When the normal circulation is compromised by damage to blood vessels, clots are formed at the site of injury. These clots seal the injury to stem the loss of blood. They are resistant to shear stress and pliant in response to pulsatile blood flow. They support clot retraction and initiate wound healing. The biomechanical properties of clots have evolved to meet these physiological functions. Thus, clots are extensible and elastic (1). When clots are formed, the plasma protein fibrinogen is converted to fibrin. Fibrin is a network of fibers. These fibers provide the supporting framework of blood clots. It is generally accepted that the properties of these fibers underlie the mechanical properties of clots in vivo (2). For example, studies with mice lacking fibrinogen have shown that clots formed in the absence of a fibrin network failed to resist shear stress (3). Such clots grow to a certain size at the site of injury, then break off and flow downstream. Because the mechanical performance of fibrin is critical to its physiological function, the mechanical properties of fibrin have been broadly investigated (4–9). Many studies have shown that clots of pure fibrin are extensible and elastic (5, 9, 10). Recent work has shown these biomechanical properties are inherent in the fibrinogen molecule (9, 11–13). Here we review the recent work and consider the molecular origins of these remarkable biomechanical properties.
Background
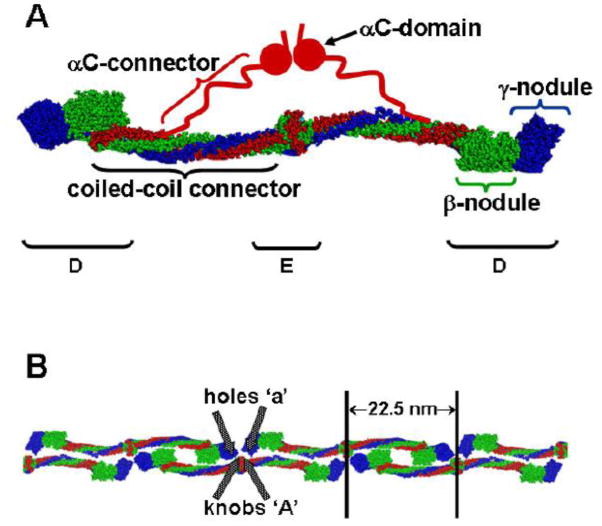
Human fibrinogen is a 340 kDa glycoprotein. It is assembled as a dimer of three polypeptide chains, called Aα, Bβ, and γ (14). The recent crystal structure(15), as well as many other studies, shows fibrinogen is an elongated molecule with a unique center and two identical symmetric distal regions (Fig. 1A). The center, or E region, is a single nodule that contains the N- termini of all six chains. The three chains extend from the center in two coiled-coils that terminate in the two distal D regions. The C-terminal segments of the Bβ and γ chains fold independently to form the β- and γ-nodules, respectively. The C-terminal segment of the Aα chain goes through the D region, and folds back to form a four chain coiled-coil. Beyond this short, fourth alpha-helix, the structure of the C-terminus of the Aα chain is not visible in the X-ray structure. Presumably this region is mobile within the crystal, and thus does not provide interpretable diffraction data (15). Other studies, including electron microscopy(16) and NMR (17, 18) analysis, indicate the C-terminal region of the Aα chain contains two regions, called the αC connector (Aα 221–391) and the αC domain (Aα 391–610) (19).
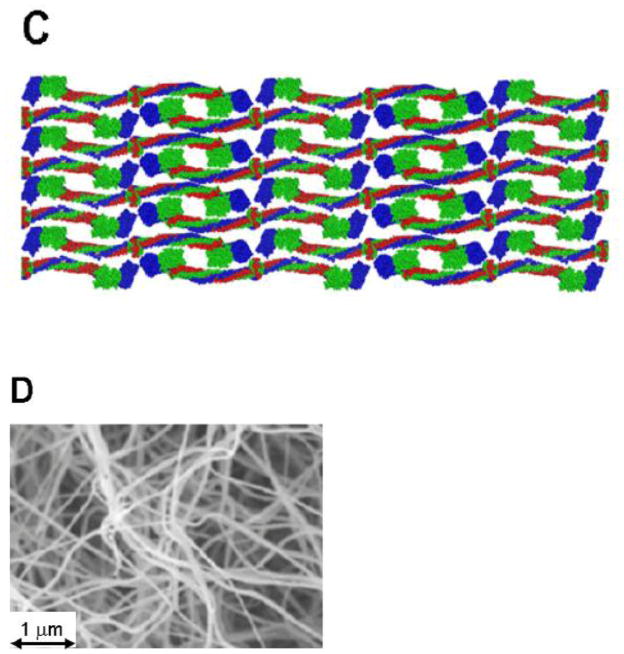
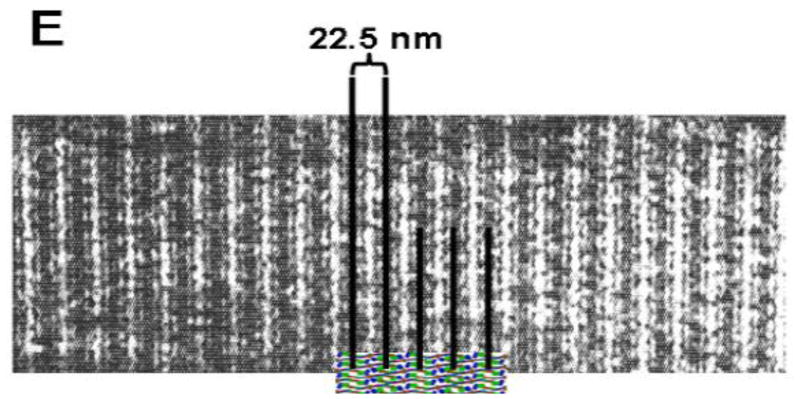
Figure 1. Structures of fibrinogen and fibrin.
(A) Model of human fibrinogen based on the crystal structure (15) modified from [19]. (B) Model of half-staggered, double-stranded fibrin protofibril, illustrating the knob-hole interactions ‘A:a’. Because the locations of the αC regions in the protofibril are unknown, they are omitted from this model. (C) Model of fibrin fiber assembled from protofibrils shown in panel B. (D) Scanning electron micrograph of a fibrin clot. (57). (E) The model in panel C superimposed on a transmission electron micrograph of a negatively-stained fibrin fiber segment (58) with alignment of the stain pattern with the assembled fiber pattern.
Following vessel injury, a series of reactions occur on the surfaces of cells at the site of injury, leading to the generation of the protease thrombin. Thrombin cleaves fibrinogen at 4 sites, two after residue 16 in the Aα chain, releasing fibrinopeptide A (FpA), and two after residue 14 in the Bβ chain, releasing fibrinopeptide B (FpB). FpA is cleaved first, exposing a new site called knob ‘A’. Knob ‘A’ binds with a site in the γ-nodule, called hole ‘a’. These ‘A:a’, knob-hole, interactions lead to formation of half-staggered, double-stranded protofibrils that have a structural periodicity of 22.5 nm (20) ( Fig. 1B). Subsequent cleavage of FpB exposes the knob ‘B’ which binds with a site in the β-nodule, called hole ‘b’. Concurrent with FpB release, the protofibrils assemble into fibers. The product is a three dimensional fibrin network or gel (Fig. 1D). The molecular interactions that promote branching and assembly of fibers are not well understood, although these processes appear to be competitive (10), as fibrin gels with many branches have thinner fibers than gels with fewer branches. The ‘B:b’ interactions, which are homologous to the ‘A:a’ interactions, were proposed to support the assembly of protofibrils into fibers. More recent studies`, however`, suggest the ‘B:b’ interactions occur between strands within the protofibril`, reinforcing the ‘A:a’ bonds (21, 22). Several studies have suggested the αC regions support the assembly of protofibrils into fibers, the step called lateral aggregation (23) Nevertheless, the αC region is not required for lateral aggregation, as nearly normal polymers are formed from fibrin monomers lacking this region (24). Fibrin polymers are stabilized by the FXIIIa-catalyzed formation of γ-glutamyl-ε-lysyl amide bonds between monomers. These bonds link γ chains to form reciprocal γ-γ dimers parallel to the long fiber axis. These bonds also link α chains to form α-polymers, linking multiple monomers across the long fiber axis (25, 26).
The vast majority of mechanical studies of fibrin have been done with macroscopic gel samples evaluated in a cone plate rheometer. John Ferry pioneered the mechanical evaluation of fibrin gels in experiments that established their macroscopic viscoelastic properties (6–8). On their own, cone plate rheometry data cannot distinguish the contributions of individual fibers, network architecture and branch point strength to the mechanical properties of fibrin clots. When coupled with optical methods (birefringence) (27) and x-ray scattering (28), however, such measurements of fibrin films provided insight into the microscopic mechanisms of deformation. These studies were the first to suggest that stress induces conformational changes within the protein monomer, attributed to either the coiled-coil or the D region of the protein. Recent studies have measured the mechanical properties of individual fibers (13, 29–32) or cylindrical fibrin clots (9). These studies showed the mechanical properties of clots depend on the mechanical properties of the individual fibrin monomers.
Considering the global structure of the fibrin monomer, there are three features that could underlie these remarkable mechanical properties: the coiled-coil connectors, the folded globular nodules and the relatively unstructured αC regions (30). Experimental data suggest that each of these structures, or some combination of them, contributes. In their recent Science paper, Weisel and colleagues proposed the forced unfolding of the coiled-coil connectors occurs at relatively low strain, exposing hydrophobic residues that interact to form fiber bundles and expel water (9). This model follows from earlier work that showed forced unfolding of the coiled-coils in single molecules or small oligomers of fibrinogen (11, 12). Two recent papers by Gorkun and colleagues have shown the ‘A:a’ interactions that mediate protofibril formation are sufficiently strong to unfold the γ-nodule prior to breaking this knob-hole bond (33, 34). These experiments suggest unfolding of the γ-nodules occurs under strain. Lastly, by comparing fibrinogens isolated from different species, Lord, Falvo and colleagues have shown that extensibility of individual fibers varies with the length of the αC region (13). They hypothesize that the unstructured repeat in the αC connector has a key role in fiber extensibility. Here we review the data that support each possibility.
The Coiled-Coil Connectors
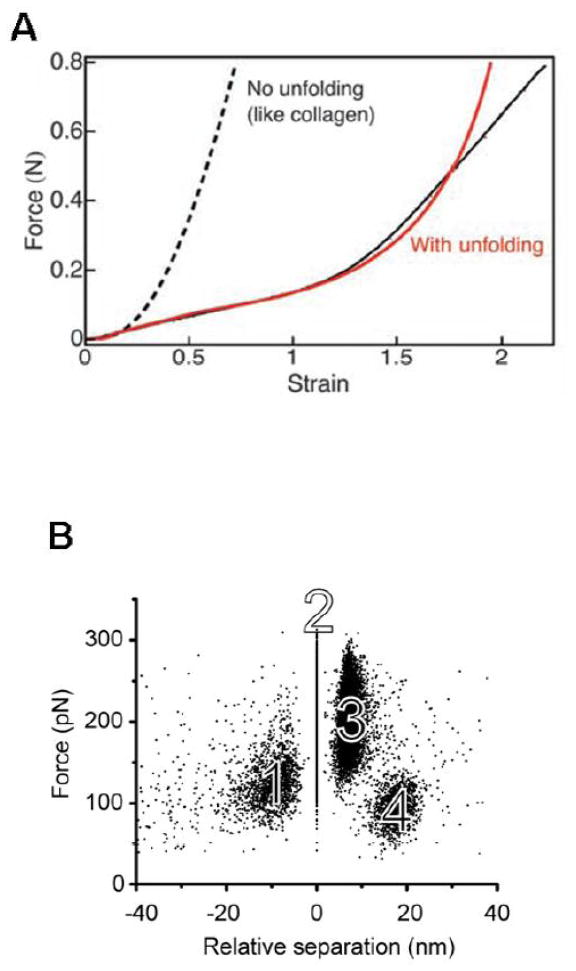
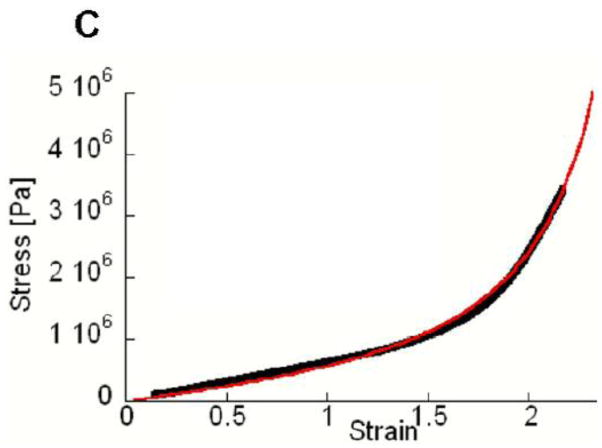
A pair of single molecule studies by Lim et al. (12) and Brown et al. (11) proposed that unfolding of the coiled-coil connectors mediates fiber extensibility. These groups used atomic force microscopy to measure the extension of single molecules within protofibrils (12) or small oligomers of fibrinogen (11) as a function of the force applied. These force-extension data were compared to simulations of forced unfolding of the coiled-coil region of the molecule. Lim et al. performed steered molecular dynamics simulations on a single fibrinogen molecular model, while Brown et al. performed Monte Carlo simulations of the force threshold for unfolding the coiled-coil region. Both studies showed some degree of quantitative agreement between AFM force-extension data and the simulations of coiled-coil unfolding. However, in the Lim study, both the experimental data and the molecular dynamics simulation show a force plateau after unfolding, which is similar to that seen in myosin coil-coil unfolding studies(35); where as in the Brown study, a worm-like-chain unfolding pattern was observed, similar to that seen in proteins such as titin (36, 37). Brown et al. also analyzed the distance between peaks in the force-extension data (11). A histogram of peak-to-peak lengths showed reasonable statistical agreement with the 22.5 nm length expected for the difference between a fully folded and stretched coiled-coil region, consistent with a sequential unfolding of coiled-coils within the fibrinogen oligomer. In these studies, the applied force was necessarily transferred through the D region, coiled-coil and E region, eliminating any contribution of the unstructured αC region. Thus, although these studies provide evidence that the coiled-coil is unfolding, it is not clear that the data are relevant to the stretching of physiological fibrin fibers, which are polymers with many parallel and series connections between monomers. In a more recent study (9), Brown et al performed force-extension measurements on specially prepared clots whose cylindrical geometry allowed tensile stress vs. strain evaluation (Fig 2A). This study included impressive small angle x-ray scattering experiments that showed a gradual disappearance of the 22.5 nm peak corresponding to the half-length of the fibrin monomer. These data are consistent with an unfolding of the coiled-coil, but may also indicate that the continuous half-stagger alignment of the protofibrils (Fig. 1E) is broken through shearing.
Figure 2. Changes of molecular dimensions with applied force.
(A) Stress-strain curves of cylindrical fibrin clots. Data are shown with solid black like, fitted model in red. [1] (B) Force-induced unfolding of the γ-nodule measured by AFM. Distances between knob ‘A’ and hole ‘a’ are plotted relative to event 2. The clusters of forces representing the four events in the complex force pattern are labeled with numbers. (34) (C) Stress-strain curves of individual fibrin fibers. Data are in black, fitted model in red (32).
To analyze their data, Brown et al. (9) developed a multi-scale mechanical model spanning six orders of spatial magnitude. In this model the mechanism of extension for the single fiber was assumed to be the unfolding and subsequent stretching of the coiled-coil connectors or perhaps another structured region of the protein. The connectors were in one of two states, folded or unfolded, and the distribution of folded and unfolded was governed thermodynamically by the unfolding energy barrier and the length difference between folded and unfolded states. The force-extension relation contained two components, a linear region representing the fiber stiffness prior to unfolding and an non-linear, stiffening region modeled with an ideal polymer chain expression (worm like chain) representing the unfolded fiber stiffness. In order to fit the low strain linear region data to this model, a value of 14.5 MPa was used as the elastic modulus of the fiber prior to unfolding of any domains of the monomer. This value was based on previous experiments that determined the single fiber modulus through analysis of thermal motion (38). It is typical of elastomeric materials and similar in magnitude to the 4 MPa stiffness measured for single fibrin fibers in AFM experiments (39). In fact, experimental determination of the elastic modulus for protofibrils yields roughly the same values as the whole fiber indicating the low modulus is an inherent molecular scale materials property (40, 41). This modulus is not consistent, however, with the known stiffness of individual α-helices, coiled-coils, or globular domains as determined in experimental and theoretical treatments. Calculations of the persistence length of a single α-helix yield values in the 100–150 nm range (42). This places the elastic modulus of an α-helix in the range ~1 GPa (persistence length can be expressed as a ratio of thermal energy to the bending modulus of the filament, see (43)). These calculated values are in close agreement with the experimental values (1–4 GPa) for α-keratin materials ((43), page142) for which the coiled-coil was shown to mediate stiffness (44, 45). Thus, the elastic modulus that Brown et al. used to fit the low strain fibrin fiber data is 100–1000 times lower than the published values for coiled-coils.
A model in which the coiled-coil is the structural component supporting the majority of the initial strain of the fibrin fiber requires connection of adjacent coiled-coils in series. This is reasonable for fibrin since the D regions are connected in this way when crosslinked with FXIIIa. However, if the extensibility is being born primarily by the coiled-coil, then the stiffness of the fiber overall must be comparable to the stiffness of the coiled-coil. One way to reconcile the GPa unfolding of known coiled-coils with the MPa scale for fibrin fibers is a zipper type mechanism in which the stress is concentrated rather than distributed throughout the fiber. Only a small fraction of the coiled-coils would bear the total load, and as they unfold, would pass the concentrated load to the next isolated cohort of coiled-coils. The global stiffness of the fiber would then appear to be much lower than expected if the load were distributed uniformly. Because there is no structural data consistent with a non-uniform distribution of the applied load, this zipper mechanism is unlikely.
The γ–nodules
Recent studies by Averett et al. provide direct evidence that the γ-nodules unfold in response to mechanical stress (33, 34). These authors developed a novel method to measure the forced dissociation of ‘A:a’ interaction using AFM. Molecules containing holes ‘a’ were bound to the AFM chip and molecules containing knobs ‘A’ were bound to the cantilever tip. They measured the distribution of interaction forces and found a complex pattern consisting of several rupture events where the last event represented the bond dissociation (Fig. 2B). Careful control experiments showed the complex force pattern was associated with an interaction between single molecules. The specific interactions that contributed to this pattern were elucidated using fibrinogen and fibrin fragments, and recombinant fibrinogen variants. These experiments showed the nature of the knob-bearing molecule did not contribute to the complex pattern. Rather, the pattern reflected only the nature of hole ‘a’. The same force pattern was observed when knobs ‘A’ were present in the central region of fibrin, specifically the fragment desAB-NDSK, or the isolated chain of fibrin. No substantive differences were seen between holes ‘a’ in fibrinogen and in fragment D, demonstrating that the coiled-coil and αC regions did not contribute to force dissociation pattern. In contrast, no interactions were seen with fibrinogen γ D364H, a variant where hole ‘a’ is changed and ‘A:a’ interactions are disrupted. The force pattern seen with fibrinogen BβD432A, which disrupts ‘B:b’ interactions, was not different from normal fibrinogen. Thus, the complex force pattern was specifically associated with the ‘A:a’ interaction and the γ-nodule.
In subsequent studies of multi-step ‘A:a’ bond rupture, Averett et al. demonstrated that the force applied to hole ‘a’ through bound knob ‘A’ induced stepwise unfolding of the γ-nodule prior to rupture of the ‘A:a’ bond (33, 34). Remarkably, the strength of the ‘A:a’ bond was sufficient to survive through the unfolding events. That is, the ‘A:a’ bond was sufficiently stable under tension to allow propagation of the applied forces to other regions of fibrinogen. By comparing the probability distributions of rupture forces, it was found that the unfolding events represent a rupture of sequential bonds whereby each protects the next from breaking. This stepwise unfolding of the γ-nodule would create fibrin intermediates of different lengths. These intermediates may be considered mechanically stable as they resist further unfolding for short periods of time (the time between ruptures) but they exist only while under tension. The analyses show each fibrin ‘A:a’ interaction can be maintained for strains up to 23 nm prior to rupture. With two symmetric ‘A:a’ interactions in one molecule, this strain would double the normal fibrin monomer length.
These studies clearly show the γ-nodule can be extended without breaking the bonds between monomers. Nevertheless, it is not obvious that unfolding of the γ-nodule is relevant to the extensibility of fibrin fibers. Some unfolding forces for the ‘A:a’ interaction are quite high: upwards of 220 pN per molecule. To reach forces this high per monomer, the stress in the fiber (the force divided by the cross sectional area) needs to be in excess of 10 MPa (220pN divided by the cross sectional area a monomer with a 5nm diameter). In single fiber measurements, 10 MPa is higher than the average fiber strength. High stress, sufficient to unfold the ‘A:a’, has been observed only in rare cases at very high strain. Thus, unfolding the γ-nodule is not a likely source of extension when force is applied directly through the ‘A:a’ interaction. Moreover, when adjacent fibrin monomers are ligated by FXIIIa, the strength of this covalent interaction may circumvent the role of the ‘A:a’ bond during fiber stretching. It is possible that γ-nodule unfolding occurs in the ligated fiber. One might expect, however, a different unfolding when force is applied through the ligated C-terminus of the γ chain rather than through the ‘A:a’ interaction, because the energy landscape is highly dependent on the direction of force application (46) In other words, the energy barriers to bond dissociation in force-induced unfolding are direction dependent, so force applied through the ligated bond will encounter different energy barriers than force applied though the ‘A:a’ bonds. If this is the case, unfolding would occur if the barriers were sufficiently low along the direction of applied force. If γ-nodule unfolding occurs in both ligated and unligated fibers, this difference in the direction of force may contribute to the difference between the extensibilities of these two fibers.
The αC regions
Recent studies probing the mechanical properties of individual fibrin fibers have shown that the fiber itself is highly extensible, possesses tensile elastic modulus in the few MPa range (between 1–10 MPa) and exhibits strain stiffening behavior (Fig. 2C) (13, 29–32, 38). All three properties are characteristic of other elastomeric protein fibers such as elastin, resilin or spider silk (47–49) and suggest that, like these protein fibers, fibrin’s mechanical properties lie in the straightening of unstructured polypeptides. Protein assemblies whose stiffness is mediated by solid folded proteins possess moduli in the GPa range and extensibilities typically well under 50% (50, 51). For these reasons, proponents of all of the extensibility models described in this review are in agreement on one point: the extensibility of fibrin must be mediated by an unstructured length of polypeptide. The question is whether this unstructured segment is an unfolded part of the structured portion of the fibrin molecule or whether it is the “natively unfolded” region of the monomer. Extensibility models based on the coiled-coil or gamma region require a transition from a folded to an unfolded state during stretching. In the transition from mostly folded monomers to mostly unfolded, one expects a measurable change (lowering) in overall fiber stiffness as the stiff folded monomers are transformed into random coil polypeptides. This lowering of stiffness is seen in single molecule studies on globular proteins such as titin (36, 37) and coiled coils such as myosin (35), as well in protein assemblies such as keratin (44) in which stretching induced protein unfolding is known to occur. However, the stress vs. strain data collected on single fibers show a constant (elastomeric-like) stiffness from very low strain up to ~100% strain where strain stiffening begins.
These arguments point to the αC region, the only natively unstructured part of fibrinogen, as the source of fiber extensibility. Indeed, studies of clots made with recombinant fibrinogen lacking the αC region provide definitive evidence that this region has an important role in the mechanical properties of fibrin clots (52). Moreover, experiments with fibrinogens from different species suggest a role of this unstructured region. This region is the least conserved through evolution (53, 54). In particular the length of the repeat in the αC connector region varies from a low in chicken fibrinogen, which has no repeat, to a high in lamprey fibrinogen, which has over 20 repeats of 18 residues (55). Of special note, the amino acid content of the tandem repeat within all αC connectors is intriguingly reminiscent of those in the repeat sequences in elastin, resilin, and spider-silk (56). If the αC region or the repeat region is important, then the fibrin fibers from different species would have different mechanical properties. Indeed, this is the case. Falvo et al. used their integrated nanomanipulation system to examine the extensibility and elasticity of fibers made of human, mouse and chicken fibrinogen (13). In human fibrinogen, the tandem repeat segment is 128 amino acids long (10 repeats of mostly 13); in mouse it is 60 (five repeats of mostly 13) (53). Consequently, the contour length of the loosely tethered αC region is roughly 22 nm longer in mouse and 50 nm longer in human than in chicken fibrinogen. With two α chains per monomer, this would be a total difference in contour length of 44 and 100 nm per monomer, respectively. These experiments showed extensibility was correlated with the lengths of the tandem repeat segments. Extensibility for chicken fibers was 47 ± 23% (n = 42); for mouse: 187 ± 44% (n = 89); and for human: 217 ± 47% (n = 75). The differences were significant for each pairing (mouse/human, P = 0.0004; chicken/human, P < 0.0001; mouse/chicken, P < 0.0001). Thus, the relatively low extensibility of chicken fibrin, and the intermediate values for mouse as compared with human fibers, correlate with the length of their corresponding connector regions.
Although these findings clearly support the hypothesis that the unstructured αC connector regions provide the structural basis for fiber extensibility, interpretation of this data is limited by the nature of the fibrins. The structures of the fibrinogens from these three species differ in ways outside the tandem repeats. In particular, their C-terminal αC domains are markedly different in sequence. Thus, these data point to the potential importance of the entire αC regions, not just the tandem repeats.
Summary
Recent studies have examined the molecular basis for the extensibility of fibrin fibers. Indirect studies, by analogy to the coiled-coils in keratin, implicate unfolding of the coiled-coils in fibrin (9, 11, 12). Direct studies have shown the feasibility of maintaining ‘A:a’ interactions between fibrin monomers with partial unfolding of the γ-nodule (33, 34). Direct studies have demonstrated that the αC regions influence the extensibility of fibers (13). As described here, there are substantive limitations for each of these studies. Because there are pros and cons for each, it appears reasonable to conclude that all three contribute to the mechanical properties of fibrin fibers. Indeed, Guthold and his colleagues recently provided a scheme where the unfolding of the coiled-coils would accommodate approximately 100% strain, while partial unfolding of the γ-nodules could accommodate strains up to 320%; extension of the αC regions would parallel both unfolding events (30, 31). In our view of the fibrin fiber, the coiled-coil and the γ-nodule are relatively stiff elastic elements in series with much softer unstructured regions. When under stress, the softer αC regions will bear the majority of the strain. A look at the magnitudes of fibrin’s stiffness relative to other elastomeric proteins as well as the known stiffness of α-helix coiled coils supports this view.
We believe that the current data present a conundrum. The conventional understanding of fibrin structure, shown in Fig 1C-E, is consistent with unfolding the coiled-coils and the γ-nodules. But, it is hard to reconcile these extensions with the force data. The fibers are too soft and the force vs. strain curves are too smooth. In contrast, stretching the unstructured αC region makes sense in the force context. It is hard, however, to reconcile a model of extensibility that relies completely on the αC region with the long covalently crosslinked protofibrils depicted in the conventional fiber structure. Thus, when we favor the view that the αC regions bear the strain, we bring forward the conclusion that the conventional understanding of fibrin structure is incomplete or incorrect. We are obliged to examine this possibility. Similarly, when others favor the view that extension ensues from unfolding structured regions like the coiled-coils and the γ-nodule, they are compelled to reconcile the known stress of unfolding with the stress strain curves for individual fibers and fiber bundles.
The success of our model supporting a natively unfolded origin to fibrin elasticity, the feasibility of unfolding the γ-nodule while retaining ‘A:a’ interaction, and the impressive set of SAXS data suggesting unfolding of some region of the molecule point to the need for direct evidence of the molecular origins of fibrin fiber extensibility and define the challenge for future work.
Research Highlights.
The mechanical properties of fibrin depend on the mechanical properties of the individual fibrin monomers.
Experimental data suggest three structures could contribute to the monomer properties: the coiled-coil connectors, the folded globular nodules and the relatively unstructured αC regions.
Review with a focus on the molecular origins of the remarkable biomechanical properties of fibrin clots.
Acknowledgments
The authors’ research is support by funding from the United States National Institutes of Health (HL031508, P41-EB002025), the National Science Foundation (0705977) and the American Heart Association (0855252E). The authors thank Laurel E. Averett and Nathan E. Hudson for their helpful comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dempfle CE, Kalsch T, Elmas E, Suvajac N, Lucke T, Munch E, Borggrefe M. Impact of fibrinogen concentration in severely ill patients on mechanical properties of whole blood clots. Blood Coagul Fibrinolysis. 2008;19(8):765–70. doi: 10.1097/MBC.0b013e32830f1b68. [DOI] [PubMed] [Google Scholar]
- 2.Weisel JW. The mechanical properties of fibrin for basic scientists and clinicians. Biophys Chem. 2004;112(2–3):267–76. doi: 10.1016/j.bpc.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Ni H, Denis CV, Subbarao S, Degen JL, Sato TN, Hynes RO, Wagner DD. Persistence of platelet thrombus formation in arterioles of mice lacking both von Willebrand factor and fibrinogen. J Clin Invest. 2000;106(3):385–92. doi: 10.1172/JCI9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janmey PA, Bale MD, Ferry JD. Polymerization of fibrin: analysis of light-scattering data and relation to a peptide release. Biopolymers. 1983;22(9):2017–9. doi: 10.1002/bip.360220902. [DOI] [PubMed] [Google Scholar]
- 5.Janmey PA, Amis EJ, Ferry JD. Rheology of Fibrin Clots .6. Stress-Relaxation, Creep, and Differential Dynamic Modulus of Fine Clots in Large Shearing Deformations. Journal of Rheology. 1983;27(2):135–153. [Google Scholar]
- 6.Nelb GW, Gerth C, Ferry JD. Rheology of fibrin clots. III. Shear creep and creep recovery of fine ligated and coarse unligated closts. Biophys Chem. 1976;5(3):377–87. doi: 10.1016/0301-4622(76)80050-6. [DOI] [PubMed] [Google Scholar]
- 7.Roberts WW, Kramer O, Rosser RW, Nestler FH, Ferry JD. Rheology of fibrin clots. I. Dynamic viscoelastic properties and fluid permeation. Biophys Chem. 1974;1(3):152–60. doi: 10.1016/0301-4622(74)80002-5. [DOI] [PubMed] [Google Scholar]
- 8.Gerth C, Roberts WW, Ferry JD. Rheology of fibrin clots. II. Linear viscoelastic behavior in shear creep. Biophys Chem. 1974;2(3):208–17. doi: 10.1016/0301-4622(74)80046-3. [DOI] [PubMed] [Google Scholar]
- 9.Brown AE, Litvinov RI, Discher DE, Purohit PK, Weisel JW. Multiscale mechanics of fibrin polymer: gel stretching with protein unfolding and loss of water. Science. 2009;325(5941):741–4. doi: 10.1126/science.1172484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan EA, Mockros LF, Weisel JW, Lorand L. Structural origins of fibrin clot rheology. Biophys J. 1999;77(5):2813–26. doi: 10.1016/S0006-3495(99)77113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown AE, Litvinov RI, Discher DE, Weisel JW. Forced Unfolding of Coiled-Coils in Fibrinogen by Single-Molecule AFM. Biophys J. 2007;92(5):L39–41. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim BB, Lee EH, Sotomayor M, Schulten K. Molecular basis of fibrin clot elasticity. Structure. 2008;16(3):449–59. doi: 10.1016/j.str.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 13.Falvo MR, Millard D, O’Brien ET, 3rd, Superfine R, Lord ST. Length of tandem repeats in fibrin’s. alphaC region correlates with fiber extensibility. J Thromb Haemost. 2008;6(11):1991–3. doi: 10.1111/j.1538-7836.2008.03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–99. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- 15.Kollman JM, Pandi L, Sawaya MR, Riley M, Doolittle RF. Crystal structure of human fibrinogen. Biochemistry. 2009;48(18):3877–86. doi: 10.1021/bi802205g. [DOI] [PubMed] [Google Scholar]
- 16.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W, Weisel JW. Carboxyl-terminal portions of the alpha chains of fibrinogen and fibrin. Localization by electron microscopy and the effects of isolated alpha C fragments on polymerization. J Biol Chem. 1993;268(18):13577–85. [PubMed] [Google Scholar]
- 17.Burton RA, Tsurupa G, Hantgan RR, Tjandra N, Medved L. NMR solution structure, stability, and interaction of the recombinant bovine fibrinogen alphaC-domain fragment. Biochemistry. 2007;46(29):8550–60. doi: 10.1021/bi700606v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton RA, Tsurupa G, Medved L, Tjandra N. Identification of an ordered compact structure within the recombinant bovine fibrinogen alphaC-domain fragment by NMR. Biochemistry. 2006;45(7):2257–66. doi: 10.1021/bi052380c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medved L, Weisel JW. Recommendations for nomenclature on fibrinogen and fibrin. J Thromb Haemost. 2009;7(2):355–9. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weisel JW. The electron microscope band pattern of human fibrin: various stains, lateral order, and carbohydrate localization. J Ultrastruct Mol Struct Res. 1986;96(1–3):176–88. doi: 10.1016/0889-1605(86)90019-4. [DOI] [PubMed] [Google Scholar]
- 21.Lounes KC, Ping L, Gorkun OV, Lord ST. Analysis of engineered fibrinogen variants suggests that an additional site mediates platelet aggregation and that “B-b” interactions have a role in protofibril formation. Biochemistry. 2002;41(16):5291–9. doi: 10.1021/bi011988s. [DOI] [PubMed] [Google Scholar]
- 22.Okumura N, Terasawa F, Haneishi A, Fujihara N, Hirota-Kawadobora M, Yamauchi K, Ota H, Lord ST. B:b interactions are essential for polymerization of variant fibrinogens with impaired holes ‘a’. J Thromb Haemost. 2007;5(12):2352–9. doi: 10.1111/j.1538-7836.2007.02793.x. [DOI] [PubMed] [Google Scholar]
- 23.Gorkun OV, Veklich YI, Medved LV, Henschen AH, Weisel JW. Role of the alpha C domains of fibrin in clot formation. Biochemistry. 1994;33(22):6986–97. doi: 10.1021/bi00188a031. [DOI] [PubMed] [Google Scholar]
- 24.Gorkun OV, Henschen-Edman AH, Ping LF, Lord ST. Analysis of A alpha 251 fibrinogen: the alpha C domain has a role in polymerization, albeit more subtle than anticipated from the analogous proteolytic fragment X. Biochemistry. 1998;37(44):15434–41. doi: 10.1021/bi981551t. [DOI] [PubMed] [Google Scholar]
- 25.Francis CW, V, Marder J. Rapid formation of large molecular weight alpha-polymers in cross-linked fibrin induced by high factor XIII concentrations. Role of platelet factor XIII. J Clin Invest. 1987;80(5):1459–65. doi: 10.1172/JCI113226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz ML, Pizzo SV, Hill RL, McKee PA. The effect of fibrin-stabilizing factor on the subunit structure of human fibrin. J Clin Invest. 1971;50(7):1506–13. doi: 10.1172/JCI106636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roska FJ, Ferry JD. Studies of fibrin film. I. Stress relaxation and birefringence. Biopolymers. 1982;21(9):1811–32. doi: 10.1002/bip.360210910. [DOI] [PubMed] [Google Scholar]
- 28.Roska FJ, Ferry JD, Lin JS, Anderegg JW. Studies of fibrin film. II. Small-angle x-ray scattering. Biopolymers. 1982;21(9):1833–45. doi: 10.1002/bip.360210911. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Jawerth LM, Sparks EA, Falvo MR, Hantgan RR, Superfine R, Lord ST, Guthold M. Fibrin fibers have extraordinary extensibility and elasticity. Science. 2006;313(5787):634. doi: 10.1126/science.1127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guthold M, Liu W, Sparks EA, Jawerth LM, Peng L, Falvo M, Superfine R, Hantgan RR, Lord ST. A comparison of the mechanical and structural properties of fibrin fibers with other protein fibers. Cell Biochem Biophys. 2007;49(3):165–81. doi: 10.1007/s12013-007-9001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Carlisle CR, Sparks EA, Guthold M. The mechanical properties of single fibrin fibers. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.03745.x. (Epub ahead of print. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson NE, Houser JR, OBET, Taylor RM, II, Superfine R, Lord ST, Falvo MR. Stiffening of Individual Fibrin Fibers Equitably Distributes Strain and Strengthens Networks. Biophysical Journal. 2010;98(8):1632–1640. doi: 10.1016/j.bpj.2009.12.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Averett LE, Schoenfisch MH, Akhremitchev BB, Gorkun OV. Kinetics of the multistep rupture of fibrin ‘A-a’ polymerization interactions measured using atomic force microscopy. Biophys J. 2009;97(10):2820–8. doi: 10.1016/j.bpj.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Averett LE, Geer CB, Fuierer RR, Akhremitchev BB, Gorkun OV, Schoenfisch MH. Complexity of “A-a” knob-hole fibrin interaction revealed by atomic force spectroscopy. Langmuir. 2008;24(9):4979–88. doi: 10.1021/la703264x. [DOI] [PubMed] [Google Scholar]
- 35.Schwaiger I, Sattler C, Hostetter DR, Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat Mater. 2002;1(4):232–5. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 36.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–12. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 37.Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Mechanical unfolding intermediates in titin modules. Nature. 1999;402(6757):100–3. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 38.Collet JP, Shuman H, Ledger RE, Lee S, Weisel JW. The elasticity of an individual fibrin fiber in a clot. Proc Natl Acad Sci U S A. 2005;102(26):9133–7. doi: 10.1073/pnas.0504120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falvo MR, Houser JR, Hudson NE. Unpublished Results. [Google Scholar]
- 40.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435(7039):191–4. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 41.Janmey PA, Winer JP, Weisel JW. Fibrin gels and their clinical and bioengineering applications. J R Soc Interface. 2009;6(30):1–10. doi: 10.1098/rsif.2008.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe S, Sun SX. The elasticity of alpha-helices. Journal of Chemical Physics. 2005;122(24) doi: 10.1063/1.1940048. [DOI] [PubMed] [Google Scholar]
- 43.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates, Inc; 2001. [Google Scholar]
- 44.Hearle JW. A critical review of the structural mechanics of wool and hair fibres. Int J Biol Macromol. 2000;27(2):123–38. doi: 10.1016/s0141-8130(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 45.Kreplak L, Doucet J, Dumas P, Briki F. New aspects of the alpha-helix to beta-sheet transition in stretched hard alpha-keratin fibers. Biophys J. 2004;87(1):640–7. doi: 10.1529/biophysj.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ke C, Jiang Y, Rivera M, Clark RL, Marszalek PE. Pulling geometry-induced errors in single molecule force spectroscopy measurements. Biophys J. 2007;92(9):L76–8. doi: 10.1529/biophysj.107.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Elastic proteins: biological roles and mechanical properties. Philos Trans R Soc Lond B Biol Sci. 2002;357(1418):121–32. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: from fibroin sequence to mechanical function. J Exp Biol. 1999;202(Pt 23):3295–303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 49.Fudge DS, Levy N, Chiu S, Gosline JM. Composition, morphology and mechanics of hagfish slime. J Exp Biol. 2005;208(Pt 24):4613–25. doi: 10.1242/jeb.01963. [DOI] [PubMed] [Google Scholar]
- 50.Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318(5858):1900–3. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 51.Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci U S A. 2006;103(33):12285–90. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collet JP, Moen JL, Veklich YI, Gorkun OV, Lord ST, Montalescot G, Weisel JW. The alphaC domains of fibrinogen affect the structure of the fibrin clot, its physical properties, and its susceptibility to fibrinolysis. Blood. 2005;106(12):3824–30. doi: 10.1182/blood-2005-05-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakawa M, Okamura T, Kamura T, Shibuya T, Harada M, Niho Y. Diversity of primary structures of the carboxy-terminal regions of mammalian fibrinogen A alpha-chains. Characterization of the partial nucleotide and deduced amino acid sequences in five mammalian species; rhesus monkey, pig, dog, mouse and Syrian hamster. Thromb Haemost. 1993;69(4):351–60. [PubMed] [Google Scholar]
- 54.Doolittle RF, Kollman JM. Natively unfolded regions of the vertebrate fibrinogen molecule. Proteins. 2006;63(2):391–7. doi: 10.1002/prot.20758. [DOI] [PubMed] [Google Scholar]
- 55.Wang YZ, Patterson J, Gray JE, Yu C, Cottrell BA, Shimizu A, Graham D, Riley M, Doolittle RF. Complete sequence of the lamprey fibrinogen alpha chain. Biochemistry. 1989;28(25):9801–6. doi: 10.1021/bi00451a039. [DOI] [PubMed] [Google Scholar]
- 56.Tatham AS, Shewry PR. Comparative structures and properties of elastic proteins. Philos Trans R Soc Lond B Biol Sci. 2002;357(1418):229–34. doi: 10.1098/rstb.2001.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowley SR, Okumura N, Lord ST. Impaired protofibril formation in fibrinogen gamma N308K is due to altered D:D and “A:a” interactions. Biochemistry. 2009;48(36):8656–63. doi: 10.1021/bi900239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams RC. Band patterns seen by electron microscopy in ordered arrays of bovine and human fibrinogen and fibrin after negative staining. Proc Natl Acad Sci U S A. 1983;80(6):1570–3. doi: 10.1073/pnas.80.6.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]