Abstract
The Krüppel-like factor (KLF) family of transcription factors regulates diverse biological processes that include proliferation, differentiation, growth, development, survival, and responses to external stress. Seventeen mammalian KLFs have been identified, and numerous studies have been published that describe their basic biology and contribution to human diseases. KLF proteins have received much attention because of their involvement in the development and homeostasis of numerous organ systems. KLFs are critical regulators of physiological systems that include the cardiovascular, digestive, respiratory, hematological, and immune systems and are involved in disorders such as obesity, cardiovascular disease, cancer, and inflammatory conditions. Furthermore, KLFs play an important role in reprogramming somatic cells into induced pluripotent stem (iPS) cells and maintaining the pluripotent state of embryonic stem cells. As research on KLF proteins progresses, additional KLF functions and associations with disease are likely to be discovered. Here, we review the current knowledge of KLF proteins and describe common attributes of their biochemical and physiological functions and their pathophysiological roles.
Keywords: Krüppel-Like Factor, Proliferation, Differentiation, Inflammation, Cardiovascular Diseases, Tumorigenesis, Fat Metabolism, Induced Pluripotent Stem Cell
Chapter I: Background
A. Introduction
Krüppel-like factors (KLFs) are zinc finger-containing transcription factors that regulate proliferation, differentiation, development, and programmed cell death. Alterations in their functions have been associated with the pathobiology of numerous human diseases, including cardiovascular disease, metabolic disorders, and cancer. KLF family members have homology to the Drosophila melanogaster Krüppel protein, a member of the ‘gap’ class of segmentation gene products that regulates body segmentation in the thorax and anterior abdomen of the Drosophila embryo (361). KLFs also share homology with the transcription factor Sp1, one of the first mammalian transcription factors to be identified and characterized (205). Sp1 binds GC-rich regions in DNA via three C2H2-type zinc fingers. Because KLF proteins also contain this zinc-finger structure, they are classified as part of the Sp1/KLF family. Although Sp1 was initially viewed as a general transcription factor that regulates basal expression of housekeeping genes, it was later discovered that Sp1/KLF family members regulate a complex set of genes that have distinct roles in development and homeostasis of many tissue types. KLF proteins share common mechanisms of regulation, recruiting transcriptional regulatory proteins that include transcriptional co-activators and co-repressors, and other chromatin remodeling proteins. Together, KLFs function in the physiology and pathophysiology of many organ systems, including cardiovascular, respiratory, digestive, hematological, and immune systems. Many KLFs are also involved in tumor biology, in reprogramming somatic cells into inducible pluripotent stem (iPS) cells, and maintaining the pluripotent state of embryonic stem (ES) cells (198, 300, 415, 416). As the study of KLF proteins progresses, new biological and pathobiological roles for these factors are constantly being discovered. This review addresses the current understanding of biochemical, biological, and pathophysiological functions of KLF family members.
B. Conservation and Phylogenetic Analysis of KLFs
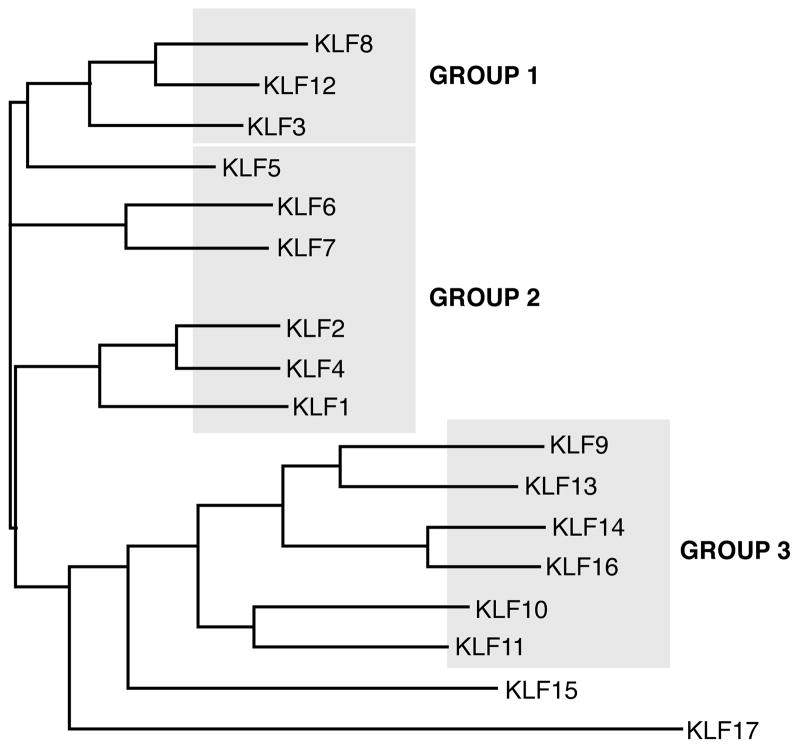
KLF proteins are conserved among mammals from human to rat, with many KLFs also having homologs in Gallus gallus (chicken), Danio rerio (zebrafish) and Xenopus laevis (frog). In addition, the Caenorhabditis elegans genome contains 3 predicted KLF homologs, klf-1, klf-2 and klf-3 (42). Tissue expression of the KLFs varies—some family members are expressed ubiquitously (e.g. KLFs 6, 10, and 11) whereas others are expressed in specific tissues—KLF1 is expressed predominantly in erythroid cells, KLF2 is highly expressed in lung, and KLFs 4 and 5 are very abundant in the gastrointestinal tract (331). KLF proteins exhibit homology in their carboxyl-terminal zinc finger domains that allow KLFs to bind GC-rich sites in promoter and enhancer regions of the genes they regulate. These structural similarities create overlap in their transcriptional targets. For example, in ES cells, KLFs 2, 4, and 5 can all bind and activate Esrrb, Fbxo15, Nanog, and Tcl1 (198). However, KLF proteins have distinct amino-terminal sequences that provide unique regions for interaction with specific binding partners. Phylogenetic analysis of protein sequences of the 17 human KLFs defines evolutionary distances of individual family members (Figure 1). Structural homologies of KLFs correlate with functional similarities; this connection is likely due to homologous protein interaction motifs in amino-terminal domains. Based on functional characteristics, KLF proteins can be divided into three distinct groups. KLFs in Group 1 (KLFs 3, 8, and 12) serve as transcriptional repressors through their interaction with the C-terminal binding protein (CtBP). Family members in Group 2 (KLFs 1, 2, 4, 5, 6, and 7) function predominantly as transcriptional activators. KLFs in Group 3 (KLFs 9, 10, 11, 13, 14, and 16) have repressor activity through their interaction with the common transcriptional co-repressor, Sin3A. KLFs 15 and 17 are more distantly related based on phylogenetic analysis and contain no defined protein interaction motifs.
Figure 1. Phylogenetic tree of human KLFs.
Multiple sequence alignment and phylogenetic analysis were performed using the ClustalW tool, Version 2.0.12. Analysis was conducted on full-length protein sequences of the 17 human KLF proteins. Structural analysis corresponded with the division of KLFs into distinct groups that have functional similarities.
C. Protein Structure of the KLFs
1. The Zinc Finger Domain
Zinc finger domains are common motifs in transcription factors. The most frequently encountered zinc finger motif is the C2H2 type, in which a zinc atom is tetrahedrally coordinated by two conserved cysteine and histidine residues that allow the domain to fold into a ββα structure (40). All members of the KLF family have three zinc finger motifs at the carboxyl-terminal ends of the proteins that are highly conserved. Their location within KLF protein structures are shown in Figure 2. The first and second zinc fingers contain 25 amino acids and the third contains 23 amino acids. Each zinc finger recognizes three base pairs in the DNA sequence and interacts with nine base pairs in total (294). Several studies have examined the preferred DNA binding motifs for a number of KLFs, based on binding studies of promoter regions and oligonucleotide screens (279, 376). DNA binding sites are similar among the KLF proteins; they include GC-rich sequences with a preference for the 5′-CACCC-3′ core motif, which is present in the β-globin gene promoter recognized by KLF1 (279). A number of other KLF proteins also bind this motif (431, 432, 464).
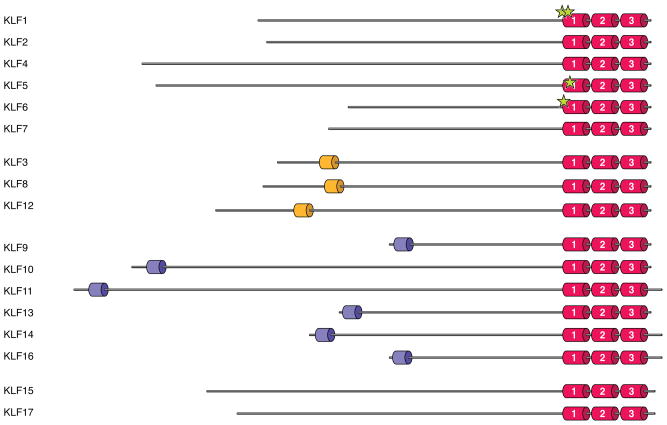
Figure 2. Protein structure of human KLF family members.
KLF proteins are grouped according to common structural and functional domains. KLFs are highly homologous in their carboxyl-terminal DNA-binding regions, which contain three C2H2 zinc finger motifs. The family members were grouped based on: (1) the ability to bind acetylases (KLFs 1, 2, 4, 5, 6, and 7); (2) the presence of a CtBP-binding site (KLFs 3, 8, and 12); or (3) the presence of a Sin3A-binding site (KLFs 9, 10, 11, 13, 14, and 16). Established sites of acetylation are marked by stars.
In addition to its role in DNA binding, the zinc finger region may be important for nuclear import. Nuclear localization signals have been identified in the zinc finger domains of KLFs 1, 4, 8, and 11 (275, 325, 377, 400) and in a basic region of KLF4 immediately amino-terminal to the zinc fingers (377).
2. Functional Binding Domains
The amino-terminal regions of KLFs vary significantly and allow them to bind different co-activators, co-repressors, and modifiers, resulting in functional diversity and specificity. Through the identification of KLF binding partners, several protein interaction domains have been characterized for subgroups of the KLFs that help define their function. Figure 2 shows the protein-binding domains within the protein structures of the KLFs.
C-Terminal Binding Protein (CtBP)-Binding Site
Although KLF3 was initially thought to function as a transcriptional activator, it was later shown to be a strong repressor, with its activity localized to a 74 amino-acid sequence in the amino-terminal region (427). In a yeast two-hybrid screen, KLF3 interacted with the transcriptional co-repressor CtBP. KLF3, as well as KLFs 8 and 12, bind CtBP via the consensus sequence PXDLS (362, 427, 432). This interaction mediates the repressor activities of KLFs 3 and 8, and the ability of KLF12 to repress expression of the AP-2α gene.
Sin3A-Binding Site
KLFs 10 and 11 also act as transcriptional repressors (90). This activity maps to three distinct repression sites in amino-terminal regions of the proteins designated R1, R2 and R3. The R1 domain was later shown to contain a Sin3-interacting domain (SID), a hydrophobic-rich motif that forms a α-helical structure to support interaction with Sin3 proteins, which are histone deacetylase-dependent co-repressors (488). KLFs 9, 10, 11, 13, and 16 share a conserved α-helical motif AA/VXXL that mediates their binding to Sin3A and their activities as transcriptional repressors (488). Whereas the protein sequence of KLF14 contains a putative SID, physical interaction between KLF14 and Sin3A has not been established. Surprisingly, KLF1, which does not contain a SID, binds and recruits Sin3A to function as a transcriptional repressor (65). However, this interaction was mediated through the carboxyl-terminal zinc finger domain of KLF1 rather than an amino-terminal hydrophobic consensus site.
Chapter II: Biochemical Mechanisms of KLFs
A. Common Interacting Proteins
1. Histone Acetyltransferases (HATs)
Sequence-specific DNA-binding factors like KLFs regulate transcription by recruiting chromatin modifiers, cofactors, and transcription machinery to promoters of specific genes. A number of KLFs from Group 1 of the phylogenetic analysis bind to co-regulators that have acetyltransferase activity, such as cAMP response element binding-binding protein (CBP), p300, and p300/CBP-associated factor (P/CAF) (127, 246, 283, 395, 491). KLF1 binds CBP/p300 and P/CAF in vivo and is subsequently acetylated at K288 and K302 (494) (Figure 2). Whereas acetylation of KLF1 at K288 is associated with its transactivation (370), acetylation at K302 is required for its interaction with the transcriptional repressor, Sin3A (64). KLFs 5 and 6 also bind CBP/p300 and are acetylated at defined sites (163, 246). KLFs 2 and 4 interact with CBP/p300 (127, 147, 369) and have putative acetylation sites that are conserved with the K288 site of KLF1, but these sites have not been empirically determined.
2. CtBP
KLFs 3, 8, and 12 interact with the transcriptional regulator CtBP through a consensus binding sequence in their amino-terminal regions (362, 427, 432). CtBP1 was originally characterized for its ability to bind the adenovirus E1A protein (39); and vertebrate CtBP1 and CtBP2 are established transcriptional repressors. One mechanism by which CtBPs promote gene silencing is through recruitment of histone deacetylases (HDACs) and histone methyl transferases to transcriptional complexes. These proteins deacetylate and methylate histones, respectively, to cause chromatin compaction and transcriptional silencing (78). CtBPs also have HDAC-independent mechanisms of action; CtBP1 and CtBP2 bind and inhibit HAT co-activators such as p300/CBP (78) and recruit other repressors that promote chromosome silencing, such as Ikaros (223) and members of the polycomb group (371). Therefore, the primary mechanism by which CtBP proteins repress transcription is through the recruitment of proteins that affect chromatin remodeling.
KLF3 is a negative regulator of adipogenesis and thereby a regulator of fat metabolism (409). Adipocyte differentiation is normally accompanied by decreased expression of KLF3, and overexpression of KLF3 blocks differentiation of 3T3-L1 cells in a CtBP-dependent manner. Given that CtBP binds NADH, CtBP might be a metabolic sensor for KLF3-related repressor function.
3. Sin3A
KLFs 9, 10, 11, 13, 14, and 16 contain a hydrophobic consensus sequence in their amino terminus that recruits the transcriptional repressor Sin3A (488). Mammalian Sin3 proteins (Sin3A and Sin3B) are large, multi-domain proteins made up of four highly conserved imperfect repeats, each of which fold into two amphipathic helices (221). These proteins bind HDAC1 and HDAC2 and other proteins, including Mad, Ume6, MeCP2, N-CoR, silencing mediator of retinoid and thyroid receptor (SMRT), and Ikaros (96). Given their size and various protein interaction sites, Sin3 proteins are likely to provide a scaffold for assembly of multi-unit complexes that modify chromatin conformation (221). HDACs, as part of these complexes, are essential for mediating repressor activity, as mutation of the HDAC binding sites in Sin3A or use of HDAC inhibitors abrogates repressor activity (206, 392).
B. Post-translational Modifications
Co-regulatory proteins modify KLF family members via acetylation, phosphorylation, ubiquitination, and sumoylation to refine their transcriptional activity.
1. Acetylation
A role for acetylation in regulating KLFs first came to light with the modulation of KLF1 activity by histone acetyltransferases (HATs) (491). Acetylation of KLF1 at K288 is required for binding of KLF1 to the β-globin locus, for recruitment of CBP to the locus, and for changes in chromatin structure that activate transcription (370). The zinc finger domain of KLF1 interacts with the amino terminus of histone H3 to coordinate this process. KLFs 4, 5, 6, and 13 are also acetylated, resulting in enhanced transcriptional activity (127, 246, 283, 395). In contrast, interaction of KLF5 with HDAC1 blocks binding of KLF5 to p300, reducing KLF5 binding and activation of transcriptional targets (269).
The acetylation of KLFs is regulated by signaling pathways that affect the association of HATs and HDACs with KLF proteins. For example, treatment of vascular smooth muscle cells (SMCs) with all-trans retinoic acid (ATRA) induces phosphorylation of HDAC2, which disrupts its interaction with KLF4 and allows KLF4 to become acetylated and bind the SM22α promoter (276). Furthermore, treatment of HaCaT epidermal cells with transforming growth factor-β (TGF-β induces an interaction between KLF5 and p300 that results in KLF5 acetylation (166). This modification not only affects the cofactors recruited by KLF5 to the promoter of the gene encoding the cell cycle inhibitor p15 (CDKN2B), but also alters KLF5’s regulation of CDKN2B, resulting in transcriptional activation rather than repression (166).
2. Phosphorylation
The transcriptional activity of several KLFs is regulated by phosphorylation. KLF1 is phosphorylated at serine and threonine residues within its transactivation region by casein kinase II, which increases transcription of KLF1 target genes (323). Phosphorylation and dephosphorylation of KLF5 have been reported to affect its binding to various effector proteins, including c-Jun (176), CBP (499), retinoic acid receptor-α (496), and the ubiquitin ligase, F-box and WD40 domain protein (Fbw7/hCDC4) (254). In T cells, phosphorylation of KLF13 by the serine/threonine kinase PRP4 increases KLF13 nuclear localization and transcriptional activation of chemokine C-C motif ligand 5 (CCL5) (187). In contrast, phosphorylation of KLF11 by extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) disrupts the interaction of KLF11 with Sin3A to prevent repression of Smad7 (122).
3. Ubiquitination
Ubiquitination is a multi-step protein modification process mediated by an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase (460). E3 ubiquitin ligases bind to the E2 ubiquitin conjugase and a substrate-specific domain in the target protein. Polyubiquitination provides a signal for the degradation of substrate proteins by the 26S proteasomal complex whereas monoubiquitination usually alters the activity of a protein (227). KLFs 1, 2, 4, 5, 6, and 10 are regulated by ubiquitination. KLF1 is ubiquitinated in vivo, and inhibition of the 26S proteasomal complex results in accumulation of KLF1 protein (341). Thus, ubiquitin-targeted degradation of KLF1 functions to maintain cellular levels of the protein. Levels of KLFs 4 and 6 are also regulated by degradation via ubiquitination, depending on physiological requirements. Following serum stimulation of quiescent HCT116 cells, KLF4 undergoes ubiquitin modification and rapid degradation through the proteasome pathway (74). Degradation of KLF4 eliminates its cell cycle inhibitory effects, allowing the cells to re-enter the cell cycle. KLF6 is ubiquitinated and undergoes proteasomal degradation in cultured cells following exposure to DNA damaging agents (17). In this study, KLF6 is degraded only with high levels of DNA damage. Lower levels of damage actually increase KLF6 levels, resulting in cell cycle arrest. Degradation of KLF6 might therefore regulate cell fate decisions between cell cycle arrest and death, depending on the extent of DNA damage. A splice variant of KLF6, KLF6-SV1, is ubiquitinated and subject to rapid degradation (112). In cancer cells that overexpress KLF6-SV1, which is often associated with poor survival, KLF6-SV1 binds to the pro-apoptotic protein NOXA, resulting in ubiquitination and degradation of both proteins, promoting cancer cell survival.
KLFs 2 and 5 bind the ubiquitin ligase WWP1, and ubiquitination of these KLFs promotes their rapid degradation by the proteasomal complex (60, 495). In some prostate and breast cancer cell lines, overexpression of WWP1 has been reported to increase degradation and loss of KLF5 (59). Similarly, KLF5 interacts with and is ubiquitinated by the E3 ubiquitin ligase and tumor suppressor, Fbw7/hCDC4, in a CDC4 phosphodegron (CPD)-dependent manner (254). However, deficiency of Fbw7/hCDC4 from some cancer cells delays turnover of KLF5, leading to the accumulation of KLF5.
KLF10 provides an example of regulation by mono-ubiquitination. KLF10 is a target of the E3 ligase Itch, which mediates both mono- and poly-ubiquitination in response to TGF-β signaling in naïve T cells (438). Mono-ubiquitination of KLF10 by Itch promotes transcriptional activation of the KLF10 target Foxp3.
4. Sumoylation
Many transcription factors and co-regulators are modified by the small ubiquitin-like modifier (SUMO) peptide, resulting in enhancement or suppression of their transcriptional activity. KLF3, which normally functions as a transcriptional repressor, interacts with the E2 SUMO-conjugating enzyme Ubc9 and is covalently modified by SUMO-1 in vitro and in vivo (332). KLF3 is sumoylated at lysines K10 and K197, and mutations at these sites compromise the transcriptional repressor activities of KLF3. KLF8 promotes cell cycle progression through positive regulation of the CCDN1 (cyclin D1) promoter. KLF8 can be sumoylated through interaction with several SUMO E3 ligase family members including protein inhibitor of activated STAT1 (PIAS1), PIASy and PIASxα (455). Overexpression of SUMO-1 suppresses the cell-cycle promoting effects of KLF8, whereas mutation of the primary sumoylation site in KLF8 increases its effects on cell cycle progression. KLFs 4 and 5 also interact with the SUMO E3 ligase, PIAS1. PIAS1 promotes KLF4 sumoylation and subsequent degradation so it no longer represses α-smooth muscle actin (α-SMA) in SMCs (213). Alternatively, sumoylation of KLF5 increases its nuclear localization and promotes its activation of the cell cycle genes cyclin D1 and Cdc2 (119, 120).
The physiological roles of KLFs 1 and 5 are modulated by sumoylation. In erythroid progenitor cells, KLF1 acts as a transcriptional repressor of megakaryocyte differentiation by blocking expression of the transcription factor FLI-1, which is required for megakaryopoiesis (144). This activity depends on sumoylation of KLF1 at K74—mutation of this site attenuates its activity as a repressor (380). KLF5 has an important role in lipid metabolism through peroxisome proliferator-activated receptor-δ(PPAR-δ) signaling (317). Under basal conditions, KLF5 is sumoylated and associates with transcriptional repressor complexes that regulate genes associated with lipid oxidation, including CPt1b, Ucp2 and Ucp3. When PPARδ interacts with an agonist, KLF5 is de-sumoylated and binds to transcriptional activation complexes that drive expression of genes that control lipid metabolism (317). Thus, KLF5 can “switch” from transcriptional repression to activation, depending on its sumoylation status.
Chapter III: Cell-Based Functions of KLFs in Normal Biological Processes
A. Proliferation
Many KLF family members function as regulators of cell growth. KLFs 4 and 5, which are highly expressed in intestinal tissues, have been studied extensively in regulating proliferation. KLF5 is localized to actively proliferating cells at the base of intestinal crypts and promotes proliferation of different types of cultured cells, including fibroblasts and epithelial cells (54, 410, 473). KLF5 is upregulated in proliferating vascular SMCs and is activated by serum stimulation of quiescent NIH3T3 cells. KLF5 participates in several growth factor signaling pathways, including the RAS/MAPK, protein kinase C, and phosphatidyloinositol-3-kinase (PI3K) pathways (115). KLF5 promotes proliferation by accelerating cells through G1/S and G2/M phases of the cell cycle. A number of transcriptional targets of KFL5 promote cell cycle progression, including cyclin D1 (301), cyclin B1 and Cdc2 (297). KLF5 also represses expression of the cell cycle inhibitory proteins p27 and p15 (56). However, the ability of KLF5 to promote proliferation can be downregulated or reversed in cultured cells through activation of signaling pathways that suppress cellular proliferation, including retinoic acid receptor signaling and TGF-β signaling (54, 163, 164).
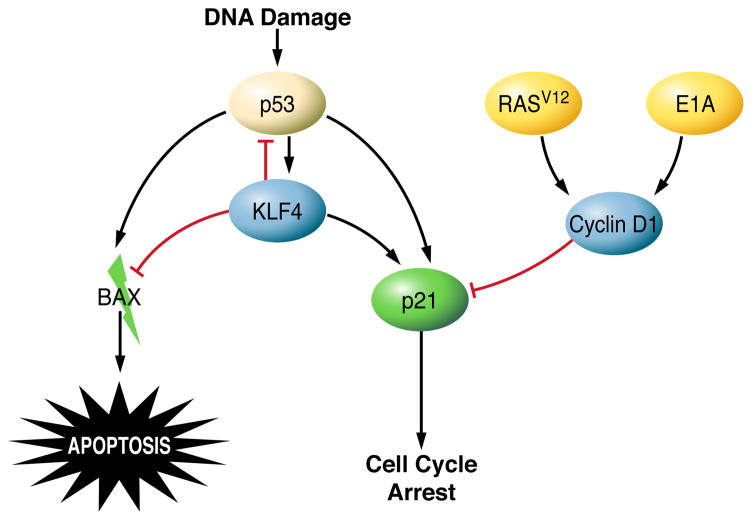
In contrast to KLF5, KLF4 inhibits cell cycle progression. In the intestinal mucosa, KLF4 is expressed in differentiated cells at the luminal surface that have undergone growth arrest (272, 375). Studies in NIH3T3 cells indicate that KLF4 is expressed at very low levels in actively proliferating cells but is induced as cells enter quiescence following serum-starvation or contact inhibition (375). Furthermore, transcriptional profiling studies reveal a global inhibitory function of KLF4 in regulating genes that promote transcription and the biosynthesis of proteins and cholesterol (458). The primary mechanism by which KLF4 contributes to cell cycle arrest has been determined from studies of KLF4 in DNA damage-induced growth arrest. Exposure of cultured cells to DNA damaging agents or γ-irradiation increases the levels of KLF4 in a p53-dependent manner (479, 493). KLF4 binds directly to the promoter of the gene that encodes the cell cycle inhibitor p21Cip1/Waf1 and recruits p53 to activate expression of p21Cip1/Waf1 (493). KLF4 also inhibits expression of the cell cycle-promoting genes CCND1 (cyclin D1) (373) and CCNB1 (cyclin B1) (481). Furthermore, in response to DNA damage, KLF4 suppresses transcription of the gene that encodes cyclin E and prevents chromosomal amplification (480). Thus, KLF4 activates cell cycle checkpoints to prevent inappropriate cell cycle progression and maintain DNA integrity.
KLFs 6 and 8, which are ubiquitously expressed, also regulate cell cycle progression. KLF6 induces G1 cell cycle arrest by upregulating expression of p21Cip1/Waf1 and repressing CCND1, thus disrupting formation of cyclin D1/cyclin-dependent kinase 4 (CDK4) complexes (27, 373). However, KLF8, as a mediator of focal adhesion kinase signaling, activates the CCND1 promoter, and ectopic expression of KLF8 promotes cell cycle progression (500).
KLFs 10 and 11, initially identified as TGF-β-inducible genes, play significant roles in TGF-β mediated cell growth control and differentiation. Ectopic expression of KLF10 can mimic many of the effects of TGF-β signaling including suppression of proliferation (92, 177). KLF10 mediates TGF-β signaling by blocking expression of Smad7, a negative regulator of TGF-β (202), and activating expression of the positive effector, Smad 2 (203). These events promote expression of p21Cip1/Waf1 and thereby inhibit cell cycle progression (202). KLF11 mediates TGF-β/Smad signaling through downregulation of Smad7 by recruiting the transcriptional repressor Sin3A to the Smad7 promoter (122). KLF11 also suppresses cell growth through TGF-β dependent regulation of c-Myc (122). Upon TGF-β stimulation of epithelial cells, KLF11 interacts with Smad3 to bind a TGF-β-inhibitory element (TIE) within the c-Myc promoter and blocks its expression (43).
As another example of regulating cell growth, KLFs participate in the maintenance of quiescence in lymphocytes. Naïve lymphocytes are held in a non-cycling, G0 growth phase until they are activated by specific antigens. Quiescence requires negative regulation of cell cycle progression and expression of genes that maintain small cell size and low metabolic activity. Overexpression of KLF2 in Jurkat leukemia T cells inhibits growth and DNA synthesis. KLF2 promotes a quiescent phenotype by blocking expression of the growth-promoting c-Myc (44) and upregulating the cell cycle inhibitor p21Cip1/Waf1 (462). Similar activities have been ascribed to KLF4 in B lymphocytes. KLF4 levels are decreased upon B cell activation, and ectopic expression of KLF4 induces G1 cell cycle arrest (484). As with KLF2, the arrest is associated with increased expression of p21Cip1/Waf1 and decreased expression of c-Myc, as well as reduced expression of CCND1.
KLFs therefore regulate proliferation in a variety of cell types through transcriptional control of cell cycle regulatory components. Transcriptional targets include cyclins D1, D2, B1, E, and cyclin-dependent kinase inhibitors p21Cip1/Waf1, p15 and p27, as well as the proliferative factor c-Myc.
B. Differentiation
KLFs play critical roles in differentiation during development and in maintenance of tissue homeostasis. As a primary example, KLF1 regulates differentiation during erythropoiesis. The development of red blood cells requires carefully regulated changes in cell morphology, globin expression, and heme synthesis. KLF1 mediates the switch from expression of fetal γ-globin to adult β-globin and regulates transcription of genes that encode cytoskeletal proteins, heme synthesis enzymes, and blood group antigens (118, 183, 339). Binding of KLF1 to the CACCC consensus site at nucleotide position −90 in the β-globin promoter initiates the recruitment of large transcriptional complexes, including the mammalian SWI/SNF chromatin-remodeling proteins, BRG1, BAF155, and CBP/p300 (204). KLF1 therefore regulates the maturation of erythroid cells by allowing the chromatin structure to open and become transcriptionally active.
KLF1 also regulates the lineage progression of megakaryocyte-erythroid progenitor (MEP) cells. While promoting erythroid maturation, KLF1 simultaneously suppresses megakaryocyte differentiation by antagonizing the transcription factor FLI-1. FLI-1 is an ETS-related factor that is normally expressed in the MEP and its activity is required for megakaryopoiesis. Expression of FLI-1 is negatively regulated in the MEP by a SUMO-modified form of KLF1 (380). Upon sumyolation, KLF1 interacts with the Mi-2β component of the NuRD repression complex and recruits HDAC, indicating that Mi-2β/HDAC activity is involved in the SUMO-dependent repression of FLI-1.
In the intestinal epithelium, KLF4 expression is restricted to terminally differentiated epithelial cells of the mucosa, where it promotes differentiation (272). Transcriptional targets of KLF4 include Lama1, which encodes the basement membrane component Laminin-1, and the gene that encodes the enterocyte differentiation marker intestinal alkaline phosphatase (181, 336). A specific role for KLF4 in goblet cell differentiation is demonstrated in studies of Klf4−/−mice. In these mice, the number of colonic goblet cells is significantly reduced and goblet cells have aberrant morphology, with low levels of the cell-specific marker MUC2 (211).
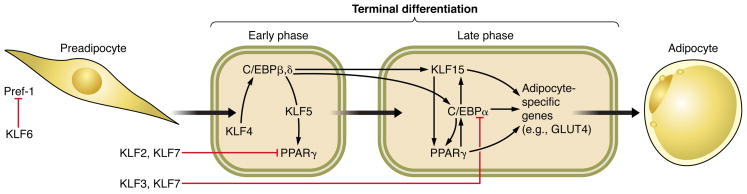
A number of KLFs participate in differentiation during adipogenesis. KLFs 2, 3, 4, 5, 6, 7, 11, and 15 have all been reported to function as positive or negative regulators of adipocyte differentiation, and their roles are described in detail in Chapter IV, Section F, “The Role of KLFs in Metabolic Regulation.” Other tissues in which KLFs regulate differentiation include KLFs 2, 4, and 5 in vascular SMCs (1, 234, 378), KLF2 in thymocytes (51), KLF13 in cardiomyocytes (307), KLF7 in olfactory sensory neurons (207), KLF4 in corneal epithelial cells (413), KLF4 in monocytes (6), KLF5 in lung respiratory epithelial cells (440), and KLF9 in hippocampal adult born neurons (363).
C. Apoptosis
Many of the studies examining the role of KLFs in apoptosis have arisen from observations of up or downregulation of KLFs in cancer. The most highly characterized KLFs in regard to apoptosis are KLFs 4, 5, and 6. KLF4 is generally regarded as a tumor suppressor due to its ability to induce cell cycle arrest. However, when the ability of KLF4 to induce growth arrest is inactivated (351), it can actually contribute to tumor progression because it also inhibits apoptosis. KLF4 regulates apoptosis following DNA damage, when cells must either activate cell cycle checkpoints and repair machinery or commit to apoptosis. In γ-irradiated RKO colon cancer cells, ectopic expression of KLF4 significantly reduces the percentage of apoptotic cells, causing them to instead undergo growth arrest (149). In this study, upregulation of KLF4 blocks expression of the pro-apoptotic protein BAX by inhibiting p53-dependent transactivation of the BAX promoter. In another study of MDA-MB-134 breast cancer cells, KLF4 modulates apoptosis by binding directly to the promoter of p53 and suppressing its transcription (352). KLF4 may be a determining factor in the outcome of p53 responses to DNA damage, depending on the extent of damage (507). Following low, cytostatic doses of adriamycin, KLF4 is induced and promotes cell cycle arrest. However, when cells are exposed to high levels of adriamycin, KLF4 expression is inhibited and p53-dependent apoptosis proceeds. Ectopic expression of KLF4 prevents cell death in response to high levels of adriamycin. KLF4 is therefore an important determinant of cell cycle arrest or death induction by p53.
KLF6 is similar to KLF4 in that it regulates growth arrest and can function as a tumor suppressor. KLF6 is often mutated or deleted in human prostate tumors (305), and ectopic expression of KLF6 in prostate cancer cells or non-small cell lung cancer induces apoptosis (189, 193). However, silencing of KLF6 in HepG2 and COS-7 cells sensitizes these cells to apoptosis and increases the sub-G1 population (98, 388). Situations in which KLF6 appears to suppress apoptosis may be attributable to specific splice variants of the KLF6 gene that can act in a manner distinct from full-length KLF6. For example, the KLF6-SV1 isoform is overexpressed in ovarian cancer and binds the BH3-only protein NOXA to block apoptosis (112). Downregulation of this splice variant induces spontaneous apoptosis in ovarian, lung, and prostate cancer cell lines (111, 112, 302, 358), suggesting that KLF6-SV1 might be a therapeutic target for cancer.
Although the exact role of KLF6 in apoptosis is controversial, KLF5 is clearly a suppressor of apoptosis. In EU-8 leukemia cells that have low levels of KLF5, introduction of ectopic KLF5 induces the expression of survivin, an inhibitor-of-apoptosis (IAP) protein (509). Conversely, blocking KLF5 expression with small interfering RNA (siRNA) downregulates survivin and sensitizes leukemia cells to chemotherapeutic-induced apoptosis. As a mechanism of action, KLF5 binds directly to p53 and blocks p53-regulated repression of survivin (509). In HCT116 colon cancer cells, KLF5 is activated following exposure to 5-fluorouracil and ultraviolet irradiation (502). Depletion of KLF5 from these cells increases their sensitivity to apoptosis in response to DNA-damaging agents. In this study, KLF5’s pro-survival activity does not depend on the activity of p53, but is instead associated with regulation of Pim1—a kinase that negatively regulates the pro-apoptotic protein BAD. Finally, Suzuki et al. have shown that KLF5 inhibits apoptosis of SMCs in vascular lesions by interacting with poly(ADP-ribose) polymerase-1, a nuclear enzyme that controls DNA repair and apoptosis (411).
Chapter IV: Organ-Based Functions in Physiology and Pathophysiology
KLF family members regulate key events in development, maintenance of homeostasis, and adaptive responses to physiological or pathobiological stimuli in mammalian tissues. The following sections describe the functions of KLF proteins in various organs and ways in which their dysregulation contributes to the pathogenesis of diseases. Table I summarizes the expression, function, and pathobiological roles of mammalian KLFs.
Table I.
Expression, Function, and Pathobiology of Mammalian KLF Family Proteins
| Name | Also known as | Expression pattern | Tissue function/disease association | Gene knockout phenotypes | References |
|---|---|---|---|---|---|
| KLF1 | EKLF (Erythroid Krüppel-like factor) | Erythroid cells, fetal liver, adult bone marrow and spleen | Erythropoiesis; megakaryocyte differentiation | Defective hematopoiesis and lethal β-thalassemia at E14.5 | (144, 311, 335) |
| KLF2 | LKLF (Lung Krüppel-like factor) | Lungs, erythroid cells, vascular endothelial cells, T lymphocytes, white adipose tissue | Erythropoiesis; regulator of hemodynamics; lung development; T cell survival, migration and trafficking; inhibitor of adipogenesis; important in generating induced-pluripotent stem (iPS) cells | Embryonic lethality due to compromised vessel integrity/rupture; delayed lung development; defective T-lymphocyte and adipocyte differentiation | (21, 51, 234, 235, 243, 295, 448, 464) |
| KLF3 | BKLF (Basic Krüppel-like factor) | Widely expressed; abundant in erythroid cells | Inhibits adipocyte differentiation | Decreased white adipose tissue | (95, 409) |
| KLF4 | GKLF (Gut-enriched Krüppel-like factor); EZF (Endothelial zinc finger protein) | Epithelia of gut, skin and lungs; endothelial cells; early precursor B cells | Intestinal epithelial homeostasis; tumor suppressor in colon cancer; oncogene in breast cancer and squamous cell carcinoma; promotes adipogenesis; critical factor in generating iPS cells | Perinatal lethal due to dehydration with skin barrier defect; abnormal differentiation of goblet cells in the colon | (151, 211, 272, 367, 415, 416, 433) |
| KLF5 | IKLF (Intestinal-enriched Krüppel-like factor); BTEB2 (Basic transcription element binding protein-2) | Enriched in gut epithelia; expressed in epidermis, vascular smooth muscle and white adipose tissue; also developing/perinatal skeleton and lung | Intestinal homeostasis; cardiac remodeling; lung maturation and morphogenesis; promotes adipogenesis; important in generating iPS cells; dysregulated expression in colon, breast, prostate cancer and other solid tumors | Embryonic lethal at E8.5; heterozygous and tissue-specific deletions show: reduced cardiac fibrosis and hypertrophy in response to stress, deficiencies in white adipose tissue development, skeletal growth retardation, defective perinatal lung morphogenesis; heterozygous deletion drastically reduces intestinal tumor formation in ApcMin mice and ApcMin/KRASV12 mice | (52, 57, 58, 89, 271, 272, 295, 298, 299, 318, 378, 379, 417, 423, 440) |
| KLF6 | ZF9 (Zinc-finger transcription factor-9); CPBP (Core promoter binding protein); GBF (G-box binding factor) | Strongly expressed in placenta and developing hindgut, heart, lung and kidney; adult endothelial cells, heart, liver, lung, kidney, intestine | Promotes vascular remodeling; stimulates adipogenesis; tumor suppressor in prostate cancer; silenced or mutated in a variety of other cancers | Embryonic lethal at E12.5, with reduced hematopoiesis and poor yolk sac vascularization | (13, 113, 135, 240, 267, 268, 305, 389) |
| KLF7 | UKLF (Ubiquitous Krüppel-like factor) | High expression in brain and spinal cord; expressed in developing central and peripheral nervous systems; | Inhibits adipocyte differentiation; single-nucleotide polymorphisms (SNPs) associated with type 2 diabetes | Neonatal lethality by P3 associated with defects in neurite outgrowth and axonal misprojection | (209, 239, 241, 268) |
| KLF8 | - | Low, ubiquitous expression | Elevated in ovarian and other human cancers | - | (432, 445) |
| KLF9 | BTEB (Basic transcription element-binding protein) | Broadly expressed; high in developing brain, thymus, epithelia and smooth muscle of gut and bladder | Endocrine-responsive cancers, including endometrial cancer | Normal lifespan, but specific behavioral abnormalities; subfertility and parturition defects in females; shorter small intestinal villi | (265, 290, 383–385) |
| KLF10 | TIEG1 (TGF-inducible gene-1); mGIF (murine glial cell-derived neurotrophic factor (GDNF)-inducible factor) | Broadly expressed, including pancreas, kidney, lung, brain, liver, heart and testis | T eg cell differentiation and T cell activation; bone development; SNPs associated with volumetric bone mineral density; implicated in breast cancer | Normal lifespan, but osteopenia in females; cardiac hypertrophy in males; defects in structure and healing of tendons | (26, 50, 175, 343, 405, 406, 468, 476) |
| KLF11 | FKLF (Fetal-like globin gene-activating Krüppel-like factor); TIEG2 (TGF- -inducible gene-2); MODY7 (Maturity-onset diabetes of the young-7) | Erythroid cells in fetal liver; ubiquitous in adult including pancreas | Tumor suppressor in pancreatic cancer; variants associated with early-onset type 2 diabetes | Normal development, lifespan, fertility | (12, 43, 91, 122, 308, 396) |
| KLF12 | AP-2rep (Activator protein-2α repressor) | Expression in developing brain and kidney; adult kidney, very low in liver and lung | Implicated in breast and gastric cancer | - | (192, 296, 353, 408) |
| KFL13 | BTEB3 (Basic transcription element-binding protein-3); FKLF2 (Fetal -like globin gene-activating Krüppel-like factor-2); RFLAT-1 (RANTES factor of late-activated T lymphocytes 1) | Broadly expressed; high temporal expression in developing heart, brain, thymus, epidermis, gut and bladder epithelia | Cardiac development; B and T cell development | Splenomegaly and aberrant erythroblast differentiation; abnormal T and B cell development; increased T cell survival | (157, 242, 265, 322, 506) |
| KFL14 | - | Ubiquitous | Variants associated with basal cell carcinoma | - | (328, 364, 401) |
| KLF15 | KKLF (Kidney-enriched Krüppel-like factor) | Low cardiac expression during development; adult kidney, liver, pancreas, heart, skeletal muscle, lung, ovary | Regulator of gluconeogenesis; negative regulator of cardiac hypertrophy and fibrosis | Normal viability, fertility; exaggerated cardiac fibrosis and hypertrophy in response to stress; defective amino acid metabolism resulting in hypoglycemia | (134, 159, 428, 441) |
| KLF16 | BTEB4 (Basic transcription element-binding protein-4); DRRF (Dopamine receptor regulating factor) | Highly expressed in regions of developing brain; lower expression in thymus, duodenum, kidney, liver, heart, bladder and lung | - | - | (99, 191) |
| KLF17 | Zfp393 (Zinc finger protein 393) | Spermatids and oocytes | - | - | (431, 471) |
A. The Cardiovascular System
1. Development of the Cardiovascular System
KLF13 is required for cardiac development—it is expressed in a variety of tissues in the mouse embryo, including the developing heart (265). At E10.5, Klf13 is present predominantly in the atrial myocardium and endocardial layer (242). By E12.5, the atria and ventricles both contain Klf13 (242). KLF13 functionally and physically interacts with GATA-4, which regulates cardiac-specific genes during embryonic and postnatal heart development (55, 284). Knockdown of KLF13 in Xenopus leads to atrial septal defects and hypotrabeculation similar to those observed in humans or mice with hypomorphic GATA-4 alleles (242). One of the transcriptional targets of KLF13 is cyclin D1, through which KLF13 regulates cardiac cell proliferation (307). Klf13−/− mice have enlarged hearts and increased susceptibility to cardiac vacuolar lesions (157); patients with microdeletion of chromosome 15q13, which includes KLF13, have cardiac defects (429).
During embryonic development, KLF2 is expressed in vascular endothelial cells (10). Klf2−/−mice die in utero from intra-embryonic and intra-amniotic hemorrhage, despite normal vasculogenesis, angiogenesis, and cardiogenesis (234, 447). However, the recruitment of pericytes and vascular SMCs to the tunica media of Klf2−/− embryos is defective, resulting in compromised vessel integrity that is manifested by aneurysmal dilatation of arteries and veins and subsequent rupture (234). A subsequent study demonstrated the failure of mural SMCs to migrate around normally developed endothelial cells in the aorta of Klf2−/− embryos (461). Interestingly, in mouse embryos, endothelial expression of Klf2 correlates with the rise of fluid shear forces; conditional deletion of Klf2 from the endothelium results in embryonic lethality from high-output heart failure (243). In contrast to Klf2−/− mice, mice with endothelial-specific loss of Klf2 do not suffer from primary vascular abnormality or hemorrhage; instead, increased cardiac output is a result of loss of peripheral vascular resistance and reduced vessel tone (243). KLF2 is therefore an important in vivo regulator of hemodynamics whose regulation in response to fluid shear stress is necessary for normal cardiovascular development.
2. Pathobiology of the Heart
Whereas KLF13 is highly expressed in fetal heart, expression of KLF15 in the heart increases significantly following birth and is highest in the adult heart (134). KLF15 levels are reduced in hypertrophic hearts of rodents following pressure overload and in biopsy samples from patients with left ventricular hypertrophy due to chronic valvular aortic stenosis (134). Similarly, expression of KLF15 in neonatal rat ventricular muscle cells is reduced by prohypertrophic stimuli such as phenylephrine and endothelin-1 (134). Overexpression of KLF15 in cadiomyocytes reduces cell size and expression of atrial natriuretic factor and B-type natriuretic peptide—both are expressed in the fetal heart and associated with cardiac hypertrophy (134). Klf15−/− mice are viable but have exaggerated, hypertrophic remodeling of the heart in response to pressure overload, manifested by increased heart weight, left ventricular cavity enlargement, impaired left ventricular systolic function, and increased expression of hypertrophic genes (134). KLF15 might inhibit cardiac hypertrophy by attenuating the functions of GATA-4 and myocyte enhancer factor 2 (MEF2)—transcription factors that are critical effectors of cardiac hypertrophy (97, 338).
Following pressure overload, Klf15−/− mice also develop fibrosis and deposition of excessive amounts of collagen in the heart (441). This phenotype is associated with increased expression of connective tissue growth factor (CTGF), which has been implicated in the pathogenesis of fibrotic diseases of the heart (62, 312). CTGF expression is regulated by diverse stimuli, including TGF-β1, which promotes fibrosis (62). Incubation of neonatal rat ventricular fibroblasts with TGF-β1 reduces expression of KLF15 and increases that of CTGF (441). Conversely, overexpression of KLF15 suppresses basal and TGF-β1-induced expression of CTGF (441). These studies demonstrate that KLF15 is a negative regulator of cardiac fibrosis.
In contrast to the anti-fibrotic action of KLF15 in the heart, KLF5 promotes fibrosis. Following infusion of angiotensin II, a potent mediator of cardiac hypertrophy (270), Klf5+/− mice have less cardiac hypertrophy and interstitial fibrosis than wild-type mice given angiotensin II (378). In addition, the level of TGF-β in the hearts of Klf5+/− mice given angiotensin II is significantly lower than that of wild-type mice, suggesting that TGF-β lies downstream from KLF5 (378). In cultured cardiac fibroblasts, angiotensin II increases expression of KLF5 and platelet-derived growth factor (PDGF)-A, which controls tissue remodeling (340). Moreover, KLF5 is directly responsible for the induction of PDGF-A expression in response to angiotensin II stimulation (378). The essential role of cardiac fibroblasts in mediating the adaptive response to pressure overload was recently confirmed in mice with cardiac fibroblast-specific deletion of Klf5 (417). An attempt to identify compounds that regulate KLF5 activity yielded several retinoid acid receptor (RAR) ligands. Am80, a synthetic retinoid agonist, reduces PDGF-A promoter activity in cells that co-express KLF5 and RAR-α (378). Wild-type mice given Am80 have reduced angiotensin II-induced cardiac hypertrophy, which approximates that observed in Klf5+/− mice given angiotensin II (378). Reagents that alter KLF5 activity might therefore be developed to control cardiac remodeling.
KLF10 is important during development of the heart. Male Klf10−/− mice develop cardiac hypertrophy by 16 months (343); other phenotypes include asymmetric septal hypertrophy and increased ventricular size, wall thickness, and heart weight (343). The hearts of Klf10−/− mice have evidence of myocyte disarray and myofibroblast fibrosis (343), but the mechanisms by which KLF10 prevents cardiac hypertrophy are not known.
3. Endothelial Biology and Pathobiology
KLFs 2, 4, and 6 are expressed in endothelial cells (36, 226, 477), where they have important roles in cell function and pathobiology (13). Expression of KLF4 in endothelial cells is induced by shear stress (274) and pro-inflammatory stimuli (172). Overexpression of KLF4 in endothelial cells activates expression of anti-inflammatory and anti-thrombotic genes, such as those that encode endothelial nitric-oxide synthase (eNOS) and thrombomodulin, whereas reduction of KLF4 levels increases tumor necrosis factor-α (TNF-α)-induced expression of vascular cell adhesion molecule-1 (VCAM-1) and tissue factor (172). In one study, overexpression of KLF4 in endothelial cells significantly reduces TNF-α-induced E-selectin and VCAM-1 expression (277). Moreover, KLF4 significantly reduces adhesion of inflammatory cells to endothelial cells and prolongs clotting time under inflammatory conditions. These findings support an anti-inflammatory function for KLF4 in endothelial cells (172).
Endothelial expression of KLF6 is induced after vascular injury and is responsible for the transcriptional activation of several genes involved in vascular remodeling, including urokinase plasminogen activator, endoglin, collagen α1(I), TGF-β1, and TGF-β receptor type I (36, 226). This transactivation requires the participation and interaction between KLF6 and Sp1 (36). KLF6 was shown to regulate endothelial cell motility by negatively regulating, in conjunction with Sp2, transcription of the gene encoding matrix metalloproteinase-9 (MMP-9) (105). Disruption of the KLF6/Sp2 repressor complex by small heterodimeric partner, which is activated by farnesoid X receptor, results in upregulation of MMP-9 and increased cell motility. KLF6 is therefore an important mediator of vascular remodeling and response to injury.
Most information on the function of KLFs in endothelial cell biology and pathobiology have come from studies of KLF2 (13). KLF2 expression is highly upregulated in cultured endothelial cells subjected to prolonged laminar flow shear stress (108, 190, 369). These results were supported by the in vivo observation that KLF2 is restricted to the endothelium of healthy human aorta, in regions predicted to be exposed to laminar shear stress (108). In contrast, KLF2 expression is decreased or absent from regions of vessels exposed to non-laminar shear stress, such as bifurcations of the aorta to the iliac and carotid arteries (109)—these bifurcation areas are also susceptible to atherosclerosis. KLF2 is one of several differentially expressed genes in endothelial cells subjected to ‘atheroprotective’ waveforms, relative to ‘atheroprone’ waveforms, which are modeled after the flow patterns in arteries from healthy human subjects (100). Consequences of flow-induced KLF2 expression in endothelial cells include activated expression of eNOS (107, 109, 329, 369) and repressed expression of angiotensin-converting enzyme, endothelin-1, and adrenomodullin—all of which are involved in the control of vascular tone in response to flow (107, 109). KLF2 therefore has flow-dependent, atheroprotective functions in endothelial cells.
The atheroprotective activity of KLF2 was demonstrated by its induction in response to 3-hydroxy-3-methyblutaryl coenzyme A inhibitors (statins) (195, 330, 368, 426), which protect against atherosclerosis (24, 249). Importantly, upregulation of KLF2 is required for many of the transcriptional effects of statins in endothelial cells, so KLF2 might mediate their atheroprotective effects (330). In a background of apolipoprotein E (ApoE−/−) deficiency, Klf2+/−mice have increased diet-induced atherosclerosis, compared with ApoE−/− mice (14). Interestingly, peritoneal macrophages isolated from Klf2+/− mice have increased lipid uptake, compared with wild-type macrophages; this increase might be mediated by the ability of KLF2 to activate expression of aP2, a lipid chaperon involved in macrophage lipid accumulation and atherogenesis (35, 263).
Contrary to the upregulation of KLF2 by shear stress and statins, expression of KLF2 in endothelial cells is suppressed by pro-inflammatory cytokines such as TNF-α and interleukin (IL)-1β, which are important in the pathogenesis of atherosclerosis (233, 369). TNF-α inhibits KLF2 via NF-κB and HDAC4, which cooperatively inhibit MEF2—an essential transcriptional activator of KLF2 (190). Conversely, overexpression of KLF2 in endothelial cells inhibits IL-1β-dependent induction of the pro-inflammatory molecules VCAM-1 and E-selectin (369). Consistent with these findings, T-cell rolling and attachment are significantly reduced in endothelial monolayers transduced with KLF2 (369). Similarly, IL-1β-induced leukocyte-endothelial cell interactions are inhibited by ectopic expression of KLF2 (329). Importantly, KLF2 inhibits endothelial activation by many pro-inflammatory stimuli, including IL-1β, TNF-α, lipopolysaccharide (LPS), and thrombin (13, 329, 369). The mechanisms by which KLF2 achieve its anti-inflammatory function are multiple and include nhibition of NF-κB (369), activator protein-1 (AP-1) (34), and activating transcription factor 2 (137). Thus, KLF2 is an important regulator of pro-inflammatory cytokine-mediated activation of endothelial cells.
In addition to its anti-inflammatory activity, KLF2 regulates endothelial thrombotic function by controlling expression of factors that maintain an anti-thrombotic endothelial surface. Overexpression of KLF2 in endothelial cells induces thrombomodulin and eNOS (253), which inhibit blood coagulation and platelet aggregation, respectively (125). In contrast, KLF2 inhibits expression of the pro-coagulant factors plasminogen activator inhibitor-1 (PAI-1) and cytokine-mediated induction of tissue factor (253). KLF2 also inhibits thrombin-mediated activation of endothelial cells by inhibiting expression of the thrombin receptor protease activated receptor-1 (329). Consequently, KLF2 overexpression increases blood clotting time and flow rates under basal and inflammatory conditions (253). These studies demonstrate that KLF2 is an important transcriptional regulator of endothelial thrombotic function.
KLF2 regulates vascular remodeling through inhibition of angiogenesis, proliferation, and cell migration (13). In vivo, KLF2 overexpression inhibits vascular endothelial growth factor (VEGF)-mediated angiogenesis and tissue edema (28). In vitro, ectopic KLF2 expression slows VEGF-mediated activation of endothelial cells, resulting in reduced intracellular calcium influx, proliferation, and expression of pro-inflammatory genes (28). KLF2 exerts these anti-angiogenic effects via its ability to inhibit expression of VEGF receptor 2 (VEGFR2) (28). Furthermore, KLF2 attenuates endothelial migration by regulating expression of genes that control cell migration, including VEGFR2 and semaphorin-3 (107). Endothelial expression of KLF2 has paracrine effects, reducing migration of co-cultured SMCs (261); this is similar to the failure of SMCs to properly migrate to the developing aorta in Klf2−/− mouse embryos (461). These findings reveal the important link between endothelial KLF2 expression and SMC function.
4. Vascular Smooth Muscle Biology and Pathobiology
Several KLFs have important roles in the biology and pathobiology of vascular SMCs, which contribute to blood vessel walls; KLFs 4 and 5 are the best characterized (170). In rabbit SMCs, KLF5 regulates transcription of embryonic smooth muscle myosin heavy chain by binding to a specific sequence in the gene’s promoter (451). Expression of KLF5 in SMC is developmentally regulated—it is abundant in fetal but not in adult aortic SMCs of humans and rabbits (184, 451). However, under pathological conditions, such as coronary atherosclerosis, vein graft hyperplasia, and response to vascular injury, KLF5 expression is reactivated (15, 184, 313, 314). Following aortic balloon injury, KLF5 becomes expressed in the neointimal layer of the blood vessels (184). KLF5 is also highly expressed in most human coronary lesion samples (both primary and restenotic) collected during atherectomy (184)—the presence of KLF5-positive lesions has been correlated with the incidence of restenosis (184). KLF5 is more frequently expressed in SMCs cultured from human coronary atherectomy samples that have the ability for outgrowth; the ability for outgrowth correlates with a shorter time to restenosis in patients (357). In SMCs, KLF5 expression is induced by various proliferative or inflammatory stimuli, including angiotensin II, TNF-α, survivin, and MAPKs. Overexpression of KLF5 induces factors that regulate the vascular injury response such as inducible nitric oxide synthase (iNOS), PAI-1, PDGF-A, and VEGFRs (15, 214, 293, 378). Together, these findings indicate that KLF5 induces SMC proliferation in response to injury.
The role of KLF5 in blood vessel pathobiology was demonstrated by the vascular phenotype of Klf5+/− mice, which also have a cardiac phenotype (see section A2. Pathobiology of the Heart) (378). The medial and advential layers of the aortas from Klf5+/− mice are thinner than those of wild-type mice. The femoral arteries of Klf5+/− mice have impaired response to cuff-induced injury, with reduced activation and proliferation of SMCs and fibroblasts, inflammatory responses, and angiogenesis (378). These defects result in smaller areas of neointimas, and reduced granulation tissues and angiogenesis around the cuffs of femoral arteries (378). Importantly, Am80, a synthetic retinoid agonist that attenuates KLF5’s transcriptional activity, reduces neointima and granulation tissues in cuff-injured femoral arteries of wild-type mice, compared with Klf5+/− mice (378). KLF5 is therefore involved in several facets of vascular remodeling, including mesenchymal-cell activation, development of interstitial fibrosis, and angiogenesis.
The mechanisms by which KLF5 promotes vascular SMC proliferation have been examined. Angiotensin II stimulates proliferation of vascular SMCs and increases the levels of KLF5 mRNA and protein (15, 146). Incubation of vascular SMC with angiotensin II stimulates phosphorylation of KLF5, which increases its interaction with c-Jun and subsequent repression of the cell cycle inhibitor p21Cip1/Waf1 (176). The phosphorylation of KLF5 in response to angiotensin II is mediated in part by the MAPK/extracellular signal-regulated kinase (ERK) kinase-1 (MEK1); incubation of vascular SMCs with the MEK1 inhibitor PD98059 inhibits KLF5 phosphorylation and its interaction with c-Jun (176). Overexpression of KLF5 stimulates SMC proliferation, increases cyclin D1 expression, and inhibits apoptosis (412). Moreover, KLF5 interacts with RAR-α to regulate the smooth muscle phenotype; this interaction is interrupted by the synthetic retinoid Am80 (145). Am80 inhibits the interaction between KLF5 and RAR-α by inducing dephosphorylation of KLF5 that is mediated by PI3K–AKT signaling in vascular SMCs (496). These studies delineated the signaling pathways by which KLF5 is regulated by various stimuli.
In addition to its physiological function in endothelial cells (see section A3. Endothelial Biology and Pathobiology), KLF4 regulates vascular smooth muscle function. KLF4 binds to the promoter of the smooth muscle gene SM22α, in a region identified as the TGF-β control element (TCE), and suppresses the TGF-β-dependent increase in transcription of SM22α and α-SMA (1). KLF4 also represses serum response factor/myocardin-induced activation of α-SMA and mediates the repressive effect of PDGF-BB on this gene (256). Expression of KLF4 in SMCs is activated in response to members of the TGF-β superfamily, including TGF-β1 and several bone morphogenetic proteins (BMPs), which regulate the vascular SMC phenotype (219). These results indicate that KLF4 is involved in determining the SMC phenotype by repressing expression of specific genes in response to external stimuli.
Similar to KLF5, expression of KLF4 is very low in normal blood vessels but is increased in the medial layer following vascular injury (1, 482, 483). Following injury, SMC differentiation markers are repressed—this repression is transiently delayed in mice with conditional deletion of Klf4 (483). Moreover, neointimal formation increases in Klf4 mutant mice in response to vascular injury, primarily from an increase in cell proliferation (483). KLF4 therefore regulates expression of differentiation markers and proliferation in SMCs following injury. Furthermore, KLF4 expression in SMCs is induced by oxidized phospholipids, including 1-palmytoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine (POVPC), that accumulate in atherosclerotic lesions (337). POVPC strongly inhibits expression of SMC differentiation marker such as α-SMA and SM-myosin heavy chain, but activates those of inflammatory mediators, including MCP-1 and TGF-β (337). The inhibitory effect of POVPC on SMC gene expression is coordinated by KLF4, the transcription factor Elk-1, and several HDACs (482). Moreover, in SMCs, POVPC induces expression of several extracellular matrix proteins (including type VIII collagen α1 chain) and cell migration—both of these are reduced in the absence of KLF4 (77). Thus, there is much evidence for the role of KLF4 in the pathogenesis of atherosclerotic vascular diseases.
Overexpression of KLF4 in vascular SMCs inhibits proliferation via a process that depends on p53 (450) and is antagonized by the inhibitor of DNA binding 3 (Id3), which promotes proliferation (449). KLF4 is also required for ATRA to inhibit vascular SMC proliferation (442); ATRA upregulates KLF4 at transcriptional and post-transcriptional levels (276, 442). KLF4 inhibits SMC proliferation by inhibiting expression of several genes that encode growth factor receptors, including PDGF receptor β (503). However, KLF4 appears to have pleiotropic effects on expression of smooth muscle genes—although KLF4 induces expression of SM22α and α-SMA in response to ATRA (442), it inhibits that of the same genes in response to TGF-β and PDGF-BB (1, 256). This context-dependent nature of KLF4 activity has also been described in other systems.
Therefore, KLF4 and KLF5 have opposite effects in regulating proliferation of vascular SMCs. Similar findings have been observed in other cell or tissue types (151). Induction of a combination of KLF4 and KLF5 in vascular SMCs after injury might provide the balanced gene expression required to achieve homeostasis. MicroRNA (miR)-145, a non-coding RNA that regulates expression of KLFs 4 and 5, is highly expressed in normal vascular walls and freshly isolated vascular SMCs (76, 93). Expression of miR-145 is significantly downregulated in vessel walls with neointimal lesion formation and in vascular SMCs that have de-differentiated in response to PDGF. MiR-145 controls SMC fate and expression of SMC markers by regulating transcription factors that include KLFs 4 and 5 (76, 93). MicroRNAs can therefore direct SMC fate and regulate vascular SMCs proliferation under pathological conditions, in part by influencing expression of KLFs 4 and 5.
B. The Respiratory System
Although there is much information about the functions of KLFs in the cardiovascular system, few studies have investigated KLFs in the respiratory system; these have focus on the roles of KLFs in lung development and morphogenesis. KLF2 is expressed at a high level in the lung (10). Klf2−/− mice die in utero between embryonic days (E) 11.5 and 13.5, making it difficult to study the function of KLF2 in lung development (447). However, in vitro cultures of lung bud tissues removed from E11.5 Klf2−/− embryos form normal tracheobronchial trees (448). To examine lung development at a later stage, chimeric mice were generated by injection of Klf2−/− ES cells into blastocysts of wild-type mice (448). In mice that survived to adulthood, Klf2−/− ES cells contribute to all internal organs except lung. In contrast, animals with high levels of chimerism die at birth and have abnormal lung development and function (448). The lungs of these animals do not expand, appear to stop growing in the late canalicular stage, and have undilated acinar tubules and buds in peripheral regions (448). These results indicate that KLF2 is an important regulator of late-stage lung development.
Klf5 expression is developmentally regulated in mice (315). At E16.5, Klf5 is expressed at high levels in the bronchiolar epithelium and epithelial lining of the trachea (315). Klf5−/− mice die at E8.5 (378), so the exact role of KLF5 in lung development cannot be determined. Mice with conditional, lung-specific deletion of Klf5 (Klf5Δ/Δ) survive until birth but die shortly afterward from respiratory failure (440). They have abnormalities in lung maturation and morphogenesis in the respiratory epithelium, bronchiolar smooth muscle, and pulmonary vasculature; respiratory epithelial cells of the conducting and peripheral airways are immature. Levels of surfactant phospholipids are reduced and lamellar bodies, the storage form of surfactant, are rarely found. Expression profiling studies demonstrated that KLF5 regulates genes involved in surfactant lipid and protein homeostasis, vasculogenesis (including VEGF-A), and SMC differentiation (440). These observations indicate that KLF5 is required for perinatal maturation of lung morphology and function.
KLF4 suppresses TGF-β-dependent induction of α-SMA in vascular SMCs via interaction with the TCE (1) (see section A4, Vascular Smooth Muscle Biology and Pathobiology). A similar effect was observed in fibroblasts isolated from rat lungs (185), although the ability of KLF4 to inhibit α-SMA transcription does not soely depend on the TCE (185). Instead, KLF4 physically interacts with Smad3, which mediates the stimulatory effect of TGF-β on α-SMA transcription through an upstream element called Smad-binding element (SBE) (185). Interaction between KLF4 and Smad3 therefore prevents binding of Smad3 to the SBE and TGF-β-dependent, de novo differentiation of lung myofibroblasts (185).
KLF4 might also be involved in the inflammatory response of airway smooth muscle (ASM) cells, via a post-transcriptional mechanism (231). Several microRNAs are repressed in human ASM cells following stimulation with inflammatory cytokines such as IL-1β, TNF-α, or interferon-γ (IFN-γ) (231). One of these, miR-25, targets KLF4, which is upregulated in ASM cells following cytokine stimulation (231). KLF4 might therefore have a role in the pathophysiology of lung diseases such as pulmonary fibrosis and inflammatory airway disease.
C. The Hematopoietic System
KLFs regulate erythropoiesis, lymphopoiesis, and formation and functions of monocytes and macrophages. Erythropoiesis is a complex physiologic process in which pluripotent hematopoietic stem cells undergo several stages of commitment to precursor and progenitor cells with gradually restricted potential; this process is controlled by lineage-specific transcription factors (for review, see (48)). Erythropoiesis requires the highly regulated expression of β-globin and related proteins that constitute an essential subunit of hemoglobin. The β-globin gene locus contains a cluster of several related genes on chromosome 11, arranged in the order in which they are expressed during development: 5′-ε-γG-γA-δ-β-3′ (245). Thus, hemoglobin ε is the first globin gene expressed (in the yolk sac), followed by fetal hemoglobins Aγ and Gγ (in the liver), and eventually β-globin at birth (in the bone marrow). Each globin gene is controlled by specific cis-acting DNA elements in its own promoter. In addition, the differentiation- and development-specific expression of the β-globin gene locus depends on a distal regulatory sequence called the locus control region (LCR), which is located approximately 6 kb upstream from the ε-globin gene (245). The LCR interacts with multiple, sequence-specific DNA binding transcription factors and is required for high levels of expression of the β-globin genes at all developmental stages (180).
To understand the regulation of expression of the globin genes, Miller and Bieker performed subtraction hybridization studies using a Friend mouse eythroleukemia cell line. They identified a cDNA clone they named erythroid KLF or EKLF (279). Since EKLF was the first mammalian KLF identified, it was given the name KLF1. KLF1 binds the CACCC element in the β-globin gene promoter and LCR enhancer and activates a heterologous promoter that contained the CACCC sequence, as well as the endogenous β-globin promoter (31, 279). Mutations in the CACCC element of the β-globin gene have been associated with β-thalassemia (232, 320, 321)—KLF1 cannot transactivate a reporter gene that contains the mutated elements (132). Expression of Klf1 is restricted to two hematopoietic organs of the adult—mouse the bone marrow and spleen—and to cell lines of the erythroid lineage (279). KLF1 therefore has erythroid-specific distribution and function.
The developmental expression pattern of KLF1 indicates its importance in erythropoiesis. Klf1 expression begins on E7.5 in mice, within primitive erythroid cells at the beginning of blood island formation in the yolk sac (399). By E9.0, Klf1 is expressed in the hepatic primordia and liver, which becomes the sole source of Klf1 mRNA in an E14.5 fetus (399). In the adult spleen, which is an erythropoietic organ in mice, Klf1 is exclusively expressed in the red pulp (399). This is consistent with data from cell line studies showing that KLF1 is expressed in erythroid but not lymphoid cell lines (279). The physiological function of KLF1 in erythropoiesis was demonstrated in experiments with Klf1−/− mice; these mice have normal embryonic hematopoiesis but develop fatal anemia during early fetal life (E14.5), when hematopoiesis starts to occur in the liver (311, 335). Klf1−/− mice form enucleated erythrocytes that are deficient in β-globin, similar to patients with β-thalassemia (311, 335). In contrast, expression of embryonic globin genes during the early stages of embryogenesis is not affected. These findings indicate that KLF1 is not required for yolk sac erythropoiesis or erythroid commitment but is essential for the final stage of definitive erythropoiesis in the fetal liver. The stage-specific and β-globin-specific requirement for KLF1 indicates that it is involved in the switch from fetal-to-adult hemoglobin (γ to β) in humans. This concept is supported by the finding that KLF1 binds with higher affinity to the CACCC element from the adult globin gene promoter than to a similar element in the mouse embryonic (and human fetal) globin gene promoter (116). The reduction in β-globin transcription in the fetal livers of Klf1−/− mice that have a single copy of the entire human β-globin locus is accompanied by increased transcription of the human γ-globin gene (333, 459). KLF1 is therefore involved in the fetal-to-adult hemoglobin switch, mediating an adult stage-specific interaction between the β-globin gene promoter and the LCR that excludes the γ-globin gene (333, 459). Studies in which KLF1 was expressed in immortalized cell lines from Klf1−/− fetal liver progenitor cells indicated that KLF1 is involved in coordinating differentiation, hemoglobinization, and erythroid-cell proliferation (88); erythroid cells from Klf1−/− have defects in cell cycle regulation and terminal differentiation (339).
ES cells isolated from Klf1−/− mice differentiate into definitive erythroid colonies that contain reduced levels of hemoglobin. When the Klf1−/− ES cells were injected into blastocysts, they did not contribute to the mature erythrocyte compartment of the resulting chimeric mice, indicating that Klf1−/− erythrocytes are short-lived (251). Fetal expression of a human γ-globin transgene restored globin gene expression to Klf1−/− embryos, but mice do not survive due to hemolysis (334). Other KLF1 gene targets, beyond the globin genes, therefore appear to be required for erythrocyte development—several studies have shown that KLF1 regulates expression of proteins involved in membrane and cytoskeletal stability (118, 183, 310), Loss of their regulation might contribute to the severity of the Klf1−/− phenotype of disrupted erythropoiesis.
In addition to regulating erythrocyte maturation, KLF1 controls the development of megakaryocytes. Megakaryocytes and erythrocytes are derived from a common progenitor called the MEP (326). Studies indicate that KLF1 is directly involved in determining the bi-potential fate of MEP—KLF1 inhibits the formation of megakaryocytes while stimulating erythroid differentiation (144). Expression of KLF1 is uniquely downregulated in megakaryocytes after formation of the MEP (144). KLF1 might inhibit megakaryocyte differentiation by repressing expression of FLI-1 (37, 144), which is required for their development (173, 212). Conversely, FLI-1 represses KLF1-dependent transcription (403), so cross-antagonism between KLF1 and FLI-1 controls erythrocytic versus megakaryocytic differentiation (403). Lastly, sumoylation of KLF1 is required for it to suppress megakaryocyte differentiation (380).
The developmental- and erythroid-specific patterns expression patterns of KLF1 prompted investigations of the mechanisms that regulate KLF1. An enhancer element in the distal promoter of KLF1, approximately 700 bp upstream of the transcription start-site, and a proximal promoter element are required for its erythrocyte-specific expression (67). The erythroid-specific transcription factor GATA-1 binds the enhancer and promoter elements and transactivates the KLF1 promoter (9, 94). Studies in transgenic mice showed that 950 bp of the KLF1 5′-flanking region is sufficient to induce erythroid-specific expression of a reporter gene and requires the GATA-1-binding sites (8, 466). Using an in vitro system in which ES cells differentiate into embryoid bodies (EB), it was shown that the BMP-4/SMAD pathway controls differentiation-dependent expression of KLF1 (2). This study also showed that KLF1 expression is activated before terminal erythroid differentiation in two stages: it is expressed at low levels in progenitor cells (before erythroid commitment), which depends on GATA-2 and SMAD5, and then at high levels by committed erythroid cells, which depends on GATA-1 (258). The stage- and lineage-specific dependent control of KLF1 expression indicates that it is a regulator of lineage fate decisions during hematopoiesis (258).
Although KLF1 has important functions in erythropoiesis, other KLFs are also involved. A functional screen for KLFs that regulate the human γ-globin promoter through the CACCC element revealed the presence of 8 KLFs aside from KLF1 in human erythroid cell lines (490). Levels of KLFs 2, 4, 5, and 12 increase significantly during erythroid differentiation. Co-transfection studies indicated that KLFs 2, 4, and 13 activate, whereas KLF8 represses, the γ-globin promoter through its CACCC element (490). The roles of KLF2, and to some extent, KLF6, in erythropoiesis have been examined. Klf2−/− mice die in utero between E12.5 and E14.5 from intra-embryonic hemorrhage (234, 447). Compared with wild-type mice, E10.5 Klf2−/−embryos are anemic and have significant reductions in expression of mouse embryonic Ey- and βh1-globin but not ζ-globin genes (21). Expression of the adult βmaj- and βmin-globin genes is unaffected in the fetal livers of E12.5 Klf2−/− embryos. In mice that carry the entire human globin locus, KLF2 also regulates the expression of the human embryonic ε-globin gene but not the adult β-globin gene, suggesting that this developmental stage-specific role is evolutionarily conserved. KLF2 also regulates maturation and/or stability of erythroid cells in the yolk sac; Klf2−/− embryos have a significant increase in numbers of primitive erythroid cells that undergo apoptotic cell death (21).
KLF6 also regulates hematopoiesis (267). Klf6−/− ES cells have significant hematopoietic defects following differentiation into EBs (267), prolongation of epiblast-like cells, delays in mesoderm induction, and delayed expression of Klf1 and Gata1 (267). Ectopic expression of KLF6 increases the hematopoietic potential of wild-type EBs (267), so KLF6 appears to be an important regulator of ES cell differentiation and hematopoiesis.
KLF1 and KLF2 have compensatory roles in controlling embryonic β-globin gene expression and primitive erythropoiesis (20). Klf1−/−/Klf2−/− double mutant mice are anemic at E10.5, with greatly reduced levels of Ey- and βh1-globin mRNAs. Examination of the E9.5 Klf1−/−/Klf2−/− yolk sacs indicated that erythroid and endothelial cells are more abnormal than in either Klf1−/− or Klf2−/− mice (20). KLF1 and KLF2 might therefore have redundant functions in regulating expression of the embryonic β-like globin gene, primitive erythropoiesis, and endothelial development (20).
D. The Immune System and the Inflammatory Responses
1. T Lymphocytes
Kuo et al. were the first to identify a KLF that regulates T cell functions (235)—they found that KLF2 is expressed in lymphoid organs, including the thymus, and exclusively in the medulla, which contains mature, single-positive (SP; CD4+ or CD8+) T cells (235). In addition, KLF2 is expressed in SP splenocytes but not in the less mature double-positive (CD4+CD8+) T cells. SP T cells circulate through the blood and peripheral lymphoid organs in a quiescent state and are activated when the T-cell receptor binds antigen. When T cells are activated, KLF2 mRNA and protein are rapidly degraded (124, 235)—potentially via WWP1 E3-ubiquitin ligase-mediated degradation (100). Klf2−/− T cells have a spontaneously activated phenotype and die in the spleen and lymph nodes from Fas ligand-induced apoptosis (235). KLF2 is therefore required for the quiescent state of mature T cells and for maintenance of viability in the peripheral lymph organs and blood.
Additional evidence supports a role for KLF2 in T cell quiescence. Ectopic expression of KLF2 in Jurkat T cells induces quiescence, characterized by decreased proliferation, reduced cell size and protein synthesis, and decreased expression of surface markers of activation (44). Conversely, Klf2−/− T cells have increased proliferation, cell size, and cell-surface markers of activation (44). The mechanism by which KLF2 inhibits T cell activation, growth, and proliferation might include its ability to inhibit expression of c-Myc (44, 168) and stimulate expression of p21Cip1/Waf1 (462).
Although studies had demonstrated that KLF2 is downregulated upon T cell activation, its expression appears to be dynamic. For example, expression of KLF2 is re-induced in antigen-activated CD8+ T cells in the presence of certain cytokines, including IL-7 (124, 360). The ability of IL-7 to induce KLF2 expression in activated T cells is mediated by the MEK5/ERK5 pathway and depends on the transcription factor MEF2 (124, 391). Prolonged culture of activated CD8+ T cells in the presence of IL-7 causes them to acquire characteristics of memory CD8+ T cells, which maintain stable levels of KLF2 (360); stable expression of KLF2 has been reported in memory T cells (160, 360). In activated T cells, KLF2 might upregulate transcription of IL-2 (160, 360, 463). Specific cytokines might therefore induce expression of KLF2 in activated T cells, possibly to regulate factors required for long-term survival and development of the memory T-cell phenotype.
In addition to regulating T cell proliferation and survival, KLF2 controls T cell migration and trafficking. Compared to Klf2+/− cells, Klf2−/− thymocytes have reduced expression of several receptors required for emigration and peripheral trafficking, including the sphingosine-1-phosphate receptor (S1P1), CD62L (L-selectin), CCR7, and integrin subunit β7 (51). As a consequence, Klf2−/− thymocytes have abnormal tissue distribution following adoptive transfer; they are absent from blood and lymphoid organs and accumulate in the spleen (51). Pulse-labeling experiments showed impaired emigration of Klf2−/− thymocytes into the periphery (51). Conversely, primary T cells that express ectopic Klf2 are more efficient in homing to lymphoid organs (16). KLF2 accomplishes these activities by directly activating expression of CD62L (L-selectin) (16), a receptor required for T cells to enter lymphoid nodes (439); and SIP1 (51), which allows T cells to exit the thymus (266). Interestingly, KLF2 and S1P1 have similar patterns of expression in T cells—they are upregulated in mature thymocytes, downregulated upon T cell activation, and re-expressed in memory T cells (51). The PI3K signaling pathway regulates KLF2. PI3K and its downstream nutrient sensor, the mammalian target of rapamycin (mTOR), determine the repertoire of adhesion and chemokine receptors expressed by T cells and are required for T-cell trafficking (128, 387). PI3K regulates the transcription factor forkhead box O1 (FOXO1), which regulates KLF2 expression (128). KLF2 then directly regulates the expression patterns of chemokine receptors that induce T cell migration (366, 457); KLF2 thereby controls the regulatory network that controls T cell migration and trafficking.
KLF10 has an important role in the development of CD4+CD25+ regulatory T (Treg) cells, which maintain self-tolerance and immune suppression (138). The transcription factor Foxp3 regulates the Treg cell lineage (138) and TGF-β converts CD4+CD25− naïve T cells into Foxp3+CD4+CD25+ Treg cells in vitro and maintains Treg cells in vivo (63, 129, 247). KLF10 induces expression of Foxp3 in T cells following exposure to TGF-β (50, 438). KLF10 cooperates with Itch to induce Foxp3 expression (438); both Klf10−/− and Itch−/− T cells are resistant to the effects of TGF-β and have reduced TGF-β (438). Following incubation with TGF-β converted Klf10−/− Treg cells are unable to suppress airway inflammation in mice (438). Moreover, CD4+CD25− T cells that overexpress KLF10 increase TGF-β1 expression, compared with those that do not overexpress KLF10, and have reduced expression of Th1 and Th2 markers (50). Conversely, Klf10−/− CD4+CD25− T cells increase expression of Th1 and Th2 cytokines and cannot be suppressed by wild-type Treg cells (50). Because KLF10 transactivates TGF-β1 and Foxp3 in response to TGF-β, it is involved in a positive feedback loop that controls T cell activation (50).
KLF13 was initially identified as a transcription factor that regulates expression of regulated upon activation normal T cell expressed and secreted (RANTES or CCL5), a chemokine that generates inflammatory infiltrates and is expressed late after T cell activation (393, 394). Although levels of KLF13 mRNA are similar between naive and activated T cells, only KLF13 protein is abundant in the late stage of T cell activation, so KLF13 is likely to be regulated at the level of translation (393). Studies have shown that KLF13 expression is translationally regulated through the 5′-untranslated region of its mRNA in a cell type-specific manner (309). Overexpression of the translation initiation factor eIF4E and Mnk1 (which phosphorylates eIF4E) increase KLF13 protein levels (309). These events are regulated by ERK1/2 and p38 MAPKs and allow T cells to rapidly adjust levels of RANTES expression in response to changes in the cellular environment, such as stress or exposure to growth factors (309). The ability of KLF13 to activate RANTES transcription requires the recruitment of several transcription co-activators and the serine/threonine protein kinase PRP4, which phosphorylates KLF13 (4, 187). Klf13−/−mice have enlarged thymuses and spleens because of decreased thymocyte apoptosis; KLF13 inhibits expression of the anti-apoptotic factor BCL-XL (506). In addition, KLF13 influences multiple stages of B- and T-cell development in vivo (322). KLF13 therefore appears to regulate T cell activation, survival, and development.
2. B Lymphocytes
The most extensively studied KLF in B cell function is KLF4. KLF4 is expressed in early-stage B cell precursors, at the time of immunoglobulin gene rearrangement (433); its expression continuously increases until B cells are mature (220). Upon B cell activation, levels of KLF4 are rapidly reduced. Klf4−/− mice have modest reductions in numbers of pre-B cells and mature B cells in the bone marrow and spleen, respectively. B cells isolated from Klf4−/− mice have reduced DNA synthesis and delayed entry into the cell cycle in response to B-cell receptor (BCR) activation, a result of decreased cyclin D2 expression (220). KLF4 binds directly to the cyclin D2 promoter and activates cyclin D2 expression (220). KLF4 is therefore a positive regulator of BCR-mediated B cell proliferation. However, overexpression of KLF4 in pre-B cells transformed by the ABL oncogene induces arrest in the G1 phase of the cell cycle and apoptosis (216). Transformed B cells that overexpress KLF4 increase expression of p21Cip1/Waf1 and reduce expression of c-Myc and cyclin D2 (216). Moreover, the anti-proliferative effect of KLF4 on B cells requires its activation by FOXO1 (484).
KLF4 also mediates the proliferative response of memory B cells. Ectopic expression of KLF4 in memory B cells delays cell division and thereby reduces the number of proliferating cells (156). It is not clear what accounts for the discrepancies observed in the effects of KLF4 on B cell proliferation but differences could arise from variations in physiologic contexts, genetic backgrounds, or experimental methodologies.
3. Monocytes and Macrophages
KLFs regulate inflammatory responses in both endothelial and SMCs (172, 233, 369). KLFs regulate signaling following activation of macrophages, and could thereby mediate the development of acute and chronic inflammatory disorders. KLF4 mediates macrophages signaling in response to inflammatory cytokines, such as IFN-γ, LPS, or TNF-α, and anti-inflammatory factors such as TGF-β1 (130). KLF4 is rapidly induced in macrophages incubated with IFN-γ, LPS, or TNF-α and reduced in response to TGF-β1 (130). In combination with the p65 (RelA) subunit of NF-κB, overexpression of KLF4 in macrophages induces iNOS, a marker of cell activation (130). However, KLF4 overexpression also inhibits PAI-1 (a TGF-β1-regulated gene) without binding to it (130).
KLF4 regulates monocyte differentiation in vitro and in vivo (6, 131). Although overexpression of KLF4 in HL-60 cells confers characteristics of mature monocytes, KLF4 inhibition blocks phorbol ester-induced monocyte differentiation (6, 131). Ectopic expression of KLF4 in common myeloid progenitors or hematopoietic stem cells induces differentiation of only monocytes in clonogenic assays, whereas inhibition of KLF4 increases differentiation of granulocytes, at the expense of monocytes (131). Hematopoietic Klf4−/− chimeras (generated by transplantation of Klf4−/− fetal liver cells into irradiated wild-type mice) completely lack circulating inflammatory cytokines (6). KLF4 is therefore an important promoter of monocyte differentiation.
Whereas KLF4 activates macrophages to promote inflammation, KLF2 has the opposite effect (106); its expression is reduced upon monocyte activation and differentiation into macrophages. Ectopic expression of KLF2 inhibits monocyte activation and their phagocytic capacity, whereas KLF2 inhibition increases expression of inflammation genes. Reconstitution of immunodeficient mice with KLF2-overexpressing monocytes significantly reduces carrageenan-induced acute paw edema formation (106). KLF2 inhibits the transcriptional activity of both NF-κB and AP-1 by recruitment of co-activator P/CAF, thereby preventing activation of inflammatory genes (106). These results demonstrate that KLF2, as opposed to KLF4, is a negative regulator of monocytic activation.
E. The Digestive System
1. Small Intestine and Colon
The mammalian intestinal epithelium is a continuously renewing system in which cell proliferation, differentiation, migration, and apoptosis are carefully orchestrated to achieve homeostasis. The epithelium of the small and large intestines consist of a crypt/villus and crypt/surface epithelium compartment, respectively. The bulk of the villus and surface epithelium of the small and large intestine, respectively, is composed of differentiated columnar epithelial cells that are divided into absorptive (primarily enterocytes) and secretory (including goblet, enteroendocrine and Paneth cells) classes. Paneth cells are unique to the small intestine under normal conditions. The differentiated epithelial cells are descendants of the crypt progenitor cells, which themselves are derived from the multi-potent stem cells, also located in the crypt compartment (19, 342, 365). Intestinal epithelial homeostasis is regulated by signaling mechanisms that include the Wnt, Notch, Hedgehog, BMP, and PI3K pathways (19, 342, 365).
Both KLFs 4 and 5 control and maintain intestinal epithelial homeostasis (151, 272). Expression of KLFs 4 and 5 in the intestines is developmentally regulated, with higher levels of expression occurring towards the later stage of fetal development and adulthood (315, 422). The temporal pattern of expression of Klf4 during fetal development correlates with the transition of a pseudo-stratified epithelial cell layer into a simple columnar monolayer at approximately E15–E17 of development (422). In the adult, KLF4 and KLF5 are highly expressed in the intestinal epithelial cells but have different distribution—KLF4 is found primarily in the terminally differentiated cells in the villi and surface epithelium of the small intestine and colon, respectively, whereas KLF5 is present primarily in the proliferating crypt epithelial compartment (89, 272, 375). This is demonstrated in Figure 3, which shows the distribution of Klf4 and Klf5 in the mouse colon in conjunction with the proliferation marker, Ki67. The biological activities of KLFs 4 and 5 are reflective of their cellular distribution; KLF4, when overexpressed, inhibits cell proliferation, whereas KLF5 promotes it (54, 151, 272, 375). The cytostatic effect of KLF4 is mediated by induction of the gene that encodes the cell cycle inhibitor p21Cip1/Waf1 (66) and by inhibition of the genes that encode cyclin D1 and ornithine decarboxylase (ODC) (71, 373); this combination blocks the G1/S transition of the cell cycle. KLF4 also mediates the cytostatic effect of p53 following DNA damage and by blocking the G1/S and G2/M phases of the cell cycle (479, 481, 493). Transcriptional profiling studies have shown that many of the genes upregulated by KLF4 inhibit the cell cycle and many of those that are downregulated promote the cell cycle (68).
Figure 3. Localization of Klf4 and Klf5 in the mouse colon.
Immunofluorescence staining of Klf 4 or 5 (green) with the proliferation marker, Ki67 (red), was conducted on mouse colon. Klf4 (green) is present in the nuclei of terminally differentiated epithelial cells in the upper regions of colonic crypts. In contrast, Klf5 is localized to nuclei of proliferating epithelial cells at the base of the crypts. Ki67 highlights regions of active proliferation. Although Klf4 and Ki67 staining patterns do not overlap, Klf5 and Ki67 exhibit considerable co-localization, indicated by yellow staining.
In non-transformed cells, such as NIH3T3 and several intestinal epithelial cell lines, overexpression of KLF5 stimulates proliferation and in some cases, leads to anchorage-independent growth (22, 54, 410). This is consistent with the findings that KLF5 expression is upregulated by mitogens such as serum, phorbol ester, and the oncogenic HRAS (214, 297, 301, 410). A common mechanism by which these stimuli induce KLF5 expression is through activation of the MAPK/early growth response gene (EGR)-1 signaling pathway (214, 301), which activates KLF5 transcription (214). KLF5 subsequently stimulates expression of cyclin D1 and cyclin B1/Cdc2, accelerating cell cycle progression (297, 301). Conversely, KLF5 expression in intestinal epithelial cells is inhibited by ATRA, an inhibitor of epithelial cell proliferation; constitutive expression of KLF5 blocks the growth inhibitory effect of ATRA (54). KLF5 also mediates the induction of proliferation of colon cancer cells by lysophosphatidic acid (LPA) (486). The effects of KLF5 on cell proliferation require post-translational regulation including protein protein interaction and modifications. Interaction with PIAS1 and modification by SUMO increase KLF5’s pro-proliferative activity (119, 120). Interestingly, whereas KLF4 activates its own promoter, KLF5 inhibits the KLF4 promoter (104, 262). This result is reminescen to the opposing activity and cellular distribution of these KLFs in the intestinal epithelium.
The in vivo functions of KLFs 4 and 5 in the intestine have been partially determined through studies of knockout mice. Klf4−/− mice die shortly after birth with a defect in skin barrier function (367). Newborn Klf4−/− mice also have a 90% reduction in the number of goblet cells in their colon, compared to wild-type controls (211), so KLF4 might regulate intestinal epithelial differentiation. This finding is consistent with results of in vitro studies demonstrating that KLF4 regulates expression of intestinal epithelial markers, including intestinal alkaline phosphatase and keratins (68, 181, 381). Expression of KLF4 increases with differentiation of colon cancer cell lines of goblet and absorptive cell lineages (136). Short-chain fatty acids such as sodium butyrate, which regulates colonic epithelial differentiation, induce KLF4 expression (70, 136), whereas Notch signaling inhibits KLF4 expression. Inhibition of Notch induces KLF4 expression and increases the number of goblet cells in the intestine (148, 345, 504). These results indicate that in the intestinal tract, KLF4 controls epithelial differentiation.
Klf5−/− mice die in utero whereas Klf5+/− mice are born with intestinal phenotypes such as misshapen villi and intestinal fibrosis (378). When Klf5+/− mice are infected with Citrobacter rodentium (a murine bacterial pathogen that causes colonic epithelial hyperplasia), they have significantly reduced colonic hyperproliferation, compared with wild-type mice (273). These results indicate that KLF5 mediates colonic crypt cell proliferation in response to bacterial infection (Figure 4). Expression of KLF5 in intestinal epithelial cells is induced by the bacterial endotoxin, LPS. LPS-mediated induction of KLF5 increases the transcriptional activity of NF-κB, leading to induction of inflammatory cytokines such as TNF-α and IL-6, as well as intercellular adhesion molecule-1 (ICAM-1) (53). Klf5+/− mice are more susceptible to dextran sodium sulfate (DSS)-induced colitis than wild-type mice—additional evidence that KLF5 regulates epithelial restitution following injury (McConnell and Yang, unpublished observations).
Figure 4. Reduced mucosal hyperplasia in Klf5+/− mice following infection with the enteric pathogen, Citrobacter rodentium.
In these experiments, C57BL/6 (WT) and Klf5+/− mice were infected with the rodent-specific pathogen, Citrobacter rodentium. Colonic tissues from uninfected and C. rodentium-infected mice were isolated at 14 days post-infection (p.i.) and were subjected to histological staining with hematoxylin and eosin (H&E) or immunofluorescence staining for expression of Klf5 (green) or the proliferation marker, Ki67 (red). Hyperplasia induced by infection was significantly reduced in the Klf5+/− mice [from McConnell, et al. (273)].
KLF9 also regulates intestinal morphogenesis. In contrast to KLFs 4 and 5, KLF9 is predominantly expressed in the SMCs of the small intestine and colon (383). The jejunum of Klf9−/− mice have shorter villi, reduced proliferation of crypt cells, and alterations in lineage determination (383). KLF9 therefore directs the production of mediators of crypt cell proliferation, lineage determination, and migration from intestinal SMCs.
2. Esophagus and Stomach
Several KLFs function in the esophagus and stomach. KLF4 is expressed in the squamous epithelial cells of the esophagus, where it activates the promoters of two genes that are highly expressed in the esophageal squamous epithelium: the Epstein-Barr virus ED-L2 and keratin 4 (196). Keratin 4 is also regulated by KLF9 (319). Together, Sp1 and KLF4 activate transcription of keratin 19 in esophageal epithelial-derived cell lines (41).
The esophageal squamous epithelium contains a proliferating layer and a differentiated suprabasal layer. KLF4 is expressed in the suprabasal layer while KLF5 is expressed in the basal layer. Overexpression of KLF4 or KLF5 modulates proliferation, apoptosis, and invasion of human esophageal squamous cancer cell line TE2 (472). Expression of KLF5 coincides with that of the epidermal growth factor (EGF) receptor (EGFR) (473). Overexpression of KLF5 in primary esophageal keratinocytes increases proliferation, EGFR levels and MAPK signaling (473). KLF5 activates EGFR expression by directly binding to a regulatory element of the EGFR promoter (473). Overexpression of KLF5 also increases migration of primary esophageal keratinocytes, via its upregulation off integrin-linked kinase (ILK) (474) and increases proliferation of the basal, but not suprabasal, esophageal epithelial cells (155). KLF5 therefore regulates proliferation and migration of squamous esophageal epithelial cells.
Conditional deletion of Klf4 from mouse stomach (using Cre recombinase, regulated by the Foxa3 promoter) (210) results in mice that survive to adulthood but have increased proliferation in the gastric epithelium without evidence of dysplasia or cancer. The Klf4 mutant mice also have altered differentiation of gastric epithelia, including a dramatic increase in the number of trefoil factor 2-positive cells—a characteristic of premalignant conditions of the stomach (210). KLF4 therefore also regulates gastric epithelial homeostasis.
3. Liver and Pancreas
KLF6 regulates the early stages of the fibrotic response during liver injury. Hepatic stellate cells (HSCs) are the major source of extracellular matrix during liver fibrosis; they are often activated upon liver injury and during subsequent wound healing (142). Using a rat model of liver fibrosis, Friedman and colleagues found that KLF6 is induced during the early phase of HSC activation (238, 344). In fibrotic liver, KLF6 transactivates the promoters of several genes involved in fibrotic injury and repair, such as collagen α1(I) and TGF-β receptors (218, 344). In vivo, KLF6 slows proliferation; mice that overexpress KLF6 specifically in liver have reduced liver size, whereas Klf6+/− mice have increased liver size (306). Several alternatively spliced forms of KLF6 exist in human and rodents that have dominant-negative activities (303). A specific, germ-line, single-nucleotide DNA polymorphism results in a splice variant of KLF6 that is associated with increased risk of prostate cancer and advanced fibrosis in patients with nonalcoholic fatty liver disease (278, 303).
KLFs 10 and 11 are expressed in the pancreas. KLF10 is expressed in the acinar and ductular epithelial cell populations of the exocrine pancreas (414). KLFs 10 and 11 belong to the TGF-β-induced early gene subfamily and mediate the anti-proliferative effect of TGF-β on pancreatic epithelial cells—a process that involves its downstream effector Smad4 (91, 92). Overexpression of KLF10 in TGF-β-sensitive pancreatic epithelial cells causes apoptosis (414). KLFs 10 and 11 share an amino-terminal transcription repressive domain called the SID, which is also required for the growth suppressive effect of KLF11 (90, 133, 488). KLF11 potentiates TGF-β signaling and transcription in normal epithelial cells by suppressing expression of Smad7, which normally inhibits TGF-β signaling (122). Transgenic mice that express KLF11 under the control of a pancreas-specific promoter have smaller pancreases, compared with the non-transgenic control mice, and down-regulate expression of the oxidative stress scavengers SOD2 and catalase 1 (133). KLF11 expression renders cells more sensitive to oxidative drugs, an effect that is rescued by overexpression of SOD and catalase 1 (133).
KLF11 regulates β cell function in the pancreas. It is expressed in human pancreatic islets and in pancreatic β cell lines (308). β cells cultured in high levels of glucose have higher levels of KLF11, which activates the insulin promoter (308). KLF11 regulates expression of pancreatic-duodenal homeobox-1, which is required for pancreatic organogenesis and activity of insulin-secreting β cells in adults (434). Genetic analysis of KLF11 among North European populations revealed the presence of several rare variants that impair KLF11’s transcriptional activity and are associated with early onset of type 2 diabetes (308). A variant of the promoter of KLF11 has also been linked to insulin sensitivity in a Danish population (167). However, similar studies failed to demonstrate an association of any genetic variants of KLF11 with type 2 diabetes in Japanese or native American populations (236, 260, 418).
F. The Role of KLFs in Metabolic Regulation
No fewer than eight KLFs are involved in metabolism, primarily by regulating differentiation of fat cells (adipocytes) (see (42) for a recent review). The adipose tissue is the major site of energy storage and determines energy homeostasis, the perturbation of which can lead to pathological conditions including obesity and diabetes. The differentiation of precursor adipocytes (pre-adipocytes) into mature adipocytes is regulated by an intricate network that is best modeled in cell culture systems (348–350). Genes that mediate adipocyte differentiation are controlled by the transcription factor families C/EBP, PPAR, and sterol regulatory element binding proteins (SREBP)/adipocyte differentiation and determination factor (349, 350). C/EBPs β and δ are activated in the early phase of adipogenesis, followed by sustained expression of C/EBPα and PPARγin the late phase of differentiation and the eventual expression of adipocyte-specific genes (349, 350). KLFs are an important part of the regulatory cascade that leads to adipogenesis (42). The roles of various KLFs as positive and negative regulators of adipocyte differentiation are summarized in Figure 5.
Figure 5. Contribution of KLFs in regulating adipocyte differentiation.
Pre-adipocytes progress through an early stage of differentiation in which they become committed to their fate, followed by a late stage of terminal differentiation into mature adipocytes. Various members of the KLF family contribute to transcriptional control of this process and have positive and negative effects on adipogenesis [Adapted from Nagai, Friedman and Kasuga, Editors (294)].
1. KLFs 2 and 3
KLFs 2 and 3 inhibit adipocyte differentiation. In a well-characterized 3T3-L1 model of differentiation (161), levels of KLFs 2 and 3 are high in undifferentiated pre-adipocytes and reduced upon differentiation into adipocytes (18, 409, 464). Overexpression of KLF2 or 3 blocks adipocyte differentiation (18, 409, 464), partly through KLF2’s ability to repress PPARγ expression (18) and KLF3’s ability to inhibit C/EBPα expression (409).
Mouse embryonic fibroblasts (MEFs) that lack Klf2 (Klf2−/−) or Klf3 (Klf3−/−) are more prone to differentiate into adipocytes, indicating that KLFs 2 and 3 inhibit adipogenesis in vivo (409, 464). Recent studies in Caenorhabditis elegans have implicated a role for the C. elegans Klf3 homolog in regulation of fat metabolism (487).
2. KLFs 4 and 5
KLFs 4 and 5 promote adipogenesis. KLF4 is expressed very early in the course of 3T3-L1 differentiation with a pattern similar to that of previously described transcription factors, Krox20, C/EBPβ, and C/EBPδ (32). KLF4, in conjunction with Krox20, activates expression of the C/EBPβ gene (32). Importantly, knockdown of KLF4 inhibits adipogenesis and reduces C/EBPβ levels (32). Interestingly, knockdown of C/EBPβ increases levels of KLF4 and Krox20, indicating that C/EBPβ normally controls KLF4 and Krox20 expression via a negative feedback loop (32).
Similar to KLF4, KLF5 is also induced at an early stage of 3T3-L1 differentiation that requires C/EBPs β and δ (318). Then, KLF5 activates expression of PPARγ2 (318). Overexpression KLF5 spontaneously induces adipocyte differentiation, whereas expression of a dominant-negative form of KLF5 inhibits adipogenesis (318). Klf5+/− have defects in development of white adipose tissue and MEFs obtained from Klf5+/− mice have attenuated adipocyte differentiation (318). These results indicate that KLF5 is a key regulator of adipocyte differentiation in vitro and in vivo. Furthermore, Klf5+/− mice are resistant to high fat diet-induced obesity, hypercholesterolemia, and glucose intolerance, despite their consumption of more food than wild-type mice (317). Instead, Klf5+/− mice have increased energy expenditure, in part from increased expression genes that encode lipid oxidation in the soleus muscle such as carnitine-palmitoyl transferase-1b (Cpt1b) and uncoupling proteins 2 and 3 (Ucp2 and Ucp3) (317). KLF5 must be sumoylated to repress these oxidizing genes, which allows it to interact with co-repressors such as nuclear receptor co-repressor and SMRT, as well as unliganded PPARδ (317). PPARδ agonists stimulate desumoylation of KLF5; KLF5 then associates with transcriptional activation complexes containing ligand-bound PPARδ and CBP (317). This activation complex subsequently activates expression of the lipid oxidizing genes (317). Sumoylation is therefore a molecular switch that controls KLF5 function and the transcriptional machinery that governs lipid metabolism.
3. KLF6
Delta-like 1 (Dlk1), also called pre-adipocyte factor-1 (Pref-1), regulates adipogenesis by preventing differentiation of 3T3-L1 cells into adipocytes (390). KLF6 represses Dlk-1 expression and thereby stimulates 3T3-L1 differentiation (390). Conversely, knockdown of KLF6 prevents adipogenesis (390). The repressive effect of KLF6 on the Dlk-1 promoter requires HDAC3 activity (390).
4. KLF7
A search for single nucleotide polymorphisms (SNPs) in 12 KLF genes identified a SNP in KLF7 that was significantly associated with type 2 diabetes in Japanese subjects (209). Although the same SNP is not associated with diabetes in a Danish population, a minor allele of a different KLF7 locus protects against obesity in the same population (510). These studies provided evidence that KLF7 is involved in fat metabolism. Expression of KLF7 is decreased upon 3T3-L1 adipocyte differentiation and overexpression of KLF7 inhibits adipogenesis (209). Similarly, overexpression of KLF7 in human pre-adipocytes inhibited their differentiation into adipocytes and reduced expression of genes involved in adipogenesis, including C/EBPα and PPARγ (215). Overexpression of KLF7 in differentiated adipocytes significantly reduces the levels of adipocytokines such as leptin and adiponectin (215). When overexpressed in an insulin-secreting pancreatic β cell line, KLF7 inhibits expression of insulin and glucose-induced secretion of insulin (215). In addition, KLF7 inhibits expression of hexokinase 2 in a skeletal muscle cell line and of glucose transporter 2 in a liver-derived cell line (215). KLF7 might therefore contribute to the pathogenesis of diabetes by impairing insulin biosynthesis and secretion in pancreatic β cells and reducing insulin sensitivity in peripheral tissues. These results are consistent with the observation that (-)-catechin, a component of the green tea polyphenols, promotes adipogenesis by inhibiting expression of KLF7 and activating that of adiponectin in 3T3-L1 cells (81)
5. KLF11
The role for KLF11 in regulating pancreatic β cell functions was addressed in section E3. Liver and Pancreas. KLF11 also regulates cholesterol-mediated gene expression. Following exposure of vascular endothelial cells to cholesterol, KLF11 represses the gene that encodes caveolin-1 gene, in which is involved in cholesterol homeostasis (49). Upon depletion of cholesterol, KLF11 is displaced from a Sp1-site that flanks a sterol-response element in the caveolin-1 promoter, allowing the binding of SREBP and Sp1 and activation of the caveolin-1 promoter (49). In response to cholesterol, KLF11 therefore represses genes that SREBP/Sp1 otherwise activate.
6. KLF15
KLF15 also regulates adipogenesis; its expression is highly upregulated during differentiation of 3T3-L1 pre-adipocytes into adipocytes (288). Dominant-negative forms or RNA interference of KLF15 reduce expression of PPARγand block adipogenesis of 3T3-L1 pre-adipocytes (288). KLF15 and C/EBPα act synergistically to increase the activity of PPARγ2 in 3T3-L1 adipocytes (288). In non-adipocyte cell lines such as NIH3T3 and C2C12, ectopic expression of KLF15 increases PPARγ levels and lipid accumulation (288). Moreover, KLF15 expression is increased by ectopic expression of C/EBPα, β, or γ in NIH3T3 cells (288). KLF15 therefore plays an essential role in regulating adipocyte differentiation through its regulation of PPARγ expression.
In addition to its role in adipogenesis, KLF15 is involved in energy metabolism in the liver, skeletal muscle, and adipocytes. KLF15 regulates expression of the insulin-sensitive glucose transporter GLUT4 in both adipose and muscle tissues (158). KLF15 also contributes to the transcriptional activation of mitochondrial acetyl-CoA synthetase 2 in skeletal muscle during fasting (469). In the liver, expression of KLF15 is increased by fasting and reduced by refeeding (421). In cultured hepatocytes, KLF15 expression is increased by cAMP and decreased by insulin (421). Ectopic expression of KLF15 in cultured hepatocytes increases expression of phophoenolpyruvate carboxykinase (PEPCK), which is required for gluconeogenesis (421). Consistent with a role for KLF15 in gluconeogenesis, Klf15−/− mice have severe hypoglycemia after an overnight fast (159). Interestingly, instead of a reduction in PEPCK levels, Klf15−/− mice have defects in amino acid catabolism that arise from reduced expression of enzymes required for amino acid degradation in the liver and skeletal muscle; reductions of these enzymes limit the substrates available for gluconeogenesis (159). The activity of alanine aminotransferase (ALT), which helps convert the amino acid alanine into the gluconeogenic substrate pyruvate, is reduced by 50% in the livers of Klf15−/− mice (159). These studies implicate KLF15 as a critical regulator of gluconeogenesis.
G. KLFs and Bone Metabolism
KLF 10 is the only family member known to be associated with bone metabolism. It was initially identified as a TGF-β-induced cDNA from a human fetal osteoblast (hFOB) library (406). Expression of KLF10 is rapidly and significantly induced in hFOB cells incubated with TGF-β1 and BMP-2, and to a less extent, EGF (406). Other TGF-β superfamily members such as BMP-4, BMP-6, and activin, also induce KLF10 in hFOB cells but are less potent than TGF-β1 (178). In addition, KLF10 is rapidly induced by the estrogen 17β-estradiol (E2), in estrogen receptor (ER)-positive hFOB cells—this induction is antagonized by parathyroid hormone (419). E2-induced KLF10 inhibits DNA synthesis (419) and proliferation of hFOB cells incubated with TGF-β (177). In fact, human osteosarcoma cells (MG-63) that have been stably transfected with a vector that encodes KLF10 display changes similar to those following incubation with TGF-β (177). KLF10 increases TGF-β signaling through transcriptional activation of Smad2 and inhibition of Smad7, (202, 203).
KLF10’s role in bone physiology has been studied in knockout mice. Klf10−/− mice have a greater number of osteoblasts, but not osteoclasts, during bone formation than wild-type mice (405). Following exposure to BMP-2, osteoblasts differentiate, but osteoblasts from Klf10−/− mice fail to mineralize of support differentiation of osteoclasts (405). As a consequence, Klf10−/− mice have defects in bone and tendon strength and microarchitecture (25, 26). The defects in bone mineral content, density, and area in Klf10−/− mice appear to be sex-specific (175). KLF10 was one of five genes whose expression was associated with volumetric bone density in a SNP study of bone metabolism genes (476).
H. The Nervous System and Neuronal Morphogenesis
Several KLFs participate in neuronal morphogenesis. Klf7 exhibits three phases of expression in the nervous system of the developing mouse (239). During embryogenesis, Klf7 is highly expressed in the spinal cord, dorsal root ganglia, and sympathetic ganglia. During early postnatal development, Klf7 is expressed in the cerebral cortex and is subsequently downregulated. In adult mice, Klf7 is localized to the cerebellum and dorsal root ganglia. These phases of expression correspond with establishment of the neuronal phenotype in the embryonic spinal cord, development of synaptic circuitry in the postnatal cerebral cortex, and survival and function of adult sensory neurons and cerebellar granule cells. Deletion of Klf7 in mice results in neonatal lethality which is associated with deficits in neurite outgrowth and axonal misprojection (241). Axonal pathways affected include the olfactory and optic systems, the cerebral cortex and the hippocampus.
Klf9 is expressed in dentate granule neurons of the dentate gyrus (DG), a region of the mammalian brain in which neurogenesis occurs in adulthood (363). Klf9 is upregulated during the early postnatal period and is expressed in dentate granule neurons during the late stage of maturation, when the cells are integrated into the hippocampal network. Dentate granule neurons from Klf9−/− mice show delayed maturation, and adult Klf9−/− mice exhibit impaired differentiation of adult-born neurons. Thus, Klf9 is necessary for late-phase maturation of dentate granule neurons both in DG development and during adult hippocampal neurogenesis.
KLF4 was recently identified as an important factor regulating the regeneration potential of neurons in the central nervous system (CNS) (286). Gene expression profiling was used to identify factors that are upregulated in retinal ganglion cells (RGCs) during postnatal loss of axon growth ability. Of the candidate genes identified, KLF4 is the most effective suppressor of neurite outgrowth when overexpressed in RGCs. Conversely, RGCs lacking KLF4 exhibit increased axon growth both in vitro and after optic nerve injury in vivo. Other KLFs are also developmentally regulated in postnatal RGCs—KLFs 6 and 7 which have growth-enhancing effects are downregulated, whereas KLF9 which is growth-suppressive is highly upregulated. In agreement with this study, Klfs 6 and 7 are required for regeneration of RGCs in zebrafish (437). Thus, KLFs that act as positive and negative regulators of axon outgrowth are coordinated to control the regenerative capacity of CNS neurons.
I. The Role of KLFs in Tumor Biology
Numerous KLFs are involved in the pathobiology of cancer (33, 46, 121, 126, 152, 355, 384, 453). Here we review the role of representative KLF family members in tumor biology.
1. KLF4
As discussed in Section E.1. Small Intestine and Colon, KLF4 was initially identified as a growth arrest-associated gene in the intestinal epithelium that suppresses DNA synthesis when expressed ectopically (375). In RKO colon cancer cells, induction of KLF4 leads to G1/S cell cycle arrest, which correlates with an increase in the level of p21Cip1/Waf1 (66). Using cell culture models in which DNA damage is elicited by γ irradiation or methyl methanesulfonate, KLF4 is transcriptionally activated by the tumor suppressor p53 and mediates subsequent G1/S and G2/M cell cycle checkpoints by activating transcription of p21Cip1/Waf1 (479, 481, 493). KLF4 is necessary for and sufficient to prevent centrosome amplification following γ-irradiation–induced DNA damage; it does so by inhibiting expression of the gene encoding cyclin E, which promotes centrosome amplification when overexpressed (480). KLF4 also represses expression of cell cycle-promoting genes such as cyclin D1 and ODC in the human colon cancer cell line HT29 (71, 373). Agents that have anti-proliferative agents in colon cancer cells, including IFN-γ (72), sulforaphane (424, 425), sodium butyrate (70), 15-deoxy-Δ12,14-prostaglandin J2 (73), and certain phytochemicals (79, 80), increase levels of KLF4.
The checkpoint function of KLF4 in colon cancer-derived cell lines suggests that it is a tumor suppressor. Induced KLF4 expression in RKO cells, which do not express endogenous KLF4, reduces colony formation, cell migration, invasion, and in vivo tumorigenecity (102). KLF4 is regulated by adenomatous polyposis coli (APC), the most frequently mutated tumor suppressor gene in colon cancer (103). APC is part of the Wnt signaling pathway responsible for maintaining intestinal epithelial homeostasis by modulating cellular levels and localization of β-catenin (19, 342, 365). Either the presence of Wnt ligands or mutational inactivation of APC leads to nuclear translocation of β-catenin; formation of nuclear β-catenin/T cell factor 4 (TCF4) transcriptional complex increases cell proliferation (228, 289). KLF4 inhibits Wnt signaling by downregulating β-catenin protein and mRNA levels (404), interacting physically with β-catenin, and repressing β-catenin/TCF4 transcriptional activity (492). KLF4 therefore assists the tumor suppressive activity of APC via regulation of the Wnt/β-catenin signaling pathway in colon cancer cells.
The tumor suppressor activity of KLF4 in colon cancer has been investigated in primary tumor samples. The levels of KLF4 mRNA are reduced, compared with control tissues, in the respective intestinal and colonic adenomas from the ApcMin mice and patients with familial adenomatous polyposis—both carrying germ-line APC mutations (101). Similarly, KLF4 mRNA levels are lower in adenocarcinoma of the colon compared to normal colon (374). In a panel of 30 colorectal cancer specimens, the mean level of KLF4 mRNA was reduced by 50% compared with paired normal colon samples (501). KLF4 mRNA levels are also reduced in cultured colon cancer cell lines, compared to non-transformed colonic epithelial cells (501). Reductions in KLF4 expression arise through several mechanisms often involved with tumor suppressor inhibition, including loss of heterozygosity (LOH), promoter hypermethylation, and mutations that disrupt protein activity (501). Several studies have validated the loss of KLF4 expression in colorectal cancer, including those showing that KLF4 mRNA levels are inversely correlated with Duke stages (86, 356, 465). Moreover, KLF4 has tumor suppressive activity in cancers of other regions of the gastrointestinal tract, including the esophagus (259, 443), stomach (85, 208, 452), and pancreas (454). Expression of KLF4 is also reduced in tumors that arise outside the gastrointestinal tract, such as in lung (30, 186), prostate (139), and urinary bladder (316).
The tumor suppressive function of KLF4 has been examined in vivo. Between 10 and 20 weeks of age, ApcMin/Klf4+/− mice develop on average 55% more intestinal adenomas than ApcMin mice (150). The levels of Klf4 protein in the normal-appearing mucosa of the ApcMin/Klf4+/−mice are lower than in that of wild-type, ApcMin, or Klf4+/− mice. In contrast, the levels of β-catenin and cyclin D1 are higher in the intestinal tissues of ApcMin/Klf4+/− mice, compared with the other genotypes (150). Klf4 levels are further decreased in adenomas derived from ApcMin/Klf4+/− and ApcMin mice compared to their respective normal surrounding mucosa (150). There is an inverse correlation between the size of adenoma and Klf4 transcript levels and a correlation between LOH at Apc and adenoma size (150). Haploinsufficiency of Klf4 therefore promotes Apc-dependent intestinal tumorigenesis, consistent with the observations that KLF4 is regulated by APC (103) and that KLF4 inhibits Wnt/β-catenin signaling (404, 492). MEFs isolated from Klf4−/−embryos and maintained in cultured are genetically unstable—they show aneuploidy, DNA damage, chromosome aberrations, and centrosome amplification (169). These results support a role for KLF4 as a tumor suppressor that maintains genetic stability (169).
An additional mechanism by which KLF4 suppresses intestinal tumor formation involves Notch signaling, which suppresses goblet cell differentiation in the intestinal epithelium and is upregulated in intestinal tumors (197, 430). KLF4 is required for goblet cell differentiation (211); overexpression of Notch in HT29 cells suppresses KLF4 expression and increases proliferation (148). Conversely, inhibition of Notch by pharmacological or genetic means increases KLF4 expression and reduces proliferation (148). ApcMin mice given the Notch inhibitor dibenzazepine (DBZ) have a 50% reduction in numbers of intestinal adenomas, compared with mice given vehicle alone (controls) (148). Importantly, the normal-appearing intestinal mucosa and adenomas from DBZ-treated ApcMin mice have increased levels of Klf4 and numbers of goblet cells, as well as reduced cyclin D1 and Ki67 levels, compared with controls (148). These results were subsequently replicated (504) and suggest that KLF4 mediates the anti-tumor effect of Notch inhibitors such as DBZ (148).
Although there is much evidence that KLF4 is a tumor suppressor, contradictory data exist. For example, KLF4 mRNA and protein are present in a relatively high proportion of breast tumor samples, compared with the adjacent uninvolved epithelium (139). Furthermore, nuclear localization of KLF4 in early-stage ductal carcinoma of the breast is associated with an aggressive phenotype that includes shortened survival time (324). The mechanism by which KLF4 promotes breast carcinogenesis might involve its ability to repress transcription of p53 and Mucin 1, which are often overexpressed in breast tumors (456). KLF4 activates transcription of Notch1 in mammary epithelial cells and promotes signaling through a non-canonical Notch1 pathway (257). However, the role of KLF4 as a putative oncoprotein in breast tissue is controversial. A search of gene expression databases showed that KLF4 mRNA levels are lower in breast tumor tissues, compared with normal tissues, in 9 of 11 data sets and that the levels are inversely correlated with tumor grade (5). KLF4 mRNA levels are also correlated with ER-positive breast tumors (5). In breast cancer cells, KLF4 physically interacts with ER to inhibit its transcriptional activity and ER-dependent proliferation (5). Exposure of breast cancer cells to okadaic acid (OA) induces c-Myc and subsequent c-Myc-mediated apoptosis (489). KLF4 is also induced by OA and is responsible for OA-dependent induction of c-Myc (489). KLF4 also regulates transcription of the gene encoding laminin α3A (LAMA3A) chain, a component of the extracellular matrix protein laminin-5, which is produced by normal mammary epithelial cells but markedly down-regulated in breast cancer cells (280). This loss of laminin-5 expression is associated with progression of breast cancer (179, 264). Notably, KLF4 protein and DNA binding at the LAMA3A promoter are found in normal mammary epithelial cells but not in any of the breast cancer cell lines analyzed (280). Further studies are required to determine whether KLF4 is oncogenic or tumor suppressive in the breast.
KLF4 is also involved in the pathogenesis of squamous cell carcinoma. KLF4 is expressed in the normal squamous cell epithelium of the skin and oral cavity, in the differentiated suprabasal layer but not the basal layer (139). KLF4 staining is increased in dysplastic epithelium or squamous cell carcinoma of the oral cavity (69, 139). Conditional expression of KLF4 in basal keratinocytes of transgenic mice blocks the proliferation-differentiation switch between the basal and parabasal epithelial cells and leads to hyperplasia, dysplasia, and squamous cell carcinoma in situ (140). In normal squamous epithelium or hyperplastic epithelium derived from the skin of KLF4 transgenic mice, expression of KLF4 and the proliferation marker PCNA are mutually exclusive (188). In contrast, KLF4 and PCNA co-localize in dysplastic or carcinoma-like lesions epithelium (188). These results indicate that successive increases of KLF4 in the nuclei of basal keratinocytes increases cell turnover and progression through the different stages of squamous cell tumorigenesis (188). KLF4 also activates expression of several retinoic acid receptors, including retinoid acid receptor-γ (RAR-γ) and retinoid X receptor-α (RXR-α). Exposure of mice to RXR-selective agonists such as 9-cis UAB-30 or rexinoid prevents formation of skin tumors that arise via induction of KLF4 in the basal keratinocytes and the appearance of hyperplastic, dysplastic, and squamous cell carcinoma-like lesions (200).
The mechanisms by which KLF4 could function as a tumor suppressor or oncoprotein are presented in Figure 6. In untransformed cells, KLF4 inhibits cell proliferation, but KLF4-induced arrest is bypassed by oncogenic RASV12 due to the upregulation of cyclin D1 (351). Inactivation of p21Cip1/Waf1-induced cell cycle arrest by the upregulated cyclin D1 overcomes the suppressive effects of KLF4 and contributes to cell transformation (351). Overexpression of KLF4 suppresses expression of p53 at the promoter level, allows RASV12-mediated transformation, and prevents DNA damage-induced apoptosis (351). In a similar study, KLF4 was identified in a functional screen as a transforming protein in adenovirus E1A-immortalized rat kidney RK3E cells (141). Cells that lack KLF4 exhibit a greater degree of apoptosis following γ irradiation (149). KLF4’s anti-apoptotic effects are mediated through activation of p21Cip1/Waf1 and inhibition of p53 activation of the pro-apoptotic gene, BAX (149). KLF4 levels are determined by the extent of DNA damage and different levels of KLF4 determine the outcome of p53 response to DNA damage (507). Thus, KLF4 is activated after cytostatic, low-level DNA damage, leading to cell cycle arrest, but is repressed after severe DNA damage, leading to apoptosis (507). These studies indicate that KLF4 regulates whether a cell will undergo cell cycle arrest or apoptosis depending on the level of genetic damage.
Figure 6. A model for the role of KLF4 in tumor suppression and oncogenesis.
Following DNA damage, expression of KLF4 is activated in a p53-dependent manner. Increased levels of KLF4 lead to increased expression of p21Cip1/Waf1 and decreased expression of BAX, with the net effect being to tip the balance away from apoptosis towards cell cycle arrest. However, in the presence of activated RAS or E1A, the growth inhibitory effects of p21Cip1/Waf1 are negated by high levels of cyclin D1. The combination of high levels of KLF4 expression in the presence of oncogenic RAS or E1A lead to active proliferation and suppressed apoptosis, which contribute to carcinogenesis. KLF6 likewise acts as a tumor suppressor through induction of p21Cip1/Waf1. The growth-suppressing activity of KLF6 can be offset by oncogenic RAS which induces alternative splicing of KLF6, generating a mutant form that promotes cell proliferation and suppresses apoptosis [Adapted from Ghaleb, et al. (149)].
2. KLF5
KLF5 promotes proliferation of different cell types. In intestinal epithelial cells, expression of KLF5 coincides with that of Ki67, a marker of proliferation (273). In vivo, KLF5 mediates the hyperproliferative response of the colonic epithelium following pathogenic infection by Citrobacter rodentium (273) and the regenerative response in chemical colitis caused by DSS (McConnell and Yang, unpublished observations). Similarly, nanoparticles that contain siRNA against KLF5 have anti-tumor activity (467), and ATRA inhibits proliferation of untransformed intestinal epithelial cells, IEC6, and several colon cancer cell lines by inhibiting expression of KLF5 (54). Conversely, LPA facilitates proliferation of intestinal epithelial cells and colon cancer cells by inducing KLF5 (252, 486). A recent high-throughput screening approach identified several novel and potent small molecular inhibitors of KLF5 that inhibit proliferation of several colon cancer cell lines (29). Moreover, overexpression of KLF5 in NIH3T3 fibroblasts (410), COS-7 (120), intestinal epithelial cells (22, 54), and colon cancer cells (119) increases rates of proliferation, and in some cases, anchorage-independent growth. Finally, KLF5 mediates the transforming activity of oncogenic HRAS and KRAS in NIH3T3 fibroblasts (297, 301) and IEC6 cells (299), respectively. Consistent with these findings, intestinal tumors derived from KRASV12-expressing transgenic mice and human colon cancer samples with oncogenic KRAS mutations contain increased levels of KLF5, determined by immunohistochemical analysis (299).
ApcMin/Klf5+/− mice have a 96% reduction in the number of intestinal adenomas compared with ApcMin mice (271). This reduction in tumor formation correlates with decreased nuclear localization of β-catenin and reduced expression of the β-catenin targets cyclin D1 and c-Myc (271). In cultured cells, KLF5 physically interacts with β-catenin, facilitates its nuclear localization, and modulates β-catenin’s transcriptional activity (271). KLF5 is therefore required for the tumor-initiating activity of β-catenin during intestinal tumorigenesis in ApcMin mice. The tumor-promoting effects of KLF5 are supported by the observation that Klf5 haploinsufficiency reduces intestinal burden in ApcMin mice with the KRASV12 mutation (298).
The involvement of KLF5 in cancers of the extra-intestinal tissues of the gastrointestinal tract has been investigated. As addressed in Section E.2. Esophagus and Stomach, overexpression of KLF5 in esophagus squamous epithelial cells (keratinocytes) regulates proliferation, migration, and signal transduction (155, 473, 474). In tumor samples from 247 patients with gastric cancer, KLF5 expression is correlated with early-stage cancers that are small in size and have not metastasized to the lymph nodes (237). In several pancreatic cell lines, KLF5 levels are increased independently of MAPK signaling, but increased KLF5 expression do depend on IL-1β and hypoxia-inducible factor 1-α (HIF1-α) (287). Downregulation of KLF5 expression in pancreatic cancer cells by siRNA reduced expression of the KLF5 targets survivin and PDGF-A, which promote pancreatic tumor growth (287). The chromosome region that contains KLF5, 13q22.1-22.2, is frequently amplified in salivary gland tumors, so KLF5 might also be involved in the pathogenesis of this tumor type (287).
The role of KLF5 in the pathogenesis of breast cancer is somewhat unclear—with some studies showing that it promotes breast tumor formation. KLF5 frequently undergoes hemizygous deletion and loss of expression in breast tumors, indicating a possible tumor suppressive role (57). In addition, KLF5 is actively degraded by the hyperactive E3 ubiquitin ligase, WWP1, in some breast cancer cell lines (59, 74). Conversely, several studies demonstrate that KLF5 promotes breast cancer cell proliferation and survival by upregulating transcription of fibroblast growth factor binding protein 1 and stabilizing the dual-specificity phosphatase, MAPK phosphatase 1 (255, 505). KLF5 is also induced upon overexpression of ERBB2 (also known as HER1 or NEU), which is amplified in some breast tumors (23). In tumor samples from 90 patients with breast cancer, those that express higher levels of KLF5 have shorter disease-free survival and overall survival times, compared to those that express lower levels of KLF5 (423). KLF5 expression is correlated with that of HER2 and the proliferation marker Ki67 (423). It is not clear why KLF5 is upregulated in some breast tumors and downregulated in others, but the status of the ER might be a factor—KLF5 inhibits the proliferative action of estrogen in ER-positive, but not ER-negative, cells (165).
The role of KLF5 in prostate cancer pathogenesis is also unclear. The KLF5 locus is frequently deleted and downregulated in prostate cancer and overexpression of KLF5 in prostate cancer cell lines inhibits their growth (58). KLF5 is often degraded by ubiquitin-mediated proteolysis in prostate cancer cells, as in breast cancer cells (59). However, gene expression profiling experiments showed that KLF5 transcript levels were consistently increased in prostate cancer samples, relative to normal prostate epithelium (52). Furthermore, KLF5 increases the expression of fatty acid synthase and the chemokine receptor, CXCR4 (143, 244), which are involved in prostate cancer proliferation and migration. Further studies are needed to identify the role of KLF5 in prostate cancer pathogenesis.
The studies described here indicate that KLF5 has a context-dependent proliferative or anti-proliferative function, sometimes in the same cell types. A number of recent studies explored the mechanism by which this can be achieved. In an in vitro epidermal epithelial cell system, proliferative KLF5 becomes anti-proliferative, in the presence of TGF-β (163). KLF5 inhibits the expression of the cell cycle inhibitor, p15, in the absence of TGF-β, but when TGF-β is added to cells, KLF5 becomes acetylated and co-activates p15 transcription (163). In a similar manner, although KLF5 activates c-Myc expression in the absence of TGF-β, it inhibits c-Myc in the presence of TGF-β (164). These findings suggest that KLF5 activates cell proliferation in response to TGF-β, which has well-recoginzed pleiotropic effects on cell proliferation.
3. KLF6
An association between KLF6 and cancer was made by Narla et al. who found that one allele of KLF6 is deleted and the remaining allele is mutated in most primary prostate tumors (305). Wild-type KLF6 upregulates p21Cip1/Waf1 and inhibits prostate cell proliferation but mutant forms of KLF6 do not (305), indicating that KLF6 is a prostate tumor suppressor. LOH and somatic mutations in KLF6 are detected in another study of prostate cancer specimens, although at a lower frequency than that of Narla et al. (58). More recent studies confirmed that overexpression of KLF6 in prostate cancer cells reduces proliferation and induces apoptosis (75, 189). Conversely, reduced expression of KLF6 is associated with poor prognosis in gene expression profiling studies of patients with prostate cancer (154). Expression of KLF6 is decreased in aggressive, androgen-independent, metastatic prostate tumors (402). Other studies, however, report that KLF6 mutations are rare events in sporadic or inherited-forms of prostate cancer (3, 224, 291).
Numerous studies demonstrated that loss of KLF6 expression or activity, due to LOH, mutation, or promoter methylation, occur in many other types of cancer, including hepatocellular (45, 182, 229, 230, 306), colorectal (82, 83, 282, 292, 346), gastric (84, 359), lung (193, 508), head and neck (420), ovarian (114), and brain (47, 478) cancers. However, other studies failed to establish an association between KLF6 and some of these cancer types (38, 222, 225, 250, 285, 397).
A large cohort study of men with prostate cancer identified a germ-line SNP in KLF6 that increases the relative risk of familial and sporadic prostate cancer (303). This polymorphism occurs in the first intron of KLF6 (denoted as the intervening sequence (IVS) 1–27 G>A or IVSΔA allele) and generates a functional binding site for the splicing factor SRp40 that increases transcription of three alternatively spliced forms of KLF6: KLF6-SV1, -SV2 and -SV3 (303). The KLF6-SV1 and -SV2 splice variants mislocalize to the cytoplasm, antagonize wild-type KLF6, and decrease p21Cip1/Waf1 expression (303); expression of these variants is increased in prostate tumors compared with normal prostate tissues (303) and targeted inhibition of the KLF6-SV1 variant suppresses prostate cancer cell proliferation, colony formation, and invasion (304). Conversely, overexpression of KLF6-SV1 accelerates prostate tumor progression and metastasis in humans and mice (302). These results demonstrate that dominant-negative KLF6 splice variants contribute to prostate cancer pathogenesis.
KLF6-SV1 has been detected in hepatocellular (475), ovarian (112, 114), lung (111, 358), pancreatic (174), and gastric tumors (359), and glioblastoma (47). In hepatocellular carcinoma, oncogenic RAS signaling stimulates alternative splicing of KLF6 and increases proliferation (475). In ovarian cancer cells, KLF6-SV1 promotes degradation of the anti-apoptotic protein NOXA, increasing cell survival (112). KLF6-SV1 also has anti-apoptotic effects in lung cancer cells (111). Indeed, KLF6-SV1 represents the first therapeutically targeted KLF and inhibitors of KLF6-SV1 slow tumor growth and progression in several model systems (111–113, 359).
4. KLF8
Tumors acquire invasive phenotypes via the epithelial–mesenchymal transition (EMT) (162), which is mediated by many factors, including focal adhesion kinase (FAK) (87, 354). FAK regulates expression of KLF8, which controls the cell cycle by activating cyclin D1 expression (500). In human ovarian epithelial and cancer cells, FAK regulates transcription of KLF8 by activating the PI3K–AKT pathway (444). KLF8 expression is increased in several types of human cancer cells and tissues and ectopic expression of KLF8 induces transformation (445). Stable expression of KLF8 in immortalized normal human breast epithelial cells induces the EMT, repressing transcription of E-cadherin and thereby increasing the motility and invasiveness of cancer cells (446). (446)
5. KLF9
KLF9 is associated with endocrine-responsive cancers of the female reproductive tissues such as endometrial cancer (384). Stable expression of KLF9 in the human endometrial carcinoma cell line Hec-1-A increases DNA synthesis and cell cycle kinetics by inducing genes involved in cell cycle control (386, 497). KLF9 physically interacts with the progesterone receptor; together, they co-regulate expression of progesterone-responsive target genes (435, 485, 498). KLF9 is a negative regulator of ligand-dependent ERα signaling in endometrial carcinoma cells (436). KLF9 might therefore function in the progesterone and ER signaling pathways to control endometrial cell proliferation. Human endometrial cancer is associated with unopposed estrogen activity. Levels of KLF9 mRNA are reduced in endometrial tumor of higher grades, compared with normal endometrium and low-grade endometrial tumors (382).
6. KLFs 10 and 11
KLF10 is a TGF-β-inducible early gene that is normally expressed in the acinar and ductular epithelial cells of the exocrine pancreas; its over-expression causes apoptosis in pancreatic epithelial cell lines, so KLF10 has tumor suppressive effects (414). The induction of KLF10-mediated apoptosis increases the sensitivity of pancreatic cancer cells to gemcitabine; this effect is mediated by suppression of stathmin expression (199). However, a mutational screen of the KLF10 gene in 22 pancreatic cell lines revealed no sequence alterations (11).
KLF10 has anti-proliferative effects, promotes apoptosis (7, 201, 281), and is involved in the pathogenesis of cancers in addition to pancreatic cancer. Expression of KLF10 is progressively lost during the transition of normal breast epithelium into invasive carcinoma (347, 407). KLF10 is regulated by estrogen in breast cancer and is part of a gene expression signature that can be used to discriminate ER-positive from ER-negative tumors (398). In colon cancer cells, KLF10 expression is increased by 15-hydroxy-eicosatetraenoic acid (15S-HETE), an endogenous ligand for PPARγ, and induces apoptosis (61). Colon tumors have decreased levels of KLF10 and 15-lipoxygenase, the enzyme responsible for 15S-HETE formation, compared with normal mucosa; colonic tumorigenesis is therefore associated with decreased 15S-HETE levels (61). In acute lymphoblastic leukemia (ALL), KLF10 inhibits proliferation in response to stimulation by TGF-β or BMP-6 derived from bone marrow stromal cells (117). However, the anti-proliferative effect of KLF10 protects the ALL cells against chemotherapy-induced cell death (117). KLF10 might therefore mediate signals from the microenvironment to leukemia cells. KLF10 is upregulated in renal clear cell carcinoma as a result of mutation in the tumor suppressor von-Hippel Lindau protein (194), but further studies are required to determine the exact role of KLF10 in cancer pathogenesis.
Like KLF10, KLF11 mediates the effect of TGF-β on cell growth and has a role in pancreatic tumor progression. KLF11 potentiates TGF-β signaling by binding to the mSin3A co-repressor and terminating the inhibitory Smad7 loop (122). These events are inhibited in pancreatic cancer cells with oncogenic KRAS mutations—ERK/MAPK phosphorylates KLF11, leading to disruption of the interaction between KLF11 and mSin3A (122). Expression of an ERK-insensitive, mutant form of KLF11 restores mSin3A binding and Smad7 repression and increases TGF-β signaling in pancreatic cancer cells (122). In response to TGF-β stimulation, KLF11 and Smad3 bind the TIE and inhibit transcription of c-Myc; this silencing is required for TGF-β-mediated inhibition of epithelial cell proliferation (43). In pancreatic cancer cells with oncogenic mutations in KRAS, hyperactive forms of ERK counteract TGF-β–induced repression of c-Myc and induce proliferation by disrupting the KLF11/Smad3 complex and inhibiting its binding to the TIE in the c-Myc promoter (43). ERK signaling thereby antagonizes the tumor suppressor activity of TGF-β, blocking ability of KLF11 to inhibit c-Myc expression (43).
7. KLF12
KLF12 was initially identified as a repressor of the transcription factor AP-2α (192). KLF12 is located on chromosome region 13q21-q22, which houses a putative breast cancer susceptibility gene and is the site of somatic deletions in different malignant tumors (353). In contrast, this chromosome region is frequently amplified in salivary gland and gastric tumors (153, 296). Knockdown of KLF12 induces growth arrest of HGC27 gastric cancer cells (296). Overexpression of KLF12 in NIH3T3 and AZ-521 cells increases their invasive potential (296). KLF12 mRNA levels are increased in 11 of 28 samples of gastric tumors from patients, compared to normal gastric epithelium (296), and correlate with tumor size (296). These results suggest that KLF12 has an important role in the progression of gastric cancer.
J. KLFs and Somatic Cell Reprogramming
The ability to reprogram somatic cells into pluripotent, ES-like cells has many potential clinical applications in regenerative medicine. The importance of KLF proteins in somatic cell reprogramming was revealed by Takahashi and Yamanaka, who identified four transcription factors that could reset the fate of somatic cells: Klf4, Oct3, Sox2, and c-Myc (415, 416). One mechanism by which KLF4 participates in ES cell self-renewal is through its upregulation in ES cells by leukemia inhibitory factor (LIF), which maintains pluripotency of ES cells (248). ES cells that overexpress KLF4 have a greater capacity to self-renew based on secondary embryoid body (EB) formation. Furthermore, EBs transduced with KLF4 express higher levels of Oct4, consistent with the notion that KLF4 regulates ES cell self-renewal (248). Global analysis of promoter occupancy by the four somatic cell reprogramming factors (Klf4, Oct4, Sox2 and c-Myc) and five additional factors important for pluripotency (217) revealed an integral role for KLF4 in a transcriptional hierarchy regulating ES cell pluripotency. Klf4 was identified as an upstream regulator of a large, feed-forward transcription factor loop that contains Oct4, Sox2, and c-Myc, as well as other common downstream factors such as Nanog (217) (see Figure 7). The study also indicated that c-Myc has a distinct role in regulating cell proliferation and chromosomal accessibility in ES cells, based on the identification of c-Myc promoter targets that were independent of the other pluripotency-inducing factors.
Figure 7. Klf4 as part of a transcriptional regulatory circuit for somatic cell reprogramming.
A model of the transcriptional hierarchy of the reprogramming transcription factors: Klf4, Sox2, Oct4, c-Myc. Klf4 is shown as an upstream regulator of feed-forward transcription loops—it binds the promoters of Oct4, Sox2, c-Myc and the downstream target Nanog. Klfs 2 and 5 can substitute for Klf4 in somatic cell reprogramming [Adapted from Kim, et al. (217)].
The ability of KLF4 to maintain immortality of iPS cells may arise from its cooperation with c-Myc. In a manner similar to the cooperation between KLF4 and RAS to affect transformation (352) (Figure 6), KLF4 and c-Myc may cooperate to promote iPS cell self-renewal, with KLF4 suppressing apoptosis induced by c-Myc and c-Myc neutralizing the cytostatic effect of KLF4 by suppressing p21Cip1/Waf1 (470). In this way, the balance between KLF4 and c-Myc may establish the immortalized state of iPS cells.
To identify additional factors that can generate iPS cells, Nakagawa et al. substituted the different factors used in somatic reprogramming with their respective homologs (295). They reported the successful generation of iPS clones upon substitution of Klf2 for Klf4 or L-Myc for c-Myc. Other substitutions, including Klf5 for Klf4, Sox1 for Sox2, and N-Myc for c-Myc resulted in positive clones, although fewer in number. No iPS clones developed when only three factors, devoid of Klf4, were used for reprogramming. The requirement for Oct4 and Klf4 in reprogramming somatic cells has been substantiated (110, 372).
Jiang et al. examined the contribution of KLFs 2, 4, and 5 to ES cell self-renewal (198). These KLFs are downregulated in ES cells induced to differentiate with retinoic acid. Depletion of Klf2, 4, or 5 in mouse ES cells does not affect ES self-renewal, and pair-wise depletion of the three factors likewise has no effect. In contrast, depletion of all three factors results in ES cell differentiation. The authors used chromatin immunoprecipitation studies to show that Klfs 2, 4, and 5 share gene targets with Nanog, including a number of genes that regulate pluripotency such as Esrrb, Pou5f1, Sox2 and Tcf3. Furthermore, these Klfs bind two upstream regions in the Nanog promoter, including a Nanog-enhancer element, and promote Nanog expression. These findings indicate that KLFs and Nanog work in concert to provide a core circuitry that regulates ES cell self-renewal (198).
Oct4 and LIF have additive effects that induce KLFs to sustain ES cell self-renewal, providing further support for the role of KLFs in ES cell maintenance. Oct4 primarily induces Klf2, whereas LIF/STAT3 signaling selectively increases Klf4 expression (171). Induction of both KLFs is necessary to maintain ES cell pluripotency, identity, and self-renewal (171).
KLF5 also plays a key role in self-renewal of mouse ES cells (123, 327). Parisi et al. reported that Klf5 is present in undifferentiated ES cells and co-localizes with Oct3/4 and Nanog. They also found that nuclear Klf5 localization correlates with that of Oct3/4 and Nanog in the pre-implantation embryos and in blastocysts at E3.5. In vitro depletion of Klf5 results in the morphological differentiation of ES cells, with a concomitant reduction in Oct3/4 and Nanog mRNA levels. Interestingly, Klf4 or Klf2 mRNA levels are unchanged upon Klf5 depletion. Overexpression of Klf5 prevents ES cell differentiation following removal of LIF, but the cells retain expression of Oct3/4 and Nanog. Furthermore, Klf5 was shown to regulate transcription of Oct3/4 and Nanog. Parisi et al. concluded that Klf5 is important for ES cell self-renewal and cannot be completely substituted by either Klf4 or Kfl2 (327).
Ema et al. studied Klf5−/− ES cells and made similar observations (123). Klf5−/− embryos fail to develop past E6.5 because of their failure to implant, which results from trophoectoderm defects—this time frame is earlier than what had been reported previously (378). Klf5−/− ES cells carried in culture exhibit expression of several differentiation marker genes and spontaneously differentiated at a high frequency. Overexpression of Klf5 in ES cells, on the other hand, decreases expression of differentiation markers and maintained pluripotency in the absence of LIF. To investigate the redundancy of KLFs in ESC self-renewal, Ema et al. examined whether Klf4 could rescue the differentiation defects observed in the Klf5−/− ES cells. Overexpression of Klf4 maintains Klf5−/− ES cells in an undifferentiated state, but the cells proliferate more slowly. Klf4 and Klf5 therefore suppress differentiation, but have opposing effects on proliferation (123).
A great deal of work remains to fully understand the molecular mechanisms of somatic reprogramming. KLFs are clearly key factors in self-renewal and pluriopotency of ES cells, but it will be important to determine the redundant and non-redundant features of individual KLFs in promoting a self-renewing, pluripotent state. Additional work is also required to elucidate the interactions between KLF proteins and other factors that control the stepwise transition in gene expression from somatic cell to pluripotent cell.
Chapter V. Conclusion
Since the initial characterization of KLF1 over 15 years ago, a large body of knowledge has been compiled regarding the biochemical and physiological functions of KLF family members. Studies of KLF proteins in mouse model systems and human diseases have elucidated the normal biological roles of the KLFs as well as their involvement in disease processes. However, further studies are needed to identify factors that determine the specific functions of individual KLFs, given the redundancy of family members and their common transcriptional targets. One factor contributing to KLF specificity may be post-translational modifications of KLFs, such as acetylation, sumoylation, and phosphorylation. These modifications have been shown to “switch” the transactivating or repressor functions of KLFs by altering their binding partners and changing affinities for target promoters. Structural analyses of KLF proteins will also help elucidate their specific functions—especially studies of their diverse amino-terminal regions. Finally, continued identification of novel KLF-binding proteins and co-factors in transcriptional complexes will provide information about their specific actions.
In defining functional processes in which KLFs participate, mouse models with disruptions of individual Klf genes are providing significant information about Klf expression patterns and functions during embryonic development and cell-specific lineage differentiation. Analyses of mice with tissue-specific deletion or inducible disruptions of Klfs are also helping to define tissue-specific functions. Furthermore, treatment of Klf KO mice with various stressors is yielding a more complete understanding of the participation of KLFs in stress responses. Another model system, C. elegans, provides an elegant tool for studying the role of KLFs in biological processes. KLF activity can be disrupted in C. elegans by expression of mutant forms or through RNA interference. Thus, this organism has been used to explore the participation of KLFs in fat storage and energy metabolism (42). The genetics of C. elegans also makes this system useful for the investigation of conserved regulators and downstream targets of KLF proteins.
Involvement of KLFs in the pathogenesis of cardiovascular disease and cancer have been recognized for some time; however, new activities await discovery, such as the participation of KLFs in regulating fat metabolism—currently an area of intense research. The participation of KLFs in somatic cell reprogramming has also generated much excitement for their possible application to regenerative medicine. Opportunities for development of targeted therapies will arise as we continue to expand our understanding of the normal biological functions of KLFs and their contribution to disease.
Acknowledgments
The authors wish to acknowledge the grant support from the National Institutes of Health (DK52230, DK643299, DK76742, and CA84197).
Abbreviations
- ALL
Acute lymphoblastic leukemia
- APC
Adenomatous polyposis coli
- ASM
Airway smooth muscle
- ATRA
All-trans retinoic acid
- AP-1
Activator protein-1
- BCR
B cell receptor
- BMP
Bone morphogenetic protein
- BTEB
Basic transcription element binding protein
- CBP
CREB-binding protein
- CCL5
Chemokine C-C motif ligand 5
- C/EBP
CCAAT/enhancer-binding protein
- CNS
Central nervous system
- CtBP
C-terminal binding protein
- CTGF
Connective tissue growth factor
- DBZ
Dibenzazepine
- DG
Dentate gyrus
- DSS
Dextran sodium sulfate
- EB
Embryoid body
- EGF
Epidermal growth factor
- EMT
Epithelial mesenchymal transition
- eNOS
Endothelial nitric oxide synthase
- ER
Estrogen receptor
- ERK
Extracellular signal-regulated kinase
- ES
Embryonic stem
- EZF
Epithelial zinc finger
- FAK
Focal adhesion kinase
- Fbw7
F-box and WD40 domain protein 7
- FOXO1
Forkhead box O1
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- hFOB
Human fetal osteoblast
- HSC
Hepatic stellate cell
- IEC6
Intestinal epithelial cell 6 (rat)
- IFN-γ
Interferon-γ
- IL
Interleukin
- iNOS
Inducible nitric-oxide synthase
- iPS
Induced pluripotent stem
- IVS
Intervening sequence
- KLF
Krüppel-like factors
- LAMA3A
Laminin α3A
- LCR
Locus control region
- LIF
Leukemia inhibitory factor
- LOH
Loss of heterozygosity
- LPA
Lysophosphatidic acid
- LPS
Lipopolysaccharide
- MAPK
Mitogen-activated protein kinase
- MEF
Mouse embryonic fibroblast
- MEF2
Myocyte enhancer factor 2
- MEK
MAPK/ERK kinase
- MEP
Megakaryocyte erythroid progenitor
- MMP-9
Matrix metalloproteinase-9
- OA
Okadaic acid
- ODC
Ornithine decarboxylase
- PAI-1
Plaminogen activator inhibitor-1
- P/CAF
p300/CBP-associated factor
- PDGF
Platelet-derived growth factor
- PEPCK
Phophoenolpyruvate carboxykinase
- PI3K
Phosphatidyloinositol-3-kinase
- PIAS
Protein inhibitor of activated STAT1
- POVPC
1-Palmytoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine
- PPAR
Peroxisome proliferation-activated receptors
- RANTES
Regulated upon activation, normal T cell expressed and secreted
- RAR
Retinoid acid receptor
- RGC
Retinal ganglion cell
- RXR
Retinoid X receptor
- S1P1
S1P receptor
- SBE
Smad binding element
- 15S-HETE
15-Hydroxy-eicosatetraenoic acid
- SNP
Single-nucleotide polymorphism
- SID
Sin3-interacting domain
- siRNA
Small interfering RNA
- α-SMA
α-Smooth muscle actin
- SMC
Smooth muscle cell
- SMRT
Silencing mediator of retinoid and thyroid receptor
- SNP
Single nucleotide polymorphism
- SP
Single-positive
- SREBP
Sterol response element binding protein
- SUMO
Small ubiquitin-like modifier
- TCE
TGF-β control element
- TCF4
T cell factor 4
- TIE
TGF-β-inhibitory element
- TGF-β
Transforming growth factor-β
- TNF-α
Tumor necrosis factor α
- Treg
Regulatory T cells
- VCAM-1
Vascular cell adhesion molecule-1
- VEGF
Vascular endothelial growth factor
References
- 1.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 2.Adelman CA, Chattopadhyay S, Bieker JJ. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development. 2002;129:539–549. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- 3.Agell L, Hernandez S, de Muga S, Lorente JA, Juanpere N, Esgueva R, Serrano S, Gelabert A, Lloreta J. KLF6 and TP53 mutations are a rare event in prostate cancer: distinguishing between Taq polymerase artifacts and true mutations. Mod Pathol. 2008;21:1470–1478. doi: 10.1038/modpathol.2008.145. [DOI] [PubMed] [Google Scholar]
- 4.Ahn YT, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol Cell Biol. 2007;27:253–266. doi: 10.1128/MCB.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akaogi K, Nakajima Y, Ito I, Kawasaki S, Oie SH, Murayama A, Kimura K, Yanagisawa J. KLF4 suppresses estrogen-dependent breast cancer growth by inhibiting the transcriptional activity of ERalpha. Oncogene. 2009;28:2894–2902. doi: 10.1038/onc.2009.151. [DOI] [PubMed] [Google Scholar]
- 6.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Rodriguez R, Barzi M, Berenguer J, Pons S. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J Biol Chem. 2007;282:37170–37180. doi: 10.1074/jbc.M705414200. [DOI] [PubMed] [Google Scholar]
- 8.Anderson KP, Crable SC, Lingrel JB. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood. 2000;95:1652–1655. [PubMed] [Google Scholar]
- 9.Anderson KP, Crable SC, Lingrel JB. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J Biol Chem. 1998;273:14347–14354. doi: 10.1074/jbc.273.23.14347. [DOI] [PubMed] [Google Scholar]
- 10.Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonello D, Moore PS, Zamboni G, Falconi M, Scarpa A. Absence of mutations in the transforming growth factor-beta inducible early gene 1, TIEG1, in pancreatic cancer. Cancer Lett. 2002;183:179–183. doi: 10.1016/s0304-3835(01)00802-3. [DOI] [PubMed] [Google Scholar]
- 12.Asano H, Li XS, Stamatoyannopoulos G. FKLF, a novel Kruppel-like factor that activates human embryonic and fetal beta-like globin genes. Mol Cell Biol. 1999;19:3571–3579. doi: 10.1128/mcb.19.5.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 14.Atkins GB, Wang Y, Mahabeleshwar GH, Shi H, Gao H, Kawanami D, Natesan V, Lin Z, Simon DI, Jain MK. Hemizygous deficiency of Kruppel-like factor 2 augments experimental atherosclerosis. Circ Res. 2008;103:690–693. doi: 10.1161/CIRCRESAHA.108.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bafford R, Sui XX, Wang G, Conte M. Angiotensin II and tumor necrosis factor-alpha upregulate survivin and Kruppel-like factor 5 in smooth muscle cells: Potential relevance to vein graft hyperplasia. Surgery. 2006;140:289–296. doi: 10.1016/j.surg.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 17.Banck MS, Beaven SW, Narla G, Walsh MJ, Friedman SL, Beutler AS. KLF6 degradation after apoptotic DNA damage. FEBS Lett. 2006;580:6981–6986. doi: 10.1016/j.febslet.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- 19.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu P, Lung TK, Lemsaddek W, Sargent TG, Williams DC, Jr, Basu M, Redmond LC, Lingrel JB, Haar JL, Lloyd JA. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood. 2007;110:3417–3425. doi: 10.1182/blood-2006-11-057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu P, Morris PE, Haar JL, Wani MA, Lingrel JB, Gaensler KM, Lloyd JA. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood. 2005;106:2566–2571. doi: 10.1182/blood-2005-02-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 23.Beckers J, Herrmann F, Rieger S, Drobyshev AL, Horsch M, Hrabe de Angelis M, Seliger B. Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114:590–597. doi: 10.1002/ijc.20798. [DOI] [PubMed] [Google Scholar]
- 24.Bellosta S, Ferri N, Bernini F, Paoletti R, Corsini A. Non-lipid-related effects of statins. Ann Med. 2000;32:164–176. doi: 10.3109/07853890008998823. [DOI] [PubMed] [Google Scholar]
- 25.Bensamoun SF, Hawse JR, Subramaniam M, Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ, Amadio PC, An KN, Spelsberg TC. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone. 2006;39:1244–1251. doi: 10.1016/j.bone.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Bensamoun SF, Tsubone T, Subramaniam M, Hawse JR, Boumediene E, Spelsberg TC, An KN, Amadio PC. Age-dependent changes in the mechanical properties of tail tendons in TGF-beta inducible early gene-1 knockout mice. J Appl Physiol. 2006;101:1419–1424. doi: 10.1152/japplphysiol.00800.2005. [DOI] [PubMed] [Google Scholar]
- 27.Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya R, Senbanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 29.Bialkowska AB, Du Y, Fu H, Yang VW. Identification of novel small-molecule compounds that inhibit the proproliferative Kruppel-like factor 5 in colorectal cancer cells by high-throughput screening. Mol Cancer Ther. 2009;8:563–570. doi: 10.1158/1535-7163.MCT-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianchi F, Hu J, Pelosi G, Cirincione R, Ferguson M, Ratcliffe C, Di Fiore PP, Gatter K, Pezzella F, Pastorino U. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNA microarray. Clin Cancer Res. 2004;10:6023–6028. doi: 10.1158/1078-0432.CCR-04-0619. [DOI] [PubMed] [Google Scholar]
- 31.Bieker JJ, Southwood CM. The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol Cell Biol. 1995;15:852–860. doi: 10.1128/mcb.15.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 34.Boon RA, Fledderus JO, Volger OL, van Wanrooij EJ, Pardali E, Weesie F, Kuiper J, Pannekoek H, ten Dijke P, Horrevoets AJ. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler Thromb Vasc Biol. 2007;27:532–539. doi: 10.1161/01.ATV.0000256466.65450.ce. [DOI] [PubMed] [Google Scholar]
- 35.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, Hotamisligil GS. Adipocyte fatty acid-binding protein, aP2, alters late atherosclerotic lesion formation in severe hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2002;22:1686–1691. doi: 10.1161/01.atv.0000033090.81345.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botella LM, Sanchez-Elsner T, Sanz-Rodriguez F, Kojima S, Shimada J, Guerrero-Esteo M, Cooreman MP, Ratziu V, Langa C, Vary CP, Ramirez JR, Friedman S, Bernabeu C. Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood. 2002;100:4001–4010. doi: 10.1182/blood.V100.12.4001. [DOI] [PubMed] [Google Scholar]
- 37.Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W, Louache F, Morle F. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112:576–584. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 38.Boyault S, Herault A, Balabaud C, Zucman-Rossi J. Absence of KLF6 gene mutation in 71 hepatocellular carcinomas. Hepatology. 2005;41:681–682. doi: 10.1002/hep.20588. author reply 682–683. [DOI] [PubMed] [Google Scholar]
- 39.Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50:111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 41.Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275:28230–28239. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 42.Brey CW, Nelder MP, Hailemariam T, Gaugler R, Hashmi S. Kruppel-like family of transcription factors: an emerging new frontier in fat biology. Int J Biol Sci. 2009;5:622–636. doi: 10.7150/ijbs.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck A, Buchholz M, Wagner M, Adler G, Gress T, Ellenrieder V. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol Cancer Res. 2006;4:861–872. doi: 10.1158/1541-7786.MCR-06-0081. [DOI] [PubMed] [Google Scholar]
- 44.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc--dependent pathway. Nat Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 45.Bureau C, Peron JM, Bouisson M, Danjoux M, Selves J, Bioulac-Sage P, Balabaud C, Torrisani J, Cordelier P, Buscail L, Vinel JP. Expression of the transcription factor Klf6 in cirrhosis, macronodules, and hepatocellular carcinoma. J Gastroenterol Hepatol. 2008;23:78–86. doi: 10.1111/j.1440-1746.2007.05234.x. [DOI] [PubMed] [Google Scholar]
- 46.Buttar NS, Fernandez-Zapico ME, Urrutia R. Key role of Kruppel-like factor proteins in pancreatic cancer and other gastrointestinal neoplasias. Curr Opin Gastroenterol. 2006;22:505–511. doi: 10.1097/01.mog.0000239864.73962.db. [DOI] [PubMed] [Google Scholar]
- 47.Camacho-Vanegas O, Narla G, Teixeira MS, DiFeo A, Misra A, Singh G, Chan AM, Friedman SL, Feuerstein BG, Martignetti JA. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–1395. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
- 48.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 49.Cao S, Fernandez-Zapico ME, Jin D, Puri V, Cook TA, Lerman LO, Zhu XY, Urrutia R, Shah V. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J Biol Chem. 2005;280:1901–1910. doi: 10.1074/jbc.M407941200. [DOI] [PubMed] [Google Scholar]
- 50.Cao Z, Wara AK, Icli B, Sun X, Packard RR, Esen F, Stapleton CJ, Subramaniam M, Kretschmer K, Apostolou I, von Boehmer H, Hansson GK, Spelsberg TC, Libby P, Feinberg MW. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(−) T cells and T regulatory cells. J Biol Chem. 2009;284:24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 52.Chaib H, Cockrell EK, Rubin MA, Macoska JA. Profiling and verification of gene expression patterns in normal and malignant human prostate tissues by cDNA microarray analysis. Neoplasia. 2001;3:43–52. doi: 10.1038/sj.neo.7900126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chanchevalap S, Nandan MO, McConnell BB, Charrier L, Merlin D, Katz JP, Yang VW. Kruppel-like factor 5 is an important mediator for lipopolysaccharide-induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 2006;34:1216–1223. doi: 10.1093/nar/gkl014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chanchevalap S, Nandan MO, Merlin D, Yang VW. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Kruppel-like factor 5. FEBS Lett. 2004;578:99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, Zhou Z, Cheng X, Simons JW, Dong JT. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 58.Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson HF, Jr, Vessella RL, Dong JT. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol. 2003;162:1349–1354. doi: 10.1016/S0002-9440(10)63930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 60.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 61.Chen GG, Xu H, Lee JF, Subramaniam M, Leung KL, Wang SH, Chan UP, Spelsberg TC. 15-hydroxy-eicosatetraenoic acid arrests growth of colorectal cancer cells via a peroxisome proliferator-activated receptor gamma-dependent pathway. Int J Cancer. 2003;107:837–843. doi: 10.1002/ijc.11447. [DOI] [PubMed] [Google Scholar]
- 62.Chen MM, Lam A, Abraham JA, Schreiner GF, Joly AH. CTGF expression is induced by TGF-beta in cardiac fibroblasts and cardiac myocytes: a potential role in heart fibrosis. J Mol Cell Cardiol. 2000;32:1805–1819. doi: 10.1006/jmcc.2000.1215. [DOI] [PubMed] [Google Scholar]
- 63.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Bieker JJ. Stage-specific repression by the EKLF transcriptional activator. Mol Cell Biol. 2004;24:10416–10424. doi: 10.1128/MCB.24.23.10416-10424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Bieker JJ. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol Cell Biol. 2001;21:3118–3125. doi: 10.1128/MCB.21.9.3118-3125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Reitman M, Bieker JJ. Chromatin structure and transcriptional control elements of the erythroid Kruppel-like factor (EKLF) gene. J Biol Chem. 1998;273:25031–25040. doi: 10.1074/jbc.273.39.25031. [DOI] [PubMed] [Google Scholar]
- 68.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC, Chen CM. Nuclear Kruppel-like factor 4 expression is associated with human skin squamous cell carcinoma progression and metastasis. Cancer Biol Ther. 2008;7:777–782. doi: 10.4161/cbt.7.5.5768. [DOI] [PubMed] [Google Scholar]
- 70.Chen ZY, Rex S, Tseng CC. Kruppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 71.Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 72.Chen ZY, Shie JL, Tseng CC. STAT1 is required for IFN-gamma-mediated gut-enriched Kruppel-like factor expression. Exp Cell Res. 2002;281:19–27. doi: 10.1006/excr.2002.5633. [DOI] [PubMed] [Google Scholar]
- 73.Chen ZY, Tseng CC. 15-deoxy-Delta12,14 prostaglandin J2 up-regulates Kruppel-like factor 4 expression independently of peroxisome proliferator-activated receptor gamma by activating the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signal transduction pathway in HT-29 colon cancer cells. Mol Pharmacol. 2005;68:1203–1213. doi: 10.1124/mol.105.014944. [DOI] [PubMed] [Google Scholar]
- 74.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 75.Cheng XF, Li D, Zhuang M, Chen ZY, Lu DX, Hattori T. Growth inhibitory effect of Kruppel-like factor 6 on human prostatic carcinoma and renal carcinoma cell lines. Tohoku J Exp Med. 2008;216:35–45. doi: 10.1620/tjem.216.35. [DOI] [PubMed] [Google Scholar]
- 76.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cherepanova OA, Pidkovka NA, Sarmento OF, Yoshida T, Gan Q, Adiguzel E, Bendeck MP, Berliner J, Leitinger N, Owens GK. Oxidized phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ Res. 2009;104:609–618. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 79.Chintharlapalli S, Papineni S, Liu S, Jutooru I, Chadalapaka G, Cho SD, Murthy RS, You Y, Safe S. 2-cyano-lup-1-en-3-oxo-20-oic acid, a cyano derivative of betulinic acid, activates peroxisome proliferator-activated receptor gamma in colon and pancreatic cancer cells. Carcinogenesis. 2007;28:2337–2346. doi: 10.1093/carcin/bgm189. [DOI] [PubMed] [Google Scholar]
- 80.Cho SD, Chintharlapalli S, Abdelrahim M, Papineni S, Liu S, Guo J, Lei P, Abudayyeh A, Safe S. 5,5′-Dibromo-bis(3′-indolyl)methane induces Kruppel-like factor 4 and p21 in colon cancer cells. Mol Cancer Ther. 2008;7:2109–2120. doi: 10.1158/1535-7163.MCT-07-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho SY, Park PJ, Shin HJ, Kim YK, Shin DW, Shin ES, Lee HH, Lee BG, Baik JH, Lee TR. (-)-Catechin suppresses expression of Kruppel-like factor 7 and increases expression and secretion of adiponectin protein in 3T3-L1 cells. Am J Physiol Endocrinol Metab. 2007;292:E1166–1172. doi: 10.1152/ajpendo.00436.2006. [DOI] [PubMed] [Google Scholar]
- 82.Cho YG, Choi BJ, Kim CJ, Song JW, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic alterations of the KLF6 gene in colorectal cancers. APMIS. 2006;114:458–464. doi: 10.1111/j.1600-0463.2006.apm_431.x. [DOI] [PubMed] [Google Scholar]
- 83.Cho YG, Choi BJ, Song JW, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Aberrant expression of krUppel-like factor 6 protein in colorectal cancers. World J Gastroenterol. 2006;12:2250–2253. doi: 10.3748/wjg.v12.i14.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–4590. doi: 10.1038/sj.onc.1208670. [DOI] [PubMed] [Google Scholar]
- 85.Cho YG, Song JH, Kim CJ, Nam SW, Yoo NJ, Lee JY, Park WS. Genetic and epigenetic analysis of the KLF4 gene in gastric cancer. APMIS. 2007;115:802–808. doi: 10.1111/j.1600-0463.2007.apm_643.x. [DOI] [PubMed] [Google Scholar]
- 86.Choi BJ, Cho YG, Song JW, Kim CJ, Kim SY, Nam SW, Yoo NJ, Lee JY, Park WS. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202:585–589. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 87.Cicchini C, Laudadio I, Citarella F, Corazzari M, Steindler C, Conigliaro A, Fantoni A, Amicone L, Tripodi M. TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp Cell Res. 2008;314:143–152. doi: 10.1016/j.yexcr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 88.Coghill E, Eccleston S, Fox V, Cerruti L, Brown C, Cunningham J, Jane S, Perkins A. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood. 2001;97:1861–1868. doi: 10.1182/blood.v97.6.1861. [DOI] [PubMed] [Google Scholar]
- 89.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- 91.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 92.Cook T, Urrutia R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am J Physiol Gastrointest Liver Physiol. 2000;278:G513–521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- 93.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crossley M, Tsang AP, Bieker JJ, Orkin SH. Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. J Biol Chem. 1994;269:15440–15444. [PubMed] [Google Scholar]
- 95.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr Opin Genet Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Czubryt MP, Olson EN. Balancing contractility and energy production: the role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog Horm Res. 2004;59:105–124. doi: 10.1210/rp.59.1.105. [DOI] [PubMed] [Google Scholar]
- 98.D’Astolfo DS, Gehrau RC, Bocco JL, Koritschoner NP. Silencing of the transcription factor KLF6 by siRNA leads to cell cycle arrest and sensitizes cells to apoptosis induced by DNA damage. Cell Death Differ. 2008;15:613–616. doi: 10.1038/sj.cdd.4402299. [DOI] [PubMed] [Google Scholar]
- 99.D’Souza UM, Lammers CH, Hwang CK, Yajima S, Mouradian MM. Developmental expression of the zinc finger transcription factor DRRF (dopamine receptor regulating factor) Mech Dev. 2002;110:197–201. doi: 10.1016/s0925-4773(01)00564-0. [DOI] [PubMed] [Google Scholar]
- 100.Dai G, Kaazempur-Mofrad MR, Natarajan S, Zhang Y, Vaughn S, Blackman BR, Kamm RD, Garcia-Cardena G, Gimbrone MA., Jr Distinct endothelial phenotypes evoked by arterial waveforms derived from atherosclerosis-susceptible and -resistant regions of human vasculature. Proc Natl Acad Sci U S A. 2004;101:14871–14876. doi: 10.1073/pnas.0406073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, Yang VW. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dang DT, Chen X, Feng J, Torbenson M, Dang LH, Yang VW. Overexpression of Kruppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–3430. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das A, Fernandez-Zapico ME, Cao S, Yao J, Fiorucci S, Hebbel RP, Urrutia R, Shah VH. Disruption of an SP2/KLF6 repression complex by SHP is required for farnesoid X receptor-induced endothelial cell migration. J Biol Chem. 2006;281:39105–39113. doi: 10.1074/jbc.M607720200. [DOI] [PubMed] [Google Scholar]
- 106.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dekker RJ, Boon RA, Rondaij MG, Kragt A, Volger OL, Elderkamp YW, Meijers JC, Voorberg J, Pannekoek H, Horrevoets AJ. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 108.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 109.Dekker RJ, van Thienen JV, Rohlena J, de Jager SC, Elderkamp YW, Seppen J, de Vries CJ, Biessen EA, van Berkel TJ, Pannekoek H, Horrevoets AJ. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am J Pathol. 2005;167:609–618. doi: 10.1016/S0002-9440(10)63002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Di Stefano B, Prigione A, Broccoli V. Efficient genetic reprogramming of unmodified somatic neural progenitors uncovers the essential requirement of Oct4 and Klf4. Stem Cells Dev. 2008 doi: 10.1089/scd.2008.0180. [DOI] [PubMed] [Google Scholar]
- 111.DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, Narla G. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68:965–970. doi: 10.1158/0008-5472.CAN-07-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G, Martignetti JA. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res. 2009;69:4733–4741. doi: 10.1158/0008-5472.CAN-08-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.DiFeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat. 2009;12:1–7. doi: 10.1016/j.drup.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 114.DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, Kalir T, Yao S, Levine A, Birrer MJ, Bonome T, Friedman SL, Buller RE, Martignetti JA. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–3739. doi: 10.1158/1078-0432.CCR-06-0054. [DOI] [PubMed] [Google Scholar]
- 115.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Donze D, Townes TM, Bieker JJ. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 117.Dosen-Dahl G, Munthe E, Nygren MK, Stubberud H, Hystad ME, Rian E. Bone marrow stroma cells regulate TIEG1 expression in acute lymphoblastic leukemia cells: role of TGFbeta/BMP-6 and TIEG1 in chemotherapy escape. Int J Cancer. 2008;123:2759–2766. doi: 10.1002/ijc.23833. [DOI] [PubMed] [Google Scholar]
- 118.Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, Grosveld F, Philipsen S. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol Cell Biol. 2005;25:5205–5214. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Du JX, Bialkowska AB, McConnell BB, Yang VW. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J Biol Chem. 2008;283:31991–32002. doi: 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Du JX, Yun CC, Bialkowska A, Yang VW. Protein inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor Kruppel-like factor 5. J Biol Chem. 2007;282:4782–4793. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ellenrieder V. TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res. 2008;28:1531–1539. [PubMed] [Google Scholar]
- 122.Ellenrieder V, Buck A, Harth A, Jungert K, Buchholz M, Adler G, Urrutia R, Gress TM. KLF11 mediates a critical mechanism in TGF-beta signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology. 2004;127:607–620. doi: 10.1053/j.gastro.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 123.Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, Shimosato D, Uchida K, Suzuki N, Yanagisawa J, Sogawa K, Rossant J, Yamamoto M, Takahashi S, Fujii-Kuriyama Y. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 124.Endrizzi BT, Jameson SC. Differential role for IL-7 in inducing lung Kruppel-like factor (Kruppel-like factor 2) expression by naive versus activated T cells. Int Immunol. 2003;15:1341–1348. doi: 10.1093/intimm/dxg133. [DOI] [PubMed] [Google Scholar]
- 125.Esmon CT. Crosstalk between inflammation and thrombosis. Maturitas. 2004;47:305–314. doi: 10.1016/j.maturitas.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 126.Evans PM, Liu C. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008;40:554–564. doi: 10.1111/j.1745-7270.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 128.Fabre S, Carrette F, Chen J, Lang V, Semichon M, Denoyelle C, Lazar V, Cagnard N, Dubart-Kupperschmitt A, Mangeney M, Fruman DA, Bismuth G. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 129.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25− T cells through Foxp3 induction and down-regulation of Smad7. J Immunol. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 130.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 131.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Feng WC, Southwood CM, Bieker JJ. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem. 1994;269:1493–1500. [PubMed] [Google Scholar]
- 133.Fernandez-Zapico ME, Mladek A, Ellenrieder V, Folch-Puy E, Miller L, Urrutia R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003;22:4748–4758. doi: 10.1093/emboj/cdg470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fischer EA, Verpont MC, Garrett-Sinha LA, Ronco PM, Rossert JA. Klf6 is a zinc finger protein expressed in a cell-specific manner during kidney development. J Am Soc Nephrol. 2001;12:726–735. doi: 10.1681/ASN.V124726. [DOI] [PubMed] [Google Scholar]
- 136.Flandez M, Guilmeau S, Blache P, Augenlicht LH. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res. 2008;314:3712–3723. doi: 10.1016/j.yexcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fledderus JO, van Thienen JV, Boon RA, Dekker RJ, Rohlena J, Volger OL, Bijnens AP, Daemen MJ, Kuiper J, van Berkel TJ, Pannekoek H, Horrevoets AJ. Prolonged shear stress and KLF2 suppress constitutive proinflammatory transcription through inhibition of ATF2. Blood. 2007;109:4249–4257. doi: 10.1182/blood-2006-07-036020. [DOI] [PubMed] [Google Scholar]
- 138.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005;6:331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 139.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 140.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson AJ, Kudlow JE, Lobo-Ruppert SM, Ruppert JM. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT, Ruppert JM. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 142.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frigo DE, Sherk AB, Wittmann BM, Norris JD, Wang Q, Joseph JD, Toner AP, Brown M, McDonnell DP. Induction of Kruppel-like factor 5 expression by androgens results in increased CXCR4-dependent migration of prostate cancer cells in vitro. Mol Endocrinol. 2009;23:1385–1396. doi: 10.1210/me.2009-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frontelo P, Manwani D, Galdass M, Karsunky H, Lohmann F, Gallagher PG, Bieker JJ. Novel role for EKLF in megakaryocyte lineage commitment. Blood. 2007;110:3871–3880. doi: 10.1182/blood-2007-03-082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fujiu K, Manabe I, Ishihara A, Oishi Y, Iwata H, Nishimura G, Shindo T, Maemura K, Kagechika H, Shudo K, Nagai R. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting KLF5. Circ Res. 2005;97:1132–1141. doi: 10.1161/01.RES.0000190613.22565.13. [DOI] [PubMed] [Google Scholar]
- 146.Gao D, Niu X, Ning N, Hao G. Regulation of angiotensin II-Induced Kruppel-like factor 5 expression in vascular smooth muscle cells. Biol Pharm Bull. 2006;29:2004–2008. doi: 10.1248/bpb.29.2004. [DOI] [PubMed] [Google Scholar]
- 147.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ghaleb AM, Aggarwal G, Bialkowska AB, Nandan MO, Yang VW. Notch inhibits expression of the Kruppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res. 2008;6:1920–1927. doi: 10.1158/1541-7786.MCR-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26:2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ghaleb AM, McConnell BB, Nandan MO, Katz JP, Kaestner KH, Yang VW. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–7154. doi: 10.1158/0008-5472.CAN-07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ghaleb AM, Yang VW. The Pathobiology of Kruppel-like Factors in Colorectal Cancer. Curr Colorectal Cancer Rep. 2008;4:59–64. doi: 10.1007/s11888-008-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Giefing M, Wierzbicka M, Rydzanicz M, Cegla R, Kujawski M, Szyfter K. Chromosomal gains and losses indicate oncogene and tumor suppressor gene candidates in salivary gland tumors. Neoplasma. 2008;55:55–60. [PubMed] [Google Scholar]
- 154.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1784–1792. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 156.Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc Natl Acad Sci U S A. 2007;104:13420–13425. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gordon AR, Outram SV, Keramatipour M, Goddard CA, Colledge WH, Metcalfe JC, Hager-Theodorides AL, Crompton T, Kemp PR. Splenomegaly and modified erythropoiesis in KLF13−/− mice. J Biol Chem. 2008;283:11897–11904. doi: 10.1074/jbc.M709569200. [DOI] [PubMed] [Google Scholar]
- 158.Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 159.Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grayson JM, Murali-Krishna K, Altman JD, Ahmed R. Gene expression in antigen-specific CD8+ T cells during viral infection. J Immunol. 2001;166:795–799. doi: 10.4049/jimmunol.166.2.795. [DOI] [PubMed] [Google Scholar]
- 161.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 162.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 163.Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J Biol Chem. 2009;284:6071–6078. doi: 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guo P, Dong XY, Zhao K, Sun X, Li Q, Dong JT. Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor beta. J Biol Chem. 2009;284:28243–28252. doi: 10.1074/jbc.M109.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo P, Dong XY, Zhao KW, Sun X, Li Q, Dong JT. Estrogen-induced interaction between KLF5 and estrogen receptor (ER) suppresses the function of ER in ER-positive breast cancer cells. Int J Cancer. 2009 doi: 10.1002/ijc.24696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guo P, Zhao KW, Dong XY, Sun X, Dong JT. Acetylation of KLF5 alters the assembly of p15 transcription factors in transforming growth factor-beta-mediated induction in epithelial cells. J Biol Chem. 2009;284:18184–18193. doi: 10.1074/jbc.M109.007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gutierrez-Aguilar R, Froguel P, Hamid YH, Benmezroua Y, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O, Neve B. Genetic analysis of Kruppel-like zinc finger 11 variants in 5864 Danish individuals: potential effect on insulin resistance and modified signal transducer and activator of transcription-3 binding by promoter variant −1659G>C. J Clin Endocrinol Metab. 2008;93:3128–3135. doi: 10.1210/jc.2007-2504. [DOI] [PubMed] [Google Scholar]
- 168.Haaland RE, Yu W, Rice AP. Identification of LKLF-regulated genes in quiescent CD4+ T lymphocytes. Mol Immunol. 2005;42:627–641. doi: 10.1016/j.molimm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 169.Hagos EG, Ghaleb AM, Dalton WB, Bialkowska AB, Yang VW. Mouse embryonic fibroblasts null for the Kruppel-like factor 4 gene are genetically unstable. Oncogene. 2009;28:1197–1205. doi: 10.1038/onc.2008.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Haldar SM, Ibrahim OA, Jain MK. Kruppel-like Factors (KLFs) in muscle biology. J Mol Cell Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, Tomlinson S, Smith A. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 172.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 173.Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 174.Hartel M, Narla G, Wente MN, Giese NA, Martignoni ME, Martignetti JA, Friess H, Friedman SL. Increased alternative splicing of the KLF6 tumour suppressor gene correlates with prognosis and tumour grade in patients with pancreatic cancer. Eur J Cancer. 2008;44:1895–1903. doi: 10.1016/j.ejca.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B, Rajamannan NM, Oursler MJ, Turner RT, Spelsberg TC, Subramaniam M. TIEG-null mice display an osteopenic gender-specific phenotype. Bone. 2008;42:1025–1031. doi: 10.1016/j.bone.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.He M, Han M, Zheng B, Shu YN, Wen JK. Angiotensin II stimulates KLF5 phosphorylation and its interaction with c-Jun leading to suppression of p21 expression in vascular smooth muscle cells. J Biochem. 2009 doi: 10.1093/jb/mvp115. [DOI] [PubMed] [Google Scholar]
- 177.Hefferan TE, Reinholz GG, Rickard DJ, Johnsen SA, Waters KM, Subramaniam M, Spelsberg TC. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J Biol Chem. 2000;275:20255–20259. doi: 10.1074/jbc.C000135200. [DOI] [PubMed] [Google Scholar]
- 178.Hefferan TE, Subramaniam M, Khosla S, Riggs BL, Spelsberg TC. Cytokine-specific induction of the TGF-beta inducible early gene (TIEG): regulation by specific members of the TGF-beta family. J Cell Biochem. 2000;78:380–390. doi: 10.1002/1097-4644(20000901)78:3<380::aid-jcb4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 179.Henning K, Berndt A, Katenkamp D, Kosmehl H. Loss of laminin-5 in the epithelium-stroma interface: an immunohistochemical marker of malignancy in epithelial lesions of the breast. Histopathology. 1999;34:305–309. doi: 10.1046/j.1365-2559.1999.00634.x. [DOI] [PubMed] [Google Scholar]
- 180.Higgs DR. Do LCRs open chromatin domains? Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 181.Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, Athaide CP, Abedrapo MA, Chen X, Yang VW, Hodin RA. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 182.Hirasawa Y, Arai M, Imazeki F, Tada M, Mikata R, Fukai K, Miyazaki M, Ochiai T, Saisho H, Yokosuka O. Methylation status of genes upregulated by demethylating agent 5-aza-2′-deoxycytidine in hepatocellular carcinoma. Oncology. 2006;71:77–85. doi: 10.1159/000100475. [DOI] [PubMed] [Google Scholar]
- 183.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hoshino Y, Kurabayashi M, Kanda T, Hasegawa A, Sakamoto H, Okamoto E, Kowase K, Watanabe N, Manabe I, Suzuki T, Nakano A, Takase S, Wilcox JN, Nagai R. Regulated expression of the BTEB2 transcription factor in vascular smooth muscle cells: analysis of developmental and pathological expression profiles shows implications as a predictive factor for restenosis. Circulation. 2000;102:2528–2534. doi: 10.1161/01.cir.102.20.2528. [DOI] [PubMed] [Google Scholar]
- 185.Hu B, Wu Z, Liu T, Ullenbruch MR, Jin H, Phan SH. Gut-enriched Kruppel-like factor interaction with Smad3 inhibits myofibroblast differentiation. Am J Respir Cell Mol Biol. 2007;36:78–84. doi: 10.1165/rcmb.2006-0043OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hu W, Hofstetter WL, Li H, Zhou Y, He Y, Pataer A, Wang L, Xie K, Swisher SG, Fang B. Putative tumor-suppressive function of kruppel-like factor 4 in primary lung carcinoma. Clin Cancer Res. 2009;15:5688–5695. doi: 10.1158/1078-0432.CCR-09-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Huang B, Ahn YT, McPherson L, Clayberger C, Krensky AM. Interaction of PRP4 with Kruppel-like factor 13 regulates CCL5 transcription. J Immunol. 2007;178:7081–7087. doi: 10.4049/jimmunol.178.11.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Huang CC, Liu Z, Li X, Bailey SK, Nail CD, Foster KW, Frost AR, Ruppert JM, Lobo-Ruppert SM. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol Ther. 2005;4:1401–1408. doi: 10.4161/cbt.4.12.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huang X, Li X, Guo B. KLF6 induces apoptosis in prostate cancer cells through up-regulation of ATF3. J Biol Chem. 2008;283:29795–29801. doi: 10.1074/jbc.M802515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huddleson JP, Srinivasan S, Ahmad N, Lingrel JB. Fluid shear stress induces endothelial KLF2 gene expression through a defined promoter region. Biol Chem. 2004;385:723–729. doi: 10.1515/BC.2004.088. [DOI] [PubMed] [Google Scholar]
- 191.Hwang CK, D’Souza UM, Eisch AJ, Yajima S, Lammers CH, Yang Y, Lee SH, Kim YM, Nestler EJ, Mouradian MM. Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc Natl Acad Sci U S A. 2001;98:7558–7563. doi: 10.1073/pnas.121635798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, Buettner R. Transcriptional regulation of the AP-2alpha promoter by BTEB-1 and AP-2rep, a novel wt-1/egrrelated zinc finger repressor. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, Kawabe T, Hasegawa Y, Shimokata K, Sekido Y. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64:3838–3843. doi: 10.1158/0008-5472.CAN-04-0185. [DOI] [PubMed] [Google Scholar]
- 194.Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, Lerman MI. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jain MK, Ridker PM. Anti-inflammatory effects of statins: clinical evidence and basic mechanisms. Nat Rev Drug Discov. 2005;4:977–987. doi: 10.1038/nrd1901. [DOI] [PubMed] [Google Scholar]
- 196.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein-Barr virus ED-L2 promoters by gut-enriched Kruppel-like factor. J Biol Chem. 1998;273:10747–10754. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 197.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 198.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 199.Jiang L, Chen Y, Chan CY, Wang X, Lin L, He ML, Lin MC, Yew DT, Sung JJ, Li JC, Kung HF. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009;274:101–108. doi: 10.1016/j.canlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 200.Jiang W, Deng W, Bailey SK, Nail CD, Frost AR, Brouillette WJ, Muccio DD, Grubbs CJ, Ruppert JM, Lobo-Ruppert SM. Prevention of KLF4-mediated tumor initiation and malignant transformation by UAB30 rexinoid. Cancer Biol Ther. 2009;8:289–298. doi: 10.4161/cbt.8.3.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jin W, Di G, Li J, Chen Y, Li W, Wu J, Cheng T, Yao M, Shao Z. TIEG1 induces apoptosis through mitochondrial apoptotic pathway and promotes apoptosis induced by homoharringtonine and velcade. FEBS Lett. 2007;581:3826–3832. doi: 10.1016/j.febslet.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 202.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 203.Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J Cell Biochem. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- 204.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 206.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kajimura D, Dragomir C, Ramirez F, Laub F. Identification of genes regulated by transcription factor KLF7 in differentiating olfactory sensory neurons. Gene. 2007;388:34–42. doi: 10.1016/j.gene.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 208.Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, Yao J, Xie K. Loss of Kruppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 209.Kanazawa A, Kawamura Y, Sekine A, Iida A, Tsunoda T, Kashiwagi A, Tanaka Y, Babazono T, Matsuda M, Kawai K, Iiizumi T, Fujioka T, Imanishi M, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakamura Y, Maeda S. Single nucleotide polymorphisms in the gene encoding Kruppel-like factor 7 are associated with type 2 diabetes. Diabetologia. 2005;48:1315–1322. doi: 10.1007/s00125-005-1797-0. [DOI] [PubMed] [Google Scholar]
- 210.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 211.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kawada H, Ito T, Pharr PN, Spyropoulos DD, Watson DK, Ogawa M. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int J Hematol. 2001;73:463–468. doi: 10.1007/BF02994008. [DOI] [PubMed] [Google Scholar]
- 213.Kawai-Kowase K, Kumar MS, Hoofnagle MH, Yoshida T, Owens GK. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol Cell Biol. 2005;25:8009–8023. doi: 10.1128/MCB.25.18.8009-8023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 215.Kawamura Y, Tanaka Y, Kawamori R, Maeda S. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006;20:844–856. doi: 10.1210/me.2005-0138. [DOI] [PubMed] [Google Scholar]
- 216.Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS, Fruman DA. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109:747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim Y, Ratziu V, Choi SG, Lalazar A, Theiss G, Dang Q, Kim SJ, Friedman SL. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem. 1998;273:33750–33758. doi: 10.1074/jbc.273.50.33750. [DOI] [PubMed] [Google Scholar]
- 219.King KE, Iyemere VP, Weissberg PL, Shanahan CM. Kruppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor beta 1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278:11661–11669. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
- 220.Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Kruppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. 2007;179:4679–4684. doi: 10.4049/jimmunol.179.7.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 222.Kohler B, Wolter M, Blaschke B, Reifenberger G. Absence of mutations in the putative tumor suppressor gene KLF6 in glioblastomas and meningiomas. Int J Cancer. 2004;111:644–645. doi: 10.1002/ijc.20302. [DOI] [PubMed] [Google Scholar]
- 223.Koipally J, Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J Biol Chem. 2000;275:19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 224.Koivisto PA, Hyytinen ER, Matikainen M, Tammela TL, Ikonen T, Schleutker J. Kruppel-like factor 6 germ-line mutations are infrequent in Finnish hereditary prostate cancer. J Urol. 2004;172:506–507. doi: 10.1097/01.ju.0000129242.88182.e1. [DOI] [PubMed] [Google Scholar]
- 225.Koivisto PA, Zhang X, Sallinen SL, Sallinen P, Helin HJ, Dong JT, Van Meir EG, Haapasalo H, Hyytinen ER. Absence of KLF6 gene mutations in human astrocytic tumors and cell lines. Int J Cancer. 2004;111:642–643. doi: 10.1002/ijc.20301. [DOI] [PubMed] [Google Scholar]
- 226.Kojima S, Hayashi S, Shimokado K, Suzuki Y, Shimada J, Crippa MP, Friedman SL. Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-beta1 in vascular endothelial cells. Blood. 2000;95:1309–1316. [PubMed] [Google Scholar]
- 227.Komander D. The emerging complexity of protein ubiquitination. Biochemical Society Transactions. 2009;037:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 228.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 229.Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, Lee JS, Schwartz M, Thung SN, Fiel IM, Banck M, Zimran E, Thorgeirsson SS, Mazzaferro V, Bruix J, Martignetti JA, Llovet JM, Friedman SL. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645–654. doi: 10.1016/j.jhep.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P, Martignetti JA, Friedman SL. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 231.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA Expression in Human Airway Smooth Muscle Cells: Role of miR-25 in Regulation of Airway Smooth Muscle Phenotype. Am J Respir Cell Mol Biol. 2009 doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kulozik AE, Bellan-Koch A, Bail S, Kohne E, Kleihauer E. Thalassemia intermedia: moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood. 1991;77:2054–2058. [PubMed] [Google Scholar]
- 233.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol Cell Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 236.Kuroda E, Horikawa Y, Enya M, Oda N, Suzuki E, Iizuka K, Takeda J. Identification of minimal promoter and genetic variants of Kruppel-like factor 11 gene and association analysis with type 2 diabetes in Japanese. Endocr J. 2009;56:275–286. doi: 10.1507/endocrj.k08e-302. [DOI] [PubMed] [Google Scholar]
- 237.Kwak MK, Lee HJ, Hur K, Park do J, Lee HS, Kim WH, Lee KU, Choe KJ, Guilford P, Yang HK. Expression of Kruppel-like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134:163–167. doi: 10.1007/s00432-007-0265-2. [DOI] [PubMed] [Google Scholar]
- 238.Lalazar A, Wong L, Yamasaki G, Friedman SL. Early genes induced in hepatic stellate cells during wound healing. Gene. 1997;195:235–243. doi: 10.1016/s0378-1119(97)00159-5. [DOI] [PubMed] [Google Scholar]
- 239.Laub F, Aldabe R, Friedrich V, Jr, Ohnishi S, Yoshida T, Ramirez F. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev Biol. 2001;233:305–318. doi: 10.1006/dbio.2001.0243. [DOI] [PubMed] [Google Scholar]
- 240.Laub F, Aldabe R, Ramirez F, Friedman S. Embryonic expression of Kruppel-like factor 6 in neural and non-neural tissues. Mech Dev. 2001;106:167–170. doi: 10.1016/s0925-4773(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 241.Laub F, Lei L, Sumiyoshi H, Kajimura D, Dragomir C, Smaldone S, Puche AC, Petros TJ, Mason C, Parada LF, Ramirez F. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol. 2005;25:5699–5711. doi: 10.1128/MCB.25.13.5699-5711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, Nemer M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006;25:5201–5213. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, Mericko P, Stadtfeld M, Zhou D, Cheng L, Graf T, MacRae CA, Lepore JJ, Lo CW, Kahn ML. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 244.Lee MY, Moon JS, Park SW, Koh YK, Ahn YH, Kim KS. KLF5 enhances SREBP-1 action in androgen-dependent induction of fatty acid synthase in prostate cancer cells. Biochem J. 2009;417:313–322. doi: 10.1042/BJ20080762. [DOI] [PubMed] [Google Scholar]
- 245.Levings PP, Bungert J. The human beta-globin locus control region. Eur J Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 246.Li D, Yea S, Dolios G, Martignetti JA, Narla G, Wang R, Walsh MJ, Friedman SL. Regulation of Kruppel-like factor 6 tumor suppressor activity by acetylation. Cancer Res. 2005;65:9216–9225. doi: 10.1158/0008-5472.CAN-05-1040. [DOI] [PubMed] [Google Scholar]
- 247.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 248.Li Y, McClintick J, Zhong L, Edenberg HJ, Yoder MC, Chan RJ. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- 249.Liao JK, Laufs U. Pleiotropic effects of statins. Annu Rev Pharmacol Toxicol. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lievre A, Landi B, Cote JF, Veyrie N, Zucman-Rossi J, Berger A, Laurent-Puig P. Absence of mutation in the putative tumor-suppressor gene KLF6 in colorectal cancers. Oncogene. 2005;24:7253–7256. doi: 10.1038/sj.onc.1208867. [DOI] [PubMed] [Google Scholar]
- 251.Lim SK, Bieker JJ, Lin CS, Costantini F. A shortened life span of EKLF−/− adult erythrocytes, due to a deficiency of beta-globin chains, is ameliorated by human gamma-globin chains. Blood. 1997;90:1291–1299. [PubMed] [Google Scholar]
- 252.Lin S, Wang D, Iyer S, Ghaleb AM, Shim H, Yang VW, Chun J, Yun CC. The absence of LPA2 attenuates tumor formation in an experimental model of colitis-associated cancer. Gastroenterology. 2009;136:1711–1720. doi: 10.1053/j.gastro.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Jr, Balasubramanian V, Garcia-Cardena G, Jain MK. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96:e48–57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 254.Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, Wang W, Wang R, Wang Q, Sun J, Wang P. The Fbw7/hCDC4 tumor suppressor targets pro-proliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010 doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liu R, Zheng HQ, Zhou Z, Dong JT, Chen C. KLF5 promotes breast cell survival partially through fibroblast growth factor-binding protein 1-pERK-mediated dual specificity MKP-1 protein phosphorylation and stabilization. J Biol Chem. 2009;284:16791–16798. doi: 10.1074/jbc.M808919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 257.Liu Z, Teng L, Bailey SK, Frost AR, Bland KI, Lobuglio AF, Ruppert JM, Lobo-Ruppert SM. Epithelial transformation by KLF4 requires Notch1 but not canonical Notch1 signaling. Cancer Biol Ther. 2009;8 doi: 10.4161/cbt.8.19.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lohmann F, Bieker JJ. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development. 2008;135:2071–2082. doi: 10.1242/dev.018200. [DOI] [PubMed] [Google Scholar]
- 259.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, Wang T, Zhi H, Wu M, Liu Z. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–1299. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 260.Ma L, Hanson RL, Que LN, Mack JL, Franks PW, Infante AM, Kobes S, Bogardus C, Baier LJ. Association analysis of Kruppel-like factor 11 variants with type 2 diabetes in Pima Indians. J Clin Endocrinol Metab. 2008;93:3644–3649. doi: 10.1210/jc.2008-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mack PJ, Zhang Y, Chung S, Vickerman V, Kamm RD, Garcia-Cardena G. Biomechanical Regulation of Endothelium-dependent Events Critical for Adaptive Remodeling. J Biol Chem. 2009;284:8412–8420. doi: 10.1074/jbc.M804524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mahatan CS, Kaestner KH, Geiman DE, Yang VW. Characterization of the structure and regulation of the murine gene encoding gut-enriched Kruppel-like factor (Kruppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med. 2001;7:699–705. doi: 10.1038/89076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Martin KJ, Kwan CP, Nagasaki K, Zhang X, O’Hare MJ, Kaelin CM, Burgeson RE, Pardee AB, Sager R. Down-regulation of laminin-5 in breast carcinoma cells. Mol Med. 1998;4:602–613. [PMC free article] [PubMed] [Google Scholar]
- 265.Martin KM, Metcalfe JC, Kemp PR. Expression of Klf9 and Klf13 in mouse development. Mech Dev. 2001;103:149–151. doi: 10.1016/s0925-4773(01)00343-4. [DOI] [PubMed] [Google Scholar]
- 266.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 267.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Matsumoto N, Laub F, Aldabe R, Zhang W, Ramirez F, Yoshida T, Terada M. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J Biol Chem. 1998;273:28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 269.Matsumura T, Suzuki T, Aizawa K, Munemasa Y, Muto S, Horikoshi M, Nagai R. The deacetylase HDAC1 negatively regulates the cardiovascular transcription factor Kruppel-like factor 5 through direct interaction. J Biol Chem. 2005;280:12123–12129. doi: 10.1074/jbc.M410578200. [DOI] [PubMed] [Google Scholar]
- 270.Matsusaka T, Katori H, Inagami T, Fogo A, Ichikawa I. Communication between myocytes and fibroblasts in cardiac remodeling in angiotensin chimeric mice. J Clin Invest. 1999;103:1451–1458. doi: 10.1172/JCI5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.McConnell BB, Bialkowska AB, Nandan MO, Ghaleb AM, Gordon FJ, Yang VW. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69:4125–4133. doi: 10.1158/0008-5472.CAN-08-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.McConnell BB, Klapproth JM, Sasaki M, Nandan MO, Yang VW. Kruppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology. 2008;134:1007–1016. doi: 10.1053/j.gastro.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mehta TS, Lu H, Wang X, Urvalek AM, Nguyen K-HH, Monzur F, Hammond JD, Ma JQ, Zhao J. A unique sequence in the N-terminal regulatory region controls the nuclear localization of KLF8 by cooperating with the C-terminal zinc-fingers. Cell Res. 2009;19:1098–1109. doi: 10.1038/cr.2009.64. [DOI] [PubMed] [Google Scholar]
- 276.Meng F, Han M, Zheng B, Wang C, Zhang R, Zhang XH, Wen JK. All-trans retinoic acid increases KLF4 acetylation by inducing HDAC2 phosphorylation and its dissociation from KLF4 in vascular smooth muscle cells. Biochem Biophys Res Commun. 2009;387:13–18. doi: 10.1016/j.bbrc.2009.05.112. [DOI] [PubMed] [Google Scholar]
- 277.Methe H, Balcells M, Alegret Mdel C, Santacana M, Molins B, Hamik A, Jain MK, Edelman ER. Vascular bed origin dictates flow pattern regulation of endothelial adhesion molecule expression. Am J Physiol Heart Circ Physiol. 2007;292:H2167–2175. doi: 10.1152/ajpheart.00403.2006. [DOI] [PubMed] [Google Scholar]
- 278.Miele L, Beale G, Patman G, Nobili V, Leathart J, Grieco A, Abate M, Friedman SL, Narla G, Bugianesi E, Day CP, Reeves HL. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135:282–291. e281. doi: 10.1053/j.gastro.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, Weitzman SA. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J Biol Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 281.Mitsumoto M, Mitsumoto A, Demple B. Nitric oxide-mediated upregulation of the TGF-beta-inducible early response gene-1 (TIEG1) in human fibroblasts by mRNA stabilization independent of TGF-beta. Free Radic Biol Med. 2003;34:1607–1613. doi: 10.1016/s0891-5849(03)00211-9. [DOI] [PubMed] [Google Scholar]
- 282.Miyaki M, Yamaguchi T, Iijima T, Funata N, Mori T. Difference in the role of loss of heterozygosity at 10p15 (KLF6 locus) in colorectal carcinogenesis between sporadic and familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer patients. Oncology. 2006;71:131–135. doi: 10.1159/000100523. [DOI] [PubMed] [Google Scholar]
- 283.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol. 2003;23:8528–8541. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 285.Montanini L, Bissola L, Finocchiaro G. KLF6 is not the major target of chromosome 10p losses in glioblastomas. Int J Cancer. 2004;111:640–641. doi: 10.1002/ijc.20303. [DOI] [PubMed] [Google Scholar]
- 286.Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Mori A, Moser C, Lang SA, Hackl C, Gottfried E, Kreutz M, Schlitt HJ, Geissler EK, Stoeltzing O. Up-regulation of Kruppel-like factor 5 in pancreatic cancer is promoted by interleukin-1beta signaling and hypoxia-inducible factor-1alpha. Mol Cancer Res. 2009;7:1390–1398. doi: 10.1158/1541-7786.MCR-08-0525. [DOI] [PubMed] [Google Scholar]
- 288.Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 289.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 290.Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, Fujii-Kuriyama Y. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol. 2003;23:2489–2500. doi: 10.1128/MCB.23.7.2489-2500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Muhlbauer KR, Grone HJ, Ernst T, Grone E, Tschada R, Hergenhahn M, Hollstein M. Analysis of human prostate cancers and cell lines for mutations in the TP53 and KLF6 tumour suppressor genes. Br J Cancer. 2003;89:687–690. doi: 10.1038/sj.bjc.6601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mukai S, Hiyama T, Tanaka S, Yoshihara M, Arihiro K, Chayama K. Involvement of Kruppel-like factor 6 (KLF6) mutation in the development of nonpolypoid colorectal carcinoma. World J Gastroenterol. 2007;13:3932–3938. doi: 10.3748/wjg.v13.i29.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Nagai R, Shindo T, Manabe I, Suzuki T, Kurabayashi M. KLF5/BTEB2, a Kruppel-like zinc-finger type transcription factor, mediates both smooth muscle cell activation and cardiac hypertrophy. Adv Exp Med Biol. 2003;538:57–65. doi: 10.1007/978-1-4419-9029-7_5. discussion 66. [DOI] [PubMed] [Google Scholar]
- 294.Nagai RFSL, Kasuga M, editors. The Biology of Kruppel-like Factors. Tokyo, Berlin, Heidelberg, New York: Springer; 2009. [Google Scholar]
- 295.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 296.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, Imoto I, Katai H, Yamaguchi T, Inazawa J, Hirohashi S, Ishikawa Y, Shibata T. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–1867. doi: 10.1002/ijc.24538. [DOI] [PubMed] [Google Scholar]
- 297.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Nandan MO, Ghaleb AM, McConnell BB, Patel NV, Robine S, Yang VW. Kruppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Mol Cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Nandan MO, Yang VW. The role of Kruppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol Histopathol. 2009;24:1343–1355. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, Hod E, Leake D, Friedman SL, Hall SJ, Chinnaiyan AM, Gerald WL, Rubin MA, Martignetti JA. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A, McDonnell SK, Wiley KE, Jacobsen SJ, Isaacs SD, Walsh PC, Zheng SL, Chang BL, Friedrichsen DM, Stanford JL, Ostrander EA, Chinnaiyan AM, Rubin MA, Xu J, Thibodeau SN, Friedman SL, Martignetti JA. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 304.Narla G, DiFeo A, Yao S, Banno A, Hod E, Reeves HL, Qiao RF, Camacho-Vanegas O, Levine A, Kirschenbaum A, Chan AM, Friedman SL, Martignetti JA. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 305.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 306.Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, Friedman SL. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428–4434. doi: 10.1038/sj.onc.1210223. [DOI] [PubMed] [Google Scholar]
- 307.Nemer M, Horb ME. The KLF family of transcriptional regulators in cardiomyocyte proliferation and differentiation. Cell Cycle. 2007;6:117–121. doi: 10.4161/cc.6.2.3718. [DOI] [PubMed] [Google Scholar]
- 308.Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, Dina C, Hamid YH, Joly E, Vaillant E, Benmezroua Y, Durand E, Bakaher N, Delannoy V, Vaxillaire M, Cook T, Dallinga-Thie GM, Jansen H, Charles MA, Clement K, Galan P, Hercberg S, Helbecque N, Charpentier G, Prentki M, Hansen T, Pedersen O, Urrutia R, Melloul D, Froguel P. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Nikolcheva T, Pyronnet S, Chou SY, Sonenberg N, Song A, Clayberger C, Krensky AM. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J Clin Invest. 2002;110:119–126. doi: 10.1172/JCI15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Nilson DG, Sabatino DE, Bodine DM, Gallagher PG. Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp Hematol. 2006;34:705–712. doi: 10.1016/j.exphem.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 311.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 312.Oemar BS, Werner A, Garnier JM, Do DD, Godoy N, Nauck M, Marz W, Rupp J, Pech M, Luscher TF. Human connective tissue growth factor is expressed in advanced atherosclerotic lesions. Circulation. 1997;95:831–839. doi: 10.1161/01.cir.95.4.831. [DOI] [PubMed] [Google Scholar]
- 313.Ogata T, Kurabayashi M, Hoshino Y, Ishikawa S, Takeyoshi I, Morishita Y, Nagai R. Inducible expression of BTEB2, a member of the zinc-finger family of transcription factors, in cardiac allograft arteriosclerosis. Transplant Proc. 2000;32:2032–2033. doi: 10.1016/s0041-1345(00)01544-x. [DOI] [PubMed] [Google Scholar]
- 314.Ogata T, Kurabayashi M, Hoshino YI, Sekiguchi KI, Kawai-Kowase K, Ishikawa S, Morishita Y, Nagai R. Inducible expression of basic transcription factor-binding protein 2 (BTEB2), a member of zinc finger family of transcription factors, in cardiac allograft vascular disease. Transplantation. 2000;70:1653–1656. doi: 10.1097/00007890-200012150-00019. [DOI] [PubMed] [Google Scholar]
- 315.Ohnishi S, Laub F, Matsumoto N, Asaka M, Ramirez F, Yoshida T, Terada M. Developmental expression of the mouse gene coding for the Kruppel-like transcription factor KLF5. Dev Dyn. 2000;217:421–429. doi: 10.1002/(SICI)1097-0177(200004)217:4<421::AID-DVDY9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 316.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 317.Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat Med. 2008;14:656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 318.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 319.Okano J, Opitz OG, Nakagawa H, Jenkins TD, Friedman SL, Rustgi AK. The Kruppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Lett. 2000;473:95–100. doi: 10.1016/s0014-5793(00)01468-x. [DOI] [PubMed] [Google Scholar]
- 320.Orkin SH, Antonarakis SE, Kazazian HH., Jr Base substitution at position −88 in a beta-thalassemic globin gene. Further evidence for the role of distal promoter element ACACCC. J Biol Chem. 1984;259:8679–8681. [PubMed] [Google Scholar]
- 321.Orkin SH, Kazazian HH, Jr, Antonarakis SE, Goff SC, Boehm CD, Sexton JP, Waber PG, Giardina PJ. Linkage of beta-thalassaemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature. 1982;296:627–631. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- 322.Outram SV, Gordon AR, Hager-Theodorides AL, Metcalfe J, Crompton T, Kemp P. KLF13 influences multiple stages of both B and T cell development. Cell Cycle. 2008;7:2047–2055. doi: 10.4161/cc.7.13.6234. [DOI] [PubMed] [Google Scholar]
- 323.Ouyang L, Chen X, Bieker JJ. Regulation of erythroid Kruppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J Biol Chem. 1998;273:23019–23025. doi: 10.1074/jbc.273.36.23019. [DOI] [PubMed] [Google Scholar]
- 324.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, Bland KI, LoBuglio AF, Lobo-Ruppert SM, Ruppert JM. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 325.Pandya K, Townes TM. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J Biol Chem. 2002;277:16304–16312. doi: 10.1074/jbc.M200866200. [DOI] [PubMed] [Google Scholar]
- 326.Pang L, Weiss MJ, Poncz M. Megakaryocyte biology and related disorders. J Clin Invest. 2005;115:3332–3338. doi: 10.1172/JCI26720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 328.Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, Moore GE, Kaneda M, Perry GH, Stone AC, Lee C, Meguro-Horike M, Sasaki H, Kobayashi K, Nakabayashi K, Scherer SW. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, Garcia-Cardena G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Parmar KM, Nambudiri V, Dai G, Larman HB, Gimbrone MA, Jr, Garcia-Cardena G. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J Biol Chem. 2005;280:26714–26719. doi: 10.1074/jbc.C500144200. [DOI] [PubMed] [Google Scholar]
- 331.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 332.Perdomo J, Verger A, Turner J, Crossley M. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol Cell Biol. 2005;25:1549–1559. doi: 10.1128/MCB.25.4.1549-1559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Perkins AC, Gaensler KM, Orkin SH. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc Natl Acad Sci U S A. 1996;93:12267–12271. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Perkins AC, Peterson KR, Stamatoyannopoulos G, Witkowska HE, Orkin SH. Fetal expression of a human Agamma globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood. 2000;95:1827–1833. [PubMed] [Google Scholar]
- 335.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 336.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, Leitinger N, Owens GK. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 338.Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 339.Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol. 2008;28:7394–7401. doi: 10.1128/MCB.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. I. Paracrine cells important in health and disease. Am J Physiol. 1999;277:C1–9. doi: 10.1152/ajpcell.1999.277.1.C1. [DOI] [PubMed] [Google Scholar]
- 341.Quadrini KJ, Bieker JJ. EKLF/KLF1 is ubiquitinated in vivo and its stability is regulated by activation domain sequences through the 26S proteasome. FEBS Lett. 2006;580:2285–2293. doi: 10.1016/j.febslet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 342.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 343.Rajamannan NM, Subramaniam M, Abraham TP, Vasile VC, Ackerman MJ, Monroe DG, Chew TL, Spelsberg TC. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J Cell Biochem. 2007;100:315–325. doi: 10.1002/jcb.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman SL. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, Meijerink J, Aifantis I, Basso G, Cordon-Cardo C, Ai W, Ferrando A. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S, Eng FJ, Arthur MJ, Martignetti JA, Friedman SL. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 347.Reinholz MM, An MW, Johnsen SA, Subramaniam M, Suman VJ, Ingle JN, Roche PC, Spelsberg TC. Differential gene expression of TGF beta inducible early gene (TIEG), Smad7, Smad2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2004;86:75–88. doi: 10.1023/B:BREA.0000032926.74216.7d. [DOI] [PubMed] [Google Scholar]
- 348.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 350.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 351.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 352.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 353.Rozenblum E, Vahteristo P, Sandberg T, Bergthorsson JT, Syrjakoski K, Weaver D, Haraldsson K, Johannsdottir HK, Vehmanen P, Nigam S, Golberger N, Robbins C, Pak E, Dutra A, Gillander E, Stephan DA, Bailey-Wilson J, Juo SH, Kainu T, Arason A, Barkardottir RB, Nevanlinna H, Borg A, Kallioniemi OP. A genomic map of a 6-Mb region at 13q21–q22 implicated in cancer development: identification and characterization of candidate genes. Hum Genet. 2002;110:111–121. doi: 10.1007/s00439-001-0646-6. [DOI] [PubMed] [Google Scholar]
- 354.Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 355.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 356.Saiki Y, Ishimaru S, Mimori K, Takatsuno Y, Nagahara M, Ishii H, Yamada K, Mori M. Comprehensive analysis of the clinical significance of inducing pluripotent stemness-related gene expression in colorectal cancer cells. Ann Surg Oncol. 2009;16:2638–2644. doi: 10.1245/s10434-009-0567-5. [DOI] [PubMed] [Google Scholar]
- 357.Sakamoto H, Sakamaki T, Kanda T, Hoshino Y, Sawada Y, Sato M, Sato H, Oyama Y, Nakano A, Takase S, Hasegawa A, Nagai R, Kurabayashi M. Smooth muscle cell outgrowth from coronary atherectomy specimens in vitro is associated with less time to restenosis and expression of a key Transcription factor KLF5/BTEB2. Cardiology. 2003;100:80–85. doi: 10.1159/000073043. [DOI] [PubMed] [Google Scholar]
- 358.Sangodkar J, Difeo A, Feld L, Bromberg R, Schwartz R, Huang F, Terzo EA, Choudhri A, Narla G. Targeted reduction of KLF6-SV1 restores chemotherapy sensitivity in resistant lung adenocarcinoma. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Sangodkar J, Shi J, DiFeo A, Schwartz R, Bromberg R, Choudhri A, McClinch K, Hatami R, Scheer E, Kremer-Tal S, Martignetti JA, Hui A, Leung WK, Friedman SL, Narla G. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur J Cancer. 2009;45:666–676. doi: 10.1016/j.ejca.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- 361.Schuh R, Aicher W, Gaul U, Cote S, Preiss A, Maier D, Seifert E, Nauber U, Schroder C, Kemler R, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 362.Schuierer M, Hilger-Eversheim K, Dobner T, Bosserhoff AK, Moser M, Turner J, Crossley M, Buettner R. Induction of AP-2alpha expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. J Biol Chem. 2001;276:27944–27949. doi: 10.1074/jbc.M100070200. [DOI] [PubMed] [Google Scholar]
- 363.Scobie KN, Hall BJ, Wilke SA, Klemenhagen KC, Fujii-Kuriyama Y, Ghosh A, Hen R, Sahay A. Kruppel-like factor 9 is necessary for late-phase neuronal maturation in the developing dentate gyrus and during adult hippocampal neurogenesis. J Neurosci. 2009;29:9875–9887. doi: 10.1523/JNEUROSCI.2260-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Scohy S, Gabant P, Van Reeth T, Hertveldt V, Dreze PL, Van Vooren P, Riviere M, Szpirer J, Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 365.Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 366.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 367.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 368.Sen-Banerjee S, Mir S, Lin Z, Hamik A, Atkins GB, Das H, Banerjee P, Kumar A, Jain MK. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 369.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Sengupta T, Chen K, Milot E, Bieker JJ. Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the beta-globin locus. Mol Cell Biol. 2008;28:6160–6170. doi: 10.1128/MCB.00919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Sewalt RG, Gunster MJ, van der Vlag J, Satijn DP, Otte AP. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 373.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Shie JL, Chen ZY, O’Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 375.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Kruppel-like factor define a subfamily of closely related Kruppel proteins. J Biol Chem. 1997;272:18504–18507. doi: 10.1074/jbc.272.29.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 379.Shinoda Y, Ogata N, Higashikawa A, Manabe I, Shindo T, Yamada T, Kugimiya F, Ikeda T, Kawamura N, Kawasaki Y, Tsushima K, Takeda N, Nagai R, Hoshi K, Nakamura K, Chung UI, Kawaguchi H. Kruppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J Biol Chem. 2008;283:24682–24689. doi: 10.1074/jbc.M709857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Siatecka M, Xue L, Bieker JJ. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol Cell Biol. 2007;27:8547–8560. doi: 10.1128/MCB.00589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Siddique A, Malo MS, Ocuin LM, Hinnebusch BF, Abedrapo MA, Henderson JW, Zhang W, Mozumder M, Yang VW, Hodin RA. Convergence of the thyroid hormone and gut-enriched Kruppel-like factor pathways in the context of enterocyte differentiation. J Gastrointest Surg. 2003;7:1053–1061. doi: 10.1016/j.gassur.2003.09.006. discussion 1061. [DOI] [PubMed] [Google Scholar]
- 382.Simmen FA, Su Y, Xiao R, Zeng Z, Simmen RC. The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod Biol Endocrinol. 2008;6:41. doi: 10.1186/1477-7827-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Simmen FA, Xiao R, Velarde MC, Nicholson RD, Bowman MT, Fujii-Kuriyama Y, Oh SP, Simmen RC. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–1769. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- 384.Simmen R, Pabona J, Velarde M, Simmons C, Rahal O, Simmen F. The emerging role of Kruppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2009 doi: 10.1677/JOE-09-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Simmen RC, Eason RR, McQuown JR, Linz AL, Kang TJ, Chatman L, Jr, Till SR, Fujii-Kuriyama Y, Simmen FA, Oh SP. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Kruppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J Biol Chem. 2004;279:29286–29294. doi: 10.1074/jbc.M403139200. [DOI] [PubMed] [Google Scholar]
- 386.Simmen RC, Zhang XL, Michel FJ, Min SH, Zhao G, Simmen FA. Molecular markers of endometrial epithelial cell mitogenesis mediated by the Sp/Kruppel-like factor BTEB1. DNA Cell Biol. 2002;21:115–128. doi: 10.1089/104454902753604998. [DOI] [PubMed] [Google Scholar]
- 387.Sinclair LV, Finlay D, Feijoo C, Cornish GH, Gray A, Ager A, Okkenhaug K, Hagenbeek TJ, Spits H, Cantrell DA. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Sirach E, Bureau C, Peron JM, Pradayrol L, Vinel JP, Buscail L, Cordelier P. KLF6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death Differ. 2007;14:1202–1210. doi: 10.1038/sj.cdd.4402114. [DOI] [PubMed] [Google Scholar]
- 389.Slavin D, Sapin V, Lopez-Diaz F, Jacquemin P, Koritschoner N, Dastugue B, Davidson I, Chatton B, Bocco JL. The Kruppel-like core promoter binding protein gene is primarily expressed in placenta during mouse development. Biol Reprod. 1999;61:1586–1591. doi: 10.1095/biolreprod61.6.1586. [DOI] [PubMed] [Google Scholar]
- 390.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 391.Sohn SJ, Li D, Lee LK, Winoto A. Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol Cell Biol. 2005;25:8553–8566. doi: 10.1128/MCB.25.19.8553-8566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Luscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 393.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 394.Song A, Nikolcheva T, Krensky AM. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol Rev. 2000;177:236–245. doi: 10.1034/j.1600-065x.2000.17610.x. [DOI] [PubMed] [Google Scholar]
- 395.Song A, Patel A, Thamatrakoln K, Liu C, Feng D, Clayberger C, Krensky AM. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Kruppel-like transcription factor of activated T lymphocytes. J Biol Chem. 2002;277:30055–30065. doi: 10.1074/jbc.M204278200. [DOI] [PubMed] [Google Scholar]
- 396.Song CZ, Gavriilidis G, Asano H, Stamatoyannopoulos G. Functional study of transcription factor KLF11 by targeted gene inactivation. Blood Cells Mol Dis. 2005;34:53–59. doi: 10.1016/j.bcmd.2004.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Song J, Kim CJ, Cho YG, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic and epigenetic alterations of the KLF6 gene in hepatocellular carcinoma. J Gastroenterol Hepatol. 2006;21:1286–1289. doi: 10.1111/j.1440-1746.2006.04445.x. [DOI] [PubMed] [Google Scholar]
- 398.Sorbello V, Fuso L, Sfiligoi C, Scafoglio C, Ponzone R, Biglia N, Weisz A, Sismondi P, De Bortoli M. Quantitative real-time RT-PCR analysis of eight novel estrogen-regulated genes in breast cancer. Int J Biol Markers. 2003;18:123–129. doi: 10.1177/172460080301800205. [DOI] [PubMed] [Google Scholar]
- 399.Southwood CM, Downs KM, Bieker JJ. Erythroid Kruppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev Dyn. 1996;206:248–259. doi: 10.1002/(SICI)1097-0177(199607)206:3<248::AID-AJA3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 400.Spittau B, Wang Z, Boinska D, Krieglstein K. Functional domains of the TGF-beta-inducible transcription factor Tieg3 and detection of two putative nuclear localization signals within the zinc finger DNA-binding domain. Journal of Cellular Biochemistry. 2007;101:712–722. doi: 10.1002/jcb.21228. [DOI] [PubMed] [Google Scholar]
- 401.Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Hemminki K, Rudnai P, Gurzau E, Koppova K, Botella-Estrada R, Soriano V, Juberias P, Saez B, Gilaberte Y, Fuentelsaz V, Corredera C, Grasa M, Hoiom V, Lindblom A, Bonenkamp JJ, van Rossum MM, Aben KK, de Vries E, Santinami M, Di Mauro MG, Maurichi A, Wendt J, Hochleitner P, Pehamberger H, Gudmundsson J, Magnusdottir DN, Gretarsdottir S, Holm H, Steinthorsdottir V, Frigge ML, Blondal T, Saemundsdottir J, Bjarnason H, Kristjansson K, Bjornsdottir G, Okamoto I, Rivoltini L, Rodolfo M, Kiemeney LA, Hansson J, Nagore E, Mayordomo JI, Kumar R, Karagas MR, Nelson HH, Gulcher JR, Rafnar T, Thorsteinsdottir U, Olafsson JH, Kong A, Stefansson K. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 403.Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A, Verger A, Duterque-Coquillaud M, Morle F. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol. 2003;23:1390–1402. doi: 10.1128/MCB.23.4.1390-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Stone CD, Chen ZY, Tseng CC. Gut-enriched Kruppel-like factor regulates colonic cell growth through APC/beta-catenin pathway. FEBS Lett. 2002;530:147–152. doi: 10.1016/s0014-5793(02)03449-x. [DOI] [PubMed] [Google Scholar]
- 405.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- 408.Suda S, Rai T, Sohara E, Sasaki S, Uchida S. Postnatal expression of KLF12 in the inner medullary collecting ducts of kidney and its trans-activation of UT-A1 urea transporter promoter. Biochem Biophys Res Commun. 2006;344:246–252. doi: 10.1016/j.bbrc.2006.03.138. [DOI] [PubMed] [Google Scholar]
- 409.Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, Czolij R, Denyer G, Bao S, Molero-Navajas JC, Perkins A, Fujiwara Y, Orkin SH, Bell-Anderson K, Crossley M. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Suzuki T, Nishi T, Nagino T, Sasaki K, Aizawa K, Kada N, Sawaki D, Munemasa Y, Matsumura T, Muto S, Sata M, Miyagawa K, Horikoshi M, Nagai R. Functional interaction between the transcription factor Kruppel-like factor 5 and poly(ADP-ribose) polymerase-1 in cardiovascular apoptosis. J Biol Chem. 2007;282:9895–9901. doi: 10.1074/jbc.M608098200. [DOI] [PubMed] [Google Scholar]
- 412.Suzuki T, Sawaki D, Aizawa K, Munemasa Y, Matsumura T, Ishida J, Nagai R. Kruppel-like factor 5 shows proliferation-specific roles in vascular remodeling, direct stimulation of cell growth, and inhibition of apoptosis. J Biol Chem. 2009;284:9549–9557. doi: 10.1074/jbc.M806230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, Piatigorsky J. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol Cell Biol. 2007;27:182–194. doi: 10.1128/MCB.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 416.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 417.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Tanahashi T, Shinohara K, Keshavarz P, Yamaguchi Y, Miyawaki K, Kunika K, Moritani M, Nakamura N, Yoshikawa T, Shiota H, Inoue H, Itakura M. The association of genetic variants in Kruppel-like factor 11 and Type 2 diabetes in the Japanese population. Diabet Med. 2008;25:19–26. doi: 10.1111/j.1464-5491.2007.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Tau KR, Hefferan TE, Waters KM, Robinson JA, Subramaniam M, Riggs BL, Spelsberg TC. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology. 1998;139:1346–1353. doi: 10.1210/endo.139.3.5830. [DOI] [PubMed] [Google Scholar]
- 420.Teixeira MS, Camacho-Vanegas O, Fernandez Y, Narla G, DiFeo A, Lee B, Kalir T, Friedman SL, Schlecht NF, Genden EM, Urken M, Brandwein-Gensler M, Martignetti JA. KLF6 allelic loss is associated with tumor recurrence and markedly decreased survival in head and neck squamous cell carcinoma. Int J Cancer. 2007;121:1976–1983. doi: 10.1002/ijc.22926. [DOI] [PubMed] [Google Scholar]
- 421.Teshigawara K, Ogawa W, Mori T, Matsuki Y, Watanabe E, Hiramatsu R, Inoue H, Miyake K, Sakaue H, Kasuga M. Role of Kruppel-like factor 15 in PEPCK gene expression in the liver. Biochem Biophys Res Commun. 2005;327:920–926. doi: 10.1016/j.bbrc.2004.12.096. [DOI] [PubMed] [Google Scholar]
- 422.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang VW. Expression of the gut-enriched Kruppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997;419:239–243. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- 424.Traka M, Gasper AV, Smith JA, Hawkey CJ, Bao Y, Mithen RF. Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr. 2005;135:1865–1872. doi: 10.1093/jn/135.8.1865. [DOI] [PubMed] [Google Scholar]
- 425.Traka MH, Chambers KF, Lund EK, Goodlad RA, Johnson IT, Mithen RF. Involvement of KLF4 in sulforaphane- and iberin-mediated induction of p21(waf1/cip1) Nutr Cancer. 2009;61:137–145. doi: 10.1080/01635580802348641. [DOI] [PubMed] [Google Scholar]
- 426.Tuomisto TT, Lumivuori H, Kansanen E, Hakkinen SK, Turunen MP, van Thienen JV, Horrevoets AJ, Levonen AL, Yla-Herttuala S. Simvastatin has an anti-inflammatory effect on macrophages via upregulation of an atheroprotective transcription factor, Kruppel-like factor 2. Cardiovasc Res. 2008;78:175–184. doi: 10.1093/cvr/cvn007. [DOI] [PubMed] [Google Scholar]
- 427.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Kruppel-like factor, a novel zinc finger repressor. Mol Cell Biol. 2000;20:7319–7331. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.van Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, Innis JW, de Ravel TJ, Mercer CL, Fichera M, Stewart H, Connell LE, Ounap K, Lachlan K, Castle B, Van der Aa N, van Ravenswaaij C, Nobrega MA, Serra-Juhe C, Simonic I, de Leeuw N, Pfundt R, Bongers EM, Baker C, Finnemore P, Huang S, Maloney VK, Crolla JA, van Kalmthout M, Elia M, Vandeweyer G, Fryns JP, Janssens S, Foulds N, Reitano S, Smith K, Parkel S, Loeys B, Woods CG, Oostra A, Speleman F, Pereira AC, Kurg A, Willatt L, Knight SJ, Vermeesch JR, Romano C, Barber JC, Mortier G, Perez-Jurado LA, Kooy F, Brunner HG, Eichler EE, Kleefstra T, de Vries BB. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 431.van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 432.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.van Zelm MC, van der Burg M, de Ridder D, Barendregt BH, de Haas EF, Reinders MJ, Lankester AC, Revesz T, Staal FJ, van Dongen JJ. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175:5912–5922. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- 434.Vanvelkinburgh JC, Fernandez-Zapico ME, Gutierrez-Aguilar R, Neve B, Froguel P, Urrutia R, Stein R. The MODY7 gene, KLF11, is a novel p300-dependent regulator of PDX-1 (MODY4) transcription in pancreatic islet beta cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Velarde MC, Iruthayanathan M, Eason RR, Zhang D, Simmen FA, Simmen RC. Progesterone receptor transactivation of the secretory leukocyte protease inhibitor gene in Ishikawa endometrial epithelial cells involves recruitment of Kruppel-like factor 9/basic transcription element binding protein-1. Endocrinology. 2006;147:1969–1978. doi: 10.1210/en.2005-1419. [DOI] [PubMed] [Google Scholar]
- 436.Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor alpha signaling in Ishikawa endometrial adenocarcinoma cells. Mol Endocrinol. 2007;21:2988–3001. doi: 10.1210/me.2007-0242. [DOI] [PubMed] [Google Scholar]
- 437.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 438.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, Su J, Liu YC. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.von Andrian UH, Mempel TR. Homing and cellular traffic in lymph nodes. Nat Rev Immunol. 2003;3:867–878. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- 440.Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Wang B, Haldar SM, Lu Y, Ibrahim OA, Fisch S, Gray S, Leask A, Jain MK. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J Mol Cell Cardiol. 2008;45:193–197. doi: 10.1016/j.yjmcc.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Wang C, Han M, Zhao XM, Wen JK. Kruppel-like factor 4 is required for the expression of vascular smooth muscle cell differentiation marker genes induced by all-trans retinoic acid. J Biochem. 2008;144:313–321. doi: 10.1093/jb/mvn068. [DOI] [PubMed] [Google Scholar]
- 443.Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, Wu M. Down-regulation of gut-enriched Kruppel-like factor expression in esophageal cancer. World J Gastroenterol. 2002;8:966–970. doi: 10.3748/wjg.v8.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by focal adhesion kinase in human ovarian epithelial and cancer cells. J Biol Chem. 2008;283:13934–13942. doi: 10.1074/jbc.M709300200. [DOI] [PubMed] [Google Scholar]
- 445.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- 446.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- 447.Wani MA, Means RT, Jr, Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7:229–238. doi: 10.1023/a:1008809809843. [DOI] [PubMed] [Google Scholar]
- 448.Wani MA, Wert SE, Lingrel JB. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J Biol Chem. 1999;274:21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]
- 449.Wassmann K, Mueller CF, Becher UM, Werner C, Jung A, Zimmer S, Steinmetz M, Nickenig G, Wassmann S. Interaction of Inhibitor of DNA binding 3 (Id3) with Gut-enriched Kruppel-like factor (GKLF) and p53 regulates proliferation of vascular smooth muscle cells. Mol Cell Biochem. 2010 doi: 10.1007/s11010-009-0201-7. [DOI] [PubMed] [Google Scholar]
- 450.Wassmann S, Wassmann K, Jung A, Velten M, Knuefermann P, Petoumenos V, Becher U, Werner C, Mueller C, Nickenig G. Induction of p53 by GKLF is essential for inhibition of proliferation of vascular smooth muscle cells. J Mol Cell Cardiol. 2007;43:301–307. doi: 10.1016/j.yjmcc.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 451.Watanabe N, Kurabayashi M, Shimomura Y, Kawai-Kowase K, Hoshino Y, Manabe I, Watanabe M, Aikawa M, Kuro-o M, Suzuki T, Yazaki Y, Nagai R. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 452.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 453.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 454.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem. 2006;281:16664–16671. doi: 10.1074/jbc.M513135200. [DOI] [PubMed] [Google Scholar]
- 456.Wei X, Xu H, Kufe D. Human mucin 1 oncoprotein represses transcription of the p53 tumor suppressor gene. Cancer Res. 2007;67:1853–1858. doi: 10.1158/0008-5472.CAN-06-3063. [DOI] [PubMed] [Google Scholar]
- 457.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Whitney EM, Ghaleb AM, Chen X, Yang VW. Transcriptional profiling of the cell cycle checkpoint gene kruppel-like factor 4 reveals a global inhibitory function in macromolecular biosynthesis. Gene Expr. 2006;13:85–96. doi: 10.3727/000000006783991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, Fraser P. The role of EKLF in human beta-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 460.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 461.Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J Biol Chem. 2008;283:3942–3950. doi: 10.1074/jbc.M707882200. [DOI] [PubMed] [Google Scholar]
- 462.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 463.Wu J, Lingrel JB. Kruppel-like factor 2, a novel immediate-early transcriptional factor, regulates IL-2 expression in T lymphocyte activation. J Immunol. 2005;175:3060–3066. doi: 10.4049/jimmunol.175.5.3060. [DOI] [PubMed] [Google Scholar]
- 464.Wu J, Srinivasan SV, Neumann JC, Lingrel JB. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry. 2005;44:11098–11105. doi: 10.1021/bi050166i. [DOI] [PubMed] [Google Scholar]
- 465.Xu J, Lu B, Xu F, Gu H, Fang Y, Huang Q, Lai M. Dynamic down-regulation of Kruppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. 2008;134:891–898. doi: 10.1007/s00432-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 466.Xue L, Chen X, Chang Y, Bieker JJ. Regulatory elements of the EKLF gene that direct erythroid cell-specific expression during mammalian development. Blood. 2004;103:4078–4083. doi: 10.1182/blood-2003-09-3231. [DOI] [PubMed] [Google Scholar]
- 467.Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Kim JH, Nishimura S, Fujiu K, Oishi Y, Itaka K, Kato Y, Yamauchi M, Nagai R. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69:6531–6538. doi: 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
- 468.Yajima S, Lammers CH, Lee SH, Hara Y, Mizuno K, Mouradian MM. Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF) J Neurosci. 1997;17:8657–8666. doi: 10.1523/JNEUROSCI.17-22-08657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, Iwasaki S, Ioka RX, Kaneko IW, Magoori K, Takahashi S, Mori T, Sakaue H, Kodama T, Yanagisawa M, Yamamoto TT, Ito S, Sakai J. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem. 2004;279:16954–16962. doi: 10.1074/jbc.M312079200. [DOI] [PubMed] [Google Scholar]
- 470.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 471.Yan W, Burns KH, Ma L, Matzuk MM. Identification of Zfp393, a germ cell-specific gene encoding a novel zinc finger protein. Mech Dev. 2002;118:233–239. doi: 10.1016/s0925-4773(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 472.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 473.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 474.Yang Y, Tetreault MP, Yermolina YA, Goldstein BG, Katz JP. Kruppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem. 2008;283:18812–18820. doi: 10.1074/jbc.M801384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Yea S, Narla G, Zhao X, Garg R, Tal-Kremer S, Hod E, Villanueva A, Loke J, Tarocchi M, Akita K, Shirasawa S, Sasazuki T, Martignetti JA, Llovet JM, Friedman SL. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521–1531. doi: 10.1053/j.gastro.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Yerges LM, Klei L, Cauley JA, Roeder K, Kammerer CM, Ensrud KE, Nestlerode CS, Lewis C, Lang TF, Barrett-Connor E, Moffett SP, Hoffman AR, Ferrell RE, Orwoll ES, Zmuda JM. Candidate Gene Analysis of Femoral Neck Trabecular and Cortical Volumetric Bone Mineral Density in Older Men. J Bone Miner Res. 2010 doi: 10.1359/jbmr.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Yet SF, McA’Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, Perrella MA, Jain MK, Lee ME. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 478.Yin D, Komatsu N, Miller CW, Chumakov AM, Marschesky A, McKenna R, Black KL, Koeffler HP. KLF6: mutational analysis and effect on cancer cell proliferation. Int J Oncol. 2007;30:65–72. [PubMed] [Google Scholar]
- 479.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Yoon HS, Yang VW. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem. 2004;279:5035–5041. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am J Physiol Cell Physiol. 2008;295:C1175–1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, Yang VW, Kaestner KH, Fruman DA. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 485.Zhang D, Zhang XL, Michel FJ, Blum JL, Simmen FA, Simmen RC. Direct interaction of the Kruppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology. 2002;143:62–73. doi: 10.1210/endo.143.1.8590. [DOI] [PubMed] [Google Scholar]
- 486.Zhang H, Bialkowska A, Rusovici R, Chanchevalap S, Shim H, Katz JP, Yang VW, Yun CC. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Kruppel-like factor 5. J Biol Chem. 2007;282:15541–15549. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Zhang J, Yang C, Brey C, Rodriguez M, Oksov Y, Gaugler R, Dickstein E, Huang CH, Hashmi S. Mutation in Caenorhabditis elegans Kruppel-like factor, KLF-3 results in fat accumulation and alters fatty acid composition. Exp Cell Res. 2009;315:2568–2580. doi: 10.1016/j.yexcr.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 488.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Zhang L, Wali A, Ramana CV, Rishi AK. Cell growth inhibition by okadaic acid involves gut-enriched Kruppel-like factor mediated enhanced expression of c-Myc. Cancer Res. 2007;67:10198–10206. doi: 10.1158/0008-5472.CAN-07-2505. [DOI] [PubMed] [Google Scholar]
- 490.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 491.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci U S A. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, Rychahou PG, Yang VW, He X, Evers BM, Liu C. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21:2413–2422. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Kruppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316:139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 496.Zhang XH, Zheng B, Han M, Miao SB, Wen JK. Synthetic retinoid Am80 inhibits interaction of KLF5 with RAR alpha through inducing KLF5 dephosphorylation mediated by the PI3K/Akt signaling in vascular smooth muscle cells. FEBS Lett. 2009;583:1231–1236. doi: 10.1016/j.febslet.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 497.Zhang XL, Simmen FA, Michel FJ, Simmen RC. Increased expression of the Zn-finger transcription factor BTEB1 in human endometrial cells is correlated with distinct cell phenotype, gene expression patterns, and proliferative responsiveness to serum and TGF-beta1. Mol Cell Endocrinol. 2001;181:81–96. doi: 10.1016/s0303-7207(01)00536-6. [DOI] [PubMed] [Google Scholar]
- 498.Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC. Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J Biol Chem. 2003;278:21474–21482. doi: 10.1074/jbc.M212098200. [DOI] [PubMed] [Google Scholar]
- 499.Zhang Z, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196–2208. doi: 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol Cell. 2003;11:1503–1515. doi: 10.1016/s1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 501.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Zhao Y, Hamza MS, Leong HS, Lim CB, Pan YF, Cheung E, Soo KC, Iyer NG. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1–8. doi: 10.1038/sj.onc.1210625. [DOI] [PubMed] [Google Scholar]
- 503.Zheng B, Han M, Bernier M, Zhang XH, Meng F, Miao SB, He M, Zhao XM, Wen JK. Kruppel-like factor 4 inhibits proliferation by platelet-derived growth factor receptor beta-mediated, not by retinoic acid receptor alpha-mediated, phosphatidylinositol 3-kinase and ERK signaling in vascular smooth muscle cells. J Biol Chem. 2009;284:22773–22785. doi: 10.1074/jbc.M109.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C, Ai W. KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2009;296:G490–498. doi: 10.1152/ajpgi.90393.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Zheng HQ, Zhou Z, Huang J, Chaudhury L, Dong JT, Chen C. Kruppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene. 2009;28:3702–3713. doi: 10.1038/onc.2009.235. [DOI] [PubMed] [Google Scholar]
- 506.Zhou M, McPherson L, Feng D, Song A, Dong C, Lyu SC, Zhou L, Shi X, Ahn YT, Wang D, Clayberger C, Krensky AM. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178:5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Zhou Q, Hong Y, Zhan Q, Shen Y, Liu Z. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009;69:8284–8292. doi: 10.1158/0008-5472.CAN-09-1345. [DOI] [PubMed] [Google Scholar]
- 508.Zhu H, Lam DC, Han KC, Tin VP, Suen WS, Wang E, Lam WK, Cai WW, Chung LP, Wong MP. High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Lett. 2007;245:303–314. doi: 10.1016/j.canlet.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 509.Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711–14718. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- 510.Zobel DP, Andreasen CH, Burgdorf KS, Andersson EA, Sandbaek A, Lauritzen T, Borch-Johnsen K, Jorgensen T, Maeda S, Nakamura Y, Eiberg H, Pederse O, Hansen T. Variation in the gene encoding Kruppel-like factor 7 influences body fat: studies of 14 818 Danes. Eur J Endocrinol. 2009;160:603–609. doi: 10.1530/EJE-08-0688. [DOI] [PubMed] [Google Scholar]