Abstract
Chromosome translocations are gross chromosomal rearrangements that have often been associated with cancer development in mammalian cells. The feasibility of drastically reshaping the genome with a single translocation event also gives this molecular event a powerful capacity to drive evolution. Despite these implications and their role in genome instability, very little is known about the molecular mechanisms that promote and accompany these events. Here, at the molecular level, we describe 10 morphologically and physiologically different translocants ensuing from the induction of the same bridge-induced translocation (BIT) event in the budding yeast Saccharomyces cerevisiae. We have demonstrated that, despite their common origin from the integration of the same linear DNA construct, all 10 translocation mutant strains have different phenotypes and the ability to sporulate and regulate gene expression and morphology. We also provide insights into how heterogeneous phenotypic variations originate from the same initial genomic event. Here we show eight different ways in which yeast cells have dealt with a single initial event inducing translocation. Our results are in agreement with the formation of complex rearrangements and abnormal karyotypes described in many leukemia patients, thus confirming the modellistic value of the yeast BIT system for mammalian cells.
TRANSLOCATIONS between nonhomologous chromosomes are some of the most severe genomic aberrations, which in higher eukaryotes often lead to malignant transformation. In humans, they have been associated with hematological cancers such as myelogenous leukemia but also with lymphomas, solid tumors, and recently, with mesenchymal and epithelial cancers (Dalla-Favera et al. 1982; Nowell 1988; Rowley 2001, 2008; Taki and Taniwaki 2006; Gasparini et al. 2007; Nickoloff 2008). In the past few decades, molecular and cytological studies have demonstrated that different chromosomal abnormalities such as aneuploidy can be associated with a translocation event in cancer cells. For example, acute transformation of leukemia patients is often associated with the duplication of the Philadelphia chromosome, trisomy 8 and isochromosome 17q, or other complex chromosomal rearrangements (Muehleck et al. 1984; Bernstein 1988; De Braekeleer 2007). Furthermore, unbalanced translocations can be detected in older leukemia patients. Karyotypic analyses have revealed that unbalanced translocations can advance into further rearrangements of the chromosomes involved in the translocations. This can give rise to complex and abnormal karyotypes characterized by monosomy, disomy, and trisomy for different segments of the translocation participant chromosomes (Pedersen et al. 2000).
Many molecular studies have been performed to better understand the formation of such severe chromosomal aberrations (Kanaar et al. 1998; Delneri et al. 2003; Aylon and Kupiec 2004; Egli et al. 2004; Motegi et al. 2006; Weinstock et al. 2006; Motegi and Myung 2007). Nevertheless, very little is known about the mechanisms by which chromosome translocations and secondary rearrangements arising from these events occur. The occurrence of two or more double-strand breaks (DSB) and an inappropriate use of the recombination machinery of the cells are supposed to be some of the main ways in which a DNA translocation is promoted (Rowley 2001, 2008; Agarwal et al. 2006). In humans, defects in many DSB repair mechanisms such as non-allelic homologous recombination, non-homologous end-joining, and fork stalling and template switching (FoSTeS) have been reported to be involved in translocation formation (Gu et al. 2008). In yeast cells, single-strand annealing and break-induced replication (BIR) pathways have been shown to be involved in the formation of chromosome rearrangements and translocations (Bosco and Haber 1998; Haber 2006).
Recently, our group described a method that allows the generation and subsequent selection of chromosomal translocations between any two chromosomal loci in diploid yeast strains. This was done by transformation of whole yeast cells with a linear DNA construct, carrying the KANR selectable marker flanked by DNA sequences homologous to two different chromosomal loci (Tosato et al. 2005, 2009; Nikitin et al. 2008).
We used this method to investigate the multiple molecular mechanisms and pathways that might be involved during a translocation event in Saccharomyces cerevisiae. To this end, we generated a collection of 10 different mutant strains harboring a translocation between chromosomes XVI and IX (Figure 1), followed by their comparative molecular and physiological analyses. Results obtained from our experiments suggest that the induction of the same chromosomal translocation can lead to a wide variety of secondary chromosomal rearrangements, generating aneuploidy derived from partial or complete chromosome duplication or loss of genetic material.
Figure 1.—
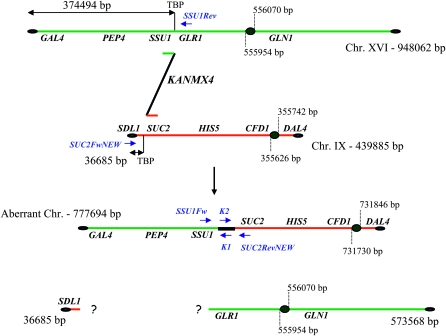
Schematic of BIT between chromosomes XVI and IX. The 777,694-bp aberrant chromosome together with the two hypothetic fragments originating after a non-reciprocal BIT translocation are shown. Genes analyzed by Southern hybridization are reported along the chromosomes together with the primers used to test the integration of the BIT construct at the level of the two targeted loci. SSU1Fw and SUc2RevNEW primers were used to perform the bridge-PCR to validate the presence of the translocant chromosome (see results). TBP, translocation breakpoint.
Physiological analyses also revealed that the 10 translocants obtained with the bridge-induced translocation (BIT) system in this work have different mutant phenotypes, which are analogous to the high variation of phenotypes characteristic of cancer cells. We demonstrated that these mutant strains exhibit altered behavior and fitness in different carbon sources for growth, different sporulation efficiencies, and ability to flocculate. Expression analyses also show that they exhibit different expression profiles of various genes located along the translocated chromosome or involved in cellular processes such as apoptosis, cell cycle regulation, and oxidative stress response. Overall, our work shows that the single integration of a linear DNA cassette with homology on two different chromosomes not only can generate a translocation, but also might be responsible for other complex chromosomal rearrangements. Such complex genomic rearrangements seen in yeast may play a role as key evolutionary forces, reshaping and remodeling genomes, followed by selection and adaptation into specialized cells or even neoplastic transformation, as observed in mammalian cancer cells.
MATERIALS AND METHODS
Strains:
The diploid S. cerevisiae strain San1 (Waghmare et al. 2003) was used as the recipient strain to induce the translocation between chromosomes XVI and IX. San1 was also used as the control strain throughout this work.
The SUSU translocants are all derivatives of San1 and were obtained using the BIT system (Tosato et al. 2005). The BIT cassette was amplified using the primers Fw(XVI) and Rev(IX) reported in the supporting information, Table S1A. Transformants were selected as kanamycin-resistant (KanR) strains, and the presence of the translocation between the SSU1 and SUC2 loci was monitored by PCR and Southern blot analysis. The two diploid mutant strains San1-DBP1/dbp1∷KANMX4 and San1-GUT2/gut2∷KANMX4 used in CHEF experiments are both derivatives of San1, and they were obtained using the standard PCR-based gene knockout technique. ΔFDBP1 (forward) and RKANDBP1 (reverse) primers were used for DBP1 deletion cassette amplification, and GUT2ΔFw (forward) and GUT2ΔrevNEW (reverse) for GUT2 deletion cassette amplification (Table S1C). Primers described in Table S1C were used to confirm the deletion in the transformants by PCR using genomic DNA. Strains I-2 and I-6 are derivatives of San1 carrying the KANMX4 insertion in chromosome XVI at the translocation breakpoint. Strains I-11 and I-17 carry the same on chromosome IX. Primers used to amplify PCR-based integration cassettes for chromosome XVI and IX are reported in Table S1A, Table S1B, and Table S1C.
Growth conditions:
All S. cerevisiae strains were grown at 30° in rich liquid or solid medium (YPD; Difco, Detroit). Geneticin G418 was added to select transformants and to grow the SUSU translocants (final concentration 200 μg/ml; Gibco, Rockville, MD). Raffinose [2% w/v, yeast extract, peptone, raffinose (YPR)] and glycerol (2% w/v, YPG) media were used to test the growth rate of the translocants. SPIII and SPII media, supplemented with amino acids, were used for sporulation tests (Kaiser et al. 1994).
The standard growth conditions to prepare samples for RT–PCR were as follows: cells were cultured in 100-ml baffled flasks with 10 ml medium in a shaking incubator at 30° and at 160 rpm. After 18 hr of preculturing in YPD, the cells were washed with sterile water and inoculated (105 cells/ml) in 250-ml flasks containing 20 ml of fresh YPD medium. After 16 hr of growth, cells were centrifuged and then resuspended in 1 ml of water. A volume corresponding to 0.3–0.5 OD600 of this suspension was then transferred to a new 250-ml flask containing 15 ml of fresh YPD or YPR medium. Cultures were further incubated for an additional 3 hr and then harvested for total RNA extraction.
Sporulation efficiency:
The sporulation efficiency of all SUSU translocants grown in sporulation medium for 4–5 days was calculated as the percentage of the four-spored asci among the total number of cells counted under the microscope and compared with the San1 sporulation efficiency considered as 100%.
Microscopy:
DAPI staining:
A single isolated colony was grown in liquid rich medium at 30°. After 16, 40, or 72 hr growth, 1 ml of the culture was harvested and cells were stained with DAPI as described (Nikitin et al. 2008). A Leica DMLB fluorescence microscope was used to monitor DAPI-stained images of yeast cells.
FUN-1 staining:
A single isolated colony was inoculated in a 100-ml flask containing 10 ml YPD medium and incubated at 30° for 18 hr to reach late log phase (107 cells/ml). A total of 200 μl of the cell suspension was then stained with FUN-1 dye diluted at an optimal concentration of 12.5 or 6.3 μm following the standard protocol suggested by the supplier (Molecular Probes, Eugene, OR). Cells were visualized under fluorescence using fluorescein and rhodamin filters. At least 300 cells were counted, and the ratio between metabolically inactive/dead cells (yellow) and metabolically active/alive cells [forming cylindrical red intravascular structures, or CIVSs (Millard et al. 1997)] was expressed as the percentage of cells that appeared yellow.
Plasmids and transformations:
Plasmid pFA6-KanMX4 containing the KANMX4 gene (Oka et al. 1981) was used as template for the amplification of the SUSU-BIT cassette. Transformation of S. cerevisiae was performed by the lithium acetate (LiAC) method following the EUROFAN protocol for PCR-based gene replacements (Wach et al. 1994) or by the spheroplast method following the instruction reported in Kaiser et al. (1994).
Molecular biology techniques:
Standard recombinant DNA techniques were carried out essentially according to Sambrook et al. (1989). Genomic DNA purification kits and plasmid mini-preparation kits were obtained from Promega (Madison, WI). Restriction enzymes and biochemicals, obtained from New England Biolabs (Beverly, MA), were used according to the manufacturer's instructions.
Semiquantitative RT–PCR analysis:
Total RNA was isolated from cells using the SV Total RNA Isolation System (Promega) or the Total Quick RNA kit (Talent, Trieste, Italy). cDNA synthesis and semiquantitative PCR based on limiting the number of cycles (20–23 cycles) were performed as described (Nikitin et al. 2008).
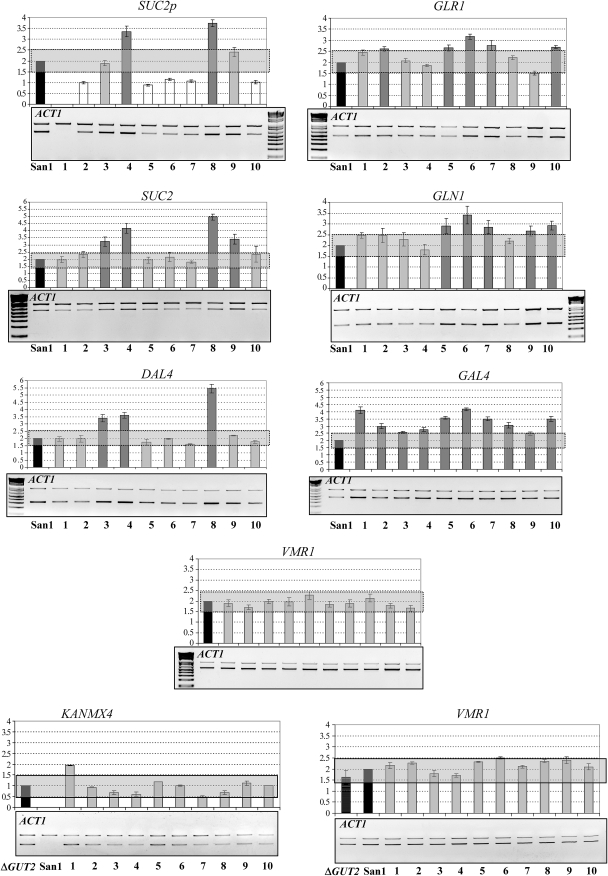
RT–PCR results were quantified using a laser-scanning densitometry (UltroScan XL, Pharmacia LKB), and the intensity of each band (area) was calculated as the average of three different measures. The expression of the housekeeping gene HSC82, located on chromosome VIII, was used to normalize the data. Data presented in Figure 8 and Figure S4 are averages with standard errors from at least three independent determinations in which the final values are the expression level of each gene normalized with respect to its expression in the San1 control strain considered as a unit.
Figure 8.—
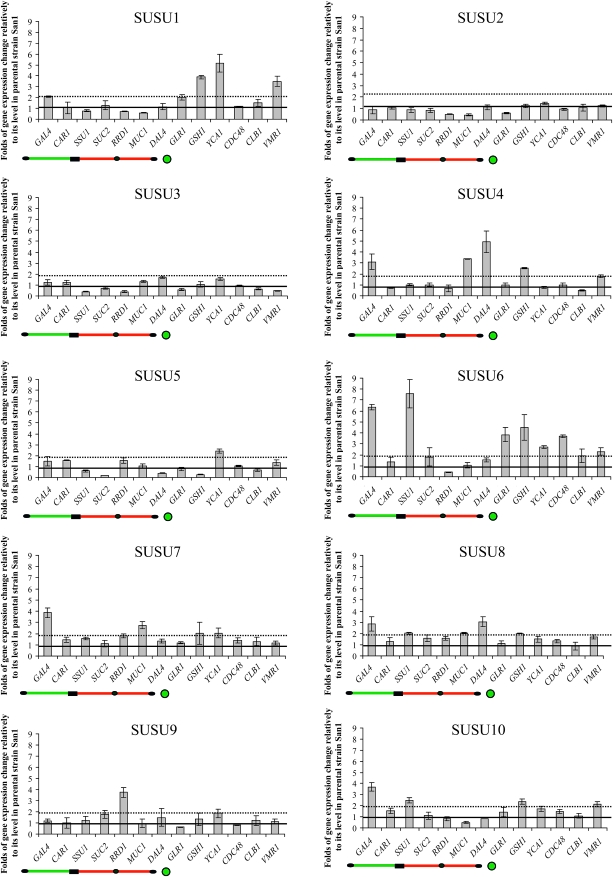
Semiquantitative RT–PCR analysis of the SUSU translocants. Graphs of the expression level of various genes located along the translocated chromosome or those involved in various cellular processes of S. cerevisiae. The names of the analyzed genes are reported below each bar (see also Table S3). Values reported in the graphs represent the expression level of each gene normalized with the expression level of the control gene HSC82 and then compared with the normalized expression of the analyzed gene in the San1 control strain considered as the unit (horizontal black line). Dotted black line represents a twofold increase in gene expression relative to the expression level in the parental strain. The scheme of the translocated chromosome shown below each graph indicates the relative position of the genes along this chromosome. The green circle below GLR1 indicates its presence on chromosome XVI but not on the translocated chromosome.
The primers used to amplify the different genes are described in Table S1A and Table S1B. YCA1 and HSC82 are located on chromosome XV, CLB1 on chromosome VII, CDC48 on chromosome IV, GSH1 on chromosome X, and VMR1 on chromosome VIII. All the other genes are located on chromosome IX or XVI (Table S3).
DNA copy-number determination:
Determination of gene copy number of the genes located on the chromosomes IX and XVI in the different SUSU translocants was made by standard quantitative PCR using a limiting number of cycles (20–22 cycles). ACT1 (on chromosome VI) was used as the control gene to normalize the data. To increase the accuracy of the experiment, ACT1 was co-amplified together with the target gene in the same reaction tube. Values presented in Figure 3 are averages and standard errors from at least three independent determinations normalized with respect to ACT1, considered as a two-copy-number gene. A control experiment was carried out in which ACT1 was amplified together with VMR1, a gene located on chromosome VIII, which is not involved in the translocation. Gel images presented are representative examples from at least four individual experiments.
Figure 3.—
Quantitative PCR copy-number analysis of genes located on chromosomes XVI and IX. In each gel, the higher band corresponds to that of the ACT1 control gene amplicon in chromosome VI, whereas the lower band corresponds to the amplicon of the analyzed gene involved in the translocation. The gene dosage of VMR1, located on chromosome VIII, but not involved in translocation, was also determined as the control for the accuracy of our experiments. Each analyzed gene and ACT1 were co-amplified in the same PCR reaction. KANMX4 quantification was performed with genomic DNA from San1-GUT2/gut2∷KANMX4 containing one copy of the KANMX4 gene. ΔGUT2, San1-GUT2/gut2::KANMX4 - control strain; San1, original parental strain; 1–10: the 10 SUSU translocants. Values presented on the graph above the gels show the gene dosage expressed as an average of at least three independent densitometric determinations from four gels. Data normalized with respect to ACT1 considered as a two-copy-number gene were then compared with the normalized values in the diploid San1 control strain. San1, control strain; 1–10: the 10 SUSU translocants. Gray rectangles highlight a ±0.5 variation in gene copy number. Variations within this interval were considered as insignificant variation in gene copy number with respect to the wild-type situation. Solid bars: San1 control strain; open bars: gene copy number lower than the control strain; lightly shaded bars: gene copy number equal to that of the control strain; darkly shaded bars: gene copy number higher than that of the control strain.
The primers used to amplify the various genes are described in Table S1A and Table S1B. For VMR1 and SUC2 in the translocant chromosome, and for GAL4 and DAL4, the same primers used in the RT–PCR experiments were utilized.
CHEF analyses:
Agarose plug preparation and CHEF electrophoresis were performed as described (Tosato et al. 2005). Hybridization and detection were performed as for a standard Southern blot. Chromosome positions were calculated from bands of the membranes in Figure 2 by polynomial equation: (a) y = −114.1 X2 − 7757 X + 1,359,000 was used to calculate the size of the SUSU9 aberrant chromosome; (b) y = −1432 X2 + 179,631 X − 300,727 was used to calculate the size of the extra band in SUSU7; and (c) y = −24,936.4 X2 + 495,490 X − 1,305,740 was used to calculate the size of the extra band in SUSU2.
Figure 2.—
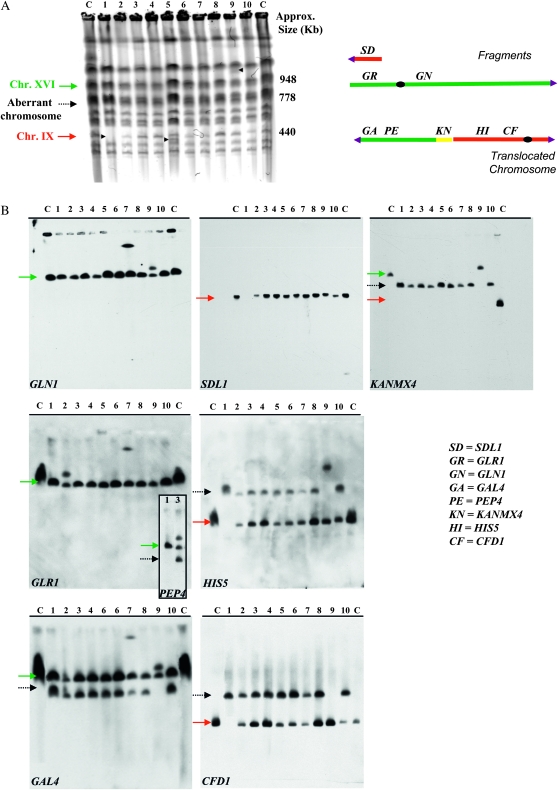
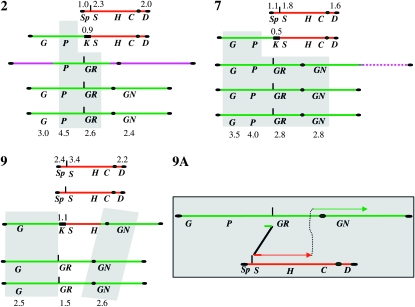
Chromosome analysis of the SUSU translocants. (A) Chromosome separation by CHEF. Arrowheads point to visible chromosomal polymorphisms in the SUSU1, SUSU5, and SUSU9 translocants. (B) Southern blot analyses. Blots were hybridized to the labeled GAL4, PEP4, GLR1, and GLN1 probes on chromosome XVI; SDL1, HIS5, and CFD1 probes are on chromosome IX; the KANMX4 probe was use to detect the aberrant chromosome. In all Southern blots, with the exception of that hybridized with KANMX4, lane C contains San1 diploid control strain DNA. In the membrane hybridized with KANMX4, the lane C contains San1-(DBP1/dbp1∷KANR) DNA; lane 12 contains San1-(GUT2/gut2∷KANR) DNA. Lanes 1–10 represent DNA derived from SUSU1 to SUSU10. The adjoining PEP4-probed membrane on the GLR1 panel shows the SUSU2 chromosomal pattern in comparison to the control strain San1. All the other translocants have the same pattern as in the GAL4 panel. Red arrow: chromosome IX; dashed arrow: translocant chromosome; green arrow: chromosome XVI.
Hybridization probes labeled using the polymerase chain reaction digoxygenin probe synthesis kit (Roche, Basel, Switzerland) were obtained from genomic DNA using primers described in Table S1A and Table S1B. For GLN1, GLR1, and GAL4, the same primers for both RT–PCR and gene copy-number experiments were used.
RESULTS
A new set of BIT translocants between chromosome XVI and IX:
The previously described BIT system (Tosato et al. 2005) was used to generate a collection of 10 translocants between chromosomes XVI and IX of S. cerevisiae. The BIT construct used in this work harbors the KANMX4 selectable marker flanked by two 65-bp sequences, one homologous to the terminator region of SSU1 (that is also the promoter region for GLR1) and the other to the promoter region of SUC2, located on chromosomes XVI and IX, respectively (Figure 1). Both these loci harbor metabolic genes that are not involved in genome regulatory processes (Perlman et al. 1982; Taussig and Carlson 1983; Lutfiyya and Johnston 1996; Avram and Bakalinsky 1997; Perez-Ortin et al. 2002; Fleming and Pennings 2007). The aberrant chromosome generated by translocation between these two loci contains 374 kb of chromosome XVI and a 403-kb fragment of chromosome IX bridged together by the selectable marker KANMX4. The centromere of the translocant chromosome belongs to chromosome IX and is located in an acrocentric position. The resulting right arm of the aberrant chromosome corresponds to the short right arm of chromosome IX (Figure 1).
The PCR-amplified linear DNA cassette was used to transform the diploid yeast strain San1, following the selection of KANR transformants on G418-containing plates. We have used two different transformation techniques to induce BIT in San1 cells. A total of 104 transformants were obtained using the standard LiAc transformation, and a collection of 138 transformants were obtained using spheroplast transformation (Table S2). All transformants were analyzed by colony PCR to check the integration of the BIT construct at both target loci in the genome, using specific primers (see Figure 1). When the transformants showed integration at both loci, the presence of the translocation was confirmed by performing a bridge-PCR from genomic DNA and further validated by sequence analysis (data not shown; see Figure 1 for more details). Among a collection of 10 translocants obtained, 9 translocants were obtained by using the LiAc transformation technique and 1 translocant (i.e., SUSU10) by using spheroplast transformation (Table S2). Our experiments showed that all the analyzed translocations are nonreciprocal (see the following paragraphs and Figures 2, 4, and 5). Furthermore, sequence analysis of the translocation breakpoints revealed no mismatches or mutations in all 10 mutant strains (data not shown).
Figure 4.—
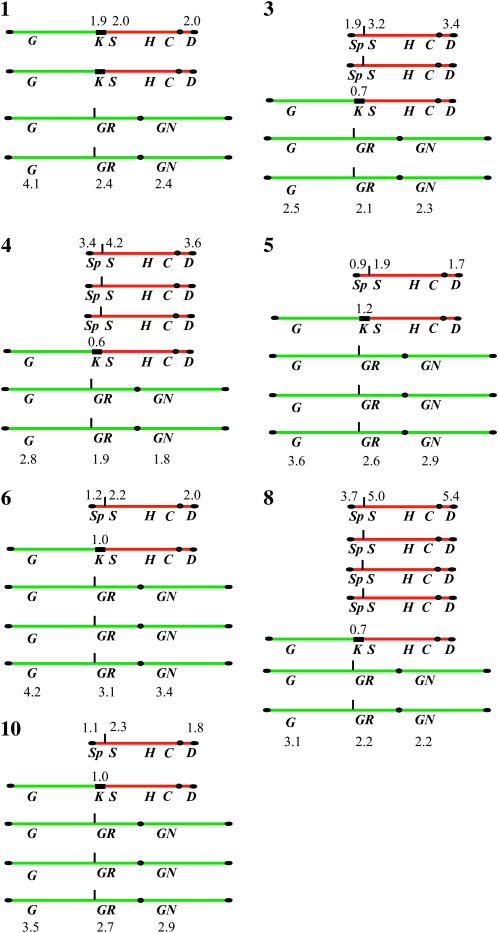
Summary of the genotypic situation of chromosomes XVI and IX in the seven SUSU translocants carrying no complex rearrangements. The genes analyzed in the Southern blot and in the gene copy-number experiments are reported. Gene copy-number values are also indicated. G, GAL4; GR, GLR1; GN, GLN1; S, SUC2; Sp, SUC2 promoter; H, HIS5; C, CFN1; D, DAL4; K, KANMX4. Numbers 1, 3, 4, 5, 6, 8, and 10 indicate SUSU1, SUSU3, SUSU4, SUSU5, SUSU6, SUSU8, and SUSU10 strains, respectively.
Figure 5.—
Summary of the genotype of chromosomes XVI and IX in three SUSU translocants harboring complex rearrangements. As in Figure 4, numbers 2, 7, and 9 indicate SUSU2, SUSU7, and SUSU9 strains, respectively. The scheme contained in panel 9A shows a putative template switching-like mechanism that might be responsible for the rearrangement observed in SUSU9 (see results and discussion for more details).
One interesting observation was that the LiAc technique was ∼10 times more efficient in inducing a BIT event in comparison to spheroplast transformation (8.6% translocation frequency with LiAc transformation, 0.7% translocation frequency with spheroplast transformation; Table S2). Moreover, we demonstrated that by using the spheroplast transformation protocol the frequency of ectopic integration was almost doubled in comparison to the LiAc transformation technique (63% vs. 37.5%; Table S2). This could be due to differential processing of the BIT cassette during its uptake into the nucleus or due to altered activation/stimulation of the DNA repair machinery by the two transformation procedures (Heale et al. 1994).
Chromosomal pattern of the SUSU translocants:
The chromosomal pattern of all the sSU1-SUc2 (i.e., SUSU) translocants named SUSU1–SUSU10 was analyzed using pulsed-field gel electrophoresis PFGE followed by Southern blot analysis. These experiments further confirm that all translocations induced by transformation with a BIT cassette between two nonhomologous chromosomes are non-reciprocal (Figure 2).
Digoxygenin-labeled probes for GAL4, PEP4, GLR1, and GLN1 were used to investigate the fate of the various parts of chromosome XVI; SDL1, HIS5, and CFD1 probes were used to investigate the same for chromosome IX. A KANMX4 probe was used to monitor the formation and occurrence of the aberrant chromosome.
Figure 2 (SDL1 panel) shows that the small acentric fragment of 36,685 bp (Figure 1), supposedly originating after the translocation, is lost or degraded. On the other hand, the 573,568-bp centromeric fragment of chromosome XVI (Figure 1) was not detected in all 10 translocants. These results, together with the gene copy-number data (the following paragraph and Figures 3, 4, and 5), suggest that this centromeric fragment was possibly copied via a BIR-like event during the integration of the BIT cassette at the SSU1 locus. This mechanism has previously been proposed to be responsible for the generation of other translocants obtained with the BIT system between chromosomes V and VIII, XV, and VIII (Tosato et al. 2005). Probes used against chromosome XVI revealed that SUSU2, SUSU7, and SUSU9 carry complex rearrangements of this chromosome (see Figure 5). In SUSU2, a fragment of chromosome XVI containing the PEP4 and GLR1 genes was rearranged elsewhere in the genome, leading to the formation of a novel chromosome (Figures 2 and 5). Using a polynomial equation to calculate chromosome size, this aberrant chromosome containing PEP4 and GLR1 was estimated to be ∼1067 kb. In SUSU7, a large fragment of chromosome XVI encompassing the centromere was fused to another chromosomal fragment, giving rise to a bigger chromosome of ∼1307 kb (see materials and methods for calculations and Figures 2 and 5). Southern blot analyses revealed that in SUSU9 the aberrant chromosome containing the kanamycin resistance gene and the sequenced BIT junction between chromosome XVI and IX was larger than expected. This chromosome of ∼1042 kb did not contain GLR1, CFD1, and DAL4 (not shown) but harbored GLN1, as detected using specific probes. Thus, it does not contain the centromere of chromosome IX but it does contain that of chromosome XVI. Moreover, it seems that it lost a portion of chromosome XVI close to the translocation breakpoint. This different aberrant chromosome that is not a true translocant may have originated by a complex Fork stalling template switching-like mechanism during synthesis (see discussion and Figure 5). Hybridization with the KANMX4 probe demonstrated the presence of the expected 778-kb translocant chromosome in all the other SUSU mutant strains.
On the basis of the fact that the parental strain San1 is diploid, at least one copy of both wild-type chromosomes XVI and IX is also present in all mutant strains, with the exception of SUSU1, in which the wild-type copy of chromosome IX was not detected. Furthermore, Figure 2A shows the presence of an additional polymorphism in SUSU5, which is clearly visible as an extra band between the parental chromosome III (370 kb) and chromosome IX (440 kb). This band did not hybridize with any of the probes located along chromosomes IX and XVI used in our experiments. Since a similar band was also observed in other translocants obtained in our laboratory (V. Tosato, personal communication), we cannot exclude that the event that leads to the formation of this polymorphism is not specifically correlated with the SUSU-BIT translocation events.
Gene copy-number determination:
CHEF and Southern blot analysis point toward the occurrence of various mechanisms that might contribute to the rearrangement of the chromosomes participating in the translocation. To better understand the mechanisms involved in the formation of the SUSU translocants and to investigate whether aneuploidy might arise in cells that undergo BIT translocation events, we determined the copy number of certain genes located along chromosomes XVI and IX by quantitative PCR. The gene dosage of VMR1, located on chromosome VIII (not involved in translocation), was also determined as the control for the accuracy of our experiments. Figure 3 shows the gene dosage of the analyzed genes in the control and in the translocant strains. The results are also summarized in Figures 4 and 5.
Gene dosage experiments suggest that the consequence of the integration of the BIT cassette might also be responsible for the generation of aneuploidy in all the translocant strains. In particular, GAL4, GLR1, and GLN1 copy-number determination showed that all the analyzed mutants have at least two copies of the wild-type chromosome XVI. In SUSU5, SUSU6, and SUSU10, the presence of three copies of this chromosome was also detected, highlighting that in the translocants a partial trisomy or tetraploidy of the fragment involved in the translocation is present. Triploid and tetraploid conditions of different parts of chromosome XVI were also detected in SUSU2, SUSU7, and SUSU9, in which Southern blot analyses revealed the presence of complex rearrangements of that chromosome. PCR data on the SUC2 promoter, the SUC2 ORF, and DAL4 located on chromosome IX pointed out that in five translocants just one copy of the wild-type chromosome IX is present, whereas in SUSU3 and SUSU9 two copies are present. Three copies were detected in SUSU4 and four copies in SUSU8. In SUSU1, the absence of the wild-type copy of chromosome IX was confirmed together with the duplication of the aberrant chromosome. With the exception of this strain, the presence of only one copy of the aberrant chromosome was noted for all the other translocants by the amplification of KANMX4, together with ACT1, by using San1-GUT2/gut2∷KANMX4 as the reference strain containing one copy of the kanamycin resistance gene.
On the basis of the above observations we suggest that SUSU translocant cells carry various types of aneuploidy. Therefore, we asked whether or not such aneuploidy was a direct consequence of targeted translocation and associated rearrangement of chromosomal fragments. To this end, we analyzed the presence of such aneuploidy in strains that carried ectopic integration of the SUSU-BIT construct and were previously obtained with both LiAc and spheroplast transformation methods. qPCR data for gene copy-number determination of SUC2 and DAL4 located on chromosome IX, and for GLR1 and GAL4 located on chromosome XVI, demonstrated that both chromosomes are present in two copies (wild-type configuration) in all the four ectopic integrants (see Figure S2). These observations suggest that the transformation process itself, or the BIT integration per se, cannot account for the rearrangements observed in the SUSU translocant strains (Figure S2).
Physiological and morphological characteristics of the SUSU translocants:
Because we noted genotypic variations among the translocants, we asked whether or not they are also expressed as physiological and morphological differences.
Growth test on carbon sources:
First, we analyzed the fitness and the ability of the SUSU translocants to grow in different carbon sources both on plates (not shown) and in liquid medium. Results plotted in Figure S1 show that, in all the carbon sources tested, the SUSU translocants reached the stationary phase at lower culture density than the parental strain San1. This phenomenon was notably evident when the mutant strains were cultured on the nonfermentable carbon source glycerol. In particular, SUSU5 was completely unable to grow on glycerol and also in other respirable-only carbon sources, such as ethanol and acetate, indicating the presence of some defect in the respiration and/or mitochondrial metabolism. The low fitness of SUSU9 on glucose and of SUSU1 on raffinose was also noteworthy. From the physiological point of view, we also noted that some strains, such as SUSU2, −6, −7, −8, and −9, were characterized by their strong ability to flocculate whereas some others were not (not shown).
Results summarized in Table 1 indicate that only 7 of 10 mutants exhibit sufficient sporulation, SUSU9 being the translocant with the highest sporulation efficiency (79% of the control strain San1) and SUSU4 the mutant strain with the lowest efficiency (2% of the control strain).
TABLE 1.
Sporulation efficiency
| Strain | % sporulation efficiency |
|---|---|
| San1 | 100 |
| SUSU1 | ND |
| SUSU2 | 20 |
| SUSU3 | 35 |
| SUSU4 | 2 |
| SUSU5 | ND |
| SUSU6 | 20 |
| SUSU7 | ND |
| SUSU8 | 22 |
| SUSU9 | 79 |
| SUSU10 | 49 |
The sporulation efficency of the mutant strains is expressed in comparison to the sporulation efficency of the control strain considered as 100%. SUSU1 and SUSU7 strains are not able to sporulate. ND, not detected.
Morphology:
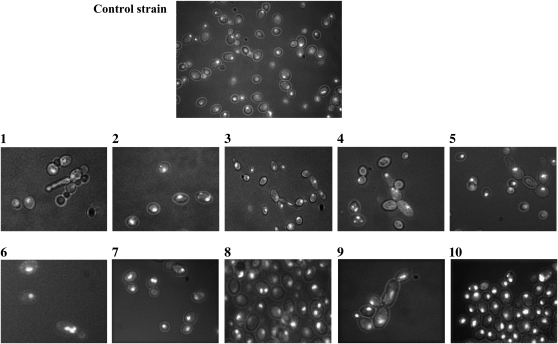
The morphology of the cells was monitored during cultivation on YPD medium and analyzed after 16, 40, and 64 hr by light fluorescence microscopy following DAPI staining. These assays highlighted that the SUSU translocant cell populations exhibit the highest percentage (2–5% of the total cell population) of morphological defects after 40 hr of growth on rich medium. After this time, the number of abnormal cells, as well as the number of actively dividing cells, decreases in all strains (data not shown). Figure 6 shows a few examples of the abnormal cell morphologies observed under the microscope, such as germination tube formation, pseudohyphal growth, multi-budded cells, and cells with elongated buds, all indicative of cell cycle defects. It is worth noting that the percentage of all these defects is much lower than that found in other translocants previously described, in which they were between 10 and 20% of the cell population (Nikitin et al. 2008). As in previous translocants obtained with the BIT system (Nikitin et al. 2008), defects in karyokinesis are also evident in most of the SUSU strains (Figure 6).
Figure 6.—
Microphotography of DAPI-stained control strain cells and SUSU translocants. (1–10) SUSU1–SUSU10 translocant cells. Various abnormal cell morphologies and karyokinetic defects are shown: germination tube formation (1), pseudohyphal growth of cells (4, 5, and 9), and multi-budded cells or cells with elongated buds (3 and 4). Nuclear fragmentation and/or defects in nuclear segregation are evident in some of the cells shown in each field (2, 4, 5, 6, 7, 8, and 10).
Viability and cell cycle regulation:
The FUN-1 stain solution is a fluorescent vital dye useful for distinguishing between metabolically active/alive yeast cells (forming CIVSs; Figure 7B) and dead/not metabolically active cells (yellow cells in Figure 7B). We used this staining method to analyze the viability of the SUSU translocants at the late exponential phase. All comparisons were made using the parental San1 strain as reference (Figure 7, A and B).
Figure 7.—
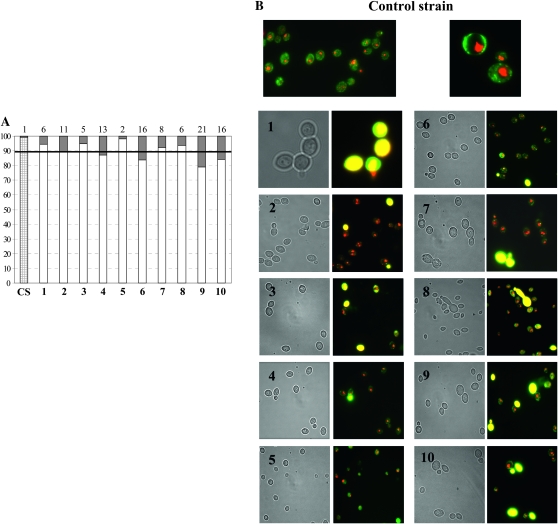
Viability test. (A) Representation of the percentage of dead cells at the late exponential phase of growth. The exact percentage of dead cells of each strain is reported on the top of the graph and is represented by shading on the bars. The dark horizontal line indicates the 10% value. CS: control strain; bars 1–10: SUSU1–SUSU10 translocants. (B) Fluorescence microphotography of FUN1-stained cells. Metabolically active/alive yeast cells containing CIVSs and dead/not metabolically active cells are shown in yellow. (1–10) SUSU1–SUSU10 translocants.
As expected, a very low percentage (1%) of cell mortality in San1 cells was reached after 18 hr of growth on YPD, whereas higher mortality was noted in all the SUSU translocants (Figure 7A). In half of the translocants analyzed, the percentage of non-metabolic active/dead cells is >10% (i.e., ∼10 times more than the control strain). The highest percentage of such cells was found in SUSU9 (21%). In agreement with these data, we observed that SUSU9 had very low fitness in YPD medium correlated to a maximum optical density of 1.36 OD600 corresponding to 1.8 × 107 cells/ml, observed for this strain. On the other hand, SUSU5 was the strain with the lowest percentage of mortality in the late exponential phase (2%). Another interesting feature characteristic of certain SUSU mutants observed by FUN-1 vital staining was the presence of some mother cells attached to a dead bud. This could be explained by the presence of defects in nuclear segregation described earlier (see Figure 6).
Expression analysis:
On the basis of the above morphological and physiological differences among the SUSU translocants, we focused on elucidating these differences at the level of gene expression. Using semiquantitative RT–PCR, we analyzed the expression of the genes located along the two chromosomes involved in the translocation. In addition, we also analyzed the expression profile of a set of genes involved in various cellular processes such as cell cycle regulation, oxidative stress response, apoptosis, and multi-drug resistance. The names of the analyzed genes are reported in Figure 8 and in Table S3. As in previous expression analyses of BIT translocants, HSC82 was chosen as a constitutive control gene (Nikitin et al. 2008; Tosato et al. 2009). For all the genes analyzed, an expression level higher than twice, or equal to or lower than half, the expression level of the control strain San1, was considered as a significant up- or downregulation of transcription, respectively. Figure 8 describes the expression patterns of all the analyzed genes in the 10 SUSU translocants grown on YPD medium. The histograms show that CLB1 and the arginase CAR1 are the only two genes with an expression level similar to that of the control strain in all 10 SUSU translocants whereas all the other genes are up- or downregulated in some of the mutant strains. Indeed, SSU1 expression data clearly demonstrate that, despite the triploid or tetraploid condition of this gene in the SUSU translocants (Figures 4 and 5), its expression was similar to the control strain in most of the SUSU strains with the exception of SUSU3, SUSU6, and SUSU10. In these strains, expression level was ∼3-fold less, and 7.5- and 2.5-fold more than in the control, respectively. The greater increase of SSU1 expression in SUSU6 and SUSU10 cannot be correlated only to the detection of four copies of the chromosomal arm harboring this gene in these two strains (Figures 4 and 5), since a similar tetraploid condition in SUSU1 and SUSU5 exhibited a wild-type expression level. Thus, other factors such as physiological conditions or the presence of unbalanced cellular components due to the effect of translocation and/or the ensuing aneuploidy condition might be responsible for its strong activation in SUSU6 and SUSU10 strains.
The expression of SUC2 was analyzed on both glucose (repressing condition; Figure 8) and raffinose (derepressing condition; see also Figure S3) (Lutfiyya and Johnston 1996; Ozcan et al. 1997) to study the effect of the translocation not only on the basal expression level of this gene but also on its transcriptional activation. In fact, the translocation breakpoints lie within a key position of the SUC2 promoter region, i.e., a few nucleotides upstream from its activation DNA-binding sites and inside a nucleosome that is lost during gene transcription activation (Fleming and Pennings 2007).
Figure 8 and Figure S3 show that the basal expression level of SUC2 is similar to that of the control strain in all the translocants with the exception of SUSU5, in which the expression of this gene is ∼5-fold less. On the other hand, the induction of SUC2 is impaired in many, but not all, translocants, in which under activating conditions the expression level of the gene is 10-fold less in SUSU1, SUSU6, and SUSU8, reduced 3-fold in SUSU7, SUSU9, and SUSU10, or halved in SUSU3. Differences in SUC2 activation levels can be explained on the basis of different genotypes and other secondary factors such as different intracellular physiological conditions. Interestingly, we found a strong reduction of the SUC2 mRNA level despite SUC2 diploid and polyploid status in various translocants. Thus, as for SSU1, gene copy number by itself cannot account for variations in gene expression patterns in the above-mentioned translocants.
Results summarized in Figure 8 also demonstrate that the increased gene expression at the translocation breakpoint and along the translocated chromosome observed in the BIT translocants previously described (Nikitin et al. 2008) was not a common feature of all the SUSU mutant strains. In fact, SUSU2 and SUSU3 did not show any significant alteration in the transcription level of the genes analyzed. The only alteration observed in these translocants was the previously described underexpression of SSU1 and of RRD1 located on chromosome IX.
Three strains, SUSU1, SUSU4, and especially SUSU6, showed strong variability in the expression of many genes, suggesting a larger alteration of their physiology and metabolism. In SUSU1, we observed a 2.1-fold increase in the mRNA level of GAL4 located on chromosome XVI and a 3.5-fold increase of the multidrug-resistant gene VMR1 transcript. In the same strain, we also found the overexpression of the caspase YCA1 gene and of GSH1, which is also involved in apoptosis and cellular response to oxidative stress. Interestingly, previous authors (Tosato et al. 2009; D. Nikitin personal communication) have revealed that VMR1 upregulation was also present in other homologous and heterologous BIT translocants.
The three genes mentioned above (GSH1, YCA1, and VMR1) are also highly overexpressed in SUSU6, which is characterized by a strong deregulation of the transcription of almost all the analyzed genes with the exceptions of CAR1, SUC2, MUC1, and CLB1. In particular, in addition to the three genes mentioned above, the two genes located on both sides of the translocation breakpoint on chromosome XVI showed a pronounced increase in their steady mRNA levels (7.6- and 3.8-fold mRNA levels increasing for SSU1 and GLR1, respectively). The overexpression of GLR1, encoding the cytosolic and mitochondrial glutathione oxidoreductase, can be correlated to the increased GSH1 transcription and could indicate oxidative stress conditions generated within SUSU6 cells. At the same time, overexpression of YCA1 suggests that an apoptotic response might be activated in this strain. The overexpression of the cell cycle-related gene CDC48 in this strain is an indication of the presence of cell cycle defects. Among other genes analyzed, only RRD1 was downregulated in SUSU6. In the SUSU5 strain, which exhibits a petite phenotype, two of the analyzed genes—SUC2 (see above) and GSH1—showed a reduced level of mRNA in comparison to the control strain, which for the latter was ∼4-fold less than that of the control.
In summary, our results show a heterogeneous situation in many SUSU translocants in which some genes are activated, others are downregulated, and others have the same expression level of the control strain. This may or may not correlate with the copy number of these genes, suggesting a more intricate adaptation of the cells to a dramatically changed karyotype.
DISCUSSION
Translocation events not only are gross chromosomal rearrangements that can be associated with cancer development in mammalian cells and acquisition of deleterious phenotypes in eukaryotic microorganisms, but also are a powerful evolutionary force capable of strongly reshaping the genome. For example, translocation events have been associated with the acquisition of advantageous antifungal drug resistance in the yeast Candida glabrata (Polakova et al. 2009). Despite their importance for the maintenance and evolution of genome organization, little is known about the molecular mechanisms and pathways involved in the formation and regulation of a translocation event.
In this work, we have used the BIT system (Tosato et al. 2005) to generate a collection of mutant yeast strains harboring the same translocation between chromosomes XVI and IX (Figure 1). Southern blot analysis and gene copy-number determination revealed that the integration of the SUSU linear DNA cassette can induce the formation of not only a non-reciprocal translocation, but also triggers other complex rearrangements generating different types of aneuploidy in all the mutant strains analyzed. This complex abnormal karyotype bearing monosomy, disomy, and trisomy for different segments of the translocation participants is strongly reminiscent of what is described (Pedersen et al. 2000) in leukemia patients carrying unbalanced translocations. Even though S. cerevisiae tolerates aneuploidy well, it is common knowledge that aneuploidy has severe effects on growth and development, as also reported in baker's yeast (Torres et al. 2007). Maintenance of euploidy is essential for survival of a species, and correlations between cancer and aneuploidy were reported in 1890 by D. P. von Hansemann and in 1902 by T. Boveri (Bignold et al. 2006). Recently, aneuploidy was proposed to be the major pre-neoplastic condition leading to chromosome instability and promoting transformation of the normal cells (Duesberg et al. 2005). Variations in chromosome numbers and aneuploidy formation are thus usually strongly inhibited by checkpoint mechanisms. In the SUSU mutant strains, such cellular surveillance seems to be bypassed, leading to the formation of gross chromosomal rearrangements possibly by various mechanisms.
One possible mechanism for recombination-associated generation of aneuploidy is BIR (Morrow et al. 1997; Jain et al. 2009). This process can well explain the partial trisomy of chromosome XVI present in SUSU3, SUSU4, and SUSU8. According to the two-step integration model of the BIT cassette proposed for other translocants previously described (Tosato et al. 2005), we might hypothesize that in these mutants the cassette initially integrates at the SUC2 locus via homologous recombination and then, in a second step, completes the integration at the SSU1 locus by strand invasion and synthesis toward the telomere. This model is also supported by the fact that the integration of the BIT cassette might mimic the condition in which homology is limited to one end of the DSB, a condition that strongly favors the repair of a break by the BIR mechanism (Malkova et al. 2005; Vanhulle et al. 2007; Jain et al. 2009). However, we cannot explain the partial trisomy of chromosome IX observed in the SUSU3 strain by a BIR-like event, as it is known that the BIR replication apparatus is blocked at the centromere (Morrow et al. 1997). One possible explanation of this situation is that during the integration of the BIT cassette an alternative replication apparatus capable of passing through the centromeric region might be involved, giving rise to a complete and functional chromosome. A striking observation of an ∼10-fold increase in the occurrence of a translocation event, upon transforming S-phase blocked yeast cells with the BIT construct, has recently been noted in our laboratory (V. Tosato, personal communication). Indeed, recent research reveals that a non-reciprocal translocation, leading to duplication of chromosomal regions, can also arise from an unconventional Pol32-independent, BIR-like mechanism (Putnam et al. 2009) This suggests that formation of complex rearrangements might occur by different mechanisms capable of repair by DNA synthesis. Furthermore, in support of this view, recent observations in our group show that deletion of POL32 does not affect the rate of BIT translocation induction (V. Tosato, unpublished data). This hypothesis, however, does not provide an exhaustive explanation of all the rearrangements depicted in Figures 4 and 5. In particular, an explanation of the multiplication of chromosome IX in SUSU4 and SUSU8 strains or of the triplication of chromosome XVI in SUSU5, SUSU6, and SUSU10 strains cannot be based on such a hypothesis. Another explanation for these phenomena could be that, in adapted cells that are able to bypass cell cycle arrest, the recombination of the BIT cassette interferes with chromosome segregation during mitosis, giving rise to the aneuploidy noted for both chromosomes XVI and IX. Missegregation due to interchromosomal association as described (Kaye et al. 2004) was also proposed to be linked to the generation of disomic cells following the integration of a DNA construct with short heterologous termini in S. cerevisiae (Svetec et al. 2007). A defect in chromosome segregation, possibly due to the size differences of chromosome IX and the newly formed aberrant chromosome, might account for the duplication or triplication of chromosome IX observed in the SUSU4 and SUSU8 strains. Indeed, perhaps unable to identify the remaining portion of chromosome IX, the cells could behave as monosomic for that chromosome, restoring a euploid situation by endo-reduplication with a mechanism similar to that already described (Waghmare and Bruschi 2005). Defects in chromosome IX segregation that lead to the loss of the wild-type copy of chromosome IX followed by the duplication of the translocant chromosome can also explain the karyotype of SUSU1, although a double translocation event leading to such a karyotype cannot be ruled out. It is noteworthy that in SUSU1 there is a loss of genetic material. In fact, it lacks all the genes contained in the 36,685-bp fragment located on the subtelomeric region upstream from the SUC2 locus. All these genes are nonessential.
Complex rearrangements involving chromosome XVI have been described in SUSU2, SUSU7, and SUSU9 (Figure 5). In particular, the SUSU9 strain does not possess the expected translocant chromosome but instead a different aberrant chromosome in which only a small part of chromosome IX was rearranged together with chromosome XVI. The configuration of this chromosome strongly suggests that a template switching-like mechanism, similar to the FoSTeS described in mammalian cells (Lee et al. 2007), might be responsible for this rearrangement. We hypothesize that in this mutant the cassette first integrates at the SSU1 locus via homologous recombination and then completes the integration at the SUC2 locus by strand invasion and synthesis (or break-induced replication). After this starting event, we suppose a stalling of the replication fork before the centromere of chromosome IX is followed by template dissociation and re-invasion of chromosome XVI and new DNA synthesis to the end of chromosome XVI, leading to the chromosome configuration described earlier (Figure 5, panel 9A).
Overall, our data also demonstrate that the integration of the linear DNA fragment into the yeast genome can be a great force for evolution. In fact, in our experiments, the integration of the same cassette at the two target loci was processed in eight different ways, generating strains different in karyotype and consequently in phenotype and physiology. This variety of morphologies and phenotypes and the variation of the expression level of various genes among the different translocants were not observed in strains carrying Kanr integration on individual chromosomes at the translocation breakpoint (see Figure S4). This observation suggests that the mere integration of KANMX4 by itself does not destabilize and deregulate gene expression or other physiological aspects such as sporulation, flocculation (not shown), or growth rate.
Some of the mutant phenotypes described in this work, such as the presence of a slow growth rate, morphological aberrations, and defects in nuclear segregation, are similar to those recently illustrated for the D10big and small mutants carrying a translocation between chromosomes VIII and XV (Nikitin et al. 2008). However, the overexpression of the genes located at the translocation breakpoints typical of the D10 mutant is not observed in most of the SUSU translocants. This suggests that the effect of a translocation on gene expression might be locus-specific, i.e., dependent on the loci and on the chromosomes involved in BIT. Moreover, in the SUSU translocants the presence of aneuploidy, previously not detected in D10big and small mutants, might also influence global gene expression. Torres et al. (2007) recently demonstrated that aneuploidy causes a transcriptional response with the doubling of gene expression along the entire length of a disomic chromosome. In SUSU strains, increased mRNA levels of genes present in a higher copy number, in comparison to the control strain, were observed in some of the mutants (Figure 8) but not in others. Perhaps in these mutants a combination of two effects, translocation and aneuploidy, is established at later time point after translocation. On the other hand, many phenotypic characteristics of the aneuploid strains described by Torres and colleagues were also found in these mutant strains, such as slow growth rate (probably also in this case because of a nonspecific disruption of the cellular homeostasis), lower OD600 values of mutants noted in the stationary phase with respect to the control strain, proliferation defects, and an increased number of G1 phase-stopped cells (data not shown).
Interestingly, some of the SUSU translocants showed the overexpression of YCA1, suggesting the activation of apoptotic pathways. The genes GSH1 and GLR1 involved in oxidative stress response were also overexpressed in some of the translocant strains together with the multi-drug resistance gene VMR1, which is homologous to the ABCC-type multi-drug transport proteins. This gene was also overexpressed in other BIT translocants between both heterologous and homologous chromosomes (Tosato et al. 2009; D. Nikitin, personal communication). The common overexpression of this gene in our translocants and the documented overexpression of the homologous human Mrp4p in some aggressive primary neuroblastomas, also characterized by unbalanced chromosomal translocations (Janoueix-Lerosey et al. 2005; Norris et al. 2005), suggest that the overexpression of this gene might be considered as a general effect of these kinds of aberrations. In this view, the detection of generally shared mutant phenotypes among translocants will have to be analyzed to clarify their dependence or independence from the specific breakpoints. It could be also interesting to know if the ectopic insertion of one of the homologies on a different chromosome could be used efficiently for a homologous repair, which would generate a reciprocal translocation. We think that the BIT system provides a valuable tool for obtaining statistically significant numbers of different translocation mutants without prior genomic engineering of cells. Results presented in this work also suggest that BIT could be considered as an experimentally reliable system to mimic the spontaneous mitotic translocation of higher eukaryotic cells. In fact, as observed by others in mammalian cells, our findings strongly support that aneuploidy arising from an initial nonreciprocal translocation event may generate a heterogeneous population of mutant cells that are phenotypically and genotypically diverse. At the same time, our laboratory recently showed that near-reciprocal translocation between homologous chromosomes does not affect gene expression at the genomic loci flanking the breakpoints (Tosato et al. 2009).
Acknowledgments
We thank Valentina Tosato and Dmitri Nikitin for help and useful discussions throughout the experimental work as well as for help with the preparation of the manuscript. We also thank Michela Ronzani for technical help. Beatrice Rossi is an International Centre for Genetic Engineering and Biotechnology (ICGEB) Post-doctoral Fellow, and Pawan Noel is a Ph.D. Fellow supported by the ICGEB doctoral program.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.120683/DC1.
References
- Agarwal, S., A. A. Tafel and R. Kanaar, 2006. DNA double-strand break repair and chromosome translocations. DNA Repair (Amst.) 5 1075–1081. [DOI] [PubMed] [Google Scholar]
- Avram, D., and A. T. Bakalinsky, 1997. SSU1 encodes a plasma membrane protein with a central role in a network of proteins conferring sulfite tolerance in Saccharomyces cerevisiae. J. Bacteriol. 179 5971–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon, Y., and M. Kupiec, 2004. New insights into the mechanism of homologous recombination in yeast. Mutat. Res. 566 231–248. [DOI] [PubMed] [Google Scholar]
- Bernstein, R., 1988. Cytogenetics of chronic myelogenous leukemia. Semin. Hematol. 25 20–34. [PubMed] [Google Scholar]
- Bignold, L. P., B. L. Coghlan and H. P. Jersmann, 2006. Hansemann, Boveri, chromosomes and the gametogenesis-related theories of tumours. Cell Biol. Int. 30 640–644. [DOI] [PubMed] [Google Scholar]
- Bosco, G., and J. E. Haber, 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera, R., M. Bregni, J. Erikson, D. Patterson, R. C. Gallo et al., 1982. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 79 7824–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Braekeleer E., N. Douet-Guilbert, M. J. Le Bris, F. Morel, M. De Braekeleer, 2007. Translocation 3;21 trisomy 8, and duplication of the Philadelphia chromosome: a rare recurrent cytogenetic pathway in the blastic phase of chronic myeloid leukemia. Cancer Genet. Cytogenet. 179 159–161. [DOI] [PubMed] [Google Scholar]
- Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis et al., 2003. Engineering evolution to study speciation in yeasts. Nature 422 68–72. [DOI] [PubMed] [Google Scholar]
- Duesberg, P., R. Li, A. Fabarius and R. Hehlmann, 2005. The chromosomal basis of cancer. Cell. Oncol. 27 293–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, D., E. Hafen and W. Schaffner, 2004. An efficient method to generate chromosomal rearrangements by targeted DNA double-strand breaks in Drosophila melanogaster. Genome Res. 14 1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A. B., and S. Pennings, 2007. Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res. 35 5520–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini, P., G. Sozzi and M. A. Pierotti, 2007. The role of chromosomal alterations in human cancer development. J. Cell Biochem. 102 320–331. [DOI] [PubMed] [Google Scholar]
- Gu, W., F. Zhang and J. R. Lupski, 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 2006. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst.) 5 998–1009. [DOI] [PubMed] [Google Scholar]
- Heale, S. M., L. I. Stateva and S. G. Oliver, 1994. Introduction of YACs into intact yeast cells by a procedure which shows low levels of recombinagenicity and co-transformation. Nucleic Acids Res. 22 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S., N. Sugawara, J. Lydeard, M. Vaze, N. Tanguy Le Gac et al., 2009. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 23 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey, I., P. Hupe, Z. Maciorowski, P. La Rosa, G. Schleiermacher et al., 2005. Preferential occurrence of chromosome breakpoints within early replicating regions in neuroblastoma. Cell Cycle 4 1842–1846. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis and A. Mitchell, 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kanaar, R., J. H. Hoeijmakers and D. C. van Gent, 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 8 483–489. [DOI] [PubMed] [Google Scholar]
- Kaye, J. A., J. A. Melo, S. K. Cheung, M. B. Vaze, J. E. Haber et al., 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14 2096–2106. [DOI] [PubMed] [Google Scholar]
- Lee, J. A., C. M. Carvalho and J. R. Lupski, 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131 1235–1247. [DOI] [PubMed] [Google Scholar]
- Lutfiyya, L. L., and M. Johnston, 1996. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 16 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova, A., M. L. Naylor, M. Yamaguchi, G. Ira and J. E. Haber, 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard, P. J., B. L. Roth, H. P. Thi, S. T. Yue and R. P. Haugland, 1997. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63 2897–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, D. M., C. Connelly and P. Hieter, 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, A., and K. Myung, 2007. Measuring the rate of gross chromosomal rearrangements in Saccharomyces cerevisiae: a practical approach to study genomic rearrangements observed in cancer. Methods 41 168–176. [DOI] [PubMed] [Google Scholar]
- Motegi, A., K. Kuntz, A. Majeed, S. Smith and K. Myung, 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehleck, S. D., R. W. McKenna, D. C. Arthur, J. L. Parkin and R. D. Brunning, 1984. Transformation of chronic myelogenous leukemia: clinical, morphologic, and cytogenetic features. Am. J. Clin. Pathol. 82 1–14. [DOI] [PubMed] [Google Scholar]
- Nickoloff, J. A., L. P. De Haro, J. Wray and R. Hromas, 2008. Mechanisms of leukemia translocations. Curr. Opin. Hematol. 15 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin, D., V. Tosato, A. B. Zavec and C. V. Bruschi, 2008. Cellular and molecular effects of nonreciprocal chromosome translocations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 105 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, M. D., J. Smith, K. Tanabe, P. Tobin, C. Flemming et al., 2005. Expression of multidrug transporter MRP4/ABCC4 is a marker of poor prognosis in neuroblastoma and confers resistance to irinotecan in vitro. Mol. Cancer Ther. 4 547–553. [DOI] [PubMed] [Google Scholar]
- Nowell, P. C., L. Jackson, A. Weiss, and R. Kurzrock, 1988. Historical communication: Philadelphia-positive chronic myelogenous leukemia followed for 27 years. Cancer Genet. Cytogenet. 34 57–61. [DOI] [PubMed] [Google Scholar]
- Oka, A., H. Sugisaki and M. Takanami, 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147 217–226. [DOI] [PubMed] [Google Scholar]
- Ozcan, S., L. G. Vallier, J. S. Flick, M. Carlson and M. Johnston, 1997. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13 127–137. [DOI] [PubMed] [Google Scholar]
- Pedersen, B., J. M. Norgaard, B. B. Pedersen, N. Clausen, I. H. Rasmussen et al., 2000. Many unbalanced translocations show duplication of a translocation participant. Clinical and cytogenetic implications in myeloid hematologic malignancies. Am. J. Hematol. 64 161–169. [DOI] [PubMed] [Google Scholar]
- Perez-Ortin, J. E., A. Querol, S. Puig and E. Barrio, 2002. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12 1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, D., H. O. Halvorson and L. E. Cannon, 1982. Presecretory and cytoplasmic invertase polypeptides encoded by distinct mRNAs derived from the same structural gene differ by a signal sequence. Proc. Natl. Acad. Sci. USA 79 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakova, S., C. Blume, J. A. Zarate, M. Mentel, D. Jorck-Ramberg et al., 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 106 2688–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam, C. D., T. K. Hayes and R. D. Kolodner, 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley, J. D., 2001. Chromosome translocations: dangerous liaisons revisited. Nat. Rev. Cancer 1 245–250. [DOI] [PubMed] [Google Scholar]
- Rowley, J. D., 2008. Chromosomal translocations: revisited yet again. Blood 112 2183–2189. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Svetec, I. K., A. Stafa and Z. Zgaga, 2007. Genetic side effects accompanying gene targeting in yeast: the influence of short heterologous termini. Yeast 24 637–652. [DOI] [PubMed] [Google Scholar]
- Taki, T., and M. Taniwaki, 2006. Chromosomal translocations in cancer and their relevance for therapy. Curr. Opin. Oncol. 18 62–68. [DOI] [PubMed] [Google Scholar]
- Taussig, R., and M. Carlson, 1983. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 11 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli et al., 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 916–924. [DOI] [PubMed] [Google Scholar]
- Tosato, V., S. K. Waghmare and C. V. Bruschi, 2005. Non-reciprocal chromosomal bridge-induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma 114 15–27. [DOI] [PubMed] [Google Scholar]
- Tosato, V., C. Nicolini and C. V. Bruschi, 2009. DNA bridging of yeast chromosomes VIII leads to near-reciprocal translocation and loss of heterozygosity with minor cellular defects. Chromosoma 118 179–191. [DOI] [PubMed] [Google Scholar]
- VanHulle, K., F. J. Lemoine, V. Narayanan, B. Downing, K. Hull et al., 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 27 2601–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Waghmare, S. K., and C. V. Bruschi, 2005. Differential chromosome control of ploidy in the yeast Saccharomyces cerevisiae. Yeast 22 625–639. [DOI] [PubMed] [Google Scholar]
- Waghmare, S. K., V. Caputo, S. Radovic and C. V. Bruschi, 2003. Specific targeted integration of kanamycin resistance-associated nonselectable DNA in the genome of the yeast Saccharomyces cerevisiae. Biotechniques 34 1024–1028, 1033. [DOI] [PubMed] [Google Scholar]
- Weinstock, D. M., C. A. Richardson, B. Elliott and M. Jasin, 2006. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst.) 5 1065–1074. [DOI] [PubMed] [Google Scholar]