Abstract
In vertebrates, including humans, individuals harbor gut microbial communities whose species composition and relative proportions of dominant microbial groups are tremendously varied. Although external and stochastic factors clearly contribute to the individuality of the microbiota, the fundamental principles dictating how environmental factors and host genetic factors combine to shape this complex ecosystem are largely unknown and require systematic study. Here we examined factors that affect microbiota composition in a large (n = 645) mouse advanced intercross line originating from a cross between C57BL/6J and an ICR-derived outbred line (HR). Quantitative pyrosequencing of the microbiota defined a core measurable microbiota (CMM) of 64 conserved taxonomic groups that varied quantitatively across most animals in the population. Although some of this variation can be explained by litter and cohort effects, individual host genotype had a measurable contribution. Testing of the CMM abundances for cosegregation with 530 fully informative SNP markers identified 18 host quantitative trait loci (QTL) that show significant or suggestive genome-wide linkage with relative abundances of specific microbial taxa. These QTL affect microbiota composition in three ways; some loci control individual microbial species, some control groups of related taxa, and some have putative pleiotropic effects on groups of distantly related organisms. These data provide clear evidence for the importance of host genetic control in shaping individual microbiome diversity in mammals, a key step toward understanding the factors that govern the assemblages of gut microbiota associated with complex diseases.
Keywords: 16S rDNA, pyrosequencing, quantitative trait loci mapping, microbiome phenotyping, population
Humans are born with a sterile gastrointestinal (GI) tract that is rapidly colonized by successive waves of microorganisms until a dense microbial population stabilizes at about the time of weaning (1). This population is dominated by thousands of bacterial species that belong to a small number of phyla (2–4). Despite conservation at the highest taxonomic ranks, the composition of the adult gut microbiota varies dramatically from individual to individual, including differences in the relative ratios of dominant phyla and variation in genera and species found in an individual host (4). Once established, these compositional features are highly resilient to perturbation (5). Although the mechanism of this homeostasis is unknown, it suggests a “top down” model for assembly of the symbiotic microbial community that is largely determined by the host.
A mechanistic insight into the assembly of the gut microbiota is immediately relevant to our understanding of complex human diseases (6). Obesity (7), coronary heart disease (8), diabetes (9), and inflammatory bowel disease (10) have all been associated with composition of gut microbiota. These diseases are well understood to be multifactorial, with both environmental and genetic components (11–13), and the contribution of the gut microbiota is currently viewed as an environmental factor (14). Although a number of studies have suggested that composition of the gut microbiota may be subject to host genetic forces, existing evidence is conflicting and confounded by the genetic diversity of vertebrate (especially human) populations and strong environmental effects (15–19).
To study the combination of environmental and host genetic factors that shape composition of the gut microbiota, we investigated a large murine intercross model in which genetic background can be systematically evaluated while environmental factors are carefully controlled. In this model, we quantified variation in taxonomic composition of gut microbiota and estimated the effects of maternal environment and host genotype. We used quantitative trait loci (QTL) analysis to test whether specific taxa cosegregate as quantitative traits with linked genomic markers. Using sophisticated methods for quantitative microbiota analysis and a suitably large number of genomic polymorphic markers, we have identified significant QTL that control variability in the abundances of different taxa in the mouse gut microbiome. We found that gut microbiota composition as a whole can be understood as a complex, polygenic trait influenced by combinations of host genomic loci and environmental factors.
Results
Core Measurable Microbiota in the G4 Intercross Population.
The availability of a large murine advanced intercross line (AIL) mapping population developed and maintained in a controlled environment (20) gave us a unique opportunity to examine the distribution of gut microbial taxa in a population of known pedigree. The random and sequential intercrossing over multiple generations in the AIL population increases the chance of recombination; as a result, AILs offer greater mapping resolution and narrower confidence intervals compared with a typical F2 mapping population (21). The breeding protocol that created the AIL used in our study effectively expanded the mapping space 3-fold from that of a standard murine map (20).
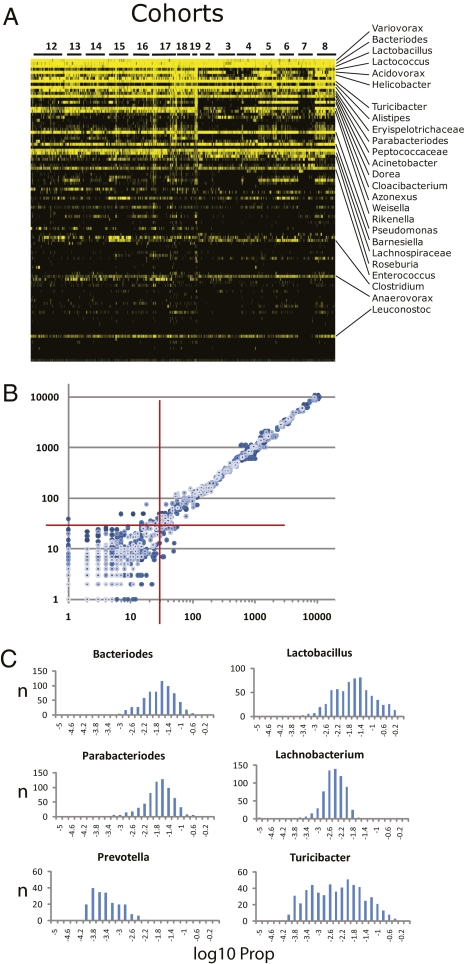
The microbiota were phenotyped by pyrosequencing of 16S rDNA, generating a detailed and quantitative estimate of the taxonomic composition of gut microbiota across the entire population of AILs. To accommodate this massive amount of data and to estimate covariation of phylogenetically related taxa up and down taxonomic ranks, we used the CLASSIFIER algorithm to predict relative abundances of organisms (22). The CLASSIFIER, which assigns taxonomic rank to sequence reads by matching distributions of nucleotide substrings to a model defined from sequences of known microorganisms, detected 420 genera, 143 families, 53 orders, 24 classes, and 16 phyla in the 645 samples sequenced. The relative abundances of the major phyla (Firmicutes, 30–70%; Bacteriodetes, 10–40%; Proteobacteria, 1–15%; Actinobacteria, Tenericutes, TM7, and Verrucomicrobia, 0.1–0.5%) were very similar to those reported for cecal sampling from murine models (7). CLASSIFIER assignments were validated by SEQMATCH (Table S1). Many genera were found in only a few animals; only a small number of genera were distributed quantitatively across most or all animals (Fig. 1A). These taxa—ones that are largely conserved and that vary quantitatively, and whose abundance can be accurately estimated from pyrosequencing data—were the focus of our analysis. Data from multiple technical repeats of five different samples (Fig. 1B) identified a minimum of 30 sequence reads for a given taxon as the threshold for quantitative repeatability. This threshold was subsequently applied as an average of 30 reads per bin across the entire mapping population. We define the resulting 19 genera and a total of 64 different taxonomic groups as a core measurable microbiota (CMM) (Table S2). Although the CMM genera represent only a small portion of the 420 total genera that we detected, they account for >90% of the sequence reads that were assigned to a genus by the CLASSIFIER, and thus define taxa that constitute a significant portion of the identifiable and quantifiable portion of the total microbiota. The CMM are log-normally distributed across the mapping population (Fig. 1C), with most genera distributed in a relatively narrow range of relative abundances and a small number of taxa, such as Turicibacter, showing a broader range (Table S2).
Fig. 1.
Characterization of the gut microbiota across the AIL population. (A) A heat map of the relative abundance of the top 100 genera identified in the G4 AIL population. Vertical columns represent individual animals; horizontal rows depict genera. Genera of interest are indicated. Black indicates absent taxa. (B) A scatterplot generated from pairwise combinations of data from technical repeats from five different samples. 16S rDNA from each sample was amplified with three different sets of bar-coded primers. Processed and filtered sequences from each barcode–sample combination were then assigned taxonomy by CLASSIFIER. Sequence counts for each taxonomic bin were log-transformed and plotted for all pairwise combinations of the three repeats for each sample. Axes are the log10-transformed values for total sequence reads of each taxon. The red crosshairs indicate the 30-read threshold. Above this number, correlation reaches >0.998: below this number, correlation dissipates rapidly. (C) Histograms of the frequency distribution of selected CMM taxa across the 645 animals. The histograms were plotted from log10-transformed values of the proportion (Prop) of sequence reads for each taxon (i.e., number of reads for that taxon/total sequence reads for a given animal). Thus, each histogram depicts the number of animals (y axis) with log10-transformed Prop values (x axis) for the given taxon.
Litter and Cohort Have Significant Effects on Gut Microbiota Composition.
If the relative abundances of the CMM are considered as complex traits, then the variation represented in their log-normal distributions would be a result of both environmental factors and host genetics. Given the well-defined nature of this large, segregating AIL population, our pyrosequencing data gave us the opportunity to evaluate systematically the relative contribution of separate apparent forces, such as the maternal environment and host genetics, a task that has not yet been accomplished in such a population.
As expected, environmental effects were readily observed by a mixed-model analysis (Table S3), which included fixed effects for parent of origin and sex along with random effects for cohort and family (nested with parent of origin) and litter (nested with cohort). On average, cohort accounted for 26% of the variation in taxa of the CMM (Table S4). Family and litter each accounted for about 5% of the variation in taxa of the CMM, with over half of the taxa showing litter effects that were significantly different from 0 (P < 0.05) (Table S3). Whereas variation between families and variation within litter include both a genetic component and an environmental component, variation between litters within a family includes only an environmental component, thereby leaving host genetics to explain significant proportions of the variation.
Composition of the Gut Microbiota Behaves as a Polygenic Trait.
We used QTL analysis to assess the degree to which host genotype contributes to the variation in CMM across the AIL mapping population. The proportion (Prop) of each CMM taxon at each taxonomic rank was treated as an individual trait and tested for cosegregation with 530 fully informative SNP markers. Although AILs enhance mapping resolution, the complex breeding history of our study population falsified the assumption of independence among individuals and made conventional mapping strategies inappropriate. To overcome this problem, we used the genome reshuffling for advanced intercross permutation (GRAIP) procedure, which estimates parental (F3 in our case) genotypes and uses a permutation scheme to simulate sets of F3 progenitors (23). From these progenitor sets, recombination and inheritance are simulated, creating randomized G4 populations (n = 50,000) that respect the original family structure while removing any association between genotype and phenotype. QTL analyses are then performed on the original and GRAIP-permuted populations. Locus-specific and genome-wide empirical P values are estimated using the distribution of P values from the permuted maps.
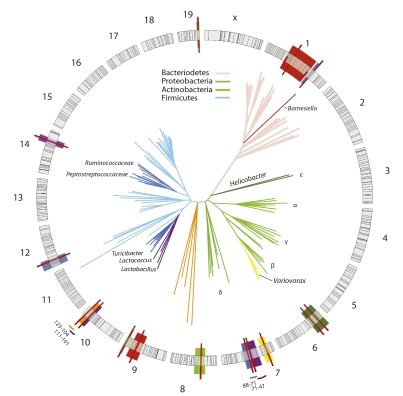
With the GRAIP procedure, 26 out of 64 taxonomic groups of organisms from the CMM showed association with 13 significant QTL (LOD ≥ 3.9; P < 0.05) and 5 additional suggestive QTL (LOD ≥ 3.5; P < 0.1). Results for significant and suggestive QTL and associated data are shown in Tables S5 and S6. QTL positions relative to the genomic markers and the phylogenetic relationships of the corresponding taxa are illustrated in Fig. 2. Each QTL individually accounted for 1.6–9.0% of the total phenotypic variation; average additive effects were frequently significant, and dominance effects were especially large for the Proteobacteria. Genetic control is exerted across the entire phylogenetic space of the gut microbiota, with at least one taxon from each of the four major phyla mapping to a significant QTL. The QTL were dispersed over eight chromosomes, with multiple QTL mapping to MMU1, MMU7, and MMU10 (Fig. 2). This pattern of cosegregation in our intercross population now provides direct evidence that composition of the gut microbiota as a whole is heritable as a complex, polygenic trait.
Fig. 2.
QTL mapping of the murine gut microbiota. The circular diagram depicts the 19 murine autosomes and X chromosome drawn to scale. Black lines mark the positions of the SNPs used for QTL mapping. QTL confidence intervals are shaded in colors that correspond to the branches of the organism(s) in the phylogenetic tree. QTL peaks are marked by solid red lines. Color-coded bars outside the circle indicate confidence intervals of adjacent QTL. Coordinates of the confidence intervals (in Mb) are also indicated. The representative phylogenetic tree was derived from 100,000 sequences randomly drawn from the total data set of 5.2. The sampled sequences were clustered with CD-Hit; representative sequences of the most abundant 200 clusters were used for phylogenetic analysis by the neighbor-joining method. Major phyla are color-coded.
Host genetic control appears to focus largely on the tips of the phylogenetic tree. This phenomenon was particularly apparent in diverse groups of organisms (e.g., Bacteriodetes, Clostridia, Bacilli) in which QTL were observed only at the genus and family levels. Phylum- or class-level QTL were apparent only in the Actinobacteria, Erysipilotrichi, and Epsilon classes of the Proteobacteria, which were each dominated numerically by a few taxa (e.g., Coriobacteriaceae within the Actinobacteria, Turicibacter within the Erysipilotrichi, Helicobacter within the Epsilon) that accounted for the QTL signal.
QTL for Host-Adapted Species of Lactobacilli.
Among the CMM organisms, only the genera Helicobacter and Lactobacillus are known to form close physical associations with host tissues, a characteristic that would be expected to be modulated by host factors. Significant QTL were detected for Helicobacter, but no QTL were identified for Lactobacillus (Table S1). Lactobacilli form dense cell layers on the murine forestomach epithelium, and its isolates’ adherence phenotypes have been shown to be host-specific (24, 25); L. reuteri even comprises host-adapted subpopulations (26). This degree of host adaptation at the species level and below, and the fact that no QTL were detected at the genus level, led us to speculate that it may be precisely at the lower taxonomic ranks that host genetic control over Lactobacilli is exerted. To test for cosegregation at the species level, we mapped as individual traits the relative abundance of three groups with 97% identity: L. reuteri, L. johnsonii/L. gasseri, and L. animalis/L. murinus (Fig. S1). Indeed, the L. johnsonii/gasseri group segregated with two significant QTL on MMU14 and MMU7 (Table S5), implying that intimate associations between the host and its microbiota are subject to heritable genetic factors.
Some QTL Have Pleiotropic Effects on the Gut Microbial Taxa.
Several QTL appear to have pleiotropic effects on multiple taxa and these effects can be divided into three groups. The first group includes QTL that affect relatively closely related organisms, such as the QTL for L. johnsonii/gasseri on MMU7 (peak at 66 Mb), which is adjacent to the QTL for Turicibacter (peak at 73 Mb), with overlapping confidence intervals. Colocalization of these QTL implies that MMU7 may encode a gene that influences both taxa, or that this region contains linked genes that, individually or in combination, affect gut microbiota composition.
The QTL for the phylum Proteobacteria exemplifies the second type of pleiotropy. Here the peak and confidence interval for a QTL on MMU6 at 28 Mb are nearly identical to those of a Helicobacter QTL. Thus, this single phylum-level QTL may have significant effects on the ability of Helicobacter to colonize the murine GI tract along with a broader effect on the entire Proteobacteria population. This finding underscores the importance of testing for cosegregation at different levels of taxonomic hierarchy. A second QTL on MMU8 was also associated with the phylum Proteobacteria, distinct from all other QTL for lower taxonomic ranks of Proteobacteria, implying that the relative abundance of an entire Phylum can be controlled by a single genomic locus.
Finally, a third type of pleiotropy can be found for the genus Lactococcus (phylum Firmicutes) and the family Coriobacteriaceae (phylum Actinobacteria). These QTL colocalize in the 104–123 Mb region of MMU10, with peaks at 107 Mb and 119 Mb, respectively. These organisms, unlike those in the first two groups of pleiotropic QTL, have a very distant phylogenetic relationship. Nonetheless, they show a positive correlation in the data set and have either shared gene action or overlapping QTL, with significant dominance effects of the C57BL/6J allele (Table S5). Thus, the effect of these colocalizing QTL was to cause positive correlation between the relative abundances of Coriobacteriaceae and Lactococcus, illustrating the significance of host genetic influence on the population structure in the gut.
Discussion
From an essentially sterile state at birth, the gut ecosystem develops rapidly as microbes successively colonize vacant niches. In humans, this period of succession persists until 18–24 mo of age, when the gut microbiota attains its “adult-like” composition and begins to behave as a highly individualized climax community (1, 27, 28). Despite tremendous diversity of the gut microbial species, many of which are sparsely distributed between individual hosts, recent work has revealed that a core of >50 taxa are found in nearly half of human subjects sampled (29, 30). This finding is consistent with the observations in our large murine population under controlled conditions (Fig. 1A). Our discovery that the CMM taxa, which are some of the most abundant organisms in the GI tract, are subject to host genetic control now supports the concept of a core microbiome as a universal feature among vertebrate hosts, with the relative abundances of CMM taxa collectively behaving as a complex polygenic trait. This glimpse of the host genetic architecture underpinning gut microbiota composition was attained under the highly controlled environmental conditions of our murine intercross population, and shows that these genetic effects are broadly distributed across the dominant CMM phyla (Fig. 2) and can influence very specific groups of organisms or have pleiotropic effects on diverse taxonomic groups.
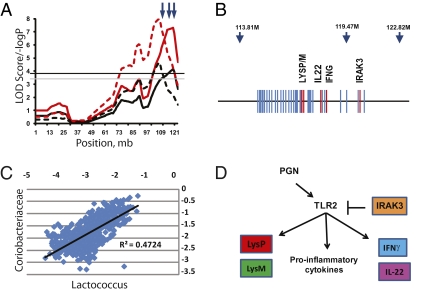
Establishment of this murine model and demonstration of heritability are important steps toward experimental paradigms that can define the mechanisms which drive the assembly of the microbiota in individuals. As an example, we again turn to the colocalized QTL for the Coriobacteriaceae and Lactococcus that span a 15-Mb region on MMU10 (Fig. 2). As shown in Fig. 3A, these QTL are closely positioned and control Gram-positive organisms, which is consistent with several genes in this region, namely Irak3, which modulates MyoD88-dependent peptidoglycan (PGN)-stimulated responses of the TLR2 pathway (31), and the two primary murine lysozyme genes, Lyz1 and Lyz2 (32). The same interval also contains genes encoding IFN-γ (Ifng) and IL-22 (Il22), which play substantial roles in mucosal immunity, where they shape T cell development and elicit antibacterial responses in intestinal epithelial cells (33, 34). Lactococci have only recently been observed in the GI tract through pyrosequencing data, but members of the Coriobacteriaceae (e.g., Eggerthella, Enterorhabdus) are associated with mouse models of inflammatory disease (35, 36). The significance of this QTL is underscored by the strong correlation of these two taxa (Fig. 3C) due, at least in part, to the QTL effect.
Fig. 3.
Fine structure of the genomic region of the significant QTL on chromosome 10. (A) The simple mapping output (red lines) and GRAIP permutation output (black lines) for QTL analysis of Coriobacteriaceae (solid lines) and Lactococcus (dashed lines). Genome-wide GRAIP-adjusted significance thresholds were generated from 50,000 permutations. Thus for the GRAIP output, a minimum possible P value with 50,000 permutations is 0.00002 (1/50,000), so the maximum −log P is 4.7. The black and gray horizontal lines represent the permuted 95% and 90% LOD thresholds, respectively. Arrows at the top show the relative positions of the three SNP markers nearest the QTL. (B) A scaled gene map of the QTL region. Arrows indicate SNP markers and their positions (in Mb). Genes are marked by blue; genes of interest are in red. (C) A scatterplot of log-transformed Prop values from the Coriobacteriaceae and the Lactococcus taxon bins of the 645 animals used in the study. (D) The combined functional pathways of the genes of interest in the QTL across multiple cell types. The bar from IRAK3 to TLR2 represents direct action. Arrows represent the relative influence of each gene and not necessarily direct gene action.
The Il22 gene is duplicated in the C57BL/6J genome, making it tempting to speculate that this duplication at least partially accounts for the MMU10 QTL effect. Indeed, in G4 progeny homozygous for the C57BL/6J allele of the JAX0030095 marker (at 119 Mb, adjacent to Il22), the Coriobacteriaceae and Lactococcus are both significantly less abundant (Fig. S3). Although this result would be anticipated, it is not clear whether the duplicated gene, which is truncated, is actually functional (37). Given the collective antimicrobial functions of genes within this cluster, an alternative explanation is that cumulative allelic variation in several candidate genes in this region accounts for the overall QTL effect, as has been previously observed for several QTL that were dissected into subregions through congenic analysis (38, 39). The mapping power of our approach will increase as we continue into later generations of the AIL (now at G10). Moreover, new genetic resource populations that will soon be available, such as the Collaborative Cross (40, 41), will increase the genomic search space, ultimately allowing the discovery of new QTL for gut microbiota and the refinement of QTL signals to fewer candidate genes.
Fundamentally, the pattern of host genetic control that we observed is consistent with the broader effects of evolutionary divergence of the gut microbiota composition across many host species (2–4). Specifically, the effects of host genetics, like those of host speciation, involve all dominant phyla and favor selection at the tips of the phylogenetic tree. Such patterns could be predicted to emerge from host speciation events that involve concerted divergence of complex sets of loci (e.g., different QTL) and corresponding stepwise changes in the microbial populations they control. This could explain the evolution of highly specialized mammalian organs (e.g., foregut, hindgut, ceca) that harness microbes for fermentation of fibrous plant materials (42). By exerting top-down selection pressure, host genetic control would subdue microbial competition within the gut ecosystem to promote microbes that benefit the host at the cost of their own competitive fitness. This view is consistent with the suggestion that the adaptive immune system has specifically evolved in vertebrates to regulate and maintain beneficial microbial communities (43). Important insights into this question will clearly emerge from QTL analyses across multiple host species.
Beyond the fundamental significance for host–microbe interactions, demonstrating that heritable traits affect the gut microbiota also may shed new light on our understanding of complex diseases. In many ways, the gut microbiota does behave as an environmental factor implicated in fat storage (14) or immune system development (44–46). However, our work shows that the gut microbiota can now be viewed as an environmental factor that itself is controlled in part by host genetic factors and potentially by interactions between host and microbial genomes. This view implies that genetic predisposition to complex diseases may be manifested in part by a predisposition to aberrant patterns of microbial colonization, which in turn contribute to disease processes. This concept is reinforced by recent studies in monogenic models showing that both aberrations in gut microbiome composition and characteristics of complex diseases can be caused by a single null mutation (9, 36, 47, 48). Moreover, it is interesting to point out that Turicibacter, Barnesiella, and members of the Coriobacteriaceae—taxa that we have now shown to be controlled by QTL—are associated with complex disease characteristics in murine models (36, 49); in each instance, the confidence intervals of our QTL overlap known QTL for complex diseases. For example, the QTL for Turicibacter of MMU7 overlaps the HCS1 QTL for susceptibility to murine hepatocellular carcinomas (50), whereas the QTL for Coriobacteriaceae on MMU10 overlaps the Scc9 locus associated with murine susceptibility to colon tumors (51). The QTL on MMU1 for Barnesiella also overlaps the conserved gene ATG16L, and this region is syntenic with the ATG16L region of the human chromosome 2 (234Mb region) recently shown to be associated with Crohn’s disease (52). Although these discoveries were made in different genetic backgrounds, and the confidence intervals of each QTL contain many genes, it will be interesting to see if any of these loci have pleiotropic effects on both microbiota abundance and disease. Conversely, for complex diseases whose genetic architecture is already well defined, such as the >200 QTL mapped for traits related to obesity (53), our discovery now begs the question of whether some of these QTL could manifest their phenotypes through their effects on gut microbiome composition and, if so, which organisms they affect.
Similarly, the CMM concept can now be translated to genome-wide association studies in humans, in which dense panels of well-defined genomic markers can be tested for association with CMM characteristics. We believe that, with highly refined data from murine models, mapping heritable genetic factors controlling gut microbiome composition will ultimately be an important tool for studying disease. This strategy is also applicable to agriculturally relevant food animals, where host genetic control is likely to be implicated in colonization by zoonotic pathogens as well as organisms important for ruminal fermentations and feed intake phenotypes.
Methods
Animal Population.
A moderately (G4) advanced intercross line (AIL) was bred from reciprocal crosses between the inbred strain C57BL/6J and the ICR-derived HR line (54). In brief, F3 breeding pairs produced multiple litters to expand (n = 815) the G4 population, with staggered mating to reduce intergroup age variation. To accommodate phenotyping constraints, G4 individuals were divided into 19 consecutive cohorts of ∼45 mice each, with approximately even numbers of both sexes. After weaning, G4 animals were group-caged by sex and provided ad libitum access to a repeatable synthetic diet (Research Diet D10001) and water. At ∼8 wk of age, mice were caged individually; the following day, fecal samples were collected and stored at −30 °C.
Deep Pyrosequencing of the Gut Microbiota.
DNA extraction from fecal pellets and pyrosequencing have been described previously (55). The V1-V2 region of the 16S rRNA gene was amplified using bar-coded fusion primers with the Roche-454 A or B Titanium sequencing adapters (see SI Methods). Of the 709 G4 animals’ samples, robust PCR products were obtained from 645 samples. Pooled and gel-purified amplicon products were sequenced using Roche-454 GS FLX Titanium chemistry.
Pyrosequencing Data Processing Pipelines.
Raw reads were filtered according to length and quality criteria (see SI Methods). Filter-pass reads were parsed into sample-barcoded bins and uploaded to a publicly accessible MySQL database (http://cage.unl.edu). More than 5.2 million quality-filtered reads were obtained from 645 samples, an average of 8,000 reads per animal. Reads were assigned taxonomic status with a parallelized version of the multi-CLASSIFIER algorithm (22), and reads in each taxonomic bin were normalized as the absolute proportion (Prop) of the total number of reads for each sample (see SI Methods). These Prop values for each taxon were used as “traits” for QTL analysis.
To confirm taxonomic assignments, we randomly sampled 40,000 sequences from genus-level bins and checked best-hits from the RDP database using SeqMatch (Table S1). In addition, we validated the quantitative nature of the pyrosequencing data by qPCR using Lactobacillus-specific primers (56), which yielded highly significant correlation (r > 0.64; Fig. S2).
QTL Analysis.
Prop values of microbial taxa were log10-transformed, and for animals for which no counts were obtained for a given taxon, a value of 0.5/total reads was log10-transformed and used. Each individual microbial “trait” was then evaluated for location and magnitude of QTL. Complete descriptions of the marker genotyping and the final set of SNPs (n = 530, with an average spacing of 4.7 Mb) used in the QTL analyses are provided elsewhere (20). To account for the G4 family structure (nonindependence of individuals), we used the GRAIP procedure (23), as described previously (20). Details of the QTL analysis are presented in SI Methods.
Supplementary Material
Acknowledgments
We thank the members of the Ribosomal Database Project at Michigan State University for kindly providing the parallelized CLASSIFIER and training data sets. We gratefully acknowledge Theodore Garland, Jr., University of California-Riverside, for providing the original HR mice that contributed to creation of the G4 population. We benefited significantly from discussions with David Threadgill (North Carolina State University). We thank Nina Murray (University of Nebraska-Lincoln) for editing the manuscript and creating the artwork. This project was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants RC1DK087346 (to A.K.B. and D.P.) and DK076050 (to D.P.). Some phenotypes were collected using the Animal Metabolism Phenotyping core facility at the University of North Carolina’s Nutrition Obesity Research Center (funded by NIDDK Grant DK056350).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007028107/-/DCSupplemental.
References
- 1.Tannock GW. What immunologists should know about bacterial communities of the human bowel. Semin Immunol. 2007;19:94–105. doi: 10.1016/j.smim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll IM, Threadgill DW, Threadgill DS. The gastrointestinal microbiome: A malleable, third genome of mammals. Mamm Genome. 2009;20:395–403. doi: 10.1007/s00335-009-9204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fava F, Lovegrove JA, Gitau R, Jackson KG, Tuohy KM. The gut microbiota and lipid metabolism: Implications for human health and coronary heart disease. Curr Med Chem. 2006;13:3005–3021. doi: 10.2174/092986706778521814. [DOI] [PubMed] [Google Scholar]
- 9.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindgren CM, et al. Wellcome Trust Case Control Consortium; Procardis Consortia; Giant Consortium. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt C, et al. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol Genomics. 2008;34:42–53. doi: 10.1152/physiolgenomics.00267.2007. [DOI] [PubMed] [Google Scholar]
- 13.van Heel DA, et al. Genome Scan Meta-Analysis Group of the IBD International Genetics Consortium. Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet. 2004;13:763–770. doi: 10.1093/hmg/ddh090. [DOI] [PubMed] [Google Scholar]
- 14.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoetendal EG, et al. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
- 16.Van de Merwe JP, Stegeman JH, Hazenberg MP. The resident faecal flora is determined by genetic characteristics of the host: Implications for Crohn’s disease? Antonie van Leeuwenhoek. 1983;49:119–124. doi: 10.1007/BF00393669. [DOI] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khachatryan ZA, et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deloris Alexander A, et al. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–1104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- 20.Kelly SA, et al. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics. 2010;42:120–200. doi: 10.1152/physiolgenomics.00028.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peirce JL, et al. Genome Reshuffling for Advanced Intercross Permutation (GRAIP): Simulation and permutation for advanced intercross population analysis. PLoS One. 2008;3:e1977. doi: 10.1371/journal.pone.0001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuller R, Brooker BE. Lactobacilli which attach to the crop epithelium of the fowl. Am J Clin Nutr. 1974;27:1305–1312. doi: 10.1093/ajcn/27.11.1305. [DOI] [PubMed] [Google Scholar]
- 25.Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127:67–76. doi: 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh PL, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 27.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 29.Qin J, et al. MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tap J, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, et al. Involvement of IRAK-M in peptidoglycan-induced tolerance in macrophages. J Biol Chem. 2004;279:6629–6634. doi: 10.1074/jbc.M308620200. [DOI] [PubMed] [Google Scholar]
- 32.Markart P, et al. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem J. 2004;380:385–392. doi: 10.1042/BJ20031810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Kimpe SJ, Kengatharan M, Thiemermann C, Vane JR. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proc Natl Acad Sci USA. 1995;92:10359–10363. doi: 10.1073/pnas.92.22.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 35.Clavel T, et al. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. J Syst Evol Microbiol. 2009;59:1805–1812. doi: 10.1099/ijs.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 36.Ye J, et al. Bacteria and bacterial rRNA genes associated with the development of colitis in IL-10(-/-) mice. Inflamm Bowel Dis. 2008;14:1041–1050. doi: 10.1002/ibd.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumoutier L, Van Roost E, Ameye G, Michaux L, Renauld JC. IL-TIF/IL-22: Genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1:488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
- 38.Farber CR, Medrano JF. Dissection of a genetically complex cluster of growth and obesity QTLs on mouse chromosome 2 using subcongenic intercrosses. Mamm Genome. 2007;18:635–645. doi: 10.1007/s00335-007-9046-0. [DOI] [PubMed] [Google Scholar]
- 39.Jerez-Timaure NC, Eisen EJ, Pomp D. Fine mapping of a QTL region with large effects on growth and fatness on mouse chromosome 2. Physiol Genomics. 2005;21:411–422. doi: 10.1152/physiolgenomics.00256.2004. [DOI] [PubMed] [Google Scholar]
- 40.Churchill GA, et al. Complex Trait Consortium. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 41.Chesler EJ, et al. The Collaborative Cross at Oak Ridge National Laboratory: Developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens CE, Hume ID. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev. 1998;78:393–427. doi: 10.1152/physrev.1998.78.2.393. [DOI] [PubMed] [Google Scholar]
- 43.McFall-Ngai M. Adaptive immunity: Care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, et al. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazmanian SKL, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 47.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 49.Presley LL, Wei B, Braun J, Borneman J. Bacteria associated with immunoregulatory cells in mice. Appl Environ Microbiol. 2010;76:936–941. doi: 10.1128/AEM.01561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gariboldi M, et al. Chromosome mapping of murine susceptibility loci to liver carcinogenesis. Cancer Res. 1993;53:209–211. [PubMed] [Google Scholar]
- 51.van Wezel T, Ruivenkamp CA, Stassen AP, Moen CJ, Demant P. Four new colon cancer susceptibility loci, Scc6 to Scc9 in the mouse. Cancer Res. 1999;59:4216–4218. [PubMed] [Google Scholar]
- 52.Parkes M, et al. Wellcome Trust Case Control Consortium. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pomp D, Allan MF, Wesolowski SR. Quantitative genomics: Exploring the genetic architecture of complex trait predisposition. J Anim Sci. 2004;82(E-Suppl):E300–312. doi: 10.2527/2004.8213_supplE300x. [DOI] [PubMed] [Google Scholar]
- 54.Kelly SA, et al. Parent-of-origin effects on voluntary exercise levels and body composition in mice. Physiol Genomics. 2010;40:111–120. doi: 10.1152/physiolgenomics.00139.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez I, et al. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walter J, et al. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.