Abstract
Class switch DNA recombination (CSR) substitutes an immunoglobulin (Ig) constant heavy chain (CH) region with a different CH region, thereby endowing an antibody with different biological effector functions. CSR requires activation-induced cytidine deaminase (AID) and occurrence of double-strand DNA breaks (DSBs) in S regions of upstream and downstream CH region genes. DSBs are critical for CSR and would be generated through deamination of dC by AID, subsequent dU deglycosylation by uracil DNA glycosylase (Ung) and nicking by apurinic/apyrimidic endonuclease (APE) of nearby abasic sites on opposite DNA strands. We show here that in human and mouse B cells, S region DSBs can be generated in an AID- and Ung-independent fashion. These DSBs are blunt and 5'-phosphorylated. In B cells undergoing CSR, blunt and 5'-phosphorylated DSBs are processed in an AID- and Ung-dependent fashion to yield staggered DNA ends. Blunt and 5'-phosphorylated DSBs can be readily detected in human and mouse AID- or Ung-deficient B cells. These B cells are CSR defective, but show evidence of intra-S region recombination. Forced expression of AID in AID-negative B cells converts blunt S region DSBs to staggered DSBs. Conversely, forced expression of dominant negative AID or inhibition of Ung by Ung inhibitor (Ugi) in switching B cells abrogates the emergence of staggered DSBs and concomitant CSR. Thus, AID and Ung generate staggered DSBs not only by cleaving intact double-strand DNA, but also by processing blunt DSB ends, whose generation is AID- and Ung-independent, thereby outlining a post-cleavage role for AID in CSR.
Keywords: activation-induced cytidine deaminase (AID), antibody, B cell, class switch DNA recombination (CSR), double-strand DNA break (DSB), immunoglobulin (Ig), switch region, knockout mice, uracil DNA glycosylase (Ung
1. Introduction
In the early stages of B cell development, RAG-dependent gene recombination selects variable (V), diversity (D) and joining (J) segments from their respective gene pools and combines them into a single Ig V(D)J exon. After a B cell encounters antigen, Ig genes undergo two types of DNA modification: class switch DNA recombination (CSR) and somatic hypermutation (SHM) (Casali et al., 2006; Chaudhuri and Alt, 2004; Di Noia and Neuberger, 2007; Diaz and Casali, 2002; Honjo et al., 2002; Kenter, 2005; Li et al., 2004; Maizels, 2005; Wu et al., 2003; Xu et al., 2005; Xu et al., 2007a; Xu et al., 2007b). CSR and SHM diversify antibodies in different ways. SHM inserts single nucleotide changes (point-mutations) in V(D)J DNA, thereby providing the substrate for selection by antigen of high-affinity mutants. CSR substitutes the IgH constant (CH) region with a downstream CH region by introducing double-strand DNA breaks (DSBs) in the switch (S) regions of an upstream and a downstream CH gene and then perfecting an intrachromosomal and inter-S-S region recombination, entailing excision of intervening genomic DNA and introduction of point-mutations around recombined S-S region junctions (Chaudhuri and Alt, 2004; Rush et al., 2004; Schrader et al., 2007; Schrader et al., 2005; Wuerffel et al., 1997). CSR is S region-specific and results in switching from IgM to IgG, IgA or IgE, without altering the antigen specificity of the expressed antibody molecule, thereby furthering the effectiveness of the immune response.
Like SHM, CSR occurs, in general, in germinal centers (GCs) in response to T cell-dependent stimuli, including, engagement of CD40 on B cells by CD154 and cytokines expressed by activated CD4+ T cells (Honjo et al., 2002; Stavnezer and Amemiya, 2004; Wu et al., 2003; Xu et al., 2005; Xu et al., 2007a; Xu et al., 2007b; Zan et al., 1999; Zan et al., 2001; Zan et al., 2000). CSR requires germline transcription of the intervening (IH), S and CH regions of the upstream (donor) and downstream (acceptor) CH loci (Nambu et al., 2003; Wuerffel et al., 2007), leading to chromosomal opening and increased accessibility to the S regions that will be targets of recombination. CSR targets DSBs to the RGYW/WRCY motif, mainly in its AGCT iteration, which accounts for more than 20% of S region sequences (Kataoka et al., 1981; Zarrin et al., 2004). A clear understanding of the mechanisms underlying CSR and SHM remains elusive, although it is now known that, like SHM, CSR requires activation-induced (cytidine) deaminase (AID) (Honjo et al., 2002; Honjo et al., 2004; Xu et al., 2007a). AID deaminates dC residues to dUs in DNA (Chaudhuri et al., 2003; Neuberger et al., 2003; Pham et al., 2003; Yu and Lieber, 2003), more efficiently when phosphorylated and coupled with replication protein A (RPA) (Chaudhuri et al., 2004; Pasqualucci et al., 2006). dUs are deglycosylated by uracil DNA glycosylase (Ung), yielding abasic sites. Abasic sites are recognized and nicked by an apurinic/apyrimidic endonuclease (APE). Nicking of abasic sites in close proximity on opposite DNA strands would give rise to DSBs to initiate CSR (Maizels, 2005; Neuberger et al., 2003; Stavnezer and Schrader, 2006).
AID-dependent DSBs were detected in S region DNA by ligation-mediated (LM)-PCR in human and mouse B cells, but “background” DSBs were also detected in the absence of AID (Catalan et al., 2003; Rush et al., 2004; Schrader et al., 2007; Schrader et al., 2005; Wu and Stavnezer, 2007). AID-independent generation of S region DSBs has been further inferred from the occurrence of intra-S region DNA recombination events in AID-deficient B cell hybridomas (Dudley et al., 2002) and the frequent translocation of c-myc into S regions in AID-deficient mice (Casali and Zan, 2004; Unniraman and Schatz, 2006; Unniraman et al., 2004b). Accordingly, uracil DNA deglycosylation activity is dispensable for the generation of DSBs (Begum et al., 2007; Begum et al., 2004), further suggesting that S region DSBs can arise independently of DNA deamination (Casali and Zan, 2004; Unniraman et al., 2004a; Unniraman and Schatz, 2006). Likewise, DSBs can be introduced in V(D)J DNA independently of AID (Bross et al., 2000; Bross and Jacobs, 2003; Bross et al., 2002; Papavasiliou and Schatz, 2000; Papavasiliou and Schatz, 2002; Zan et al., 2003). We and others have shown that such V(D)J DSBs are most abundant in the post-replicative (G2) phase of the cell cycle, they localize at mutational RGYW hotspots, are blunt and 5'-phosphorylated (Casali and Zan, 2004; Papavasiliou and Schatz, 2000; Wu et al., 2003; Xu et al., 2005; Zan et al., 2003). We also have suggested that processing of these blunt DSBs by AID and Ung leads to emergence of staggered ends, which are characteristic of B cells undergoing SHM (Casali and Zan, 2004; Wu et al., 2003; Xu et al., 2005; Zan et al., 2003). That AID is recruited to DNA breaks is suggested by the retention of AID in the nuclei of B cells treated with DNA break inducers, such as bleomycin, hydrogen peroxide or γ-rays (Brar et al., 2004).
The generation of DSBs is critical for CSR. Targeting of DSBs displays a site-specificity (Unniraman and Schatz, 2006) that contributes to the S region specificity of CSR. It has been suggested that CSR entails the recombination of blunt upstream and downstream S region DSB ends (Schrader et al., 2007; Schrader et al., 2005), but at least two important findings point at a critical role of staggered DSBs in CSR: (i) the occurrence of microhomologies, duplications, insertions of untemplated nucleotides and point-mutations in and around the recombined S-S region DNA junctions (Chen et al., 2001; Honjo et al., 2002; Wu et al., 2006), and (ii) the demonstration that I-SceI site-specific staggered DSBs mediate CSR from IgM to IgG1 in mutant B cells lacking AID expression and in which donor Sμ and accepter S□1 were replaced with yeast I-SceI endonuclease sites (Zarrin et al., 2007).
Here, we used normal and AID- or Ung-deficient human and mouse B cells to address the nature and generation of DSBs in S regions and their role in CSR. We show that staggered DSBs in S region DNA are critical for CSR to unfold. AID and Ung generate these staggered DSBs, not only by cleaving intact double-strand DNA, but also by processing blunt DSB ends, whose generation are AID- and Ung-independent, thereby outlining a post-cleavage role for AID in CSR.
2. Materials and Methods
2.1. Human and mouse B cells
The human monoclonal 4B6, 4D11 and 3G10 IgM+IgD+ cell lines were used in these studies. 4B6 B cells express significant level of AID and undergo spontaneous CSR to IgG and IgA (Kim et al., 2004; Schaffer et al., 1999; Schaffer et al., 2003; Zan et al., 2003). 4D11 B cells express germline IH-CH transcripts, but not AID (Zan et al., 2003) and cannot be induced to upregulate AID and undergo CSR. 3G10 B cells show low AID expression, but upregulate AID and undergo CSR to IgG and IgA upon induction with an agonistic anti-CD40 mAb and huIL-4 or huTGF-β (Zan et al., 2003). IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38− lymphocytes were prepared by selection of human tonsil B cells using a mouse IgG monoclonal antibody (mAb) to human IgD (Southern Biotechnology Associates Inc., Birmingham, AL), magnetic Microbeads® conjugated with goat anti-mouse IgG (Miltenyi Biotec Inc., Auburn, CA), FITC-conjugated mouse mAb to CD38 (PharMingen, BD Biosciences, San Diego, CA), anti-FITC MicroBeads® (Miltenyi Biotec Inc.), a MACS® magnetic sorter (Miltenyi Biotec Inc.) and MultiSort Release Reagent (Miltenyi Biotec Inc.) (Cerutti et al., 2000; Zan et al., 1999; Zan et al., 2001; Zan et al., 2003). The homogeneity of the separated cell fractions was assessed using a FACScalibur™ (Becton Dickinson, Immunocytometry Systems, BD Biosciences) and RT-PCR amplification of VHDJH-Cδ and VHDJH-Cγ1 transcripts. B cells from healthy humans were separated by T cell depletion of fresh PBMCs (Zan et al., 2001) from buffy coats provided by the University of California, Irvine, Blood Blank. Primary mouse B cells were isolated from RBC-depleted splenocytes using a B cell enrichment kit (StemCell Technologies Inc.).
2.2. AID- and Ung-deficient patients
AID-deficient patient YB (Dr. A. Durandy, Hopital Necker-Enfants Malades, Paris, France) carries two missense point-mutations yielding the amino acid changes Leu59Phe and Trp68Xxx together with an in-frame deletion of three amino-acids in exon 3 (Catalan et al., 2003; Revy et al., 2000). B cells from patient YB were separated from frozen PBMCs by T cell depletion. Ung-deficient patient P3 (Dr. Leman Yel, University of California, Irvine, CA) carries a homozygous deletion of two nucleotides (dAdT) in exon 2, leading to the generation of a premature stop codon (codon 159 of nuclear Ung) (Imai et al., 2003). Patient P3 has been followed at our Immunodeficiency Clinic (University of California, Irvine, CA). B cells of Ung-deficient patient P3 were separated by T cell depletion from fresh PBMCs.
2.3. aicda−/− and ung−/− mice
aicda−/− mice (Muramatsu et al., 2000) were obtained from Dr. T. Honjo (Kyoto University, Kyoto, Japan); ung−/−mice (Nilsen et al., 2000) were obtained from Dr. T. Lindahl (Cancer Research UK London Research Institute, South Mimms, UK) through Dr. U. Storb (University of Chicago, Chicago, IL). Both aicda−/− and ung−/− mice are on the C57BL background. Wild type C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME). All animal protocols were approved by the Institutional Animal Care and Use Committee of University of California, Irvine, CA.
2.4. B cell culture and induction of CSR
Lymphoblastoid and primary human B cells were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated FBS (Sigma-Aldrich, Inc., St. Louis, MO), 2 mM L-glutamine, 100 U/ml penicillin, and 100 ng/ml streptomycin (FBS-RPMI). Mouse primary B cells were cultured in FBS-RPMI containing 0.05 mM 2-ME. Human B cells were induced to undergo CSR by an agonistic anti-human CD40 mAb (IgG1 mAb G28-5, ATCC HB 9110, Manassas, VA) (2 μg/ml) and human recombinant (hu)IL-4 (100 U/ml) (Genzyme Corp., Cambridge, MA) in the presence or absence of huTGF-β(5 ng/ml) (R&D Systems Inc., Minneapolis, MN) in a three-day culture. Mouse B cells were induced to undergo CSR by lipopolysaccharide (LPS) (10 μg/ml) from E. coli (serotype 055:B5; Sigma-Aldrich, Inc.) and mouse recombinant (mo)IL-4 (2 ng/ml, R&D Systems Inc.) in a three-day culture.
2.5. Amplification of human Aid, human and mouse germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ and post-recombination Iμ-Cγ1 transcripts, mature VHDJH-CH transcripts, mouse aicda and gapdh transcripts by specific RT-PCRs
RNA was extracted from 2 × 106 B cells using the RNeasy™ Mini Kit (Qiagen Inc., Valencia, CA). mRNA was reverse transcribed using the SuperScript™ Preamplification System (Invitrogen Corp., Carlsbad, CA). RT-PCRs were made semi-quantitative by varying the number of amplification cycles and performing dilution analysis to ensure a linear relationship between the amount of cDNA used and the intensity of the PCR product. Human Aid cDNA was amplified using the forward primer huAID-F and the reverse primer huAIDR (Table 1) (Zan et al., 2001; Zan et al., 2003). Aid or dominant negative (DN) Aid transcripts, as encoded by the pcDNA3.1 (pcDNA3.1/V5-His TOPO TA Expression Kit, Invitrogen Corp.)-based expression vector, were amplified using a pcDNA3.1 vector forward primer 5'-ACGACTC ACTATAGGGAGACC-3' specific for a sequence downstream of the transcriptional start site together with the Aid reverse primer huAID-R (Zan et al., 2003). Germline Iμ-Cμ and Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cδ and VHDJH-Cγ1 and β-actin transcripts were amplified from human B cell cDNA using RTPCRs involving specific primer pairs (Cerutti et al., 1998; Zan et al., 1998) (Table 1). The germline Iμ-Cμ and Iγ1-Cγ1, circle Iγ1-Cμ, post-recombination Iμ-Cγ1, aicda and gapdh transcripts from mouse B cell cDNA were amplified using by RT-PCRs using specific primer pairs (Zan et al., 2003) (Table 1). Amplification cDNAs was performed by an initial denaturing step of 94°C for 5 min followed by 22 cycles of PCR (94°C for 30 sec, 58°C for 30 sec, 72°C for 1 min) using Taq polymerase. Positive proof of ongoing CSR was provided by amplification of circle Iγ1-Cμ transcripts (Cerutti et al., 2002; Kinoshita et al., 2001). These were amplified using an initial denaturing step of 95°C for 5 min, followed by 35 cycles of PCR (95°C for 30 sec, 58°C for 1 min, 72°C for 1 min) using Elongase Enzyme Mix (Invitrogen Corp.), in the presence of 2.0 mM Mg++ with the reverse Cμ-specific primer and the forward Iγ1 region-specific primer Iγ1F (Table 1).
Table 1.
Oligonucleotide primers for specific RT-PCR amplification of human and mouse transcripts
| Targeted transcript | Forward Primer | Reverse Primer |
|---|---|---|
| Human | ||
| Aicda | huAID-F 5'-TGCTCTTCCTCCGCTACATCTC-3' | huAID-R 5'-AACCTCATACAGGGGCAAAAGG-3' |
| β-actin | β-actin-F 5'-GTACCACTGGCATCGTGATGGACT-3' | β-actin-R 5'-ATCCACACGGAGTACTTGCGCTCA-3' |
| Germline Iμ-Cμ | huIμ-F 5'-GTGATTAAGGAGAAACACTTTGAT-3' | huCμ-R 5'-CCGAATTCAGACGAGGGGGAAAAGGGTT-3 |
| Germline Iγ1-Cγ1 | huIγ1-F 5'-GGGCTTCCAAGCCAACAGGGCAGGACA-3' | huCγ1-R 5'-GTTTTGTCACAAGATTTGGGCTC-3' |
| Circular Iγ1-Cμ | huIγ1-F 5'-GGGCTTCCAAGCCAACAGGGCAGGACA-3' | huCμ-R 5'-CCGAATTCAGACGAGGGGGAAAAGGGTT-3 |
| Mature VHDJH-Cδ | huFR3-F 5'-GACGGGCCACACCATCC-3' | huCδ-R 5'-CTGGCCAGCGGAAGATCTCCTTCTT-3 |
| Mature VHDJH-Cγ1 | huFR3-F 5'-GACGGGCCACACCATCC-3' | huCγ1-R 5'-GTTTTGTCACAAGATTTGGGCTC-3' |
| Mouse | ||
| aicda | mAID-F 5'-GAGGGAGTCAAGAAAGTCACGCTGGA-3' | mAID-R 5'-GGCTGAGGTTAGGGTTCCATCTCAG-3' |
| gapdh | GAPDH-F 5'-ATCACTGCCACCCAGAAGACTG-3' | GAPDH-R 5'-CCCTGTTGCTGTAGCCGTATTC-3' |
| Germline Iμ-Cμ | mIμ-F 5'-CTCTGGCCCTGCTTATTGTTG-3' | mCμ-R 5'-GAAGACATTTGGGAAGGACTGAC-3' |
| Germline Iγ1-Cγ1 | mIγ1-F 5'-GGCCCTTCCAGATCTTTGAG-3' | mCγ1-R 5'-GGATCCAGAGTTCCAGGTCACT-3' |
| Circular Iγ1-Cμ | mIγ1-F 5'-GGCCCTTCCAGATCTTTGAG-3' | mCμ-R 5'-GAAGACATTTGGGAAGGACTGAC-3' |
| Post-recombination | mIμ-F 5'-CTCTGGCCCTGCTTATTGTTG-3' | mCγ1-R 5'-GGATCCAGAGTTCCAGGTCACT-3' |
| Iμ-Cγ1 |
2.6. Analysis of DSBs by specific LM-PCRs
For LM-PCR, genomic DNA was prepared from 5 × 105 cells using an anion-exchange resin (QIAGEN® Genomic-tips, Qiagen Inc.) and then ligated in a 25 μl reaction volume with the double-strand anchor linker BW. For quantification of broken DNA ends, linker-ligated DNA was serially two-fold diluted (8, 4, 2, 1, 0.5, 0.25, 0.125 and 0.0625 ng of linker-ligated DNA derived from 1280, 640, 320, 160, 80, 40, 20, and 10 cells, respectively) into unligated genomic DNA and used as template in LM-PCR or serially diluted in water and used as template for β-actin DNA amplification. A first set of primers, consisting of the linker primer BW1 5'-GCGGTGACCCGGGAG ATCTGAATTC-3' and the specific forward primer (priming the “upstream” DNA end) or reverse primer (priming the “downstream” DNA end) (Table 2) was used for the first round of PCR (12 cycles with a 56°C annealing step). A second set of primers, consisting of the same BW1 oligonucleotide and a nested specific forward or reverse primer was used for the second round of PCR (25 cycles with a 60°C annealing step (1 μl of the 25 μ1 first round reaction was used to prime the 25 μl of second round reaction). Amplified DNA was fractionated through 1.5% agarose, blotted onto Hybond-N+ membranes (Amersham Biosciences Inc., Piscataway, NJ) and hybridized to [γ-32P]-ATP labeled gene-specific oligonucleotide probes. The “anion-exchange” procedure we used to process genomic DNA does not introduce detectable amounts of DSBs, as demonstrated by the analysis of Cμ DNA, and compares favorably to the agarose embedding or “agarose plug” and the “whole cell lysate” or the “direct” methods for the detection of DSBs in VHDJH regions (Zan et al., 2003). To detect total DSBs, which comprise both blunt and staggered DSBs, genomic DNA was treated with T4 pol in the presence of dNTPs (200 mM) before being used as a template for amplification of DSB upstream or “downstream” DNA ends (Zan et al., 2003) by specific LM-PCR. The frequency of staggered DSBs was measured approximately by subtracting the frequency of blunt DSBs (T4−) from that of total DSBs (T4+).
Table 2.
Oligonucleotide primers and probes for specific LM-PCR of human and mouse genomic DNA
| Targeted DNA | First round primer | Second round primer | Probe | |
|---|---|---|---|---|
| Human | ||||
| Sμ upstream end | forward | 5'-ATGGAAGCCAGCCTGGCTGT-3' | 5'-AGCCTGGCTGTGCAGGAACC-3' | 5'-TCAGAAATGGACTCAGATGG-3' |
| Sγ1 downstream end | reverse | 5'-AGTCAGCACAGTCCAGTGTCTCTAG-3' | 5'-CATCGGTGCCACCTCAGGGACGCT-3' | 5'-TGCCTGGCTTGACCAGTGGACACTGTTCTCAGATG-3' |
| Cμ upstream end | forward | 5'-GCTTCCCATCAGTCCTGAGAG-3' | 5'-CCACCTCACAGGTGCTGCTGCCTTCC-3' | 5'-CACGTGGTGTGCAAAGTCCAGCACC-3' |
| Cμ downstream end | reverse | 5'-TGCACACCACGTGTTCGTCTGTG-3' | 5'-TGTTGCCGTTGGGGTGCTGGAC-3' | 5'-TGCATGACGTCCTTGGAAGGCAGCAG-3' |
| Cγ1 upstream end | forward | 5'-CTTCTCTCTGCAGAGCCCAAATC-3' | 5'-ACTCACACATGCCCACCGTGCCCAG-3' | 5'-AGCCAGCCCAGGCCTCGCCCTCCAGCTCAAG-3' |
| Pax5 upstream end | forward | 5'-CTTCCCGTAGGTGCGCTGGCTAG-3' | 5'-AGCACTGCTGCTCTCCCGGCTTCC-3' | 5'-CTCTACTCCGGCCGGGCCGGGTCCGCCACGTCT-3' |
| Pim1 upstream end | forward | 5'-AGCCGCTCACCCCGCCGTTCTCAG-3' | 5'-TCAGTTGTCCTCCGACTCGCCCTC-3' | 5'-CCTTCCGCGCCAGCCGCAGCCACAGCCGCAACG-3' |
| Afp upstream end | forward | 5'-GGATGAATGGTTTGTATGTTTC-3' | 5'-CACTTCAATGGTATGCATATTAACTTTG-3' | 5'-TGTGAAGAAGCCAGAATTATGCTCCTTCACATAAC-3' |
| Mouse | ||||
| Sμ upstream end | forward | 5'-GTACCCCCATGGCTTCCCGGAG-3' | 5'-CCACCATCACAGACCTTTCTCCA-3' | 5'-AGCTCAGCAGAGTGAATGACAGATGGACCTC-3' |
| Sγ1 downstream end | reverse | 5'-TTGACCTGGTACCCTAGC-3' | 5'-CATCCTGTCACCTATACAGCTAAG-3' | 5'-CTGGAGCTGCTCAGCTTGGATCTCTGCACT-3' |
| Cμ upstream end | forward | 5'-TCCTTGAACTTTGGCTCCCCAG-3' | 5'-AGAATGAGCAATAGGCAGTAGAG-3' | 5'-GAGAATCAGCTGGAAGGACCAGCATCTTCCC-3' |
| Cγ1 upstream end | forward | 5'-ACGTGTGTTGTGGTAGACATCAG-3' | 5'-AGGTCCAGTTCAGCTGGTTTG-3' | 5'-GTGGAGGTGCACACAGCTCAGACGCAACCC-3' |
2.7. Analysis of intra-Sμ region recombinations
Intra-Sμ region DNA recombinations were analyzed by specific PCRs and Southern blotting of the amplified DNAs with specific Sμ probe. Sμ DNA was amplified using mouse Sμ-specific primer (forward, moSmF1 5'-AAC TCTCCAGCCACAGTAATGACC-3'; reverse, moSmR1 5'-AGGGTAGGAGGAAGGTGGGTTATG-3') from genomic DNA isolated from mouse B cells stimulated with LPS (10 μg/ml) and moIL-4 (2 ng/ml) for three days. Amplified DNA was fractionated through 1 % agarose gel, blotted onto Hybond-N+ membranes (Amersham Biosciences Inc.) and hybridized to [γ-32P]-ATP labeled Sμ specific oligonucleotide probe moSmPro1 5'-TAGTAA GCGAGGCTCTAAAAAGCAT-3'. Intra-Sμ recombined DNAs, that is, Sμ region with internal deletions, were identified by comparing the lengths of the amplified and hybridized DNAs with those derived from amplification of the respective germline Sμ regions. The amplified Sμ region DNAs were cloned into pCR-Blunt II-TOPO® vector (Qiagen Inc.) and sequenced. Each sequence was aligned with the germline Sμ sequence from C57BL/6 chromosome 12 (nucleotides 136170–140190, GeneBank access number AC073553) to identify the internal breakpoints.
2.8. Chromatin-immunoprecipitation (ChIP) assays
ChIP assays were performed as previously described (Zan et al., 2003). Briefly, spontaneously switching human 4B6 B cells (2.5 × 107) were treated with 1% formaldehyde for 10 minutes at room temperature to cross-link chromatin. After washing with cold PBS containing protease inhibitors, crude chromatin was separated by addition of nuclei-lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.5 M NaCl, 1% Triton-X-100, 0.5% sodium deoxycholate, 0.5% sarkosyl, pH 8.0), re-suspended in IP-1 buffer (20 mM Tris-HCl, 200 mM NaCl, 2 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitors) (Santa Cruz Biotechnology Inc., Santa Cruz, CA) and sonicated to yield 200–1000 bp DNA fragments. Fragmented chromatin was pre-cleared with agarose beads bearing protein A or protein G (Santa Cruz Biotechnology Inc.) and then incubated overnight at 4 °C with: purified rabbit IgG to human Rad51 (Novus Biologicals Inc., Littleton, CO), purified rabbit IgG to Rad52 (Santa Cruz Biotechnology Inc.), purified rabbit IgG to the phosphorylated H2A histone family member X (γ-H2AX) (Upstate Biotech, Inc. Waltham, MA), purified goat IgG to human Nbs1 (Santa Cruz Biotechnology Inc.), purified goat IgG to Mre11 (Santa Cruz Biotechnology Inc.) or mouse mAb to human Ku70/Ku86 (Lab Vision/NeoMarkers Corp., Fremont, CA). Immune complexes were isolated using beads bearing protein A or protein G, and eluted with elution buffer (50 mM Tris-HCl, 0.5% SDS, 200 mM NaCl, 100 μg/ml Proteinase K, pH 8.0). Eluates were heated at 65 °C overnight to reverse cross-links. DNA was recovered by phenol extraction and ethanol precipitation and was finally solubilized in TE buffer. The recovered DNA was treated with T4 pol (T4+) or nil (T4−), ligated with BW linker and then serially two-fold diluted in untreated homologous DNA. The immunoprecipitated DNA was specified by LM-PCR using the forward human Sμ or Cμ primers and the linker primer BW1.
2.9. AID, DN AID and Ugi constructs and overexpression
The full-length human Aid cDNA was amplified from Ramos cells using the forward and reverse primers AID-F2 (5'-GAGGCAAGAAGACACTCTGG-3') and AID-R2 (5'-GTGACATTCCTGGAAGTTGC-3'). The full-length human dominant negative (DN) Aid cDNA containing the H56R/E58Q mutations (Papavasiliou and Schatz, 2002; Zan et al., 2003) was generated by PCR-targeted mutagenesis. Bacteriophage PBS2 Ugi (Ung inhibitor) gene (GeneBank Access number J04434) coding region was generated by chemical synthesis. Aid, DN Aid and Ugi genes were cloned directly into the pcDNA3.1 expression vector. The empty pcDNA3.1 vector or a pcDNA3.1 vector containing an Aid, DN Aid or Ugi gene were used to transfect human 4D11, 3G10 or 4B6 B cells, which were then cultured in the presence of G418. After 21 days, the selected cells were further cultured for 3 days in the presence of nil or agonistic anti-huCD40 mAb and huIL-4, and harvested to prepare RNA, genomic DNA and whole-cell extract to analyze the expression of Aid and DN Aid by specific RT-PCRs using the appropriate primers, determine the frequency and nature of the DSBs by LM-PCR and detect Ung activity.
2.10. DNA-dU deglycosylation assays in cell extracts
To measure Ung activity, exponentially growing cells were collected, washed in PBS buffer, resuspended in 25 mM Hepes, 5 mM EDTA, 1 mM dithiothreitol and 10% glycerol at pH 7.8 (HED buffer) with complete protease inhibitors (Roche Applied Science, Roche Diagnostics, Corp., Indianapolis, IN) at about 105 cells/μl and lysed by sonication (five 15-second pulses) (Di Noia and Neuberger, 2002). After centrifugation at 10,000 g for 20 min at 4°C, the supernatant was frozen in aliquots into ethanol-dry ice bath and stored at −70°C. DNA-dU deglycosylation assays were performed by mixing 10 μg, 1.0 μg or 0.1 μg (protein) of clarified whole-cell extract with 1 pmol of [32P]-labeled double-strand oligonucleotide substrate in 10 μl of HED buffer for 2 hrs at 37° C. This double-strand oligonucleotide substrate contained a single dU:dG mismatch. It was constructed by annealing 5' [32P] labeled 5'-ATTATTATTATTCCGUGGATTTATTTATTTATTTATTTATTT-3' with the complementary oligonucleotide, 5'-AAATAAATAAATAAATAAATAAATCCGCGGAATAATAATAAT-3'. The DNA-dU deglycosylation reaction was terminated by addition of 10 μl of formamide loading dye, and the products were resolved on 15% TBE-urea polyacrylamide gels. The APE activity existing in the cell extract was more than sufficient to cleave all the deglycosylated substrate generated during the reaction (Di Noia and Neuberger, 2002).
2.11. Sequential DNA-dC deamination/DNA-dU deglycosylation assays
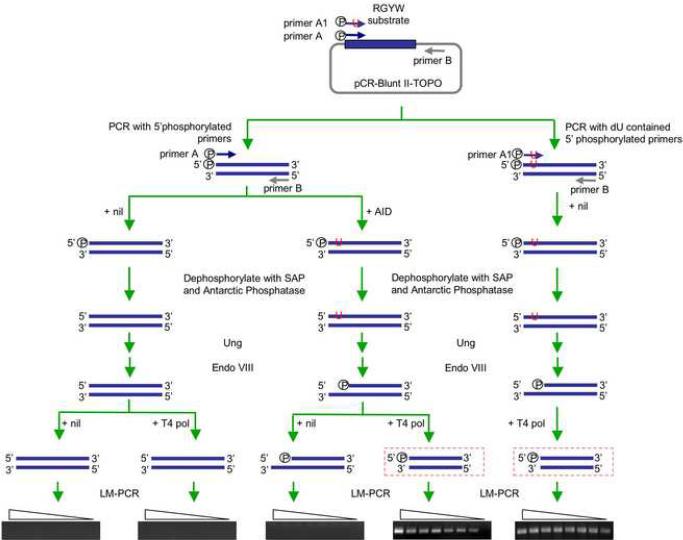
To analyze DNA-dC deamination, the 59 bp DNA fragment 5'-AGCT GGCAGGCTAGCAAGTTGGTTGGCAAGCAGGTAAGCAGGCAAGCTGGCTGAATTCC-3' (Chaudhuri et al., 2003) was cloned into pCR-Blunt II-TOPO® vector. A 191 bp 5'-phosphorylated blunt-ended linearized DNA substrate was generated by PCR amplification of the pCR-Blunt II-TOPO® vector containing the 59 bp fragment DNA, using Phusion™ high-fidelity DNA polymerase (New England Biolabs Inc., Ipswich, MA), the forward 5'-phosphorylated primer A 5'-AGCTGGCAGGCTAGCAAGTTG-3' or the forward 5'-phosphorylated primer A1 5'-AGUTGGCAGGCTAGCAAGTTG-3' (same as primer A, except for the replacement of dC at position 3 with dU), specific for the 5' region of the 59 bp DNA fragment, and the reverse primer B 5'-GTTTTCCCAGTCACGAC-3, specific for the vector sequence 449–468, 116 bp downstream of the inserted 59 bp DNA fragment (Fig. 9). The linearized DNA substrate (100 fmol) was incubated with 50 ng of purified recombinant GST-mouse AID fusion protein, a gift from Dr. Michael R. Lieber (University of Southern California, Los Angeles, CA), under conditions similar to those used by this investigator (Yu et al., 2004), followed by treatment with Shrimp Alkaline Phosphatase (SAP) and Antarctic Phosphatase (New England Biolabs Inc.). SAP and Antarctic Phosphatase catalyze the release of 5'-phosphates, yielding non-phosphorylated DNA ends, which cannot be amplified by LM-PCR. The thoroughly dephosphorylated DNA was treated with recombinant E. coli Ung (New England Biolabs Inc.) and then Endonuclease (Endo) VIII (New England Biolabs Inc.). Endo VIII possesses both N-glycosylase and APE activities, thereby cleaving 3′ and 5′ to the abasic site generated by Ung or by itself and leaving a 5′-phosphate and a 3′-phosphate. The treated DNA was LM-PCR without T4 pol pre-treatment to amplify blunt 5'-phosphorylated DSB ends or was treated with T4 pol to amplify staggered DSB ends.
Fig. 9.
AID can process blunt DSBs to generate staggered DSBs. A 59 bp DNA fragment containing 13 RGYW repeats was cloned into pCR-Blunt II-TOPO ® vector. A 191 bp 5'-phosphorylated blunt-ended linearized DNA substrate and the dU contained control DNA substrate were generated by PCR amplification of the pCR-Blunt IITOPO® vector containing the 59 bp fragment DNA using the forward 5'-phosphorylated primer A or primer A1 (containing a dU instead of a dC at position 3), specific for the 5' region of the 59 bp DNA fragment and the reverse primer B, specific for the vector sequence 116 bp downstream of the inserted 59 bp DNA fragment. The linearized DNA substrate was incubated with nil or recombinant GST-mouse AID fusion protein, then treated with SAP and Antarctic Phosphatase. These catalyze the release of 5'-phosphate groups from DNA yielding non-phosphorylated DNA ends, which cannot been amplified by LM-PCR. Incubation of DNA pretreated with nil or AID with recombinant E. coli Ung, followed by treatment with Endo VIII showed that AID could process blunt DSBs to yield staggered DSB ends. Such staggered DSB ends were amplified by specific LM-PCR after treatment with T4 pol.
3. Results
3.1. Both blunt and staggered DSBs occur in S region DNA
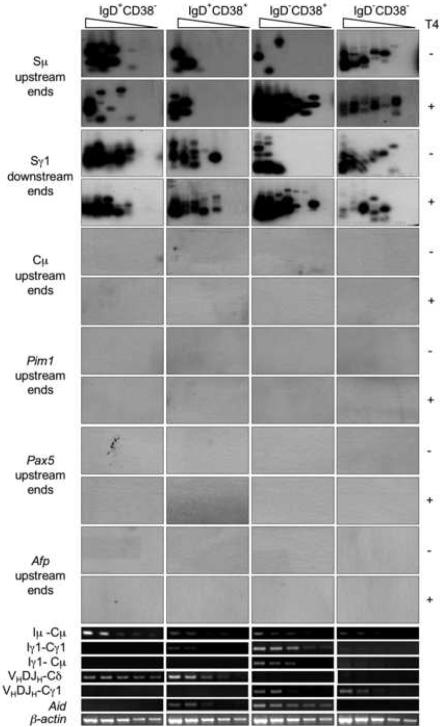
In the human, CSR is not yet active in pre-GC IgD+CD38− B cells, would initiate in IgD+CD38+ early centroblasts, unfolds in GC IgD−CD38+ centroblasts/centrocytes and extinguishes in post-GC IgD−CD38− memory B cells; SHM is negligible in IgD+CD38−, is active in IgD+CD38+ and IgD−CD38+ B cells, and extinguishes IgD−CD38− B cells (Zan et al., 2001; Zan et al., 2003). We isolated genomic DNA and RNA from human tonsil IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38− B cells after centrifugation through a Ficoll® gradient to clear debris and dead cells. DNA was ligated with the BW linker, serially two-fold diluted into unligated genomic DNA and used as template in specific LM-PCR to amplify S□ upstream and S□1 “downstream” DNA ends, and, potentially, Cμ, Pim1, Pax5 or Afp upstream DNA ends (Zan et al., 2003). Like the LM-PCR we used to detect V(D)J DSBs (Zan et al., 2003), our S region-specific LM-PCR amplifies DSB free ends that are blunt and 5'-phosphorylated; it cannot amplify staggered and/or non-phosphorylated blunt DNA ends. Using this S region-specific LM-PCR, we detected blunt DSBs in Sμ and Sγ1 DNA of IgD−CD38+ B cells, which expressed AID and underwent high-rate CSR, as shown by their expression of circle Iγ1-Cμ and mature VHDJH-Cγ1 transcripts (Fig. 1). We amplified DSB ends from as few as 320 IgD−CD38+ B cells, as LM-PCR bands disappeared at the fourth (two-fold) dilution. We also detected blunt DSBs in S regions of IgD+CD38+ B cells, which expressed AID at lower level and initiated CSR, as indicated by their expression of germline Iγ1-Cγ1 transcripts. In IgD+CD38+ B cells, Sμ or Sγ1 region DSB ends could be amplified from as few as 640 or 80 cells, respectively, as LM-PCR bands disappeared at the third (two-fold) dilution (Sμ DSB ends) and at the sixth (two-fold) dilution (Sγ1 DSB ends), respectively. We detected Sμ and Sγ1 region DSBs also in IgD+CD38− cells, in which AID was not expressed and CSR was not yet active, and IgD−CD38− B cells, in which AID expression and CSR were extinct. The specificity of these findings was further strengthened by the lack of amplification of DSB free ends in Cμ, Pim1, Pax5 or Afp DNA in the same B cells and using specific primers.
Fig. 1.
DSBs occur at high frequency in S regions of both switching and non-switching B cells. In non-switching B cells, DSBs are blunt and 5'-phosphorylated; in GC B cells undergoing CSR, they include a high proportion of staggered DNA ends. Genomic DNA from freshly isolated human tonsil IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38− B cells was treated with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as templates in LM-PCR to amplify blunt or total (blunt plus staggered) Sμ, Cμ, Pim1, Pax5 and Afp upstream DSB ends, and Sγ1 downstream DSB ends (lanes 1–8 in each panel). The amplified DNA was blotted and then probed with [γ-32P]-ATP labeled gene-specific oligonucleotide probes. Germline Iμ-Cμ and Iγ1-Cγ1 transcripts, circle Iγ1-Cμ transcripts from switch Sμ-Sγ1 circles, mature VHDJH-Cδ and VHDJH-Cγ1, as well as Aid and β-actin transcripts were detected using specific RT-PCRs.
Treatment of genomic DNA with T4 DNA polymerase (T4 pol), which trims back 3' overhangs while “filling in” 3' recessed ends, thereby yielding 5' phosphorylated blunt DNA ends (Zan et al., 2003), prior to linker ligation allowed us to analyze total (blunt plus staggered) DSBs. The differential amplification of DNA treated with nil (T4−) or T4 pol (T4+) provided a measure of the frequency of staggered DSB ends, although did not allow us to estimate the length of the overhangs. Pre-treatment of DNA with T4 pol resulted in a 16-fold higher detection level of Sμ or Sγ1 DSBs in IgD−CD38+ B cells. Indeed, we could amplify DSB ends from about 20 cells, as LM-PCR bands disappeared at the seventh (two-fold) dilution (Fig. 1), indicating that most DSBs in these switching B cells were staggered. The specificity of these findings was strengthened by the lack of staggered DSBs in non-switching IgD+CD38−, IgD+CD38+ and IgD−CD38− B cells, further showing that S region staggered DSBs characteristically occur in B cells undergoing CSR. The specificity of these findings was further strengthened by the lack of amplification of DSB free ends in Cμ, Pim1, Pax5 or Afp DNA after T4 pol treatment using specific primers.
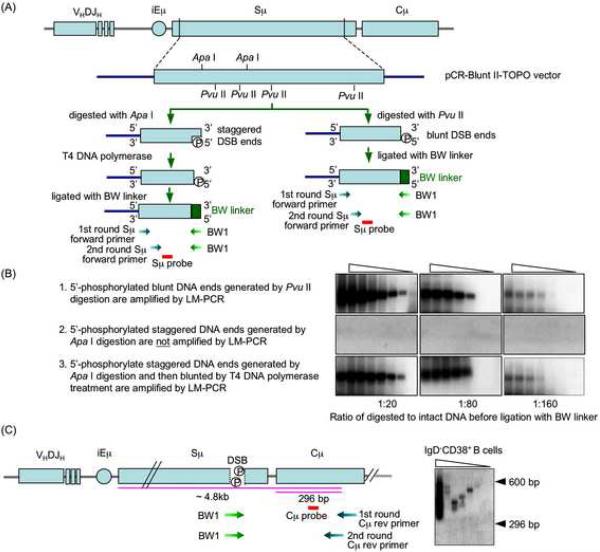
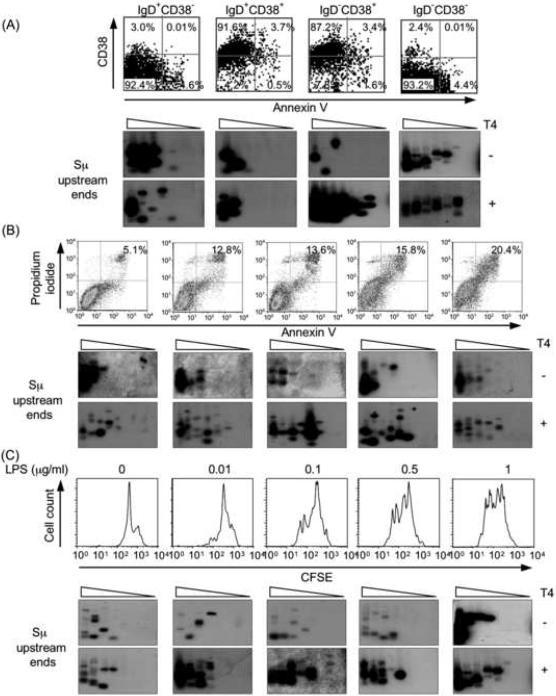
We have shown that the lack of amplification of Cμ, Pim1, Pax5 or Afp DNA by LM-PCR reflects the absence of DSBs in these DNA sequences, not marginal DNA amplification efficiency or lack of specificity of the Cμ, Pim1, Pax5 and Afp primers (Zan et al., 2003). Indeed, these effectively amplified the respective intact genomic DNA and “detected” artificially inserted free DSB blunt ends in a dose-dependent fashion, thereby emphasizing the in vivo origin of amplified free DNA ends in S regions (Zan et al., 2003). We extended these findings and proved the specificity and the sensitivity of our S region LM-PCR by amplifying free DNA ends of blunt and staggered DSBs we generated by digestion with Pvu II and Apa I, respectively, of a plasmid containing human Sμ DNA (Fig. 2A and B). The Sμ but not Cμ location of these DSBs was further confirmed by specific LM-PCR amplification using the BW1 primer, targeting the BW linker, nested reverse Cμ primers and a probe specific for Cμ DNA to amplify free ends in Sμ and the region 5' of Cμ DNA in IgD−CD38+ B cells (Fig. 2C). Like in Ig VHDJH DNA (Zan et al., 2003), amplification of free DSB ends was not influenced by “apoptotic” DSBs. The proportion of apoptotic cells was negligible in all IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38− B cell fractions (Fig. 3A); high levels of apoptotic cells or differential cell proliferation had no impact on the detection of Sμ region DSBs (Fig. 3B and C). Thus, DSBs occur in S regions of B cells that are not undergoing CSR or already completed CSR. Such DSBs are blunt and 5'-phosphorylated. Staggered DSBs emerge in B cells that express AID and undergo CSR.
Fig. 2.
The methods used to detect DSBs in S region are highly specific and allow for differential amplification of blunt and staggered DNA ends. A, Sμ plasmid DNA construct was generated by inserting the 4.3 kb human Sμ DNA into the pCR-Blunt II-TOPO® vector. This human Sμ plasmid DNA construct was digested with Pvu II to generate blunt ends or Apa I to generate staggered ends. To amplify DSB free ends, the digested DNAs were treated with nil or T4 pol, which trims back 3' overhangs while “filling in” 3'-recessed ends, thereby yielding blunt 5'-phosphorylated DNA ends, before ligation with BW linker and amplification by LM-PCR using the Sμ specific forward primers and the linker specific BW1 primer. The differential amplification of DNA treated with nil and DNA treated with T4 pol provided a measure of the occurrence of blunt 5'-phosphorylated or staggered DSB ends. B, The digested hu Sμ-pCR-Blunt II plasmid DNA was mixed with undigested plasmid DNA at ratios of 1:20, 1:80 and 1:160, and followed by ligation with BW linker and amplification by specific Sμ upstream DSB end LM-PCR, with or without pre-treatment with T4 pol. This LM-PCR amplifies only dsDNA ends that are blunt and 5'-phosphorylated. Staggered DSB ends can be amplified only after the treatment with T4 pol, which converts staggered DSB ends to blunt ends. For LM-PCR, samples consisted of 2.1 × 10−4, 1.05 × 10−4, 5.3 × 10−5, 2.6 × 10−5, 1.3 × 10−5, 6.6 × 10−6, 3.3 × 10−6 and 1.6 × 10−6 ng of plasmid DNA, which was digested and then linker-ligated; these amount of digested and linker-ligated plasmid contained Sμ DNA copies equivalent to those in genomic DNA of 1280, 640, 320, 160, 80, 40, 20 and 10 cells, respectively. C, DSBs occur in the Sμ region but not in the Cμ region of human IgD−CD38+ B cells. DSBs were detected by LM-PCR using BW1 primer targeting the BW linker sequence and reverse Cμ primers targeting 300–320 bp (first round PCR) and 274–296 bp (second round PCR) of Cμ DNA. LM-PCR products smaller than 296 bp (shorter pink line) reflected the amplification of DSB ends in Cμ DNA; LM-PCR products between 300 bp to about 4.8 kb (longer pink line) reflected the amplification of DSB ends in Sμ DNA.
Fig. 3.
Cell apoptosis or proliferation has no significant impact on specific Sμ DSB detection. A, Purified IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38− B cell fractions contain negligible proportions of apoptotic cells. Human tonsil IgD+CD38−, IgD+CD38+, IgD−CD38+ and IgD−CD38−B lymphocyte fractions were analyzed for their content in apoptotic cells (upper and lower right quadrants) using the Annexin V Apoptosis Detection Kit (Oncogene Research Products, Inc., San Diego, CA) and flow cytometry. Blunt and total (blunt plus staggered) Sμ upstream DSB ends were detected by specific LM-PCR in each B cell subset DNA after pre-treatment with nil (T4−) or T4 pol (T4+). B, Apoptosis does not affect specific Sμ DSB detection. 4B6 cells were cultured in RPMI-1640 medium supplemented with 10% FBS for 24 hr or 0% FBS for 12, 24, 48, and 72 hr (left to right panels) and then analyzed for apoptosis by Annexin V-FITC and propidium iodide staining in flow cytometry. Apoptotic cells accounted for 5.1%, 12.8%, 13.6%, 15.8% and 20.4% of each subset (left to right panels). Blunt and total (blunt plus staggered) Sμ upstream DSB ends were detected by LM-PCR after pre-treatment with nil (T4−) or T4 pol (T4+), respectively. No correlation was observed between degree of apoptosis and specific detection of DSBs in Sμ DNA. C, Differential cell proliferation does not affect specific Sμ DSB detection. Freshly isolated mouse spleen B cells were stained with 5 μg/ml CFSE at 37°C for 10 min. After washing, cells were cultured in the presence of LPS (0, 0.01, 0.1, 0.5 and 1 μg/ml) in triplicate in 24-well flat-bottom plates at the density of 1 × 106 cells/ml for 42 hr, and analyzed by flow cytometry. Blunt and total Sμ upstream DSB ends were detected by LM-PCR after pre-treatment with nil (T4−) or T4 pol (T4+). For LM-PCR, samples consisted of 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.0625 ng of linker-ligated DNA derived from 1280, 640, 320, 160, 80, 40, 20, and 10 B cells.
3.2. S region DSBs recruit γ-H2AX, Mre11, Nbs1, Ku70/Ku86, Rad52 and Rad51
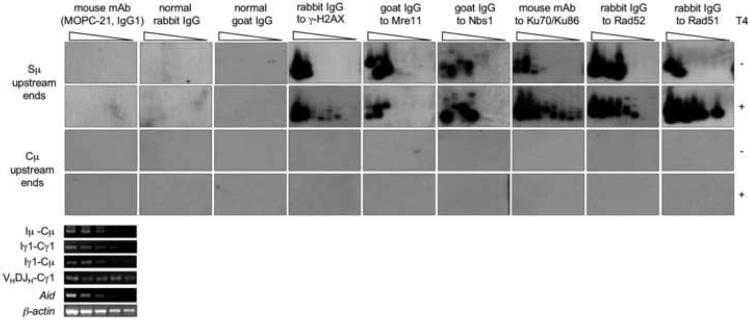
To further address the specificity of the blunt and staggered DSBs detected in S region DNA, we adapted a modified ChIP assay to “pull-down” DNA repair proteins, that are activated and/or recruited by DSBs, such as γ-H2AX, Mre11, Nbs1, Ku70/Ku86, Rad52 and Rad51 to specify by LM-PCR the co-precipitated DNA in 4B6 cells, a human lymphoblastoid IgM+IgD+ B cell subclone that undergoes spontaneous CSR (Kim et al., 2004; Schaffer et al., 2003; Zan et al., 2003).
We reasoned that if the DSBs we detected in S regions are relevant to CSR, i.e., they occur in vivo and are not resulted from in vitro artifacts, they must recruit DNA repair factors that are know to be involved in DSB repair, including γ-H2AX, Mre11, Nbs1, K70/Ku86, Rad52 and Rad51. Specific Abs to Mre11 and Nbs1 pulled down Mre11 and Nbs1, as bound mainly to blunt Sμ DNA ends (Fig. 4). In the same switching B cells, Abs to γ-H2AX, Ku70/Ku86, Rad52 and Rad51 pulled down these proteins, bound mostly to staggered Sμ DNA ends, as indicated by the significantly increased amplification of free ends when DNA was pre-treated with T4 pol. By showing that they recruit DSB-specific repair factors, these experiments further support the “in vivo” generation and specificity of the S region DSBs detected here.
Fig. 4.
S region DSB ends recruit γ-H2AX, Mre11, Nbs1, Ku70/K86, Rad52 and Rad51. Genomic DNA was precipitated from spontaneously switching 4B6 B cells using Abs and mAbs specific for human γ-H2AX, Mre11, Nbs1, Ku70/K86, Rad52 or Rad51. The precipitated DNA was treated with either nil (T4−) or T4 pol (T4+) and then ligated with the BW linker before being serially two-fold diluted into water (from 10,000, 5,000, 2,500, 1,250, 625, 313, 156 and 78 B cells, lanes 1–8) for the amplification of upstream blunt or total (blunt plus staggered) Sμ ends by LM-PCR. Control Abs were IgG from non-intentionally immunized normal rabbits or goats and mouse MOPC-21 IgG1 mAb with irrelevant binding activity. Germline Iμ-Cμ and Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cγ1, Aid and β-actin transcripts were detected by RT-PCR using serially two-fold diluted cDNAs as templates. Depicted are findings from one of four ChIP assays yielding comparable results.
3.3. AID and Ung are critical for the generation of staggered DNA ends and the unfolding of CSR
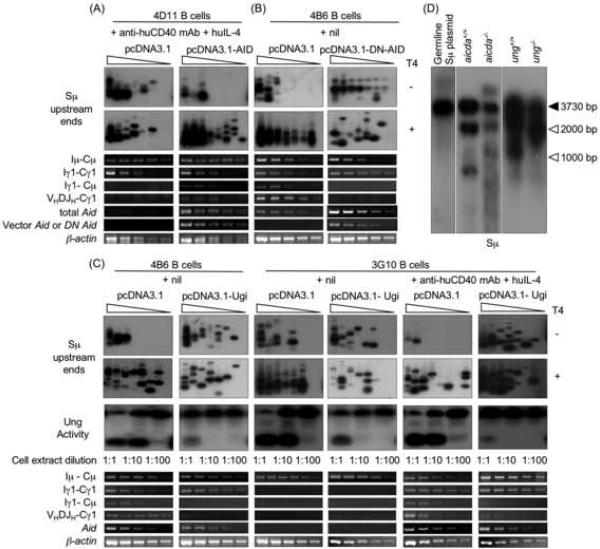
As we have argued, staggered DSBs in the Ig locus stem from blunt DSBs following processing by AID (Casali and Zan, 2004; Wu et al., 2003; Xu et al., 2005; Zan et al., 2003). To define the role of AID in the generation of staggered DSBs, we expressed AID in human monoclonal 4D11 B cells by transfection with a plasmid vector containing human AID cDNA, as driven by the CMV promoter. 4D11 B cells are IgD+IgM+. After stimulation with an agonistic anti-huCD40 mAb and huIL-4, these B cells upregulate germline Iμ-Cμ and Iγ1-Cγ1 transcription, but do not express AID and do not undergo CSR. 4D11 B cells transfected with vector containing human AID cDNA or empty vector were cultured in the presence of anti-huCD40 mAb and huIL-4 for three days, and then cleared of debris and dead cells through a Ficoll® gradient, before amplifying Sμ upstream DSB ends. Expression of AID significantly increased the overall frequency of DSBs. Such an increase stemmed mainly from increased staggered DSBs, and resulted in ongoing CSR, as revealed the increased levels of circle Iγ1-Cμ and mature VHDJH-Cγ1 transcripts (Fig. 5A). To further address the role of AID in the generation of staggered DSBs, we used a plasmid vector containing a mutated human DN Aid cDNA, as driven by the CMV promoter (Zan et al., 2003), to transfect human 4B6 cells. Expression of DN Aid in these B cells hampered the emergence of staggered DSBs and abrogated CSR (Fig. 5B), further indicating that AID processes blunt DSBs to yield staggered DSBs.
Fig. 5.
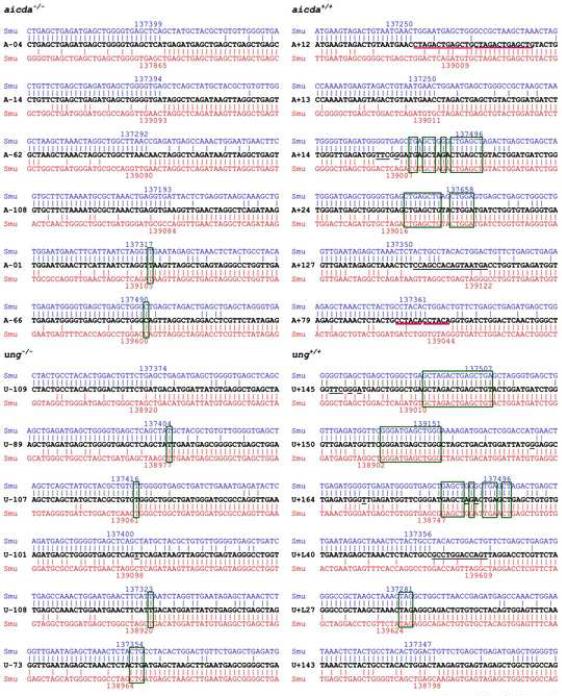
AID and Ung are critical for the generation of staggered DSBs and CSR. A, Forced expression of Aid results in occurrence of staggered DSBs. Paired samples of low Aid expression 4D11 B cells were transfected with an empty pcDNA3.1 vector or a pcDNA3.1 vector containing human Aid cDNA, and then selected in a 21-day culture in the presence of G418. The selected cells were further stimulated for three days with agonistic anti-huCD40 mAb and huIL-4, and harvested to prepare genomic DNA and RNA. To analyze DSBs, blunt or total (blunt plus staggered) Sμ upstream DSB ends were amplified by LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as template in LM-PCR (lanes 1–8 in each panel). Germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cμ1, total Aid or expression vector-encoded Aid (vector Aid), and β-actin transcripts were analyzed by RT-PCR using serially two-fold diluted cDNA as templates. B, Expression of DN Aid abrogates the generation of staggered DNA ends. Paired samples of spontaneously switching 4B6 B cells transfected with an empty pcDNA3.1 vector or a pcDNA3.1 vector containing a human DN Aid construct and then selected in a 21 days culture in the presence of G418. The selected cells were further cultured with nil for three days and then harvested to prepare genomic DNA and RNA. To analyze DSBs, blunt or total (blunt plus staggered) Sμ upstream DSB ends were amplified by LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as template in LM-PCR (lanes 1–8 in each panel). Germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cγ1, total Aid or expression vector-encoded DN Aid, and β-actin transcripts were analyzed by RT-PCR using serially two-fold diluted cDNA as templates. C, Expression of Ugi abrogates the emergence of staggered DSBs. Paired samples of spontaneously switching human 4B6 B cells or CSR-inducible human 3G10 B cells were transfected with an empty pcDNA3.1 vector or a pcDNA3.1 vector containing an Ugi construct and selected in a 21 days culture in the presence of G418. Genomic DNA, RNA and whole-cell extract were prepared from transfected 4B6 B cells cultured with nil for three days and transfected 3G10 B cells stimulated for three day with nil or agonistic anti-huCD40 mAb and huIL-4. To analyze DSBs, blunt or total (blunt plus staggered) Sμ upstream DSB ends were amplified by LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as template in LM-PCR (lanes 1–8 in each panel). Ung activity of the B cells was detected by incubate 10 μg, 1.0 μg or 0.1 μg (protein) of clarified whole-cell extract with a [32P] labeled double-stranded oligonucleotide containing a single dU/dG residue. The reaction products were resolved in 15% TBE-urea polyacrylamide gels. Germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cγ1, Aid and β-actin transcripts were analyzed by RT-PCR using serially two-fold diluted cDNA as templates. D, Intra-S region recombination occurs in the absence of AID or Ung. DNA was PCR-amplified from a mouse germline S□ region, as inserted into pCR-Blunt II-TOPO® plasmid (left panel) or genomic DNA isolated from aicda+/+ and aicda−/− (center panel) or ung+/+ and ung−/− (right panel) mouse B cells stimulated by LPS and moIL-4 for three days (left, center and right panels were from different gels), and specified by Southern-blot using a specific [32P]-labeled Sμ probe. Recombined intra-Sμ DNA resulted in Sμ DNA amplification products smaller than those expected based on germline Sμ (3730 bp) region length (closed arrowhead). Migration of molecular weight markers is indicated by open arrowheads.
To address the role of Ung in the generation of staggered DSBs, we inhibited Ung activity using Ugi (Di Noia and Neuberger, 2002), which blocks Ung binding to DNA, thereby aborting the insertion of DNA nicks resulting from APE-mediated digestion of abasic sites. In spontaneously switching 4B6 B cells, expression of Ugi significantly inhibited Ung activity, greatly reduced the level of staggered DSBs and dampened CSR (Fig. 5C). These results were extended by experiments using the human monoclonal lymphoblastoid 3G10 B cells. Upon stimulation with anti-huCD40 mAb and huIL-4, these IgD+IgM+ B cells upregulate AID expression and generate staggered DSBs in V(D)J DNA (Zan et al., 2003); they also generate staggered DSBs in Sμ region and undergo CSR to IgG, IgA or IgE (Fig. 5C). In 3G10 B cells, inhibition of Ung by Ugi inhibited the level of staggered DSBs and abrogated CSR. Thus, AID and Ung are critical for the generation of staggered DSB ends in S regions and for the unfolding of CSR.
3.4. DSBs occur in S regions of Ung- and AID-deficient B cells, and are blunt and 5'-phosphorylated
We further addressed the dispensability of AID and Ung in generating S region DSBs using B cells from patients with hyper-IgM syndrome type 2 (HIGM2) or Ung-deficient HIGM. Because of defective CSR, HIGM patients display normal or increased serum IgM concentrations with much decreased or no serum IgG, IgA and IgE. In HIGM2 patients, both CSR and SHM are impaired due to lack of AID (Revy et al., 2000); in Ung-deficient HIGM patients, CSR deficiency is usually associated with mutations in Ung, the gene encoding Ung. In these patients, Ung-deficiency leads to profoundly impaired CSR (Imai et al., 2003).
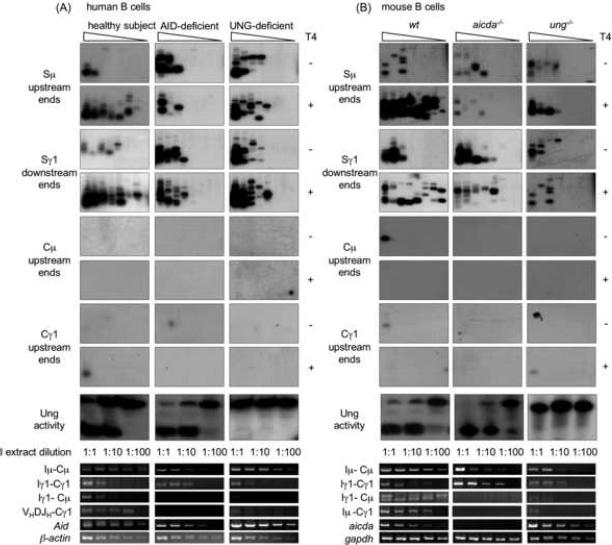
We induced peripheral blood B cells from AID-deficient HIGM2 patient YB (Catalan et al., 2003; Revy et al., 2000), Ung-deficient HIGM patient P3 (Imai et al., 2003) and a healthy subject with agonistic anti-huCD40 mAb, huIL-4 and huTGF-β. After a three-day culture, B cells were centrifuged through a Ficoll® gradient to clear debris and dead cells. Genomic DNA, RNA and whole-cell extract were isolated and used to amplify free DSB ends by LM-PCR specific for Sμ or Sγ region, analyze CSR-relevant transcripts, including Aid, and measure Ung activity by a dU excision assay on an oligonucleotide substrate. B cells from patient YB expressed Aid transcripts with the expected mutation (G203A and 175–183 deletion) and showed Ung activity comparable to healthy subject (Fig. 6A). B cells from patient P3 expressed AID, but showed no Ung activity. B cells from patients YB and P3 as well as the healthy subject showed comparable germline Iμ-Cμ and Iγ1-Cγ1 transcripts. B cells from patient YB did not undergo CSR, as neither circle Iγ1-Cμ transcripts nor mature VHDJH-Cγ1 transcripts could be detected in these lymphocytes; B cells from patient P3 underwent negligible CSR, as only minimal circle Iγ1-Cμ and mature VHDJH-Cγ1 transcripts were detected in these lymphocytes. Nevertheless, in YB and P3 B cells, Sμ and Sγ1 region DNA accumulated DSBs at levels comparable to that of B cells from the healthy subject, in the absence of DSBs in Cμ and Cγ1 regions. Virtually all S region DSBs in patient YB and P3 B cells were blunt and 5'-phosphorylated, while in normal B cells, CSR-associated DSBs were predominantly staggered, as shown by increased amplification of Sμ and Sγ1 DSB ends after T4 pol pre-treatment of genomic DNA.
Fig. 6.
S region DSBs occur at a high frequency in AID- or Ung-deficient human and mouse B cells. A, B cells from AID- or Ung-deficient HIGM patients or normal subjects were stimulated with an agonistic anti-huCD40 mAb in the presence of huIL-4 and huTGF-β, and harvested after three days of culture for preparation of genomic DNA, total RNA and whole-cell extract. Blunt or total (blunt plus staggered) Sμ upstream DSB ends were detected by LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as template in specific LM-PCR to amplify Sμ, Cμ and Cγ1 upstream or Sγ1 downstream DNA ends (lanes 1–8 in each panel). Ung activity was measured by incubating 10 μg, 1.0 μg or 0.1 μg (protein) of clarified whole-cell extract with a [32P] labeled double-strand oligonucleotide containing a single dU/dG residue. The reaction products were resolved in 15% TBE-urea polyacrylamide gels. Germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ, mature VHDJH-Cγ1, Aid and β-actin transcripts were analyzed by RT-PCR using serially two-fold diluted cDNA as templates. B, B cells isolated from spleens of aicda−/−, ung−/− or wt mice were stimulated with LPS and moIL-4, and harvested after three days of culture for preparation of genomic DNA, total RNA and whole-cell extract. Blunt or total (blunt plus staggered) Sμ upstream DSB ends were detected by LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+) before being ligated with BW linker. Linker-ligated genomic DNA (8 ng from 1280 B cells) was serially two-fold diluted into unligated homologous genomic DNA and used as template in LMPCR (lanes 1–8 in each panel). Ung activity was measured by incubating 10 μg, 1.0 μg or 0.1 μg of clarified whole-cell protein extract with a [32P] labeled double-stranded oligonucleotide containing a single dU/dG residue. The reaction products were resolved in 15% TBE-urea polyacrylamide gels. Germline Iμ-Cμ, Iγ1-Cγ1, circle Iγ1-Cμ and post-recombination Iμ-Cγ1 transcripts, and aicda and gapdh transcripts were analyzed by RT-PCR using serially two-fold diluted cDNAs as templates.
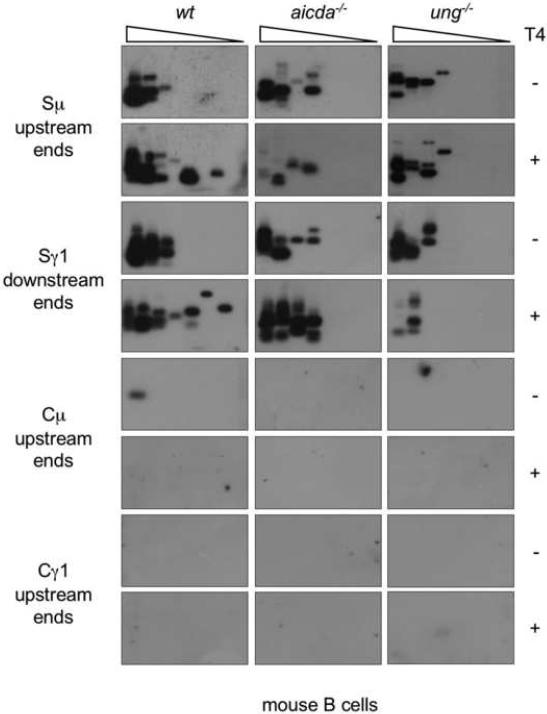
We further addressed the role of AID and Ung in the generation of DSBs in mouse B cells. In response to stimulation with LPS and moIL-4, spleen B cells isolated from aicda−/− or ung−/− mice showed a proliferation rate in a three-day culture comparable to their counterparts from wild type (wt) littermates (not shown). aicda−/− B cells did not express AID, but showed an Ung activity comparable to that of wt B cells (Fig. 6B). ung−/− B cells expressed AID, but showed no Ung activity. While aicda−/−, ung−/− and wt B cells expressed comparable levels of germline Iμ-Cμ and Iγ1-Cγ1 transcripts, aicda−/− B did not undergo CSR, as neither circle Iγ1-Cμ nor post-recombination Iμ-Cγ1 transcripts could be detected in these B cells. ung−/− B cells underwent negligible CSR, as only minimal circle Iγ1-Cμ and post-recombination Iμ-Cγ1 transcripts were detected in these cells. Consistent with what we found in human B cells, we detected virtually no DSBs in Cμ and Cγ1 regions DNA of all these knockout B cells, but readily showed blunt DSBs in Sμ and Sγ1 regions, in not only wt but also aicda−/− and ung−/− B cells (Fig. 6B: compare T4−with T4+). In aicda−/− and ung−/− B cells, the detection frequency of DSBs was not increased by T4 pol treatment, suggesting that virtually all DSBs were blunt. In contrast, in wt B cells undergoing CSR, T4 pol treatment increased the amplification frequency of DSB ends. These staggered DSBs accounted for virtually the whole DSB increase associated with ongoing CSR. To further confirm that the S region DSBs we detected were not introduced by the procedure we used to prepare genomic DNA, we analyzed blunt and total DSBs in Sμ, Sγ1, Cμ and Cγ1 regions of aicda−/−, ung−/− and wt B cells using genomic DNA prepared by the “DNA plugs” method (Papavasiliou and Schatz, 2000; Rush et al., 2004; Schrader et al., 2005). In all these B cells, we detected DSBs in Sμ and Sγ1 regions at levels comparable to those in genomic DNA prepared using our “anion-exchange” method (Fig. 7), and, likewise, found no evidence of DSBs in Cμ and Cγ1 DNA. Thus, in human and mouse B cells, AID and Ung are dispensable for generation of blunt DSBs, but are essential for generation of staggered DSBs and unfolding of CSR.
Fig. 7.
DSBs are readily detected in Sμ and Sγ1 regions but not Cμ or Cγ1 regions of genomic DNA prepared by the “agarose plug” method from aicda−/−, ung−/− or wt B cells. Spleen B cells from aicda−/−, ung−/− or wt mice were stimulated with LPS and moIL-4 and then harvested after three days of culture. Viable B cells were isolated by centrifugation over a Ficoll gradient, immediately washed and then embedded in low-melting temperature agarose. Cells in agarose plugs were used to prepare genomic DNA for double-strand DNA BW linker-ligation. Blunt or total (blunt plus staggered) Sμ upstream and Sγ1 downstream DSB ends, as well as potential Cμ and Cγ1 upstream DSB ends were detected by specific LM-PCR after pre-treatment of genomic DNA with nil (T4−) or T4 pol (T4+), before ligation with the BW linker. Linker-ligated genomic DNA (1280 B cells equivalent) was serially two-fold diluted into unligated homologous genomic DNA and used as template in LM-PCR (lanes 1–8 in each panel). The amplified DNA was blotted and then probed with [γ-32P]-ATP labeled gene-specific oligonucleotide probes.
3.5. AID and Ung are dispensable for intra-Sμ region recombination
Deletions in S regions, possibly resulting from intra-S DNA recombinations, have been detected in aicda−/−, ung−/− and ung−/−msh2−/− B cell hybridomas (Begum et al., 2007; Chaudhuri and Alt, 2004; Dudley et al., 2002). To analyze intra-S region recombination in the presence and absence of AID and/or Ung, we stimulated spleen B cells from aicda−/−, ung−/− and wt mice with LPS and moIL-4. We then PCR-amplified Sμ region DNA using specific upstream forward and downstream reverse primers, and Southern-blotted the amplified DNA with an internal Sμ oligonucleotide probe. Using this approach, we identified “conserved” germline Sμ region DNAs (3730 bp) and Sμ DNAs shorter than the germline Sμ region in not only wt, but also aicda−/− and ung−/− B cells (Fig. 5D). The shorter Sμ DNA amplification products reflected intra-Sμ region DNA recombinations involving internal Sμ deletions (Fig. 8). To further address the nature of S region DSBs in the presence and absence of AID and/or Ung, we sequenced the recombined intra-Sμ DNA junctions of in vitro stimulated B cells and compared them with the Sμ genomic templates to determine the degree of putative overlap (microhomology) of the upstream DNA ends with downstream DNA ends, DNA duplications or the presence of insertions of untemplated nucleotides between the Sμ upstream and downstream DNA ends. Only one out of twelve DNAs amplified from aicda−/− or ung−/− B cells displayed a three nucleotide microhomology, five contained only one nucleotide microhomology, one contained a single untemplated nucleotide insertion and the remaining five contained no microhomology, duplication or insertion (Fig. 8). In contrast, six out of the twelve intra-Sμ junctions in B cells from the respective aicda+/+ or ung+/+ littermates contained stretches of microhomology (three to 15 nucleotides), two included duplications and two contained insertions of untemplated nucleotides (12 and 16). Further, none of the twelve intra-Sμ junction regions from aicda−/− or ung−/− B cells contained point-mutations, while four intra-Sμ junctions from aicda+/+ or ung+/+ B cells did. Thus, AID- or Ung-deficient B cells accumulate DSBs in their Sμ regions, and such DSBs are blunt and 5'-phosphorylated. These DSBs can provide the substrate for intra-Sμ region DNA recombination, which involves blunt junctions. In contrast, in the presence of AID and Ung, Sμ region DSBs are predominantly staggered and can give rise to intra-Sμ region recombinations involving stretches of microhomologies, duplications, untemplated nucleotide insertions and point-mutations. The different nature of recombined intra-Sμ DNA junctions in aicda−/−, ung−/− and wt B cells confirmed that these sequences stemmed from intra-S DNA deletions, not from PCR artifacts.
Fig. 8.
Lack of microhomologies, duplications, insertions and point-mutations in recombined intra-Sμ DNA in aicda−/− or ung−/− B cells. Spleen B cells from aicda−/−, aicda+/+, ung−/− and ung+/+ mice were stimulated by LPS and IL-4 for three days. Intra-Sμ junctional DNAs from stimulated cells were amplified, cloned, and sequenced. Each sequence is compared with germline Sμ (nucleotides 136170–140190 of the C57/BL mouse chromosome 12 sequence: GeneBank access number AC073553). Upstream and down-stream germline Sμ sequences involved in the intra-Sμ recombination are in blue and red, respectively. The numbers on top and bottom of each aligned sequence indicate upstream and downstream breakpoints of recombined Sμ sequences. The Sμ DNA between the upstream and downstream breakpoints was deleted during intra-Sμ recombination. Microhomologies (boxed) were defined by identifying the longest region of perfect uninterrupted donor/acceptor identity at the Sμ junction. Pink lines mark duplications; untemplated nucleotide insertions between the recombined Sμ upstream and downstream DNA ends and point-mutations are underlined.
3.6. AID and Ung process blunt 5'-phosphorylated DSBs to give rise to staggered DSBs
Both single-strand DNA and double-strand DNA can be deaminated by AID (Chaudhuri et al., 2003; Neuberger et al., 2003; Pham et al., 2003; Yu and Lieber, 2003). Our present experiments and previous findings (Chaudhuri et al., 2003; Neuberger et al., 2003; Pham et al., 2003; Yu and Lieber, 2003) led us to hypothesize that AID deaminates dC nearby blunt 5'-phosphorylated DSB ends to give rise to staggered DSB ends, in the presence of Ung and APE activity. To test our hypothesis, we treated a blunt-ended 5'-phosphorylated 191 bp double-strand DNA, which contained 13 copies of seven different iterations of the RGYW motif near one end, with recombinant GST-mouse AID protein. This was followed by dephosphorylation with SAP and Antarctic Phosphatase to delete any 5'-phosphates. The thoroughly dephosphorylated DNA was then treated with recombinant E. coli Ung protein and Endo VIII. Endo VIII possesses a strong APE activity, thereby cleaving 3′ and 5′ to the abasic site generated by Ung and leaving a 5′-phosphate and a 3′-phosphate. We reasoned that, in the presence of Ung and Endo VIII, AID deamination of dC residues nearby DSB ends would result in single-strand DNA cleavages, thereby giving rise to staggered DNA ends. After treatment with T4 pol, such staggered DNA ends could be amplified by our specific LM-PCR. As expected, staggered DSB ends were detected in the DNA substrate treated with AID, Ung and Endo VIII, but were not in the DNA substrate treated with Ung and Endo VIII only (Fig. 9). The identity of the amplified staggered DSB ends was confirmed by sequencing (not shown). These experiments proved that AID can directly target and process blunt DSB ends to generate staggered DSB ends in the presence of Ung and APE activity.
4. Discussion
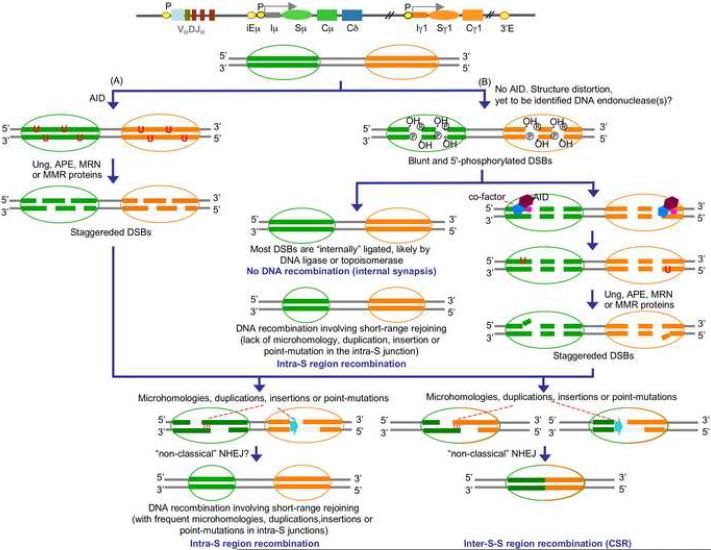
We demonstrated here that DSBs occur at a significant frequency in S regions of not only GC B cells that express AID and undergo CSR, but also in pre- and post-GC B cells, which express virtually no AID and do not undergo CSR. Such DSBs are blunt and 5'-phosphorylated, and arise independently of AID or Ung. They also occur in S regions of human and mouse B cells deficient in AID or Ung. As we showed, in the Sμ region, they are synapsed, resulting in intra-Sμ joining sequences that are conserved or display minimal or no microhomologies, duplications, untemplated nucleotide insertions or point-mutations. In B cells undergoing CSR, staggered DSBs emerge from processing of blunt 5'-phosphorylated DSBs by AID and Ung, in addition to cleavage of intact double-strand DNA by AID and Ung (Fig. 10). Such staggered DSBs give rise to inter-S-S region and intra-S region recombinations entailing occurrence of stretches of microhomology, duplications and insertions as well as point-mutations. Accordingly, forced AID expression converted blunt into staggered DSBs. Conversely, AID or Ung deficiency or expression of DN AID and Ugi, which inhibits AID and Ung, respectively, abrogated the generation of staggered DSBs and concomitant CSR in human and mouse B cells. Finally, consistent with the direct AID DNA deamination model, we showed that AID can process blunt dsDNA termini containing different RGYW iterations, including AGCT, and readily generate staggered DNA ends.
Fig. 10.
Model of DSBs and CSR. A, Pre-cleavage role of AID: staggered DSBs in S regions are generated by AID-mediated DNA deamination. AID deaminates dC in DNA converting it to dU. The dU:dG mismatch can then be processed by either base-excision repair (BER) or by mismatch-repair (MMR), which includes the mutS homologue 1 (Msh1), Msh2, Msh6, mutL homologue 1 (Mlh1), Mlh3 and postmitotic segregation (Pms) proteins, to introduce gaps or nicks on opposite strands of the S region DNA. The BER pathway is thought to generate nicks by removing AID-introduced dU by Ung, thereby creating an abasic site that is processed by APE or the Mre11/Rad50/Nbs1 (MRN) complex. B, Post-cleavage role of AID: DSBs involving blunt 5'-phosphorylated free ends in S regions are generated independently of AID, possibly due to fragility of distorted S region DNA tridimensional structures, as enhanced by transcription and/or replication, and/or S region DNA cleavage by certain nucleases, such Endo G. In the absence of AID, S region DSBs, which are blunt and 5'-phosphorylated, would be “internally” synapsed by a DNA ligase or topoisomerase, implying that in B cells not undergoing CSR, S region blunt DSBs are “open and shut”, resulting in very limited intra-S region internal deletions, microhomologies or insertions. Upon stimulation by activated CD4+ T cells, AID is upregulated, migrates into the nucleus where is recruited to DSBs, perhaps through a cofactor, to deaminate dC near the free DNA ends and generate dU:dG mismatches. Attack of dU by Ung would lead to the generation of an abasic site which becomes substrate of APEs to yield DNA nick, giving rise to staggered DNA ends (dU:dG mispairs can also be processed through the BER or MMR pathway, as shown in A. Staggered DSBs are critical for CSR and can also be involved in intra-S region recombination. CSR and the intra-S recombination involving staggered DSBs would introduce long stretched microhomologies, duplications, insertions or point-mutations in the recombinational junction (from Zan and Casali, in preparation, 2008).
γ-H2AX focus formation in the IgH locus has been used as evidence for DSBs and in support of AID-dependency in the generation of DSBs in S region DNA (Begum et al., 2004; Petersen et al., 2001; Reina-San-Martin et al., 2003). We would argue here that AID-dependent γ-H2AX foci formation does not necessarily reflect the emergence of DSBs. Rather, γ-H2AX foci would form as early intermediates in DSB repair, as γ-H2AX would hold broken chromosomal DNA ends in close proximity and function as an anchor for DNA repair proteins (Bassing and Alt, 2004). Further, H2AX phosphorylation is dispensable for the initial recognition of DNA breaks (Celeste et al., 2003). In CSR, H2AX molecules would be efficiently phosphorylated in the presence of AID, and □-H2AX likely facilitates S-S region synapsis (Chua et al., 2002; Petersen et al., 2001; Pilch et al., 2003; Ward and Chen, 2001). In the absence of H2AX, DSBs emerging physiologically during DNA replication are repaired at a steady rate, as indicated by the normal cycle and proliferation of H2AX-deficient cells (Celeste et al., 2002; Reina-San-Martin et al., 2003), S region DSBs and intra-S region recombination occur at significant levels, but inter-S-S region recombination and CSR are impaired (Franco et al., 2006; Reina-San-Martin et al., 2003). Thus, lack of γ-H2AX focus formation in the IgH locus in the absence of AID does not reflect an absence of DSBs. We unequivocally show here that blunt DSBs occur in S region DNA in AID- or Ung-deficient B cells or in normal B cells before induction of AID expression. Such blunt DSBs were likely overlooked by studies aimed at indirectly identifying DSBs through detection of γ-H2AX foci (Petersen et al., 2001; Reina-San-Martin et al., 2003).
It has been assumed that the indispensability of AID in CSR reflects the role of this enzyme in generating DSBs (Catalan et al., 2003; Rush et al., 2004; Schrader et al., 2007; Schrader et al., 2005). This assumption has been supported by the findings that S region DSBs appeared simultaneously with AID expression in mouse B cells undergoing CSR and that DSBs occurred preferentially at dC:dG base-pairs in RGYW/WRCY motif, the target of AID (Schrader et al., 2007; Schrader et al., 2005; Stavnezer and Schrader, 2006). It has been suggested that those AID-mediated DSBs are staggered and are subsequently processed to generate blunt DSBs (Schrader et al., 2007; Schrader et al., 2005; Stavnezer and Schrader, 2006). However, a post-cleavage role for AID in CSR has been indicated by the occurrence of unignorable amounts of “background” DSBs in S regions of in vitro activated AID-or Ung-deficient B cells (Begum et al., 2004; Catalan et al., 2003; Rush et al., 2004; Schrader et al., 2007; Schrader et al., 2005; Wu and Stavnezer, 2007) and the in vivo AID-independent c-myc translocation to S regions in AID-deficient mice (Casali and Zan, 2004; Unniraman and Schatz, 2006; Unniraman et al., 2004b). DSBs have been shown to occur in V(D)J DNA of AID-deficient mouse B cells (Bross and Jacobs, 2003; Bross et al., 2002; Papavasiliou and Schatz, 2002) and in human B cells, in which AID activity was inhibited by a DN AID (Zan et al., 2003). Such DSBs were blunt and 5'-phosphorylated, but were processed by AID to yield staggered DSBs in B cells undergoing SHM (Zan et al., 2003). Likewise, as we show here, AID- and Ung-independent S region DSBs are blunt and 5'-phosphorylated, but are processed in an AID-dependent fashion in B cells undergoing CSR. We would argue that, as shown in cells treated with DNA break inducers (Brar et al., 2004), blunt DSBs trigger AID translocation to the nucleus, a prerequisite for direct DNA deamination. Nuclear AID would access S regions, as facilitated by germline IH-S-CH transcription occurring in the early stages of CSR, to deaminate dC residues nearby ends, thereby giving rise to staggered DNA ends after dU deglycosylation by Ung and DNA nicking by APE. This is further supported by our demonstration that AID can effectively deaminate dC within RGYW in blunt DSB ends.
The specificity and “in vivo” origin of the Sμ and Sγ1 region DSBs identified here is emphasized by the virtual absence of DSBs in Cμ, Cγ1, Pim1, Pax5 and Afp DNA. It is further strengthened by the nature of the DNA repair factors recruited by such DSB free ends and by our demonstration of comparable DSBs in DNA prepared using two different approaches: “anion-exchange” and “DNA plugs” (Ehrenstein et al., 2001; Schrader et al., 2002; Wu et al., 2006). DSBs are heterogeneous and are repaired through different pathways according mainly to the nature of their free ends, such as the length of the overhangs or lack of thereof. A whole spectrum of overhang lengths, from none to more than 20 bases, is involved in CSR, as inferred from analysis of recombined S-S junction sequences (Ehrenstein et al., 2001; Schrader et al., 2002; Wu et al., 2006). DSBs with overhangs of less than four bases are mainly repaired by nonhomologous end-joining repair (NHEJ). Longer overhangs, particularly with high dGdC content, would be less dependent on NHEJ and skewed toward homologous recombination (HR) or single strand-annealing (SSA) (Daley and Wilson, 2005). Both NHEJ and HR pathways share γ-H2AX and the Mre11/Rad50/Nbs1 (MRN) protein complex, which are involved in early response to DSBs (Cahill et al., 2006), but also require additional components that are unique to each process (D'Amours and Jackson, 2002; van Gent et al., 2001). Namely, NHEJ requires Ku70/Ku86, DNA-PKcs, XRCC4 and DNA ligase IV, while HR or SSA uses homologues of the yeast Rad52 epitasis protein group, including Rad52 and Rad51 (Shinohara and Ogawa, 1998; Song and Sung, 2000; Valerie and Povirk, 2003). Accordingly, consistent with both their blunt and staggered nature, the S region DSBs detected here recruited Ku70/Ku86 and Rad51/Rad52, in addition to γ-H2AX and Mre11/Nbs1.
Our demonstration of a high frequency of DSBs in Sμ and Sγ1 regions of mouse and human B cells deficient in AID or Ung provides an explanation for the occurrence of intra-S region recombinations, as suggested by the internal deletions, although limited in length, found in Sμ and Sγ1 region DNA of these B cells (Begum et al., 2007; Chaudhuri and Alt, 2004; Dudley et al., 2002), and as further substantiated by our findings. In B cells expressing AID and undergoing CSR, intra-S region recombinations are more obvious, resulting in extensive internal S region deletions, likely reflecting the resection by AID of S region DNA ends that are not synapsed with other S region DNA ends (Chaudhuri and Alt, 2004; Dudley et al., 2002), and the occurrence of long stretches of microhomologies, duplications and insertions as well as point-mutations around the junction regions. These parallel the extensive microhomologies, duplications, untemplated nucleotide insertions and point-mutations occurring in and around S-S region recombinations (Chen et al., 2001; Honjo et al., 2002; Wu et al., 2006). We argue here that in the absence of AID, S region DSBs, which are blunt and 5'-phosphorylated, would be synapsed by a DNA ligase or topoisomerase, implying that in B cells not undergoing CSR, S region blunt DSBs are “open and shut”, resulting in very limited intra-S region internal deletions, microhomologies or insertions. The requirement for staggered DNA ends for S-S region synapses in CSR is emphasized by the occurrence of inter-S-S region recombinations involving long-range DNA deletions and CSR in absence of AID in ΔSμSΔγ1/I-SceI mutant B cells, in which donor Sμ and acceptor Sγ1 regions were replaced with yeast I-SceI sites, allowing for the emergence of staggered DSBs upon digestion by megaendonuclease (Zarrin et al., 2007). The different mechanistic nature of intra-S region and inter-S-S region DNA recombinations is emphasized by the demonstration that H2AX and 53BP1 are required for inter-S-S region DNA recombination but not for intra-S region recombination (Manis et al., 2004; Reina-San-Martin et al., 2007; Reina-San-Martin et al., 2003; Ward et al., 2004), and by the demonstration that C-terminal deletion of AID severely impairs inter-S-S region recombination without affecting intra-Sμ recombination (Barreto et al., 2003).
One major question arising from our findings is how are AID-independent DSBs generated. S regions are inherently prone to breakage by virtue of their structure, as increased susceptibility to breakage is a property of DNA containing repetitive sequences capable of adopting non-canonical DNA conformations. These include extruded DNA structures, such as R-loops, G-loops or stem-loops, arising as result of the high dGdC content of S regions (R- and G-loops) as well as the high occurrence of palindromic AGCT motifs, which structure the stem of stem-loops (Baar et al., 1996; Chen et al., 2001; Mussmann et al., 1997; Tashiro et al., 2001). These S region structural alterations are enhanced by transcription and block progression of the replication fork (Lobachev et al., 2007). Replication forks arrested at R-loops, G-loops or stem-loops can give rise to highly distorted, long and complex, mostly single-strand DNA, which is inherently fragile and may undergo cleavage by certain nucleases (Duquette et al., 2004; Sun et al., 2001; Tian and Alt, 2000; Yu and Lieber, 2003), such as endonuclease G (Endo G) (Irvine et al., 2005). Endo G is mitochondrial in origin, but it is likely processed into a nuclear form and exported from mitochondria to the nucleus. There, it could cleave S region DNA, primarily at dG and dC (Widlak et al., 2001), which are also the preferential sites of DSBs (Rush et al., 2004; Schrader et al., 2005; Stavnezer and Schrader, 2006). In conclusion, our study argues for a postcleavage role of AID in CSR. While defining a further role of AID in CSR, it calls for investigation into the mechanisms that underlie the AID- and Ung-independent cleavage of S region DNA.
Acknowledgments
We thank Dr. Leman Yel for blood from patient P3, Dr. Anne Durandy for blood from patient YB, Dr. Tasuku Honjo for AID-deficient mice, Dr. Tomas Lindahl for ung-deficient mice and Dr. Michael R. Lieber for GST-AID fusion protein. We thank Dr. Jinsong Zhang, Dr. Zsuzsanna Pal and Dr. Zhenming Xu and for helpful discussions.
This work was supported by N.I.H. grants AI 45011, AI 60573 and AR 40908 to P.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baar J, Pennell NM, Shulman MJ. Analysis of a hot spot for DNA insertion suggests a mechanism for Ig switch recombination. J. Immunol. 1996;157:3430–3435. [PubMed] [Google Scholar]
- Barreto VM, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- Begum NA, Izumi N, Nishikori M, Nagaoka H, Shinkura R, Honjo T. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 2007;282:731–742. doi: 10.1074/jbc.M607439200. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kinoshita K, Kakazu N, Muramatsu M, Nagaoka H, Shinkura R, Biniszkiewicz D, Boyer LA, Jaenisch R, Honjo T. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- Brar S, Watson M, Diaz M. Activation-induced cytosine deaminase, AID, is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- Bross L, Jacobs H. DNA double strand breaks occur independent of AID in hypermutating Ig genes. Clin. Dev. Immunol. 2003;10:83–89. doi: 10.1080/10446670310001626571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L, Muramatsu K, Kinoshita K, Honjo H, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA souble strand break repair. Front. Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali P, Zan H. Class switching and c-myc translocation: how does DNA break? Nat. Immunol. 2004;5:6–8. doi: 10.1038/ni1104-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan N, Selz F, Imai K, Revy P, Fischer A, Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Schaffer A, Shah S, Goodwin RG, Zan H, Ely S, Casali P. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J. Immunol. 2000;165:786–794. doi: 10.4049/jimmunol.165.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Zan H, Kim EC, Shah S, Schattner EJ, Schaffer A, Casali P. Ongoing in vivo immunoglobulin class switch DNA recombination in chronic lymphocytic leukemia B cells. J. Immunol. 2002;169:6594–6603. doi: 10.4049/jimmunol.169.11.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max EE, Casali P. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+ IgD+ B cell line. J. Immunol. 1998;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen X, Kinoshita K, Honjo T. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc. Natl. Acad. Sci. USA. 2001;98:13860–5. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KF, Alt FW, Manis JP. The function of AID in somatic mutation and class switch recombination: upstream or downstream of DNA breaks. J. Exp. Med. 2002;195:F37–F41. doi: 10.1084/jem.20020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Diaz M, Casali P. Somatic Ig hypermutation. Curr. Opin. Immunol. 2002;14:235–240. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl. Acad. Sci. USA. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair. 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–96. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Honjo T, Muramatsu M, Fagarasan S. AID: How does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- Irvine RA, Adachi N, Shibata DK, Cassell GD, Yu K, Karanjawala ZE, Hsieh CL, Lieber MR. Generation and characterization of endonuclease G null mice. Mol. Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Miyata T, Honjo T. Repetitive sequences in class switch recombination regions of immunoglobulin heavy chain genes. Cell. 1981;23:357–368. doi: 10.1016/0092-8674(81)90131-8. [DOI] [PubMed] [Google Scholar]
- Kenter AL. Class switch recombination: an emerging mechanism. Curr. Top. Microbiol. Immunol. 2005;290:171–199. doi: 10.1007/3-540-26363-2_8. [DOI] [PubMed] [Google Scholar]
- Kim EC, Edmonston CR, Wu X, Schaffer A, Casali P. The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3' hs1,2 enhancer in human B cells. Relevance to class switch DNA recombination. J. Biol. Chem. 2004;279:42258–42269. doi: 10.1074/jbc.M407496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: transcripts directed by I promoters on looped-out circular DNAs. Proc. Natl. Acad. Sci. USA. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front. Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- Maizels N. Immunoglobulin gene diversification. Annu. Rev. Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Mussmann R, Courtet M, Schwager J, Du Pasquier L. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur. J. Immunol. 1997;27:2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem. Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, Redon C, Ried T, Bonner WM, Honjo T, Nussenzweig MC, Nussenzweig A. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1(−/−) B cells. Eur. J. Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rush JS, Fugmann SD, Schatz DG. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to Smu in Ig class switch recombination. Int. Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- Schaffer A, Cerutti A, Shah S, Zan H, Casali P. The evolutionary conserved sequence upstream of the human Ig Sg3 region is an inducible promoter: Synergistic activation by CD40 ligand and IL-4 via cooperative NF-kB and STAT-6 binding sites. J. Immunol. 1999;162:5327–5336. [PubMed] [Google Scholar]
- Schaffer A, Kim E, Wu X, Zan H, Testoni L, Salamon S, Cerutti A, Casali P. Selective inhibition of class switching to IgG and IgE by recruitment of the HoxC4 and Oct-1 homeodomain proteins and Ku70/Ku86 to newly identified ATTT cis-elements. J. Biol. Chem. 2003;278:23141–23150. doi: 10.1074/jbc.M212952200. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-Induced Cytidine Deaminase-Dependent DNA Breaks in Class Switch Recombination Occur during G1 Phase of the Cell Cycle and Depend upon Mismatch Repair. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–7. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Song B, Sung P. Functional Interactions among Yeast Rad51 Recombinase, Rad52 Mediator, and Replication Protein A in DNA Strand Exchange. J. Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin. Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Sun H, Yabuki A, Maizels N. A human nuclease specific for G4 DNA. Proc. Natl. Acad. Sci. USA. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro J, Kinoshita K, Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int. Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleaseses. J. Biol. Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Fugmann SD, Schatz DG. UNGstoppable switching. Science. 2004a;305:1113–1114. doi: 10.1126/science.1102692. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair. 2006;5:1259–1264. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for IgH switch region-c-myc chromosomal translocations. Nat. Immunol. 2004b;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replication. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, Nussenzweig MC, Chen J. 53BP1 is required for class switch recombination. J. Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlak P, Li LY, Wang X, Garrard WT. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates. J. Biol. Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, Casali P. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J. Immunol. 2006;176:5426–5437. doi: 10.4049/jimmunol.176.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zan H, Komori A, Feng J, Kim EC, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J. Clin. Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel RA, Du J, Thompson RJ, Kenter AL. Ig Sgamma3 DNA-specific double strand breaks are induced in mitogen- activated B cells and are implicated in switch recombination. J. Immunol. 1997;159:4139–4144. [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Zhong Y, Evinger AJ, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann. N.Y. Acad. Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Pone EJ, Al-Qahtani A, Park SR, Zan H, Casali P. Regulation of aicda expression and AID activity: relavance to somatic hypermutation and class switch DNA recombination. Crit. Rev. Immunol. 2007a;27:367–397. doi: 10.1615/critrevimmunol.v27.i4.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pal Z, Casali P. DNA replication to aid somatic hypermutation. Adv. Exp. Med. Biol. 2007b;596:111–127. doi: 10.1007/0-387-46530-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ cell line in vitro: Definition of the requirements and the modalities of hypermutation. J. Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Schaffer A, Dramitinos P, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β. Evidence for TGF-β but not IL-10-dependent direct Sμ→Sα and sequential Sμ→Sγ, Sγ→Sα DNA recombination. J. Immunol. 1998;161:5217–5225. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–53. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. BCR engagement and T cell contact induce bcl-6 hypermutation in human B cells: association with initiation of transcription and identity with Ig hypermutation. J. Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]