Abstract
In order to segregate chromosomes properly, the cell must prevent merotelic kinetochore attachment, an error that occurs when a single kinetochore is attached to microtubules emanating from both spindle poles. Merotelic kinetochore orientation represents a major mechanism of aneuploidy in mitotic mammalian cells and it is the primary mechanism of chromosome instability in cancer cells. Fission yeast mutants defective in putative microtubule-site clamp Pcs1/Mde4 or Clr4/Swi6-dependent centromeric heterochromatin display high frequencies of lagging chromosomes during anaphase. Here, we developed an assay based on laser microsurgery to show that the stretched morphology of lagging kinetochores in pcs1Δ and clr4Δ mutant cells is due to merotelic attachment. We further show that Mde4 is regulated by Cdc2 and that Cdc2 activity prevents precocious localization of Mde4 to the metaphase spindle. Finally, we show that Pcs1/Mde4 complex shares similar features with the conserved kinetochore complex Spc24/Spc25 suggesting that these two complexes may occupy a similar functional niche.
Keywords: chromosome segregation, mitosis, kinetochore, merotely, fission yeast
Introduction
For accurate segregation of chromosomes, sister kinetochores must attach to microtubules emanating from opposite spindle poles (amphitelic attachment).1-3 Merotelic kinetochore orientation is an error in which a single kinetochore binds microtubules emanating from both spindle poles.4,5 Merotelic attachments frequently occur in the early stages of mitosis, but most are corrected before anaphase onset.6,7 However, because merotelic attachments are not detected by the mitotic checkpoint,7,8 they can persist until anaphase, causing chromatids to lag on the mitotic spindle and hindering their poleward segregation.6,9 Thus, despite correction mechanisms, merotelic kinetochore orientation represents a major mechanism of aneuploidy in mitotic mammalian cells.9,10 Moreover, recent studies have shown that merotely is the primary mechanism of chromosome instability (CIN) in cancer cells.11-15 Our previous work implicated Pcs1/Mde4 complex and Clr4/Swi6-dependent centromeric heterochromatin in preventing merotelic attachments.16,17 We proposed that Pcs1 and Mde4 may prevent merotelic kinetochore orientation by clamping together (or cross-linking) microtubule attachment sites. This model makes two important predictions. First, lagging kinetochores in cells lacking Pcs1 or Mde4 should be merotelically attached and second, Pcs1 and Mde4 should localize to kinetochores at the time when stable kinetochore-microtubule attachments are formed. Here, we use laser microsurgery to show that lagging kinetochores in pcs1Δ and clr4Δ mutant cells are merotelically attached and we use in situ chromatin binding assay and chromatin immunoprecipitation to show that Pcs1 is localized to kinetochores in metaphase-like cells. Our further analysis shows that Cdc2-dependent phosphorylation of Mde4 is important for its proper localization during metaphase and that Pcs1/Mde4 complex shares similar features with Spc24/Spc25, a conserved family of eukaryotic kinetochore proteins.
Results and Discussion
Laser microsurgery shows that lagging kinetochores in pcs1- and clr4- mutants are merotelically attached
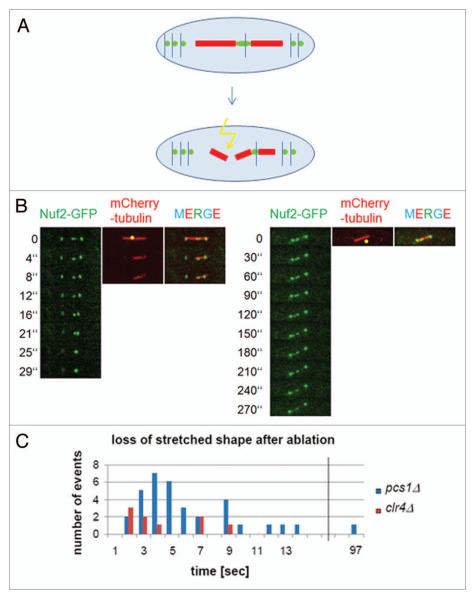
Our recent observation that lagging chromosomes in pcs1Δ, mde4Δ and clr4Δ mutants have stretched kinetochores16 is consistent with the notion that these kinetochores are merotelically attached. However, we could not exclude the possibility that chromosomes in these mutants are lagging because they are not attached to microtubules and the stretched appearance of lagging kinetochores is caused by other means than pulling forces of microtubules. We therefore decided to develop an assay that would allow us to distinguish between these two possibilities. Pulling forces of microtubules emanating from opposite spindle poles induce lateral stretching of merotelically attached kinetochores. Therefore, one of the characteristic features of merotelic kinetochores is their stretched morphology.9,16,18,19 We argued that if we sever microtubules attached to a stretched merotelic kinetochore on one side, the kinetochore should resume normal globular shape and move to the spindle pole with intact microtubules (Fig. 1A). On the other hand, if the stretched kinetochore morphology was not due to merotelic attachment, severing of microtubules should not affect kinetochore morphology. We used Nuf2-GFP to label kinetochores and mCherry-tubulin to visualize the spindle. A picosecond pulsed laser coupled to a laser scanning confocal microscope was used to sever microtubules in pcs1Δ and clr4Δ mutant cells. After laser severing, spindle poles moved toward one another suggesting that spindles were efficiently cut, as previously reported.20-23 Importantly, the lagging kinetochores recoiled and resumed their normal globular shape shortly after laser ablation (Fig. 1B and C) and invariably joined the opposite spindle pole with intact microtubules. In the control experiment, ablating a spot next to the spindle did not affect the stretched appearance of lagging kinetochore. During the course of this work, a similar strategy has been used to analyze mutants defective in sister-chromatid cohesion.24
Figure 1.
Stretched shape of lagging kinetochores is lost after severing microtubules on one side. (A) A scheme of the experiment. Pulling forces of microtubules emanating from opposite spindle poles induce lateral stretching of merotelically attached kinetochores. After severing microtubules attached to a stretched merotelic kinetochore on one side, the kinetochore should resume normal globular shape. (B) pcs1Δ mutant cells expressing mCherry-tubulin and Nuf2-GFP to label spindle microtubules and kinetochores respectively, were imaged before and after laser ablation. The laser ablated area is indicated by a yellow dot. In the control experiment on the right, laser was focused outside the spindle. (C) The loss of stretched shape of lagging kinetochores after laser ablation was quantified in pcs1Δ and clr4Δ mutant cells.
Thus, our observation that the intra-kinetochore stretching in pcs1Δ and clr4Δ mutant cells was abolished by laser severing of spindle microtubules demonstrates that stretching of lagging kinetochores is mediated by forces exerted by spindle microtubules. Importantly, it excludes the possibility that chromosomes in these mutants are lagging because they are not attached to microtubules, or attached only weakly and provides the best evidence so far kinetochores of lagging chromosomes in cells lacking Pcs1/Mde4-dependent clamps and in cells with defective centromeric heterochromatin are merotelically attached.
Importantly, we have developed an assay which is not only a tool to detect merotely, but it also provides a possibility to study structural and mechanical properties of the kinetochore in live cells. Mounting evidence suggests that the mechanical properties of the kinetochore make fundamental contributions to faithful segregation of chromosomes.25-29 However, such kinetochore properties have not been experimentally analyzed. Our observation that stretched kinetochores resumed their normal globular shape shortly after severing of the spindle suggests that the kinetochore region labeled by the Nuf2-GFP has elastic properties and paves the way for the future analysis of mechanical properties of the kinetochore in live cells.
Pcs1 localizes to the central region of centromeres and forms a complex with Mde4 in nda3-arrested cells
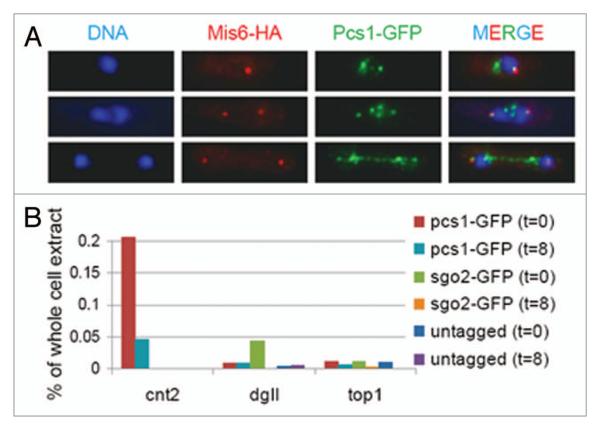
We have recently proposed that Pcs1 and Mde4 may clamp together microtubule attachment sites.16,17 If Pcs1 and Mde4 prevent merotelic kinetochore orientation by clamping together microtubule attachment sites, they should localize to kinetochores around metaphase, when stable kinetochore-microtubule attachments are formed. Both Pcs1 and Mde4 localize to kinetochores during most of the cell cycle as evidenced by co-localization of GFP-tagged Pcs1 or Mde4 with known kinetochore proteins. However, the kinetochore localization was difficult to address during early mitosis because Pcs1-GFP and Mde4-GFP signals are dispersed all over the nucleus.16,30 In order to remove the soluble fraction of Pcs1-GFP which may prevent detection of kinetochore-bound Pcs1-GFP, we used an in situ chromatin binding assay.31 Detergent extraction successfully removed soluble Pcs1-GFP from nuclei and revealed Pcs1-GFP foci in metaphase/anaphase cells. Some of the Pcs1-GFP foci co-localized with the kinetochore protein Mis6 (Fig. 2A). Interestingly, we observed co-localization of Pcs1-GFP with Mis6-HA in 90% of early anaphase cells, but only in 59% of late anaphase cells. This suggests that Pcs1 localization at kinetochores decreases during anaphase.
Figure 2.
Analysis of Pcs1 localization. (A) Cells expressing Pcs1-GFP and Mis6-HA (JG15031) were permeabilized by zymolyase digestion, extracted with detergent, fixed and stained with antibodies against HA and GFP. Nuclei were visualized by Hoechst staining. (B) Untagged cells (JG12013) or cells carrying Pcs1-GFP (JG14985) or Sgo2-GFP (JG12239) were harvested at the indicated time-points (0 and 8 minutes) after release from nda3-KM311-arrest. Chromatin binding of the GFP-tagged proteins was analyzed by chromatin-immunoprecipitation followed by quantitative PCR using oligonucleotide primers specific for the centromeric central region (cnt2), outer centromere (dgII) and chromosome arm (top1).
To determine more precisely the centromeric region to which Pcs1 binds, we arrested cells in a metaphase-like stage using a cold-sensitive tubulin allele (nda3-KM311)32 and analyzed the localization of Pcs1-GFP by chromatin immunoprecipitation. In nda3-arrested cells, Sgo2-GFP was enriched at the outer centromeric repeats, as previously reported,33 while Pcs1-GFP was enriched at the central centromeric region (Fig. 2B, t = 0). Eight minutes after release from the nda3-arrest, when 60% of cells were in anaphase, Pcs1-GFP was still enriched at the central centromeric region (Fig. 2B, t = 8), although the enrichment was lower as compared to nda3-arrested cells. We conclude that Pcs1 localizes to central region of centromeres in metaphase-like cells, and its centromeric enrichment is progressively reduced as cells undergo anaphase.
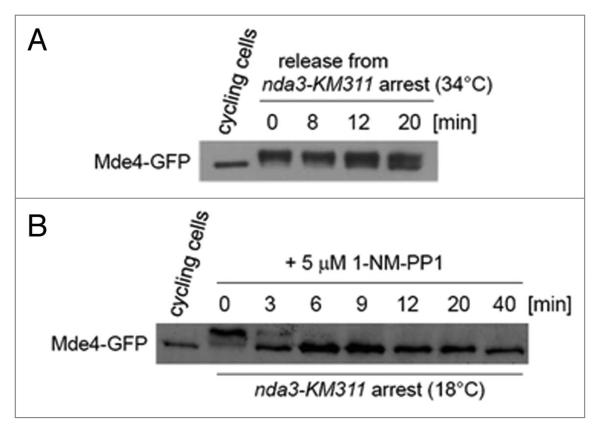
We have previously shown that Pcs1 forms a complex with Mde4 in extracts prepared from cycling cells.16 However, it is not known whether Pcs1 interacts with Mde4 during metaphase, when they are required to ensure proper attachment of kinetochores to microtubules. We used a tandem affinity purification (TAP) protocol34 to isolate Mde4-TAP together with associated proteins from both cycling and nda3-arrested cells. Mass-spectrometry analysis revealed that Mde4 associated with Pcs1 as well as with other proteins such as Net1/Cfi1-related protein Dnt1,35 and TRiC/CCT chaperonin complex36 (Fig. S1A). In addition, Mde4 co-purified with Pcs1-TAP isolated from nda3-arrested cells (Fig. S1A). While only four residues were phosphorylated on Mde4 purified from cycling cells, Mde4 was phosphorylated on 16 residues and Pcs1 was phosphorylated on serine 47 when purified from nda3-arrested cells (Fig. S1B). Five of the Mde4 phosphorylation sites have been identified during the course of this work.37-39 Western-blot analysis revealed slower-migrating forms of Mde4-GFP in nda3-arrested cells, which likely represent hyper-phosphorylated Mde4-GFP (Fig. 3A). Upon release from the nda3-arrest, when cells underwent anaphase, slower-migrating forms of Mde4 quickly decreased. We conclude that both Pcs1 and Mde4 are phosphorylated and interact with each other in metaphase-like cells.
Figure 3.
Inactivation of Cdc2 inhibits phosphorylation of Mde4. (A) nda3-KM311 cells expressing Mde4-GFP (JG15033) were grown at permissive temperature of 34°C (cycling cells) or arrested at non-permissive temperature of 18°C for 8 hours and subsequently released to permissive temperature. Samples were taken at the indicated time-points after the release and Mde4-GFP was analyzed by western blot analysis. (B) nda3-KM311 cdc2-as cells expressing Mde4-GFP (JG15356) were grown at permissive temperature of 34°C (cycling cells) or arrested at non-permissive temperature of 18°C for 8 hours. Cdc2-as was subsequently inactivated by adding 5 μM 1-NM-PP1. Samples were taken at the indicated time-points after adding 5 μM 1-NM-PP1 and Mde4-GFP was analyzed by western blot analysis.
Cdc2 activity prevents precocious localization of Mde4 to the metaphase spindle
Interestingly, eleven of the 16 residues phosphorylated on Mde4 are potential Cdk1 phosphorylation sites (SP or TP). To test whether Mde4 phosphorylation depends on the Cdk1 activity, we constructed conditional analog-sensitive allele of the fission yeast cyclin-dependent kinase Cdc2 40,41 Fig. S2). Slower-migrating forms of Mde4 rapidly disappeared upon inactivation of the Cdc2-as in nda3-arrested cells, suggesting that Cdc2 activity is necessary for Mde4 phosphorylation during mitosis (Fig. 3B). Previous observation that Cdc2 was able to phosphorylate bacterially expressed 6His-Mde4 suggests that Cdc2 may directly phosphorylate Mde4.37 Interestingly, Lrs4,16,17,42 which is the budding yeast ortholog of Mde4, was also found to be phosphorylated by Cdk1.43
In wild-type cells, Mde4 is hyper-phosphorylated during metaphase, whereas inactivation of the Cdc2-as in nda3-arrested cells creates a situation where Mde4 is hypo-phosphorylated in metaphase-like cells. We shifted nda3-arrested cells with inactivated Cdc2-as to permissive temperature to allow formation of spindles and analyzed localization of Mde4-GFP (Fig. 4A). While in wild-type cells, Mde4 localizes to spindles only in late anaphase,37,44 hypo-phosphorylated Mde4 localized to short metaphase spindles in 83% of cells (Fig. 4B). A similar phenomenon has been described in budding yeast where preventing phosphorylation of Cdk substrates such as Fin1 and Sli15/INCENP also leads to precocious localization to metaphase spindle.45,46 Chromatin immunoprecipitation showed that both hyper-phosphorylated and hypo-phosphorylated Mde4 were enriched at the central centromeric region (Fig. 4C). During the course of this work, Choi et al. showed that mutant allele of Mde4 with 12 predicted phosphorylation sites mutated to alanine (Mde4-12A) failed to localize to kinetochores and localized to metaphase spindles.37 While the premature localization of Mde4-12A to spindles is consistent with our observations (Fig. 4B), further experiments are needed to establish the role of Mde4 phosphorylation in its kinetochore localization. Thus, we conclude that Cdc2 activity prevents precocious localization of Mde4 to the metaphase spindle.
Figure 4.
Hypo-phosphorylated Mde4 localizes to metaphase spindles and kinetochores. (A) A scheme of experimental procedure. Hypo-phosphorylated Mde4 and hyper-phosphorylated Mde4 indicate experimental stages, which are referred to in other parts of the Figure. (B) nda3-KM311 cdc2-as cells expressing Mde4-GFP (JG15356) were harvested at stages indicated in the (A), fixed and stained with antibodies against tubulin and GFP. Nuclei were visualized by Hoechst staining. (C) Untagged nda3-KM311 cells (JG12013) or nda3-KM311 cdc2-as cells expressing Mde4-GFP (JG15356) were harvested at stages indicated in the (A). Chromatin binding of the Mde4-GFP was analyzed by chromatin immunoprecipitation followed by quantitative PCR using oligonucleotide primers specific for the centromeric central region (cnt2), innermost centromeric repeats (imr2), outer centromere (dgII) and chromosome arm (top1).
Pcs1/Mde4 complex shares similar features with Spc24/Spc25 complex
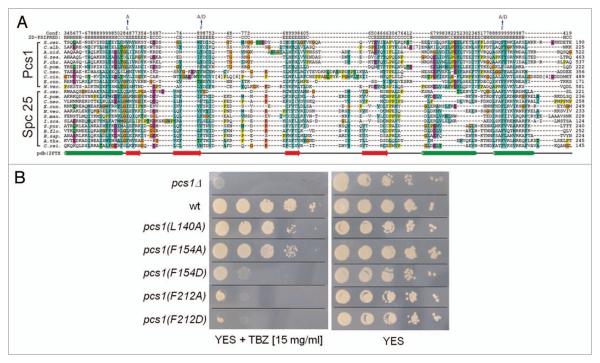
As in fission yeast, kinetochores in higher eukaryotes bind multiple microtubules and therefore must prevent merotelic attachment. However, it is not clear if higher eukaryotes employ a complex similar to Pcs1/Mde4 to prevent merotelic attachment, since Pcs1 and Mde4 orthologs have only been identified in yeast species.16 Interestingly, we noticed that PfamA Spindle_Spc25 domain was found in Pcs1 proteins (Pfam release 23.0, see Material and Methods for detailed analysis).47 Spindle_Spc25 domain is characteristic for Spc25 proteins, a conserved family of eukaryotic kinetochore proteins. Spc25 forms a dimer with Spc24 and interacts with Ndc80/Nuf2 dimer to form a four-subunit Ndc80 complex.48-54 A closer analysis revealed several lines of evidence suggesting that Pcs1/Mde4 complex is similar to Spc24/Spc25 complex. All four proteins share similar architecture, namely N-terminal coiled-coil domain followed by a globular domain. The globular part of Pcs1 and Spc25 proteins is combined in the Spindle_Spc25 domain, which aligns conserved secondary structure elements of Pcs1 to structural elements of Spc25 determined by X-ray crystallography and NMR55 (Fig. 5A). Mutating the three most conserved residues in the Spindle_Spc25 domain (L140A, F154A, F154D, F212A, F212D) diminished the Pcs1 function (Fig. 5B). Similarly as Spc24, which binds tightly to Spc25, Mde4 binds tightly to Pcs1.16,54 All four proteins localize to kinetochores, their kinetochore binding is interdependent (Fig. S3 and reviewed in ref. 37 and 50) and CENP-C is required for kinetochore binding of both Pcs1 and components of the Ndc80 complex.56-58 In addition, both Mde4 and Spc24 are phosphorylated during mitosis (Fig. S1B and reviewed in ref. 59).
Figure 5.
Pcs1 and Spc25 proteins share sequence similarity. (A) Sequence alignment of the Spindle_Spc25 domain of Pcs1 and Spc25 proteins. Mutated Pcs1 residues described in the (B) are indicated. A selected set of Pcs1 and Spc25 protein family sequences, covering a coiled coil region (the first helix shown) and the globular sequence domain, were aligned with T-coffee (default parameters, reviewed in ref. 70). The sequences used are derived from the NCBI RefSeq database71 with the following accessions for the Pcs1 sequence family: S.cer. (Saccharomyces cerevisiae, NP_010009.1), C.alb. (Candida albicans, XP_719422.1), A.nid. (Aspergillus nidulans, XP_664652.1), G.zea. (Gibberella zeae, XP_382110.1), P.chr. (Penicillium chrysogenum, XP_002559041.1), S.pom. (Schizosaccharomyces pombe, NP_001018298.1), C.neo. (Cryptococcus neoformans, XP_571257.1), C.cin. (Coprinopsis cinerea, XP_001833086.1), E.cun. (Encephalitozoon cuniculi, NP_585824.1), N.vec. (Nematostella vectensis, XP_001632729.1); the Spc25 protein family was represented by S.cer. (NP_010934.1), S.pom. (NP_588208.1), C.neo. (XP_570912.1), E.cun. (NP_586429.1), N.vec. (XP_001635865.1), S.man. (Schistosoma mansoni, XP_002580161.1), I.sca. (Ixodes scapularis, XP_002434782.1), S.pur. (Strongylocentrotus purpuratus, XP_797276.1), B.flo. (Branchiostoma floridae, XP_002592012.1), H.sap. (Homo sapiens, NP_065726.1), A.tha. (Arabidopsis thaliana, NP_566900.1), C.rei. (Chlamydomonas reinhardtii, XP_001697914.1). Stretches of unaligned sequence have been removed and the number of deleted residues is indicated in brackets. The top two lines of the alignment indicate predicted secondary structure and corresponding confidence values of S. pombe Pcs1 calculated with PSIPRED.72 The bottom line shows the secondary structure of S. cerevisiae Spc25 derived from X-ray crystallography (2FTX) as depicted from Wei et al. 2006.55 (B) Serial dilutions of wild-type cells (wt) (JG15414), pcs1Δ mutant cells (JG14821) and cells carrying the indicated pcs1 mutations pcs1(L140A) (JG15549), pcs1(F154A) (JG15550), pcs1(F154D) (JG15551), pcs1(F212A) (JG15552), pcs1(F212D) (JG15553) were spotted on YES medium or YES medium containing 15 mg/ml of thiabenda-zole (TBZ) and grown for 2 days at 32°C.
Collectively, this provides evidence that Pcs1/Mde4 complex shares similar features with the Spc24/Spc25 complex. Although a phylogenetic relationship could not be detected with sequence statistical methods, it is possible that Pcs1 and Mde4 originated by a gene duplication event in the yeast lineage, but not in higher eukaryotes. This may explain why we failed to identify orthologs of Pcs1 and Mde4 in higher eukaryotes. Alternatively, Pcs1 and Mde4 orthologs in higher eukaryotes diverged to such extend that we are not able to identify them by sequence homology. Importantly, the common features shared by Pcs1/Mde4 and Spc24/Spc25 complexes suggest that the molecular mechanism how these complexes function and their regulation may be similar.
Materials and Methods
Laser scanning confocal microscopy and laser ablation
The strains JG15161 and JG15351 were cultured in liquid EMM with appropriate supplements at 25°C. The expression of mCherry was partially repressed by the addition of 2 μM thiamine. A cover slip was coated with 2 mg/ml Lectin and glued to the round opening in the bottom of a microwell dish. Cells were mounted on the lectin spot and covered with EMM plus supplements. The microwell dish was placed in a Bachhoffer chamber to keep a constant temperature of 25°C.
Imagining of the cells was performed on an Olympus FV-1000 laser scanning confocal system. GFP and mCherry were excited at 488 nm and 561 nm, respectively, with a multi-line Argon laser (Melles Griot Bensheim, Germany). A dichroic mirror DM405/488/561/633 and a UPLSAPO 60x/1.35 oil LSM objective (Olympus) were used. Emission was detected at 500–600 nm for GFP and 600–700 nm for mCherry. During laser ablation single-plane images were taken at a free-run mode with 2 μs/pixel scanning speed. After ablation of the mitotic spindle single-plane pictures were taken using the free-run mode and Kalman filter line 2. All images have a xy-pixel size of 40 nm.
The laser setup for ablation consists of a PDL 800-B picoseconds pulsed diode laser with a LDH-P-C.405B laser head (PicoQuant, Berlin, Germany) emitting 70 picosecond pulses at 40 MHz. The laser is coupled to the bleaching port (SIM scanner) of the Olympus FV-1000 laser scanning microscope via an optical fibre. The light path of the cutting laser is different from the path of the imaging lasers. The cutting laser light is reflected onto the objective by a long pass dichroic mirror LP450 (cut on wavelength 450 nm). The laser exposure time was 4 seconds. The system was driven by Fluoview Application Software version 1.6a (Olympus).60
Chromatin immunoprecipitation
nda3-311 mutant cells were grown in YPD medium to an OD600 of 0.2–0.4 at 30°C and subsequently shifted to 18°C for 8-10 hours. Chromatin immunoprecipiation was performed essentially as described before.61-63 2.5 × 108 cells were fixed with 3% Paraformaldehyde and treated with 0.4 mg/ml Zymolyase T100. DNA was sonicated to fragments of 400 bp average size. Immunoprecipitation was performed using an anti-GFP antibody (Roche) in conjunction with ProteinA Dynabeads (Invitrogen).
Real-time PCR was performed using the IQ SYBR Green Mix (BIO-RAD) and an IQ5 Cycler (BIO-RAD). The following primers were used for qRT-PCR of S. pombe chromosome 2 loci:
Cnt2-fw AGC GCT AAC TCG TTT AAG TGA A, Cnt2-rev GGC GTG GAA AGT CAT CTG TA, Imr2-fw CTT CGG CGA CGT GAT ATA AG, Imr2-rev TTT GCA ACG ATT ACC GGT TT, DgII-fw TGC TCT GAC TTG GCT TGT CT, DgII-rev TTG CAC TCG GTT TCA GCT AT, top1-fw AGG GTT ATT TCG TGG TCG AG, top1-rev TGC CAA CCA GGT CAC TGT AT.
Strains, media and growth conditions
Tandem affinity purification
Tandem affinity purifications and mass spectrometry were performed as previously described.66-68
Sequence analysis
Iterative PSI-BLAST searches with the conserved protein sequence domain of Pcs1 family members could identify homologs in various phyla of the fungi kingdom and one animal sequence (sea anemone Nematostella vectensis) applying significant E-values below 0.01,69 (see Fig. 5A). The same approach was performed to collect the Spc25 sequence family. No significant sequence homology between Pcs1 and the Spc25 protein families could be detected and no phylogenetic relationship can be inferred. However, sequence similarity between Pcs1 and Spc25 was reported in the PfamA domain Spindle_Spc25 (PF08234, release 23.0),47 where the globular domains of both protein families were aligned. The incorporation of Pcs1 sequences into the Spindle_Spc25 domain is based on sequence searches that could not be reproduced with the latest databases. In the current Pfam release (24.0) the Pcs1 protein family is represented by the Csm1 domain (PF12539) and the Spindle_Spc25 domain contains solely sequences of the Spc25 protein family.
Supplementary Material
Acknowledgements
We thank Khodjakov A, Cimini D, McCollum D, Corbett K, Amon A, Westermann S, Cheeseman I, Riedel C, Allshire R for helpful discussions; Wang G, Benko Z and Batova M for constructing plasmids and strains; Raabe I, Kalinina I and the Light Microscopy Facility of the MPI-CBG in Dresden for help with laser ablation; Imre R and Steinmacher I for help with mass-spec analysis; Steinlein P and Stengl G for help with FACS analysis and K. Gull for the TAT1 antibody. This work was supported by Austrian Science Fund grants (P18955, P20444, F3403), HFSP grant RGY0069/2010 and the (European Community's) Seventh Framework Programme (FP7/2007-2013) under grant agreement number PIEF-GA-2008-220518. K.M. was supported by the Austrian Proteomics Platform (APP) within the Austrian Genome Research Program (GEN-AU). C.R. was supported by the F-343 stipendium from the University of Vienna.
Footnotes
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/RumpfCC9-19-sup.pdf
References
- 1.Biggins S, Walczak CE. Captivating capture: how microtubules attach to kinetochores. Curr Biol. 2003;13:449–60. doi: 10.1016/s0960-9822(03)00369-5. [DOI] [PubMed] [Google Scholar]
- 2.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–31. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–91. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 4.Cimini D. Merotelic kinetochore orientation, aneuploidy and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Cimini D. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle. 2007;6:1558–64. doi: 10.4161/cc.6.13.4452. [DOI] [PubMed] [Google Scholar]
- 6.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–25. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 7.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–55. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 8.Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–15. doi: 10.1242/jcs.115.3.507. [DOI] [PubMed] [Google Scholar]
- 9.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torosantucci L, Puzzonia Mde S, Cenciarelli C, Rens W, Degrassi F. Aneuploidy in mitosis of PtK1 cells is generated by random loss and nondisjunction of individual chromosomes. J Cell Sci. 2009;122:3455–61. doi: 10.1242/jcs.047944. [DOI] [PubMed] [Google Scholar]
- 11.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez AC, van Wely KH. Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle. 2010;9:2275–80. doi: 10.4161/cc.9.12.11865. [DOI] [PubMed] [Google Scholar]
- 16.Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, et al. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–48. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 18.Khodjakov A, Cole RW, McEwen BF, Buttle KF, Rieder CL. Chromosome fragments possessing only one kinetochore can congress to the spindle equator. J Cell Biol. 1997;136:229–40. doi: 10.1083/jcb.136.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pidoux AL, Uzawa S, Perry PE, Cande WZ, Allshire RC. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J Cell Sci. 2000;113:4177–91. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- 20.Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–6. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–40. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 22.Sacconi L, Tolic-Norrelykke IM, Antolini R, Pavone FS. Combined intracellular three-dimensional imaging and selective nanosurgery by a nonlinear microscope. J Biomed Opt. 2005;10:14002. doi: 10.1117/1.1854675. [DOI] [PubMed] [Google Scholar]
- 23.Maghelli N, Tolic-Norrelykke IM. Versatile laser-based cell manipulator. J Biophotonics. 2008;1:299–309. doi: 10.1002/jbio.200810026. [DOI] [PubMed] [Google Scholar]
- 24.Courtheoux T, Gay G, Gachet Y, Tournier S. Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J Cell Biol. 2009;187:399–412. doi: 10.1083/jcb.200902093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loncarek J, Kisurina-Evgenieva O, Vinogradova T, Hergert P, La Terra S, Kapoor TM, et al. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–9. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, et al. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 27.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–90. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–81. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 31.Kearsey SE, Brimage L, Namdar M, Ralph E, Yang X. In situ assay for analyzing the chromatin binding of proteins in fission yeast. Methods Mol Biol. 2005;296:181–8. doi: 10.1385/1-59259-857-9:181. [DOI] [PubMed] [Google Scholar]
- 32.Toda T, Umesono K, Hirata A, Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983;168:251–70. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- 33.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 34.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–2. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 35.Jin QW, Ray S, Choi SH, McCollum D. The nucleolar Net1/Cfi1-related protein Dnt1 antagonizes the septation initiation network in fission yeast. Mol Biol Cell. 2007;18:2924–34. doi: 10.1091/mbc.E06-09-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583:2647–53. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 37.Choi SH, Peli-Gulli MP, McLeod I, Sarkeshik A, Yates JR, 3rd, Simanis V, et al. Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation. Curr Biol. 2009;19:985–95. doi: 10.1016/j.cub.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–97. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 39.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregan J, Zhang C, Rumpf C, Cipak L, Li Z, Uluocak P, et al. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat Protoc. 2007;2:2996–3000. doi: 10.1038/nprot.2007.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V. Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J Cell Sci. 2008;121:843–53. doi: 10.1242/jcs.021584. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 44.Khmelinskii A, Schiebel E. Chromosome segregation: monopolin goes spindle. Curr Biol. 2009;19:482–4. doi: 10.1016/j.cub.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–12. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- 46.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–4. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 47.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, et al. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–91. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–60. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–7. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 51.Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell. 2005;16:2325–38. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 53.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 54.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–9. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function and kinetochore assembly. Mol Biol Cell. 2007;18:2155–68. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–43. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raabe I, Vogel SK, Peychl J, Tolic-Norrelykke IM. Intracellular nanosurgery and cell enucleation using a picosecond laser. J Microsc. 2009;234:1–8. doi: 10.1111/j.1365-2818.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- 61.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 62.Gregan J, Rumpf C, Li Z, Cipak L. What makes centromeric cohesion resistant to separase cleavage during meiosis I but not during meiosis II? Cell Cycle. 2008;7:10–2. doi: 10.4161/cc.7.2.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, et al. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2657–62. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rumpf C, Cipak L, Novatchkova M, Li Z, Polakova S, Dudas A, et al. High-throughput knockout screen in Schizosaccharomyces pombe identifies a novel gene required for efficient homolog disjunction during meiosis I. Cell Cycle. 2010;9:1802–8. doi: 10.4161/cc.9.9.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gregan J, Rabitsch PK, Rumpf C, Novatchkova M, Schleiffer A, Nasmyth K. High-throughput knockout screen in fission yeast. Nat Protoc. 2006;1:2457–64. doi: 10.1038/nprot.2006.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cipak L, Spirek M, Novatchkova M, Chen Z, Rumpf C, Lugmayr W, et al. An improved strategy for tandem affinity purification-tagging of Schizosaccharomyces pombe genes. Proteomics. 2009;9:4825–8. doi: 10.1002/pmic.200800948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 68.Gregan J, Riedel CG, Petronczki M, Cipak L, Rumpf C, Poser I, et al. Tandem affinity purification of functional TAP-tagged proteins from human cells. Nat Protoc. 2007;2:1145–51. doi: 10.1038/nprot.2007.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 71.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:36–8. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.