Abstract
The mammalian clock is regulated at the cellular level by a transcriptional/translational feedback loop. BMAL1/CLOCK (or NPAS2) heterodimers activate the expression of the PERIOD (PER) and CRYPTOCHROME (CRY) genes acting as transcription factors directed to the PER and CRY promoters via E-box elements. PER and CRY proteins form heterodimers and suppress the activity of the BMAL1/CLOCK (or NPAS2) completing the feedback loop. The circadian expression of BMAL1 is influenced by retinoic acid receptor-related orphan receptor α (RORα) and REV-ERBα, two nuclear receptors that target a ROR-response element in the promoter of the BMAL1 gene. Given that BMAL1 functions as an obligate heterodimer with either CLOCK or NPAS2, it is unclear how the expression of the partner is coordinated with BMAL1 expression. Here, we demonstrate that NPAS2 is also a RORα and REV-ERBα target gene. Using a ChIP/microarray screen, we identified both RORα and REV-ERBα occupancy of the NPAS2 promoter. We identified two functional ROREs within the NPAS2 promoter and also demonstrate that both RORα and REV-ERBα regulate the expression of NPAS2 mRNA. These data suggest a mechanism by which RORα and REV-ERBα coordinately regulate the expression of the positive arm of the circadian rhythm feedback loop.
Keywords: Hormone Receptors, Nuclear Receptors, Steroid Hormone Receptor, Transcription, Transcription Coactivators
Introduction
Circadian rhythms are the natural ∼24-h cycles that are conserved across a wide variety of organisms, including Arabidopsis, Drosophila, and mammals. In mammals, these rhythms are entrained by light signals and are controlled by the “master clock” in the suprachiasmatic nucleus (SCN)2 in the brain. In the periphery, semi-autonomous clocks can be entrained to signals from the SCN and signals from other cues, including nutrient status (1). The mammalian circadian clock is controlled by a transcriptional/translational feedback loop involving several key proteins. The expression of these key proteins includes brain and muscle ARNT (aryl hydrocarbon receptor nuclear translocator)-like 1 (BMAL1), circadian locomotor output kaput (CLOCK), PERIOD (PER), and CRYPTOCROME (CRY). BMAL1 and CLOCK are members of the basic helix-loop-helix-PAS family of transcription factors and form a heterodimer that targets E-box DNA elements in the promoters of the PER and CRY genes and activates their transcription. The CRY and PER proteins heterodimerize, and as the levels of this protein complex accumulate it effectively inhibits the activity of BMAL1/CLOCK limiting the expression of the PER and CRY genes. This feedback loop results in the oscillations in expression of BMAL1/CLOCK and CRY/PER that follow a circadian pattern (2, 3).
Two classes of nuclear receptors play a critical role in modulation of the mammalian circadian clock. The retinoic acid receptor-related orphan receptors (RORs) and REV-ERBs modulate expression of the BMAL1 gene through two conserved ROR-response elements (ROREs) located within the BMAL1 promoter. Expression of BMAL1 is modulated by competition between RORs and REV-ERBs for binding the BMAL1 promoter and either activation (ROR) or repression (REV-ERB) of BMAL1 (4). The expression of RORα and REV-ERBα follows a circadian pattern (with opposing phases) leading to a circadian pattern of expression of BMAL1.
Mice lacking Bmal1 completely lose circadian rhythmicity in constant darkness (5), but interestingly Clock mutant mice do not display as severe a phenotype (6, 7). This is likely due to compensatory mechanisms involving a homologue of Clock known as neuronal PAS domain protein 2 (NPAS2) (8). NPAS2 was initially discovered in the mammalian forebrain (9), but it has also been shown to be expressed in the periphery (10). NPAS2, like CLOCK, forms heterodimers with BMAL1 and effectively functions in the mammalian circadian clock (8, 9). Npas2 null mice exhibit deficiencies in circadian behavior (11), but the Npas2-Clock double knock-out phenotype is much more severe (8).
Autonomous circadian rhythms also operate in peripheral tissues such as the liver where the circadian rhythm plays an essential role in regulation of metabolic function (1). The expression of arrays of genes is a function of the molecular clock in the liver and regulate functions, including carbohydrate, fatty acid, cholesterol and bile acid metabolism. HepG2 hepatocellular carcinoma cells have been used as a model to study circadian rhythms in the liver, including the function of CLOCK and BMAL1 as well as the circadian expression of a wide number of genes, including a number of cytochrome p450s (12–19).
Based on the fact that BMAL1 and NPAS2 function as a heterodimer within the circadian clock, we found it unusual that ROR/REV-ERB would only regulate one component of this complex. In this study, we show that like BMAL1 NPAS2 is also a direct target for ROR and REV-ERB allowing for coordinated expression of these genes.
EXPERIMENTAL PROCEDURES
Reagents
The pTREX-RORα vector was a gift from Phenex Pharmaceuticals AG (Ludwigshafen, Germany). The p3×FLAG-REV-ERBα was described previously (20). The pSport6-RORα and the luciferase reporters containing the human NPAS2 promoter were a gift from the Cell-based Screening Center at The Scripps Research Institute (Jupiter, FL). All constructs were confirmed by sequencing. Mutant NPAS2:luc reporters were generated using the QuikChange II kit, according to the manufacturer's instructions (Stratagene). pG5-Luc and pGL4.73 reporters were obtained from Promega (Madison, WI). The adenoviral vector for RORα was described previously (21), and the adenoviral vector for REV-ERBα was produced in a similar manner.
Cell Culture and Cotransfection
HepG2 cells were maintained and routinely propagated in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. HepG2 cells were plated in 96-well plates at a density of 15 × 103 cells/well 24 h prior to transfection. Transfections were performed using Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen). Per well, the transfection mixture included 50 ng of Renilla luciferase as an internal control, 100 ng of the appropriate NPAS2 luciferase construct, and 100 ng of the pTREX-RORα or p3×-FLAG-REV-ERBα expression construct. The luciferase activity was measured using the Dual-Glo luciferase assay system 24 h after transfection (Promega). The luciferase readings were normalized by well to the Renilla readings. All values were normalized to the basal luciferase expression from the wild-type reporter alone to produce fold induction values.
Synchronization of Cells in Culture
HepG2 cells were plated at ∼80% confluency. The next day, cells were serum-shocked using minimal essential media with 50% horse serum media (22, 23). After 2 h, the media were changed to serum-free minimal essential media for 24 h. The time of serum-free media addition was considered t = 0, and cells were collected at various time points, as indicated in the experiments.
ChIP/Chip Screening
Either REV-ERBα or RORα was overexpressed in HepG2 cells, and ChIP/microarrays were performed as described previously by our laboratory (24–26).
ChIP
HepG2 cells were plated and serum-shocked as described. The cells were fixed, frozen on dry ice, and stored at −80 °C prior to lysis for ChIP. The ChIP-IT Express kit was used (Active Motif). At t = 4 and 16, the cells were fixed using formaldehyde added directly to the plate. Cells were thawed on ice and lysed. Sonication was used to shear the chromatin, and immunoprecipitations were incubated overnight at 4 °C. The ChIP reactions contained 25 μg of the following antibodies: IgG (Active Motif), RNA polymerase II (Active Motif), anti-hRORa (Santa Cruz Biotechnology, sr-6062X) (27), and anti-hREV-ERBα (Perseus Proteomics, PP-A8740A-00) (28). The ChIP reactions were washed, and chromatin was eluted, according to manufacturer's instructions. PCRs were performed using PCR Supermix High Fidelity (Invitrogen), 1.5 μl of each primer (10 μm) and 5 μl of eluted chromatin. The IgG, anti-RNA polymerase II, and GAPDH primers were provided in the ChIP-IT human control kit (Active Motif). The NPAS2 primers were designed with the following sequences: NPAS2 ChIP forward, 5′-CTTGGTAAAATCCTCCCTGT-3′, and NPAS2 ChIP reverse, 5′-GCAGGGGCACGTGGGGTGTG-3′; NPAS2 Control ChIP forward, 5′-TTGCATCCTTTCCCAACCTGAAGC-3′, and NPAS2 Control ChIP reverse, 5′-AAGCAGCTGTCTTCCAAGTGCT-3′.
Electrophoretic Mobility Shift Assays (EMSAs)
The RORα coding sequence was excised from pSport6-RORα, and the REV-ERBα coding sequence was excised from p3×FLAG-REV-ERBα. Both coding sequences were excised with HindIII and BamHI (Promega). The vector pcDNA3.1+ (Invitrogen) was digested with HindIII and BamHI. The digested components were gel-purified and ligated overnight at room temperature using T4 DNA ligase (Promega). The pcDNA3.1+ constructs contain a T7 promoter that allows for in vitro transcription and translation. The TnT kit for in vitro transcription and translation was used to produce protein for EMSA, according to the manufacturer's instructions (Promega). DNA containing putative response elements was annealed and then labeled with [α-32P]dATP using Klenow enzyme (Promega). Binding reactions were prepared using binding buffer (Promega), labeled probes, and equal volumes of protein. Unlabeled probes were used in 10-, 50-, and 100-fold molar excess in competition experiments. Binding reactions were loaded onto pre-cast 5% TBE gels (Bio-Rad) and analyzed by autoradiography.
mRNA Extraction, cDNA Synthesis, and Quantitative PCR
The mRNA extraction, cDNA synthesis, and quantitative PCR were performed as described previously (20). For quantitative PCR, cyclophilin B (M60857) was used as the control. All primers were designed for human genes. Primer sequences are as follows: cyclophilin B forward, 5′-GGAGATGGCACAGGAGGAAA-3′, and cyclophilin B reverse; 5′-CGTAGTGCTTCAGTTTGAAGTTCTCA-3′; RORα forward, 5′-AAACAAGCAGCGGGAGGTGA-3′, and RORα reverse, 5′-TGGCAAACTCCACCACATAC-3′; REV-ERBα forward, 5′-TTCCGCTTCGGTGGAGCAGC-3′, and REV-ERBα reverse, 5′-CCGGTTCTTCAGCACCAGAG-3′; NPAS2 forward, 5′-CCAAAGCCAATGAGAAGCTC-3′, and NPAS2 reverse. 5′-GGAAGCAGGTGTGGTCAGAT-3′.
siRNA
The siRNAs targeting human RORα and human REV-ERBα were obtained from Dharmacon (Thermo Fischer). The siRNAs were reverse-transfected into HepG2 cells using Lipofectamine RNAiMax (Invitrogen), according to the manufacturer's instructions. The HepG2 cells were incubated with siRNA for 24–48 h before being harvested for mRNA isolation.
Statistical Analysis
For cotransfection experiments, eight wells were transfected per condition per experiment. For gene expression experiments, three replicates were performed per experiment. Experiments were repeated three times, and normalized experiments are shown (mean ± S.E.). The Student's t test was used to test for significant differences between groups.
RESULTS
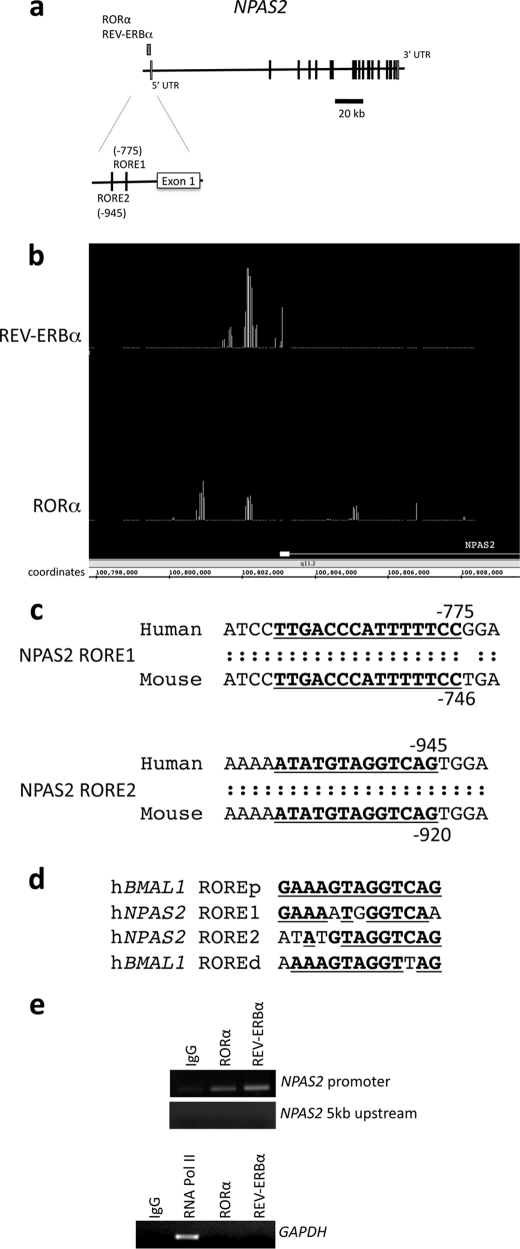
Identification of RORα and REV-ERBα Occupancy in the Human NPAS2 Promoter
A ChIP/chip screen was performed to determine RORα and REV-ERBα binding sites within the genome. RORα and REV-ERBα were overexpressed, independently, in HepG2 cells for performance of the ChIP/chip screen. HepG2 cells have been previously utilized as a model to examine circadian rhythms in peripheral tissue (liver), and we confirmed that the HepG2 cells displayed a circadian rhythm of BMAL1, NPAS2, and RORα expression following a serum shock (supplemental Fig. 1). Interestingly, although RORα displayed a robust rhythm, REV-ERBα expression remained relatively constant.
The ChIP/chip data revealed significant RORα and REV-ERBα occupancy in an identical region of the NPAS2 promoter (Fig. 1, a and b). Using the Evolutionarily Conserved Browser (29), it was determined that two putative ROREs are conserved between humans and mice (Fig. 1c). These putative ROREs were also predicted by MatInspector (30). The putative NPAS2 ROREs displayed significant similarity to the characterization of NPAS2 as a REV-ERBα/RORα target gene found in the BMAL1 gene (Fig. 1d). We performed additional ChIP experiments with primers specific for the NPAS2 promoter and confirmed occupancy of both RORα and REV-ERBα on the NPAS2 promoter (Fig. 1e). In contrast, no RORα or REV-ERBα signal was detected in regions ∼5 kb upstream of the NPAS2 promoter or in the GAPDH promoter (Fig. 1e). We focused our efforts on characterization of these putative ROREs.
FIGURE 1.
Identification of REV-ERBα and RORα binding in the NPAS2 promoter using ChIP/chip assay. Both proteins bound to a region within 1 kb of the NPAS2 gene. a, schematic detailing the position of RORα and REV-ERBα occupancy within the promoter of the NPAS2 gene, as determined by ChIP/chip. The structure of the NPAS2 gene is shown, and exons and introns are indicated. The position of the nuclear receptor occupancy is indicated above the gene structure. b, screen image of primary ChIP/chip data from Integrated Genome Browser comparing the position of positive occupancy signals from the REV-ERBα and RORα experiments. The first exon of NPAS2 is illustrated in the figure, and the coordinates within chromosome 2 are also indicated. c, analysis of RORα and REV-ERBα binding region using the Evolutionarily Conserved Region browser, which showed two conserved response elements in the region. The sequence of the two putative response elements is shown, with the conserved regions underlined. The RORE1 (proximal) and RORE2 (distal) sites are shown. d, sequences of the ROREs from the NPAS2 promoter are compared with the sequences of the ROREs (proximal and distal) from the BMAL1 promoter. The underlined boldface letters represent regions of identity with the proximal RORE derived from the BMAL1 gene. e, ChIP assay was used to verify that REV-ERBα and RORα occupancy of the NPAS2 promoter containing the ROREs. The regions encompassing the two ROREs within the NPAS2 promoter are illustrated in the top gel as well as a control region ∼5 kb upstream of this region. Additionally, a control using GAPDH primers is shown with an RNA polymerase II positive control. Numbering of the sites is relative to the transcriptional start site.
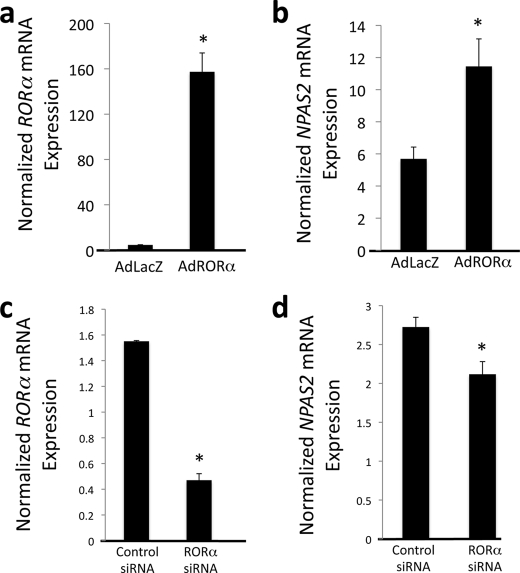
Modulation of RORα Expression Affects NPAS2 Expression
To confirm that RORα is capable of regulating NPAS2 expression, we modulated the levels of RORα and analyzed NPAS2 expression. An adenovirus encoding FLAG-tagged RORα was used to infect HepG2 cells. Forty eight h after infection, the HepG2 cells were harvested for mRNA isolation. RORα expression was significantly increased (Fig. 2a), and we noted a concomitant increase in NPAS2 mRNA expression (Fig. 2b). Regulation of NPAS2 by RORα was further examined by knockdown of RORα expression. siRNA targeting RORα resulted in a 75% decrease in RORα expression (Fig. 2c). Suppression of RORα expression resulted in a significant decrease in expression of NPAS2 (Fig. 2d). These data are consistent with RORα directly regulating the expression of NPAS2, functioning as a transcriptional activator. The degree to which an elevated level of RORα may effect NPAS2 expression may be limited by the RORα/REV-ERBα feedback loop. REV-ERB is a target gene of ROR (31, 32), and thus when ROR is massively overexpressed, REV-ERB would also be elevated by having an impact on NPAS2 expression.
FIGURE 2.
RORα regulates NPAS2 mRNA expression. a, infection of HepG2 cells with an adenovirus directing the expression of RORα results in overexpression of the receptor. b, overexpression of RORα results in induction of NPAS2 expression in HepG2 cells. c, treatment of HepG2 cells with siRNA directed at RORα suppresses expression of the receptor. d, suppression of RORα expression results in significantly lower expression of NPAS2. Data are presented as the mean ± S.E. Student's t test was used to compare the control cells to the RORα overexpressed or knocked down cells. * indicates a p value <0.05.
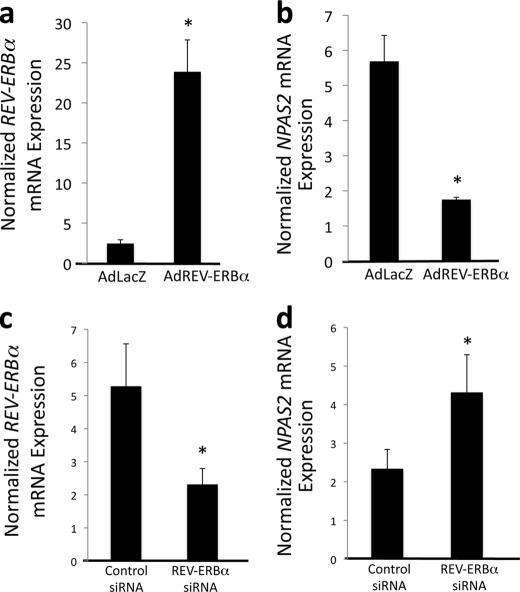
Modulation of REV-ERBα Expression Affects NPAS2 Expression
We next examined the ability of REV-ERBα to regulate NPAS2 expression. REV-ERBα was overexpressed in HepG2 cells using an adenoviral vector. Forty eight h after infection, the HepG2 cells were harvested for mRNA isolation. REV-ERBα mRNA expression was significantly increased (Fig. 3a) resulting in repression of NPAS2 (Fig. 3b). Regulation of NPAS2 by REV-ERBα was further examined by knockdown of REV-ERBα expression.
FIGURE 3.
REV-ERBα regulates NPAS2 mRNA expression. a, infection of HepG2 cells with an adenovirus directing the expression of REV-ERBα results in overexpression of the receptor. b, overexpression of REV-ERBα results in suppression of NPAS2 expression in HepG2 cells. c, treatment of HepG2 cells with siRNA directed at REV-ERBα suppresses expression of the receptor. d, suppression of REV-ERBα expression results in significantly higher expression of NPAS2. Data are presented as the mean ± S.E. Student's t test was used to compare the control cells to the REV-ERBα overexpressed or knocked down cells. * indicates a p value <0.05.
HepG2 cells were transfected with an siRNA specifically targeting REV-ERBα resulting in a 50% decrease in expression (Fig. 3c). This resulted in nearly a 2-fold increase in NPAS2 expression (Fig. 3d). These data are consistent with REV-ERBα directly regulating the expression of NPAS2 functioning as a transcriptional repressor.
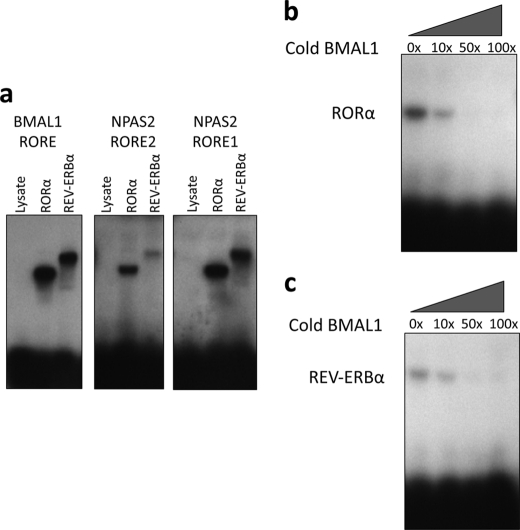
Characterization of Binding of RORα and REV-ERBα to the NPAS2 ROREs
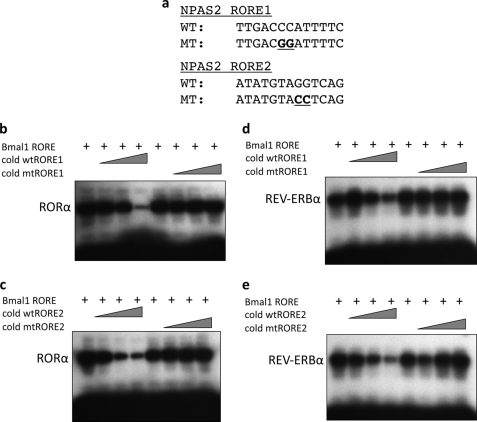
Synthetic oligonucleotides encoding the BMAL1 RORE and putative NPAS2 ROREs were utilized in an EMSA. Direct binding of REV-ERBα and RORα to each of these ROREs is shown in Fig. 4a. Competition EMSA was performed in which the BMAL1 RORE DNA was labeled and allowed to bind to RORα (Fig. 4b) or REV-ERBα (Fig. 4c). Increasing concentrations of unlabeled (cold) BMAL1 RORE DNA were added to determine the relative affinities of the nuclear receptors for the BMAL1 RORE. As shown in Fig. 4, b and c, competition was detected at a 10-fold molar excess of cold BMAL1 RORE, and the binding to the radiolabel was completely eliminated at a 50-fold molar excess of the cold DNA. Similar experiments were performed with the NPAS2 ROREs shown in Fig. 5. Both wild-type (WT) and mutant ROREs were used in this experiment to demonstrate specificity of binding to the NPAS2 ROREs (Fig. 5a). To have the ability to directly compare the relative affinities of RORα and REV-ERBα to the BMAL1 RORE and to the NPAS2 ROREs, the identical BMAL1 radiolabeled RORE used in Fig. 5, b and c, was used, but instead of unlabeled BMAL1 RORE being used as a competitor, unlabeled NPAS2 RORE1 or RORE2 was used. Fig. 5, b and c, shows results demonstrating that RORα and both the NPAS2 RORE1 (Fig. 5b) and NPAS2 RORE2 (Fig. 5c) compete for binding to the labeled BMAL1 RORE. Clearly, this interaction is less effective than if unlabeled BMAL1 RORE is used (Fig. 4b). Effective competition was not noted until a 100-fold molar excess was used for NPAS2 RORE1 or a 50-fold molar excess was used for NPAS2 RORE2. Fig. 6, d and e, shows results with REV-ERBα, and both NPAS2 RORE1 (Fig. 5d) and NPAS2 RORE2 (Fig. 5e) compete for binding to the labeled BMAL1 RORE. Again, competition is not nearly as effective compared with when the unlabeled BMAL1 RORE is used. In every case the mutant ROREs do not compete for binding indicating specificity of the NPAS2 RORE for both RORα and REV-ERBα. These data clearly demonstrate specific binding of RORα and REV-ERBα to both of the NPAS2 ROREs, but the affinity is significantly less than these NRs display for the BMAL1 RORE.
FIGURE 4.
EMSAs reveal direct binding of RORα and REV-ERBα to NPAS2 RORE1 and NPAS2 RORE2. a, EMSA displaying the ability of RORα and REV-ERBα to bind directly to radiolabeled BMAL1 RORE (proximal), NPAS2 RORE2, and NPAS2 RORE1. RORα and REV-ERBα proteins were made using rabbit reticulocyte lysate, and lysate control is also shown to demonstrate the lack of endogenous proteins within the lysate that bind to these sites. b, competition EMSA experiment using radiolabeled BMAL1 RORE (proximal) and RORα protein and competing with unlabeled (cold) BMAL1 RORE (proximal). c, competition EMSA experiment using radiolabeled BMAL1 RORE (proximal) and REV-ERBα protein and competing with unlabeled (cold) BMAL1 RORE (proximal). The numbers above the gels indicate the amount of fold molar excess of cold RORE that is included in the reactions.
FIGURE 5.
Competitive binding of RORα and REV-ERBα to ROREs. EMSAs were performed to evaluate RORα and REV-ERBα binding to the putative ROREs in the NPAS2 promoter. a, sequences of the wild-type and mutant probes are shown, with the mutated residues underlined. b, RORα binds to labeled BMAL1 RORE, and binding is competed with unlabeled NPAS2 RORE1 DNA but not mutated NPAS2 RORE1 DNA. c, RORα binds to labeled BMAL1 RORE, and binding is competed with unlabeled NPAS2 RORE2 DNA but not mutated NPAS2 RORE2 DNA. d, REV-ERBα binds to labeled BMAL1 RORE, and binding is competed with unlabeled NPAS2 RORE1 DNA but not mutated NPAS2 RORE1 DNA. e, REV-ERBα binds to labeled BMAL1 RORE, and binding is competed with unlabeled NPAS2 RORE2 DNA but not mutated NPAS2 RORE2 DNA. The triangle indicates an increasing amount of NPAS2 RORE DNA (10-, 50-, 100-fold molar excess).
FIGURE 6.
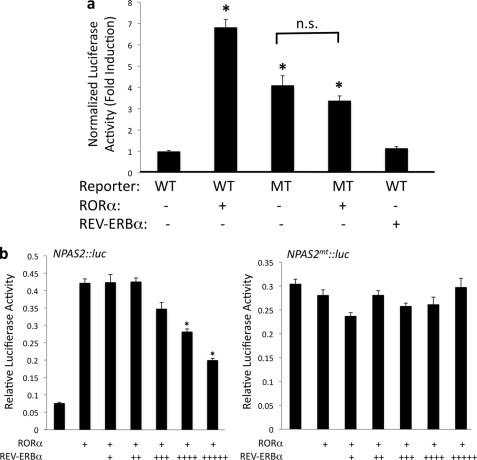
Functional characterization of ROREs within the NPAS2 promoter. a, cotransfection assay in HepG2 cells was used to analyze if RORα and REV-ERBα can regulate the expression of a luciferase reporter gene under the control of the NPAS2 promoter. The wild-type reporter contains a fragment of the NPAS2 promoter with both RORE1 and RORE2 sequences intact. The mutant reporter contains the same fragment of the NPAS2 promoter with mutations in both putative ROREs as indicated in Fig. 5a. b, ability of REV-ERBα to block the transactivation activity of RORα was examined in this cotransfection experiment. HepG2 cells were transfected with either the WT or mutant reporter as described above along with RORα. Increasing amounts of REV-ERBα were also introduced. The amounts of REV-ERBα plasmid used were 0, 10, 20, 40, 80, and 160 ng/well. Data are presented as the mean ± S.E. Student's t test was used to compare the wild-type reporter alone with the other values. * indicates a p value <0.05; n.s., not significant.
Direct Regulation of the NPAS2 Promoter by RORα and REV-ERBα
The ability of RORα and REV-ERBα to regulate the expression of a luciferase reporter driven by the NPAS2 promoter was examined using a cotransfection assay in HepG2 cells. As shown in Fig. 6a, transfection of cells with RORα along with the NPAS2:luc reporter resulted in an increase in expression (7-fold relative to the NPAS2:luc reporter alone). In the NPAS2:luc reporter in which the RORE were mutated, the RORα responsiveness was lost. Interestingly, basal luciferase expression increased in the mutant NPAS2 promoter-driven reporter suggesting that the effects of a transcriptional repressor may be lost due to this mutation. siRNA-mediated knockdown of RORα did not affect WT reporter expression suggesting that this unknown factor is not RORα (data not shown). When REV-ERBα is cotransfected into the cells along with NPAS2:luc, we note no significant effect on transcription (Fig. 6a). Clearly, REV-ERBα occupies the endogenous NPAS2 promoter as shown by the ChIP assays, and overexpression or knockdown of REV-ERBα modulates the expression of the NPAS2 mRNA in the HepG2 cells. Based on these data, it is apparent that REV-ERBα may not be directly repressing gene transcription. In fact, active repression by REV-ERB requires that two ROREs be in close proximity for this to occur. This is due to the requirement of two REV-ERB molecules for effective recruitment of corepressor (33, 34). The ROREs in the NPAS2 promoter are separated by nearly 160 bp; thus they may function as monomer binding sites. Single RORE sites can still mediate REV-ERB binding, but a REV-ERB monomer is not sufficient for active repression. However, REV-ERB can still compete for occupancy of the site for the ROR activator (33, 35, 36). Thus, REV-ERB can still block ROR-mediated activation. To test this possibility, we performed another cotransfection experiment where we added RORα to activate transcription and titrated increasing amounts of REV-ERBα. As shown in Fig. 6b, REV-ERBα effectively blocked the ability of RORα to activate transcription of the reporter in a dose-dependent manner. Additionally, this was dependent on the presence of functional ROREs because the mutant NPAS2 promoter did not respond to REV-ERBα at any concentration. Thus, it appears in the context of the NPAS2 promoter RORα functions as a transcriptional activator, but REV-ERBα may only function as an inhibitor of RORα activity by blocking binding.
DISCUSSION
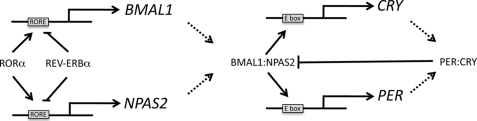
The circadian clock is essential for normal physiological function. In mammals, the master clock is located in the brain, but peripheral clocks exist in organs such as the liver. Various signals from the environment, such as light, entrain the SCN, whereas the peripheral clocks are entrained according to signals from the SCN as well as other stimuli such as nutrient status. Heterodimers of the basic helix-loop-helix transcription factors BMAL1 and CLOCK or NPAS2 play an essential role in maintaining the circadian oscillations that occur within the mammalian clock. BMAL1 is a well characterized direct target gene of RORα and REV-ERBα, and oscillations in the expression of these two nuclear receptors influence the pattern of circadian expression of BMAL1 (4, 37). It is unclear how the dimerization partner of BMAL1 is regulated in a manner that would allow coordination of the relative amounts of the partners. It is clear that coordination of expression of BMAL1 with either NPAS2 or CLOCK is essential because BMAL1 alone is not transcriptionally active. In this study, we found that NPAS2, like BMAL1, is a direct target gene of RORα and REV-ERBα. We initially identified the NPAS2 gene as a putative RORα and REV-ERBα target gene based on results from two independent ChIP/chip screens indicating both RORα and REV-ERBα occupancy of the NPAS2 promoter. We found that the NPAS2 promoter contains two functional ROREs that bind to both RORα and REV-ERBα. Our observation that NPAS2 is a target gene for RORα and REV-ERBα suggests a mechanism by which the relative levels of NPAS2 and BMAL1 can be coordinately regulated ensuring that functional heterodimer is available for maintenance of the feedback loop that is responsible for the mammalian clock (Fig. 7). However, our data suggest that this is, in fact, more complex because the ROREs found in the BMAL1 versus NPAS2 promoter display differential responsiveness to REV-ERBα. Because two REV-ERB molecules in close proximity are required to recruit corepressor, two adjacent ROREs are needed for REV-ERBα to exhibit active transcriptional repression. In the BMAL1 promoter, the proximal and distal ROREs are separated by 26 bp, and this is sufficient for REV-ERB to recruit the corepressor NCoR and actively repress BMAL1 transcription (16). In contrast, the two ROREs in the NPAS2 promoter are separated by nearly 160 bp, and our data indicate that REV-ERBα cannot effectively repress transcription in this context but can still block the action of RORα as a transcriptional activator. Additionally, our data indicate that the affinity of RORα and REV-ERBα for the NPAS2 ROREs is considerably less than for the affinity for the BMAL1 ROREp. Thus, although there is some degree of conservation of factors that regulate the expression of NPAS2 and BMAL1, there are clear distinctions in how these factors function. The biological significance of these differences is currently unclear.
FIGURE 7.
Proposed model of the circadian feedback loop illustrating coordinate regulation of BMAL1 and NPAS2 by RORα and REV-ERBα.
Our observation that NPAS2 is a REV-ERBα target gene is particularly interesting because the activities of both of these proteins have been shown to be regulated by direct binding to heme. NPAS2 contains two heme-binding domains, and heme has been shown to be necessary for binding to E-box DNA elements in target gene promoters (38, 39). Mutations in the PAS-A heme-binding domain of NPAS2 impair dimerization with BMAL1 and reduce DNA binding activity (40, 41). The heme binding domain has also been proposed to confer gas responsiveness (CO) to NPAS2. When CO is present in low micromolar concentrations, the DNA binding activity of heme-loaded NPAS2/BMAL1 heterodimers is inhibited but not for the heme-free heterodimer (42). REV-ERBα has been shown to function as a heme receptor where heme binding is necessary for the transcriptional repressor activity of the receptor (20, 43). A recent report demonstrated that REV-ERBα regulates the expression of δ-aminolevulinate synthase (ALAS1), the rate-limiting step in heme biosynthesis (44), suggesting that REV-ERB may regulate the levels of its own ligand as well as heme availability that modulates the function of one of its target genes, NPAS2. However, based on the lack of active repression of NPAS2 expression by REV-ERBα, it is unclear what the role of heme may be under these circumstances.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK080201 (to T. P. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SCN
- suprachiasmatic nucleus
- ROR
- retinoic acid receptor-related orphan receptor
- RORE
- ROR-response element
- aRTL
- Aryl hydrocarbon receptor nuclear translocator-like
- Per
- period
- Cry
- cryptochrome.
REFERENCES
- 1.Green C. B., Takahashi J. S., Bass J. (2008) Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 [DOI] [PubMed] [Google Scholar]
- 3.Reppert S. M., Weaver D. R. (2002) Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 4.Guillaumond F., Dardente H., Giguère V., Cermakian N. (2005) J. Biol. Rhythms 20, 391–403 [DOI] [PubMed] [Google Scholar]
- 5.Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A. (2000) Cell 103, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., Takahashi J. S. (1994) Science 264, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debruyne J. P., Noton E., Lambert C. M., Maywood E. S., Weaver D. R., Reppert S. M. (2006) Neuron 50, 465–477 [DOI] [PubMed] [Google Scholar]
- 8.DeBruyne J. P., Weaver D. R., Reppert S. M. (2007) Nat. Neurosci. 10, 543–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reick M., Garcia J. A., Dudley C., McKnight S. L. (2001) Science 293, 506–509 [DOI] [PubMed] [Google Scholar]
- 10.Rudic R. D., Curtis A. M., Cheng Y., FitzGerald G. (2005) Methods Enzymol. 393, 524–539 [DOI] [PubMed] [Google Scholar]
- 11.Dudley C. A., Erbel-Sieler C., Estill S. J., Reick M., Franken P., Pitts S., McKnight S. L. (2003) Science 301, 379–383 [DOI] [PubMed] [Google Scholar]
- 12.Bertolucci C., Cavallari N., Colognesi I., Aguzzi J., Chen Z., Caruso P., Foá A., Tosini G., Bernardi F., Pinotti M. (2008) Mol. Cell. Biol. 28, 3070–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunaga N., Ikeda M., Takiguchi T., Koyanagi S., Ohdo S. (2008) Hepatology 48, 240–251 [DOI] [PubMed] [Google Scholar]
- 14.Takiguchi T., Tomita M., Matsunaga N., Nakagawa H., Koyanagi S., Ohdo S. (2007) Pharmacogenet. Genomics 17, 1047–1056 [DOI] [PubMed] [Google Scholar]
- 15.Koyanagi S., Okazawa S., Kuramoto Y., Ushijima K., Shimeno H., Soeda S., Okamura H., Ohdo S. (2006) Mol. Endocrinol. 20, 573–583 [DOI] [PubMed] [Google Scholar]
- 16.Yin L., Lazar M. A. (2005) Mol. Endocrinol. 19, 1452–1459 [DOI] [PubMed] [Google Scholar]
- 17.Koyanagi S., Ohdo S. (2002) Mol. Pharmacol. 62, 1393–1399 [DOI] [PubMed] [Google Scholar]
- 18.Lavery D. J., Lopez-Molina L., Margueron R., Fleury-Olela F., Conquet F., Schibler U., Bonfils C. (1999) Mol. Cell. Biol. 19, 6488–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y. H., Alberta J. A., Gonzalez F. J., Waxman D. J. (1994) J. Biol. Chem. 269, 14681–14689 [PubMed] [Google Scholar]
- 20.Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. (2007) Nat. Struct. Mol. Biol. 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Kumar N., Solt L. A., Richardson T. I., Helvering L. M., Crumbley C., Garcia-Ordonez R. D., Stayrook K. R., Zhang X., Novick S., Chalmers M. J., Griffin P. R., Burris T. P. (2010) J. Biol. Chem. 285, 5013–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers P. M., Ying L., Burris T. P. (2008) Biochem. Biophys. Res. Commun. 368, 955–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsalobre A., Damiola F., Schibler U. (1998) Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 24.Stayrook K. R., Rogers P. M., Savkur R. S., Wang Y., Su C., Varga G., Bu X., Wei T., Nagpal S., Liu X. S., Burris T. P. (2008) Mol. Pharmacol. 73, 607–612 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Rogers P. M., Stayrook K. R., Su C., Varga G., Shen Q., Nagpal S., Burris T. P. (2008) Mol. Pharmacol. 74, 1716–1721 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Rogers P. M., Su C., Varga G., Stayrook K. R., Burris T. P. (2008) J. Biol. Chem. 283, 26332–26339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gold D. A., Baek S. H., Schork N. J., Rose D. W., Larsen D. D., Sachs B. D., Rosenfeld M. G., Hamilton B. A. (2003) Neuron 40, 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontaine C., Rigamonti E., Pourcet B., Duez H., Duhem C., Fruchart J. C., Chinetti-Gbaguidi G., Staels B. (2008) Mol. Endocrinol. 22, 1797–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ovcharenko I., Nobrega M. A., Loots G. G., Stubbs L. (2004) Nucleic Acids Res. 32, W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delerive P., Chin W. W., Suen C. S. (2002) J. Biol. Chem. 277, 35013–35018 [DOI] [PubMed] [Google Scholar]
- 32.Raspè E., Mautino G., Duval C., Fontaine C., Duez H., Barbier O., Monte D., Fruchart J., Fruchart J. C., Staels B. (2002) J. Biol. Chem. 277, 49275–49281 [DOI] [PubMed] [Google Scholar]
- 33.Harding H. P., Lazar M. A. (1995) Mol. Cell. Biol. 15, 4791–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamir I., Zhang J., Lazar M. A. (1997) Genes Dev. 11, 835–846 [DOI] [PubMed] [Google Scholar]
- 35.Forman B. M., Chen J., Blumberg B., Kliewer S. A., Henshaw R., Ong E. S., Evans R. M. (1994) Mol. Endocrinol. 8, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 36.Retnakaran R., Flock G., Giguère V. (1994) Mol. Endocrinol. 8, 1234–1244 [DOI] [PubMed] [Google Scholar]
- 37.Burris T. P. (2008) Mol. Endocrinol. 22, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukaiyama Y., Uchida T., Sato E., Sasaki A., Sato Y., Igarashi J., Kurokawa H., Sagami I., Kitagawa T., Shimizu T. (2006) FEBS J. 273, 2528–2539 [DOI] [PubMed] [Google Scholar]
- 39.Koudo R., Kurokawa H., Sato E., Igarashi J., Uchida T., Sagami I., Kitagawa T., Shimizu T. (2005) FEBS J. 272, 4153–4162 [DOI] [PubMed] [Google Scholar]
- 40.Deleted in proof
- 41.Ishida M., Ueha T., Sagami I. (2008) Biochem. Biophys. Res. Commun. 368, 292–297 [DOI] [PubMed] [Google Scholar]
- 42.Dioum E. M., Rutter J., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A., McKnight S. L. (2002) Science 298, 2385–2387 [DOI] [PubMed] [Google Scholar]
- 43.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 44.Wu N., Yin L., Hanniman E. A., Joshi S., Lazar M. A. (2009) Genes Dev. 23, 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.