Abstract
After the disappointment of clinical trials with early broad spectrum synthetic inhibitors of matrix metalloproteinases (MMPs), the field is now resurging with a new focus on the development of selective inhibitors that fully discriminate between different members of the MMP family with several therapeutic applications in perspective. Here, we report a novel class of highly selective MMP-12 inhibitors, without a phosphinic zinc-binding group, designed to plunge deeper into the S1′ cavity of the enzyme. The best inhibitor from this series, identified through a systematic chemical exploration, displays nanomolar potency toward MMP-12 and selectivity factors that range between 2 and 4 orders of magnitude toward a large set of MMPs. Comparison of the high resolution x-ray structures of MMP-12 in free state or bound to this new MMP-12 selective inhibitor reveals that this compound fits deeply within the S1′ specificity cavity, maximizing surface/volume ratios, without perturbing the S1′ loop conformation. This is in contrast with highly selective MMP-13 inhibitors that were shown to select a particular S1′ loop conformation. The search for such compounds that fit precisely to preponderant S1′ loop conformation of a particular MMP may prove to be an alternative effective strategy for developing selective inhibitors of MMPs.
Keywords: Drug Design, Matrix Metalloproteinase, Protease Inhibitor, Protein Structure, Proteolytic Enzymes, MMP-12, Macrophage Metalloelastase, Specific Inhibitor
Introduction
The association of matrix metalloproteinases (MMPs)2 with a variety of pathological states has stimulated impressive efforts over the past 20 years to develop synthetic compounds able to block efficiently (1–7) and selectively the uncontrolled activity of these enzymes (8). Extremely potent inhibitors of MMPs have been developed, but in most cases these compounds act as broad spectrum inhibitors of MMPs (9). The arguments that have been proposed to explain the difficulties in identifying inhibitors able to differentiate one MMP from another include: (a) marked sequence similarities between the catalytic domains of MMPs, (b) a well conserved enzyme active site topology (backbone RMSD between the MMP catalytic domains is 0.7–0.8 Å), and (c) the mobility of residues in the so-called S1′ specificity loop (10, 11).
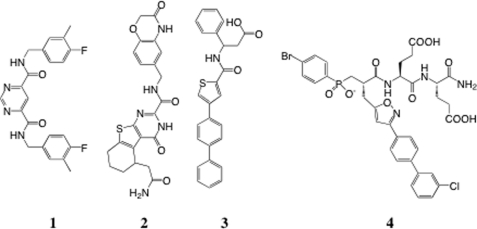
MMPs form a group of 23 proteins in humans, all of which contain a catalytic domain belonging to the zinc metalloproteinase family (12, 13). Retrospective analysis suggests that the incorporation of strong zinc-binding groups, such as hydroxamate functions, potentiates MMP inhibition but unfortunately in an indiscriminate manner affecting most members of the MMP family (7), as well as other unrelated zinc-proteinases (14). The use of a less avid zinc-binding group, like the phosphoryl group present in phosphinic peptide transition state analogs, has led to a second generation of more selective MMP inhibitors, like the selective inhibitors reported for MMP-12 (macrophage-metalloelastase) (15). The third generation MMP inhibitors possess no zinc-binding group and exploit mainly the depth of the S1′ cavity present in most MMPs (16, 17). This strategy has led to the discovery of the first extremely selective MMP-13 inhibitors, such as compound 1 (Scheme 1). The x-ray structure of MMP-13 in complex with compound 1 confirms that this compound enters deeply into the S1′ cavity of MMP-13 and involves an unusual S1′ loop conformation characterized by the presence of an additional S1′ side pocket, a feature absent from other MMP-13·inhibitor complex x-ray structures. Part of the S1′ side pocket of MMP-13 involves residues of the so-called S1′ loop, whose size and sequence varies among MMPs. The selectivity of compound 1 toward MMP-13 has been explained by the presence of Gly248, a unique feature of the S1′ loop of MMP-13 that allows it to adopt a main chain conformation that is energetically disfavored for other MMPs (16). Another selective inhibitor without zinc-binding group has also been recently reported (compound 2; Scheme 1) for the MMP-8, MMP-13 pair (18). Interestingly, the x-ray structure of 2 bound to MMP-8 again reveals the presence of an S1′ side pocket, with the distal part of 2 protruding into this pocket, in a manner similar to that observed in the MMP-13·1 complex. This suggests that the presence of an S1′ side pocket is not restricted to MMP-13 and might also occur in other MMPs through a displacement of the S1′ loop relative to the protein body. To explore this possibility, a series of compounds with no phosphinic zinc-binding group have been developed and screened against various MMPs, with the objective to identify highly selective MMP-12 inhibitors. Such attempts previously achieved only the identification of compounds with low potency toward MMP-12 and poor selectivity profile (like 3 in Scheme 1) (19).
SCHEME 1.
Chemical structures of compounds 1–4.
The drive toward the development of highly selective MMP-12 inhibitors is justified by recent studies showing the overexpression of MMP-12 in several human pathologies, such as emphysema (20), osteoarthritis (21), atherosclerosis (22), aneurisms (23), giant cell arteritis (24), and chronic obstructive pulmonary disease (25). Recently, based on a large clinical study, testing for an association between both asthma and chronic obstructive pulmonary disease and single-nucleotide polymorphisms in the gene encoding MMP-12, a detrimental role was attributed to the overexpression of MMP-12 (26). In animal models, mice deficient in MMP-12 were shown to be less susceptible to emphysema (27–29), atherosclerosis (30), and aneurisms (21). Furthermore, the need for MMP inhibitors with better selectivity profile is also justified by the poor outcomes observed in preclinical studies using broad spectrum inhibitors (31, 32) and the opposing roles that MMPs play in pathologies like cancer (33) and atherosclerosis progression (30). These studies have clearly indicated the absolute need to specifically target the MMPs involved in pathology progression and not those counteracting it. Ultimately, highly selective inhibitors are essential tools to obtain proof of principle for the efficacy of a chemical intervention in animal models, as compared with gene invalidation.
EXPERIMENTAL PROCEDURES
Chemical Synthesis
Malonic building blocks as precursors of pseudo-peptides 5–36 were first synthesized in solution and then assembled to peptide sequence on solid support. After cleavage, the resulting pseudo-peptides 5–36 were purified by preparative reverse phase HPLC, and their purity was assessed by analytical HPLC and high resolution mass spectrometry analysis. On the basis of this criteria, all of the compounds possess purity at >95%. For further details on synthesis, see supplemental “Experimental Procedures” and the analytical data of each synthesized compound (supplemental Table S2).
Enzyme Assays
All of the inhibition assays were carried out in 50 mm Tris-HCl buffer, pH 6.8, 10 mm CaCl2 at 25 °C as described previously (15, 34). ACE inhibition assays were performed using Mca-Ala-Ser-Asp-Lys-Dpa-OH as substrate (15 mm) and human somatic ACE (0.5 nm) from R & D Systems. NEP inhibition assays were performed using Mca-Arg-Pro-Pro-Gly-Phe-Ser-Pro-Dpa-OH from Enzo Life Sciences, as substrate (5 mm, Km = 2 mm) and human NEP (0.5 nm) from R & D Systems. MMPs inhibition assays were performed using Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, as substrate (13 mm, Km = 8.5 mm) and human MMPs (nanomolar range concentration) from R & D Systems (15), except for human MMP-12 that was produced and purified as described previously (35). The substrate and enzyme concentrations for the experiments were chosen so as to remain well below 10% of substrate utilization and to observe the initial rates. For each inhibitor, the percentage of inhibition was determined in triplicate experiments at five inhibitor concentrations, chosen to observe a 20–80% range of inhibition. Ki values were determined using the method proposed by Horovitz and Leviski (36) (supplemental Table S3). Continuous assays were performed by recording the fluorescence increase induced by the cleavage of fluorogenic substrates, using black, flat-bottomed, 96-well nonbinding surface plates (Corning-Costar, Schiphol-Rijk, The Netherlands). Fluorescence signals were monitored using a Fluoroscan Ascent photon counter spectrophotometer (Thermo-Labsystems, Courtaboeuf, France) equipped with a temperature control device and a plate shaker.
Crystallization
The protein inhibitor solution for crystallization consisted of 0.53 mm of the catalytic domain of the F67D mutant of human MMP12 residues 106–263 with 100 mm acetohydroxamic acid (AHA) to prevent self-degradation of the proteinase prior to crystallization in 3 mm CaCl2, 200 mm NaCl with 20 mm Tris-HCl at pH 7.5 and 10 mm of one of the four selective inhibitors (compounds 5, 9, 14, and 36 lyophilized). This solution was mixed in a ratio 1:1 with reservoir solution containing the precipitant to give drops of 2 μl (initial volume). Initial crystallization tests made use of previously published conditions and of ab initio screening with the “Stura” screens (37) (MD1–20 from Molecular Dimensions; condition 12; three-dimensional: 22.5% PEG 10,000, 200 mm imidazole malate, pH 8.5). The drops were equilibrated by sitting drop vapor diffusion for 1–4 days at 20 °C in a cooled incubator before streak seeding (38). Optimization for each inhibitor consisted of small variations in the PEG concentration and in some cases changes in ionic strength, buffer (Tris-HCl, imidazole, cacodylate, glycine) or in the molecular weight of polyethylene glycol (supplemental Table 1S). For data collection, the crystals were transferred to a cryo-protecting solution consisting of 27% PEG 8000, 15% monomethyl-PEG 550, 10% glycerol, 90 mm Tris-HCl, pH 8.0, then picked up with a loop, and cryo-cooled in liquid nitrogen.
Data Structure Determination and Refinement
The data for the MMP-12-inhibitor complexes were collected at the European Synchrotron Radiation Facility, beam lines ID14-2 and ID23-1 (Grenoble, France) or at the Soleil Synchrotron Facility (St. Aubin, France) at 100 K from single crystals. Crystals diffracted to high resolution (1.9–1.3 Å). Data reduction was carried out with MOSFLM (39). The crystals belong to the tetragonal space group P21212 with one molecule in the asymmetric unit with cell parameters similar to Protein Data Bank entry 1RMZ (11). This entry with the inhibitor removed was used for molecular replacement carried out with MOLREP (40). Subsequently structure solutions have been carried out by rigid body refinement with REFMAC (41), in all cases where lattice parameters have not varied substantially. The constraint files for the inhibitor have been generated with Monomer Library Sketcher from the CCP4 suite of programs (42). The refined ligand was then fitted in the difference electron density maps (omit σA-weighted Fo − Fc) calculated and displayed using XFIT from the XtalView suite of programs (43). In all cases a single conformation was seen for the inhibitor except for compound 5 where the chloride atom can be found in two distinct positions. The second position refined with occupancy below 0.2 was omitted from the final deposited file. Final fitting and stereochemical analysis of the refined model was carried out with COOT (44). The figures were made with PyMOL from DeLano Scientific (45).
Protein Data Bank Accession Number
The coordinates and structure factors for the catalytic subunit of human MMP12-inhibitor complexes have been deposited in the RCSB Protein Data Bank with the following codes: compound 5, code 3LIL; compound 9, code 3LIR; compound 14, code 3LJG; and compound 36, code 3LIK (supplemental Table S1).
Molecular Modeling
The molecular models of the interaction of compound 36 with MMPs was achieved with program CHARMM (version 27), using version 22. The initial position of compound 36 in MMP active sites was obtained by superimposition of the main chain atoms of the MMP-12·36 complex crystal structure onto that of other crystal structures of MMPs (Protein Data Bank code for MMP-2 (code 1QIB), MMP-3 (code 1G49), MMP-8 (code 1I76), MMP-9 (code 2OVZ), and MMP-13 (code 3I7G), respectively. The geometrical and nonbonded parameters for compound 36 were derived from ab initio quantum calculations with the program GAUSSIAN98. These calculations were done at the MP2 level of theory using a 6–31+G(d,p) basis set. The starting complex structures were then refined by energy minimization and molecular dynamics with CHARMM (15).
RESULTS
From Phosphinic Inhibitors to Non-zinc Binding Inhibitors
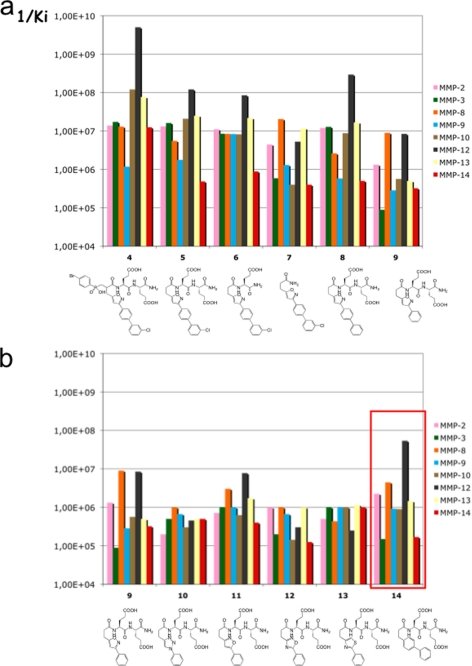
We previously reported compound 4 (RXP470; Scheme 1) as a potent and selective inhibitor of MMP-12, in which the isoxazole side chain in conjunction with the Glu-Glu motif was shown to play a key role in the selectivity of the inhibitor (15). In 4, the phosphoryl group (PO2−) is thought to interact with the zinc ion of the MMP-12 active site, as previously observed in the x-ray structure of complexes between phosphinic peptide inhibitors and other MMP members (46, 47). Starting from compound 4, a series of new compounds was generated by removal of the phosphinic moiety (Fig. 1a) as in compound 5, resulting in a concomitant loss in potency and selectivity. Similar trends were observed when either the Glu-Glu motif (compounds 6 and 7) or the isoxazole side chain (compounds 8 and 9) were modified. These first results led us to evaluate the role of the isoxazole ring trough its substitution by other five-member rings (compounds 10–13). As shown in Fig. 1b, with the exception of compound 11, a loss of potency was observed highlighting the role played by hetero-atoms in these five-member rings. However, moving to a six-member phenyl ring resulted in compound 14 with a better selectivity profile in favor of MMP-12.
FIGURE 1.
a, effects of the RXP470 deconstruction; b, the nature of proximal ring on the MMP selectivity profile. For each MMP, the 1/Ki values are reported.
From Isoxazole to Phenyl Ring in the P1′ Position
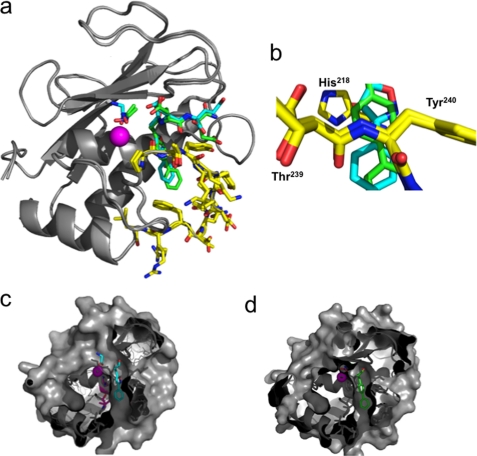
To gain some insights on the molecular factors contributing to the change in selectivity profile between compounds 9 and 14, x-ray structures of their corresponding complexes with the MMP-12 catalytic domain were determined at high resolution, 1.9 and 1.31 Å, respectively (supplemental Table S1 and text). In these structures (Fig. 2a), the zinc atom is chelated respectively by a glycine (compound 9) or an acetohydroxamic acid (AHA) molecule (compound 14) (the presence of these small zinc-binding molecules originates from the co-crystallization buffer in the case of glycine or the addition of AHA to prevent MMP-12 autolysis, see “Experimental Procedures”). Overall, these structures confirm that the P1′, P2′, and P3′ side chains in these inhibitors point toward their corresponding enzyme subsites (S1′, S2′, and S3′), with no direct interaction between the zinc ion and inhibitor atoms. This observation is sustained by the lack of electron density between glycine or AHA and compounds 9 and 14, as well as the distances between the P1′ Ca inhibitor atoms and the zinc ion (4.4 and 4.5 Å, respectively). Residues of the S1′ loop in both structures adopted almost similar orientations (Fig. 2a, yellow stick). However, because of the change in ring size, in 14 the P1′ side chain enters more deeply into the S1′ cavity, compared with 9 (Fig. 2, b–d). As a result of the phenyl shift, a better stacking is achieved between the first phenyl moiety of inhibitor 14 and His218 side chain on one side and on the other side with the peptide bond located between Thr239 and Tyr240 (Fig. 2b). As expected, the S1′ cavity in these structures appears to be partially empty, suggesting that the bottom part of the S1′ cavity could be further probed by adding groups to the para or meta position on the second phenyl group in 14 (Fig. 2d).
FIGURE 2.
a, overlay of MMP-12 complexes with compounds 9 and 14. MMP-12 is shown in ribbon representation (gray) with the active site zinc in purple. Compounds 9 and 14 and AHA are shown as sticks with nitrogen (blue) and oxygen (red), and carbon atoms are colored according to the compound (9, cyan; 14, green). S1′ loop residues are highlighted as sticks with carbon atoms in yellow. b, detailed view at the S1′ cavity entrance, showing the respective positioning of inhibitor side chain in respect to His218 and the peptide bond located between Thr239 and Tyr240. Cross-section of MMP-12 molecular surface with compound 9 (c) and 14 (d) showing the positioning of the inhibitor side chain into the S1′ cavity.
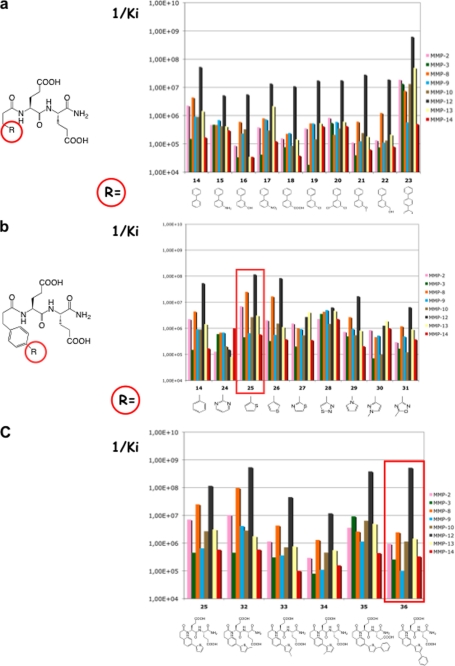
Based on the above considerations, another set of compounds was designed by introducing various small substituents on the distal phenyl group of compound 14. This series did not lead to major improvements in inhibitory potency (Fig. 3a). Some compounds (compounds 18 and 21) displayed an improved selectivity profile, but their potency toward MMP-12 remained too low (Ki = ∼100 nm). Further elongation of the P1′ side chain by adding a third phenyl strongly increased the inhibitor potency (compound 23) but at the expense of inhibitor selectivity. These results led us to explore alternative cycles in the distal part of the P1′ side chain (Fig. 3b). With the exception of a thiophene ring (compounds 25 and 26), most compounds in this series exhibited lower potency compared with 14.
FIGURE 3.
a, effects of the distal phenyl modification. b, nature of distal ring. c, thiophene modification on the MMP selectivity profile.
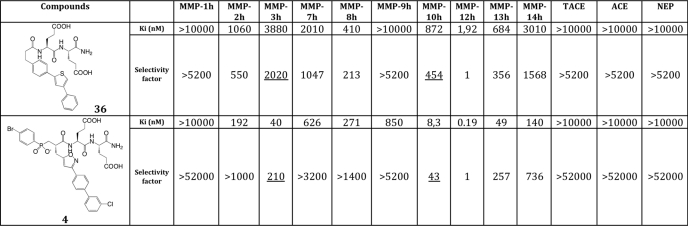
For chemistry considerations, compound 25 was selected for further modification (Fig. 3c). The introduction of a methyl at the α position relative to the sulfur atom in the thiophene ring led to marked changes both in potency and selectivity toward MMP-12 (compound 32). In contrast, the same modification at β or γ position of the thiophene sulfur atom decreased potency (compounds 33 and 34). With a phenyl, the β substitution (compound 36) provided better results than the α one in terms of selectivity (compound 35) (the γ position was discarded in this case to avoid steric clash within the S1′ cavity). As compared with our starting compound (compound 4, RXP470), compound 36 is slightly less potent toward MMP-12 and displays overall a similar selectivity profile toward a set of nine MMPs (Table 1), except for MMP-3 and MMP-10, where 36 is 10-fold less potent as compared with 4. No inhibition of TACE (TNF-α-converting enzyme), ACE, and NEP was observed when 36 was tested up to a 10 mm concentration.
TABLE 1.
Comparison of compounds 36 and 4 potency towards MMPs and their selectivity factors towards MMP-12
Ki values were determined in 50 mm Tris/HCl buffer, pH 6.8, 10 mm CaCl2 at 25 °C.
Crystal Structure of Compound 36 Bound to MMP-12
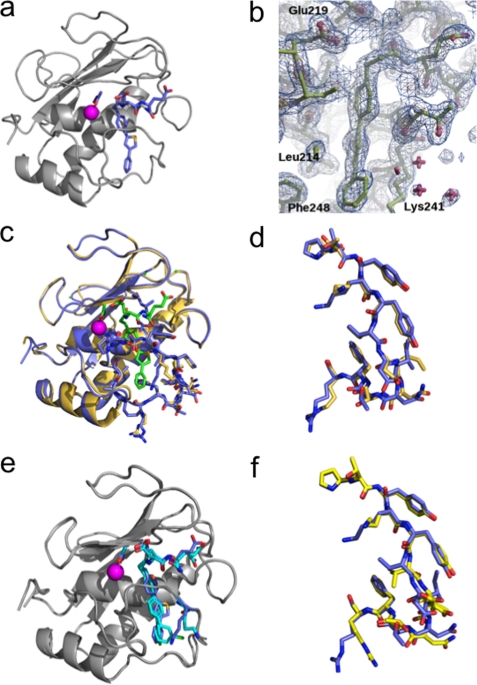
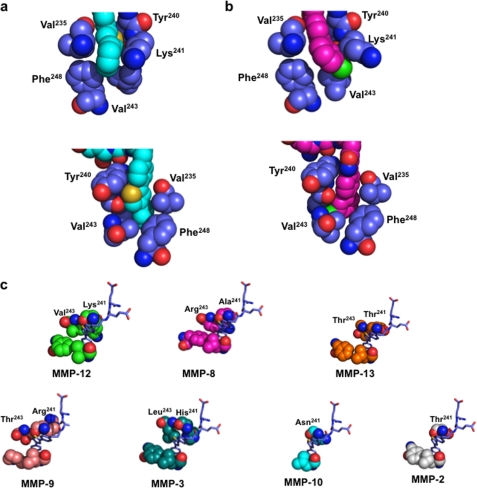
A high resolution x-ray structure of MMP-12 in complex with inhibitor 36 was solved in the presence of AHA (1.8 Å). In this complex, 36 adopts a very well defined structure (Fig. 4, a and b), with improved complementary fit between the P1′ side chain of the inhibitor and the S1′ cavity. Compounds 14 and 36 possess in common the Glu-Glu motif and the first phenyl ring. Comparison of their x-ray structures in interaction with MMP-12 revealed that these parts shared by the two inhibitors adopt almost the same binding mode. However, 36 achieves a larger number of Van der Waal's contacts between the distal part of the inhibitor (thiophene-phenyl) and several MMP-12 residues (Val235, Tyr240, Lys241, Val243, and Phe248) (Fig. 5a). This optimal accommodation relies on the bent structure of the inhibitor P1′ side chain induced by the thiophene ring. The above observations led us to wonder whether the inhibitor P1′ side chain position in the S1′ cavity might induce the concomitant conformational shift of the S1′ loop. To address this issue, we compared the crystal structures of MMP-12 in complex with 36 in the presence of AHA and MMP-12 complexed to AHA (Protein Data Bank code 1Y93, 1.03 Å) (11). The two MMP-12 structures overlay with only minor differences in their S1′ loop conformations, with the exception of Lys241 side chain (Fig. 4, c and d; RMSD in figure legend). In presence of 36, the Lys241 side chain appears to fold back over the distal phenyl of 36 (Figs. 4c and 5a), with the ϵ CH2 of Lys241 making a CH-π interaction with the phenyl ring of the inhibitor, whereas in the MMP-12·AHA complex, the lysine side chain points toward the solvent (Fig. 4c). Overall, this comparison supports the view that the accommodation of the 36 P1′ side chain into the S1′ cavity can be realized without major rearrangement of the MMP-12 S1′ loop canonical conformation.
FIGURE 4.
a, complex of MMP-12 with compound 36 in presence of AHA. Carbon atoms of 36 and AHA are shown as sticks colored light blue. b, 2Fobs − Fcalc electron density map (1σ, blue; 4σ, brown) of the inhibitor binding region. c, overlay between complexes of MMP-12 with AHA (beige) or with 36 in presence of AHA (blue). AHA and 36 are shown as stick models with their carbon atoms in green. The S1′ loop of each complex is emphasized in sticks for each complex: AHA/MMP12 (beige; Protein Data Bank code 1Y93) and 36·MMP-12 in presence of AHA (blue). The RMSD on Cα for this superposition is 0.43 Å. d, a close-up view of S1′ loop (residues 236–249) as in c (RMSD for this segment: 0.31 Å on Cα; 1.03Å for all atoms). e, overlay between complexes of MMP-12 with 5 (cyan) or with 36 (blue) in presence of AHA. The RMSD on Cα for the superposition is 0.35Å. f, a close-up view of the S1′ loop in complexes between MMP-12 and 5 (yellow stick) or with 36 (blue stick). The RMSD for the superposition of residues 236–249 is 0.61 Å on Cα and 1.44 Å for all atoms).
FIGURE 5.
a and b, close-up views of the S1′ bottom part of MMP-12 in complex with 36 in front (top) and back (bottom) view (a) or in complex with 5 in front (top panels) and back (bottom panels) view (b). 36 is colored in cyan, 5 is in magenta, and MMP-12 residues are in blue (ball-stick representation). c, a close-up view of MMP·36 models showing the S1′ bottom part of MMPs and the packing between the P1′ side chain of 36 and the MMP S1′ loop residues.
Compound 5 versus 36
Compounds 5 and 36 incorporate P1′ side chains of similar size but display markedly different selectivity profiles. To gain some insights into the molecular determinants that may explain this difference, x-ray structures of 5 in complex with MMP-12 were also solved in the presence of AHA (1.8 Å). Overall, in these two MMP-12 crystal structures, the protein adopts almost the same structure (Fig. 4, e and f), but on closer inspection the differences are observed between the S1′ loop residue conformation (Fig. 4f) and in the packing of the P1′ side chains of compounds 5 and 36 and residues within the MMP-12 S1′ cavity (Fig. 5, a and b). In the MMP12·5 complex, the distal chloro-phenyl group points toward Lys241, pushing the lysine side chain toward the solvent (Fig. 5b). In contrast, in the MMP12·36 complex, the distal phenyl points in the opposite direction, toward Phe248 (Fig. 5a). Overall, the presence of a five-member ring in 36 allows better packing between the inhibitor P1′ side chain and S1′ residues (Tyr240 and Lys241), compared with the P1′ side chain of 5, so that the void volume between the inhibitor P1′ side chain and the S1′ cavity appears lower with 36, as compared with 5 (Fig. 5, a and b). These differences may explain the higher potency of 36 toward MMP-12 and its better selectivity profile. To support this contention, we tested whether the structure adopted by 36 when bound to MMP-12 might fit in the active sites of other MMPs. Strikingly, 36 can be easily docked into other MMP active sites, without steric clash. However, these simple models reveal that the optimal close contacts observed between the distal part of the P1′ side chain of 36 and the five residues of the MMP-12 delineating the bottom part of the S1′ cavity (Val235, Tyr240, Lys241, Val243, and Phe248), with the exception of MMP-8, are not observed in other MMPs (Fig. 5c) (we selected MMPs for which the inhibitor selectivity has been improved between compound 5 and 36). Besides differences in the S1′ loop conformation between MMPs, the above observations also arise from the variability in the nature of the residues at positions 241 and 243 in various MMPs (Fig. 5c). From the above considerations, the poor selectivity displayed by compound 5 may arise from the linear shape of its P1′ side chain, as compared with the bent P1′ side chain structure of 36.
DISCUSSION
The development of most MMP inhibitors for the last 15 years has relied on the use of a strong zinc-binding group, such as hydroxamate, and by targeting the entrance of the S1′ cavity. This strategy was selected to obtain potent inhibition through tight hydroxamate/zinc ion interaction and maintaining the inhibitor in a low molecular weight range by limiting the size of the P1′ side chain. Unfortunately, although providing extremely potent MMP inhibitors, all of the compounds obtained through this strategy displayed poor selectivity toward MMPs and also targeted other unrelated zinc-metalloproteinases like NEP (14). The use of a weaker zinc-binding group (carboxylate or phosphoryl group) led to more selective inhibitors, in particular for MMP-12 (15, 48, 49). First, the present study demonstrates that pseudo-peptides with no phosphinic zinc-binding group in their structures can behave as potent MMP-12 inhibitors. Second, this study shows that exploring the bottom part of the S1′ cavity of MMP-12 by using long and bent P1′ side chain resulted in inhibitors showing even better selectivity profile than the best selective MMP-12 reported so far (compound 4) (15). Third, this study indicates that the inhibitor P1′ side chain accommodation into the S1′ cavity occurs without major conformational shift of the S1′ loop, in contrast to what was observed with MMP-13 selective inhibitors (16) and mixed MMP13/MMP-8 inhibitors (18). Selective MMP-13 inhibitors have been proposed to select a particular/minor conformation of MMP-13 from a conformational ensemble. Even if this concept seems to apply also to MMP-8, noncanonical S1′ loop conformation in MMPs is poorly documented; thus translation of this approach for developing selective inhibitor toward other MMPs approach remains challenging. In contrast a lot of three-dimensional structures (x-ray or NMR) of MMPs are available in which a preferred S1′ loop conformation can be potentially targeted by inhibitors probing the bottom part of the S1′ cavity for developing more selective MMP inhibitors. Although this strategy is appealing, the present study shows that subtle variations in the inhibitor structure entail profound selectivity and potency variations. Potency and selectivity are in fact a function of several linked and complex parameters. Thus, the exact positioning of the P1′ side chain inside the S1′ cavity seems to be critical. Such positioning depends on the nature of the ring (isoxazole versus phenyl) in the inhibitor P1′ side chain at the entrance of S1′ cavity. The shape of the P1′ side chain is also important. A long P1′ linear side chain like in 23 strongly increases the inhibitor potency but yields a poor selectivity profile. Only with a bent side chain have the best results in terms of selectivity been achieved. Compounds 5 and 36 highlight how very subtle structural differences act in synchrony to yield a different selectivity profile. The small adjustments of the residues lining the S1′ cavity in response to inhibitor binding are difficult to predict, yet such knowledge could potentiate the use of structure-based design approach to develop highly selective inhibitors. These limitations explain why the development of selective MMP inhibitors has been so disappointing in the past 20 years.
In summary, the present study reports how the systematic modification of the P1′ side chain has resulted in the identification of selective inhibitors. The absence of a zinc-binding group has facilitated the discovery process by strengthening the role played by the P1′ side chain and by removing the positional constraints associated with the zinc interaction. Considering the previous successes obtained with MMP-13, it appears extremely likely that more non-zinc-binding inhibitors of MMPs with high selectivity profile will be reported in the future. Whether such compounds will select highly or less populated S1′ loop conformations of MMPs is more difficult to foresee.
Supplementary Material
This work was supported by funds from the Commissariat à l'Energie Atomique.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, and Schemes 1S and 2S.
- MMP
- matrix metalloproteinase
- ACE
- angiotensin-converting enzyme
- NEP
- neprilysin
- AHA
- acetohydroxamic acid
- RMSD
- root mean square deviation.
REFERENCES
- 1.Babine R. E., Bender S. L. (1997) Chem. Rev. 97, 1359–1472 [DOI] [PubMed] [Google Scholar]
- 2.Whittaker M., Floyd C. D., Brown P., Gearing A. J. (1999) Chem. Rev. 99, 2735–2776 [DOI] [PubMed] [Google Scholar]
- 3.Cuniasse P., Devel L., Makaritis A., Beau F., Georgiadis D., Matziari M., Yiotakis A., Dive V. (2005) Biochimie 87, 393–402 [DOI] [PubMed] [Google Scholar]
- 4.Fisher J. F., Mobashery S. (2006) Cancer Metastasis Rev. 25, 115–136 [DOI] [PubMed] [Google Scholar]
- 5.Nuti E., Tuccinardi T., Rossello A. (2007) Curr. Pharm. Des. 13, 2087–2100 [DOI] [PubMed] [Google Scholar]
- 6.Tu G., Xu W., Huang H., Li S. (2008) Curr. Med. Chem. 15, 1388–1395 [DOI] [PubMed] [Google Scholar]
- 7.Yiotakis A., Dive V. (2008) Mol. Aspects Med. 29, 329–338 [DOI] [PubMed] [Google Scholar]
- 8.Fingleton B. (2007) Curr. Pharm. Des. 13, 333–346 [DOI] [PubMed] [Google Scholar]
- 9.Brown S., Meroueh S. O., Fridman R., Mobashery S. (2004) Curr. Top. Med. Chem. 4, 1227–1238 [DOI] [PubMed] [Google Scholar]
- 10.Moy F. J., Chanda P. K., Chen J., Cosmi S., Edris W., Levin J. I., Rush T. S., Wilhelm J., Powers R. (2002) J. Am. Chem. Soc. 124, 12658–12659 [DOI] [PubMed] [Google Scholar]
- 11.Bertini I., Calderone V., Cosenza M., Fragai M., Lee Y. M., Luchinat C., Mangani S., Terni B., Turano P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5334–5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bode W., Maskos K. (2003) Biol. Chem. 384, 863–872 [DOI] [PubMed] [Google Scholar]
- 13.Tallant C., Marrero A., Gomis-Rüth F. X. (2010) Biochim. Biophys. Acta 1803, 20–28 [DOI] [PubMed] [Google Scholar]
- 14.Saghatelian A., Jessani N., Joseph A., Humphrey M., Cravatt B. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devel L., Rogakos V., David A., Makaritis A., Beau F., Cuniasse P., Yiotakis A., Dive V. (2006) J. Biol. Chem. 281, 11152–11160 [DOI] [PubMed] [Google Scholar]
- 16.Engel C. K., Pirard B., Schimanski S., Kirsch R., Habermann J., Klingler O., Schlotte V., Weithmann K. U., Wendt K. U. (2005) Chem. Biol. 12, 181–189 [DOI] [PubMed] [Google Scholar]
- 17.Johnson A. R., Pavlovsky A. G., Ortwine D. F., Prior F., Man C. F., Bornemeier D. A., Banotai C. A., Mueller W. T., McConnell P., Yan C., Baragi V., Lesch C., Roark W. H., Wilson M., Datta K., Guzman R., Han H. K., Dyer R. D. (2007) J. Biol. Chem. 282, 27781–27791 [DOI] [PubMed] [Google Scholar]
- 18.Pochetti G., Montanari R., Gege C., Chevrier C., Taveras A. G., Mazza F. (2009) J. Med. Chem. 52, 1040–1049 [DOI] [PubMed] [Google Scholar]
- 19.Morales R., Perrier S., Florent J. M., Beltra J., Dufour S., De Mendez I., Manceau P., Tertre A., Moreau F., Compere D., Dublanchet A. C., O'Gara M. (2004) J. Mol. Biol. 341, 1063–1076 [DOI] [PubMed] [Google Scholar]
- 20.Lagente V., Le Quement C., Boichot E. (2009) Expert Opin. Ther. Targets 13, 287–295 [DOI] [PubMed] [Google Scholar]
- 21.Liu M., Sun H., Wang X., Koike T., Mishima H., Ikeda K., Watanabe T., Ochiai N., Fan J. (2004) Arthritis Rheum. 50, 3112–3117 [DOI] [PubMed] [Google Scholar]
- 22.Halpert I., Sires U. I., Roby J. D., Potter-Perigo S., Wight T. N., Shapiro S. D., Welgus H. G., Wickline S. A., Parks W. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9748–9753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curci J. A., Liao S., Huffman M. D., Shapiro S. D., Thompson R. W. (1998) J. Clin. Invest. 102, 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Pla A., Martínez-Murillo F., Savino P. J., Eagle R. C., Jr., Seo P., Soloski M. J. (2009) Rheumatology 48, 1460–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demedts I. K., Morel-Montero A., Lebecque S., Pacheco Y., Cataldo D., Joos G. F., Pauwels R. A., Brusselle G. G. (2006) Thorax 61, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunninghake G. M., Cho M. H., Tesfaigzi Y., Soto-Quiros M. E., Avila L., Lasky-Su J., Stidley C., Melén E., Söderhäll C., Hallberg J., Kull I., Kere J., Svartengren M., Pershagen G., Wickman M., Lange C., Demeo D. L., Hersh C. P., Klanderman B. J., Raby B. A., Sparrow D., Shapiro S. D., Silverman E. K., Litonjua A. A., Weiss S. T., Celedón J. C. (2009) N. Engl. J. Med. 361, 2599–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hautamaki R. D., Kobayashi D. K., Senior R. M., Shapiro S. D. (1997) Science 277, 2002–2004 [DOI] [PubMed] [Google Scholar]
- 28.Morris D. G., Huang X., Kaminski N., Wang Y., Shapiro S. D., Dolganov G., Glick A., Sheppard D. (2003) Nature 422, 169–173 [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., Mizuno-Horikawa Y., Nakano M., Asahi M., Takahashi M., Uozumi N., Ihara S., Lee S. H., Ikeda Y., Yamaguchi Y., Aze Y., Tomiyama Y., Fujii J., Suzuki K., Kondo A., Shapiro S. D., Lopez-Otin C., Kuwaki T., Okabe M., Honke K., Taniguchi N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15791–15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson J. L., George S. J., Newby A. C., Jackson C. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15575–15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coussens L. M., Fingleton B., Matrisian L. M. (2002) Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 32.Overall C. M., Kleifeld O. (2006) Br. J. Cancer 94, 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Otín C., Palavalli L. H., Samuels Y. (2009) Cell Cycle 8, 3657–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jullien N., Makritis A., Georgiadis D., Beau F., Yiotakis A., Dive V. (2010) J. Med. Chem. 53, 208–220 [DOI] [PubMed] [Google Scholar]
- 35.Dabert-Gay A. S., Czarny B., Devel L., Beau F., Lajeunesse E., Bregant S., Thai R., Yiotakis A., Dive V. (2008) J. Biol. Chem. 283, 31058–31067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horovitz A., Levitzki A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6654–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stura E. A., Nemerow G. R., Wilson I. A. (1992) J. Cryst. Growth 122, 273–285 [Google Scholar]
- 38.Stura E. A., Wilson I. A. (1991) J. Cryst. Growth 110, 270–282 [Google Scholar]
- 39.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 40.Vagin A. A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 41.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 43.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 46.Gall A. L., Ruff M., Kannan R., Cuniasse P., Yiotakis A., Dive V., Rio M. C., Basset P., Moras D. (2001) J. Mol. Biol. 307, 577–586 [DOI] [PubMed] [Google Scholar]
- 47.Tochowicz A., Maskos K., Huber R., Oltenfreiter R., Dive V., Yiotakis A., Zanda M., Pourmotabbed T., Bode W., Goettig P. (2007) J. Mol. Biol. 371, 989–1006 [DOI] [PubMed] [Google Scholar]
- 48.Li W., Li J., Wu Y., Rancati F., Vallese S., Raveglia L., Wu J., Hotchandani R., Fuller N., Cunningham K., Morgan P., Fish S., Krykbaev R., Xu X., Tam S., Goldman S. J., Abraham W., Williams C., Sypek J., Mansour T. S. (2009) J. Med. Chem. 52, 5408–5419 [DOI] [PubMed] [Google Scholar]
- 49.Li W., Li J., Wu Y., Wu J., Hotchandani R., Cunningham K., McFadyen I., Bard J., Morgan P., Schlerman F., Xu X., Tam S., Goldman S. J., Williams C., Sypek J., Mansour T. S. (2009) J. Med. Chem. 52, 1799–1802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.