Abstract
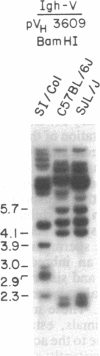
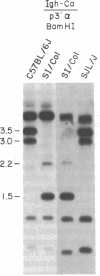
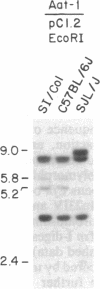
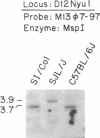
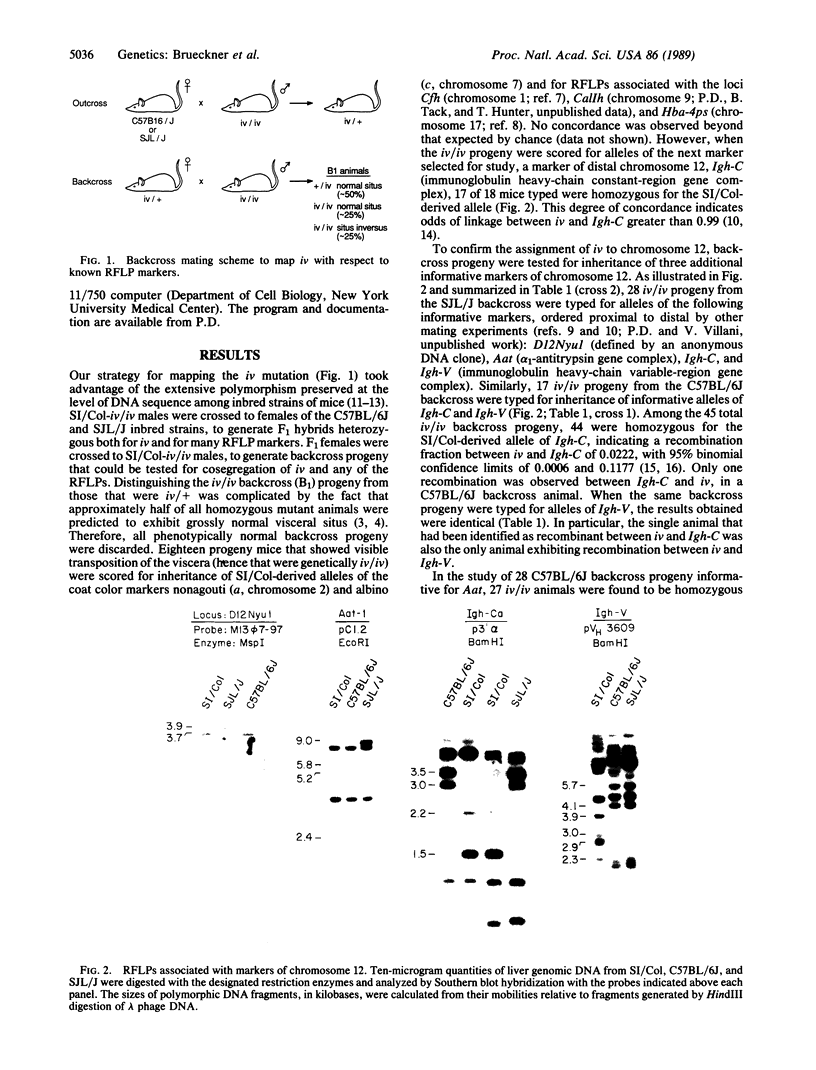
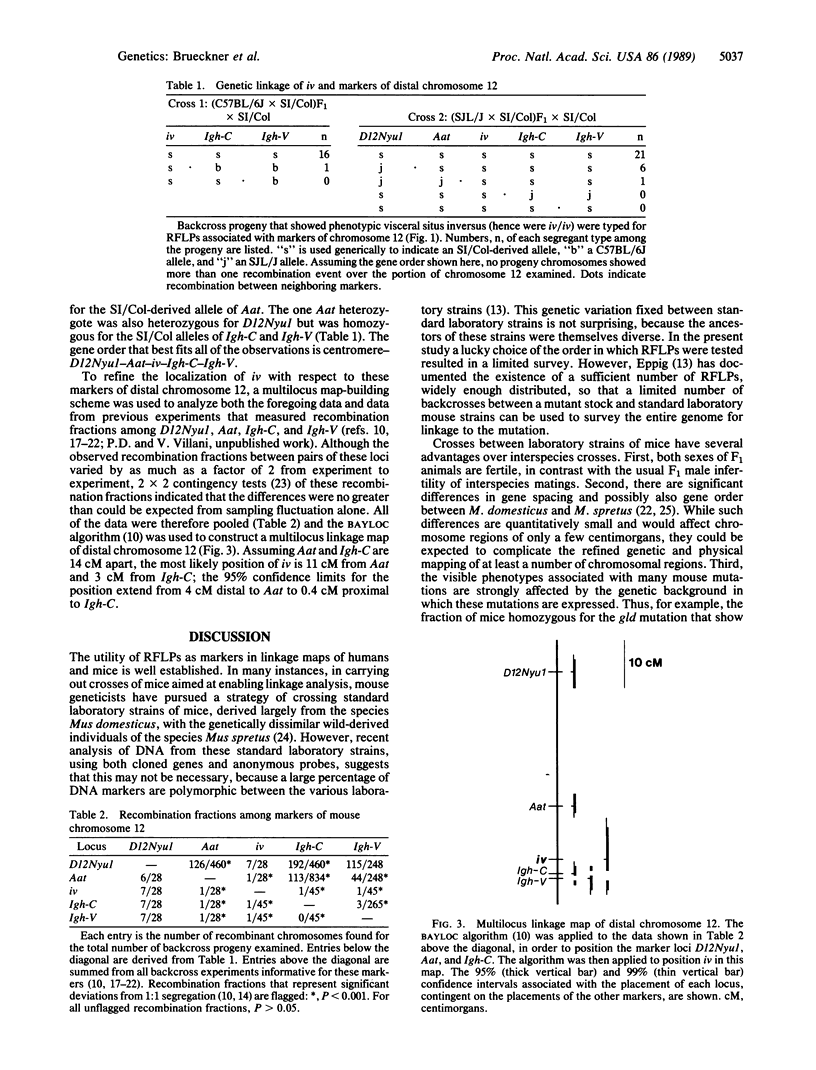
Inherited single gene defects have been identified in both humans and mice that lead to loss of developmental control over the left-right asymmetry of the heart and viscera. In mice the recessively inherited mutation iv leads to such apparent loss of control over situs: 50% of iv/iv mice exhibit situs inversus and 50% exhibit normal situs. The affected gene product has not been identified in these animals. To study the normal function of iv, we have taken an approach directed to the gene itself. As a first step, we have mapped iv genetically, by examining its segregation in backcrosses with respect to markers defined by restriction fragment length polymorphisms. The iv locus lies 3 centimorgans (cM) from the immunoglobulin heavy-chain constant-region gene complex (Igh-C) on chromosome 12. A multilocus map of the region suggests the gene order centromere-Aat (alpha 1-antitrypsin gene complex)-(11 cM)-iv-(3 cM)-Igh-C-(1 cM)-Igh-V (immunoglobulin heavy-chain variable-region gene complex).
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Afzelius B. A. A human syndrome caused by immotile cilia. Science. 1976 Jul 23;193(4250):317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- Arnold G. L., Bixler D., Girod D. Probable autosomal recessive inheritance of polysplenia, situs inversus and cardiac defects in an Amish family. Am J Med Genet. 1983 Sep;16(1):35–42. doi: 10.1002/ajmg.1320160107. [DOI] [PubMed] [Google Scholar]
- Avner P., Amar L., Dandolo L., Guénet J. L. Genetic analysis of the mouse using interspecific crosses. Trends Genet. 1988 Jan;4(1):18–23. doi: 10.1016/0168-9525(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Birkenmeier C. S., McFarland-Starr E. C., Barker J. E. Chromosomal location of three spectrin genes: relationship to the inherited hemolytic anemias of mouse and man. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8121–8125. doi: 10.1073/pnas.85.21.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank R. D., Campbell G. R., Calabro A., D'Eustachio P. A linkage map of mouse chromosome 12: localization of Igh and effects of sex and interference on recombination. Genetics. 1988 Dec;120(4):1073–1083. doi: 10.1093/genetics/120.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank R. D., Campbell G. R., D'Eustachio P. Possible derivation of the laboratory mouse genome from multiple wild Mus species. Genetics. 1986 Dec;114(4):1257–1269. doi: 10.1093/genetics/114.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur P. H., Osman G. E., Mackle J. J., Lalor T. M. The organization of the mouse Igh-V locus. Dispersion, interspersion, and the evolution of VH gene family clusters. J Exp Med. 1988 Dec 1;168(6):2261–2278. doi: 10.1084/jem.168.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Eustachio P. A genetic map of mouse chromosome 12 composed of polymorphic DNA fragments. J Exp Med. 1984 Sep 1;160(3):827–838. doi: 10.1084/jem.160.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Eustachio P., Fein B., Michaelson J., Taylor B. A. The alpha-globin pseudogene on mouse chromosome 17 is closely linked to H-2. J Exp Med. 1984 Mar 1;159(3):958–963. doi: 10.1084/jem.159.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Eustachio P., Kristensen T., Wetsel R. A., Riblet R., Taylor B. A., Tack B. F. Chromosomal location of the genes encoding complement components C5 and factor H in the mouse. J Immunol. 1986 Dec 15;137(12):3990–3995. [PubMed] [Google Scholar]
- Fitch W. M., Atchley W. R. Evolution in inbred strains of mice appears rapid. Science. 1985 Jun 7;228(4704):1169–1175. doi: 10.1126/science.4001935. [DOI] [PubMed] [Google Scholar]
- Handel M. A., Kennedy J. R. Situs inversus in homozygous mice without immotile cilia. J Hered. 1984 Nov-Dec;75(6):498–498. doi: 10.1093/oxfordjournals.jhered.a109995. [DOI] [PubMed] [Google Scholar]
- Hill R. E., Shaw P. H., Barth R. K., Hastie N. D. A genetic locus closely linked to a protease inhibitor gene complex controls the level of multiple RNA transcripts. Mol Cell Biol. 1985 Aug;5(8):2114–2122. doi: 10.1128/mcb.5.8.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Sato H., Odaka T. Mapping of the Fv-4 mouse gene controlling resistance to murine leukemia viruses. Int J Cancer. 1981 Aug 15;28(2):237–240. doi: 10.1002/ijc.2910280218. [DOI] [PubMed] [Google Scholar]
- Krauter K. S., Citron B. A., Hsu M. T., Powell D., Darnell J. E., Jr Isolation and characterization of the alpha 1-antitrypsin gene of mice. DNA. 1986 Feb;5(1):29–36. doi: 10.1089/dna.1986.5.29. [DOI] [PubMed] [Google Scholar]
- Layton W. M., Jr Random determination of a developmental process: reversal of normal visceral asymmetry in the mouse. J Hered. 1976 Nov-Dec;67(6):336–338. doi: 10.1093/oxfordjournals.jhered.a108749. [DOI] [PubMed] [Google Scholar]
- Nadeau J. H., Berger F. G., Kelley K. A., Pitha P. M., Sidman C. L., Worrall N. Rearrangement of genes located on homologous chromosomal segments in mouse and man: the location of genes for alpha- and beta-interferon, alpha-1 acid glycoprotein-1 and -2, and aminolevulinate dehydratase on mouse chromosome 4. Genetics. 1986 Dec;114(4):1239–1255. doi: 10.1093/genetics/114.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Taylor B. A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci U S A. 1984 Feb;81(3):814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Lathrop M., O'Connell P., Leppert M., Kamboh M. I., Lalouel J. M., White R. Frequent recombination is observed in the distal end of the long arm of chromosome 14. Genomics. 1989 Jan;4(1):76–81. doi: 10.1016/0888-7543(89)90317-0. [DOI] [PubMed] [Google Scholar]
- Odaka T., Ikeda H., Yoshikura H., Moriwaki K., Suzuki S. Fv-4: gene controlling resistance to NB-tropic Friend murine leukemia virus. Distribution in wild mice, introduction into genetic background of BALB/c mice, and mapping of chromosomes. J Natl Cancer Inst. 1981 Nov;67(5):1123–1127. [PubMed] [Google Scholar]
- Searle A. G., Peters J., Lyon M. F., Evans E. P., Edwards J. H., Buckle V. J. Chromosome maps of man and mouse, III. Genomics. 1987 Sep;1(1):3–18. doi: 10.1016/0888-7543(87)90099-1. [DOI] [PubMed] [Google Scholar]
- Seldin M. F., Morse H. C., 3rd, Reeves J. P., Scribner C. L., LeBoeuf R. C., Steinberg A. D. Genetic analysis of autoimmune gld mice. I. Identification of a restriction fragment length polymorphism closely linked to the gld mutation within a conserved linkage group. J Exp Med. 1988 Feb 1;167(2):688–693. doi: 10.1084/jem.167.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J., Buckler C. E. Statistical considerations for linkage analysis using recombinant inbred strains and backcrosses. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1423–1427. doi: 10.1073/pnas.83.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. Confidence limits for estimates of gene linkage based on analysis of recombinant inbred strains. J Hered. 1985 Nov-Dec;76(6):436–440. doi: 10.1093/oxfordjournals.jhered.a110140. [DOI] [PubMed] [Google Scholar]
- Taylor B. A., Bailey D. W., Cherry M., Riblet R., Weigert M. Genes for immunoglobulin heavy chain and serum prealbumin protein are linked in mouse. Nature. 1975 Aug 21;256(5519):644–646. doi: 10.1038/256644a0. [DOI] [PubMed] [Google Scholar]