Abstract
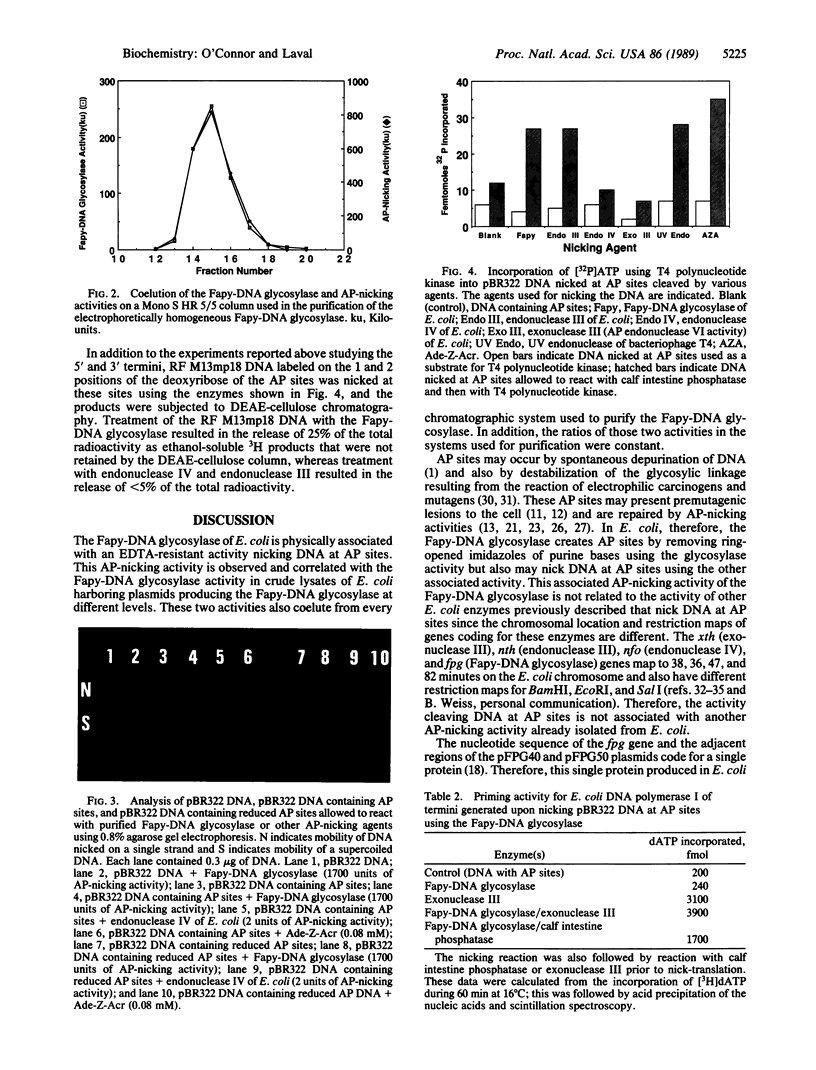
The 2,6-diamino-4-hydroxy-5N-formamidopyrimidine (Fapy)-DNA glycosylase of Escherichia coli, which is coded for by the fpg gene, excises purine bases with ring-opened imidazoles. In addition to the DNA glycosylase activity, we report that the Fapy-DNA glycosylase of E. coli has an associated activity, resistant to EDTA, that nicks DNA at apurinic/apyrimidinic (AP) sites. The levels of Fapy-DNA glycosylase and AP-nicking activity were parallel in crude lysates of E. coli HB101 harboring different plasmids constructed from the fpg gene. The fpg gene is different from the xth, nth, and nfo genes of E. coli, whose gene products also cleave DNA at AP sites. The Fapy-DNA glycosylase was purified to electrophoretic homogeneity. During this purification, the Fapy-DNA glycosylase copurified with an AP-nicking activity using chromatographic separations based on ion-exchange, molecular weight exclusion, and hydrophobicity. The cleavage at AP sites by the Fapy-DNA glycosylase left a 5'-phosphomonoester nucleotide at one terminus. In addition, DNA containing reduced AP sites was not nicked by the Fapy-DNA glycosylase. These data suggest that the mechanism of cleavage involved beta elimination. Therefore, this activity of the Fapy-DNA glycosylase nicking DNA at AP sites should be referred to as an AP lyase. The 3' terminus did not prime nick-translation by E. coli DNA polymerase I. However, the 3' terminus becomes a substrate for nick-translation if first allowed to react with calf intestine phosphatase or the E. coli exonuclease III. These data suggest that the repair of the Fapy lesion at least to some extent results in the formation of both 5'- and 3'-phosphomonoester nucleotides and the release of the deoxyribose.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Verly W. G. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987 Mar 1;242(2):565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Belleney J., Roques B. P., Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984 Jul 11;12(13):5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Huisman O. Isolation of a formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli K12. Mol Gen Genet. 1989 Jan;215(2):300–305. doi: 10.1007/BF00339732. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Huisman O., Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982 Dec 21;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochem Biophys Res Commun. 1983 Jan 27;110(2):552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987 Oct;6(10):3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breimer L. H. Enzymatic excision from gamma-irradiated polydeoxyribonucleotides of adenine residues whose imidazole rings have been ruptured. Nucleic Acids Res. 1984 Aug 24;12(16):6359–6367. doi: 10.1093/nar/12.16.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Chetsanga C. J., Frenette G. P. Excision of aflatoxin B1-imidazole ring opened guanine adducts from DNA by formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1983 Aug;4(8):997–1000. doi: 10.1093/carcin/4.8.997. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga C. J., Polidori G., Mainwaring M. Analysis and excision of ring-opened phosphoramide mustard-deoxyguanine adducts in DNA. Cancer Res. 1982 Jul;42(7):2616–2621. [PubMed] [Google Scholar]
- Clements J. E., Rogers S. G., Weiss B. A DNase for apurinic/apyrimidinic sites associated with exonuclease III of Hemophilus influenzae. J Biol Chem. 1978 May 10;253(9):2990–2999. [PubMed] [Google Scholar]
- Constant J. F., O'Connor T. R., Lhomme J., Laval J. 9-[(10-(aden-9-yl)-4,8-diazadecyl)amino]-6-chloro-2-methoxy-acridine incises DNA at apurinic sites. Nucleic Acids Res. 1988 Mar 25;16(6):2691–2703. doi: 10.1093/nar/16.6.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Saporito S. M., Spitzer S. G., Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986 Dec;168(3):1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater N. R., Miller E. C., Miller J. A. Estimation of apurinic/apyrimidinic sites and phosphotriesters in deoxyribonucleic acid treated with electrophilic carcinogens and mutagens. Biochemistry. 1980 Oct 28;19(22):5087–5092. doi: 10.1021/bi00563a023. [DOI] [PubMed] [Google Scholar]
- Franklin W. A., Lindahl T. DNA deoxyribophosphodiesterase. EMBO J. 1988 Nov;7(11):3617–3622. doi: 10.1002/j.1460-2075.1988.tb03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L. K., Haseltine W. A. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980 Dec 25;255(24):12047–12050. [PubMed] [Google Scholar]
- Haseltine W. A., Gordon L. K., Lindan C. P., Grafstrom R. H., Shaper N. L., Grossman L. Cleavage of pyrimidine dimers in specific DNA sequences by a pyrimidine dimer DNA-glycosylase of M. luteus. Nature. 1980 Jun 26;285(5767):634–641. doi: 10.1038/285634a0. [DOI] [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Kim J., Linn S. The mechanisms of action of E. coli endonuclease III and T4 UV endonuclease (endonuclease V) at AP sites. Nucleic Acids Res. 1988 Feb 11;16(3):1135–1141. doi: 10.1093/nar/16.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Laval J. Two enzymes are required from strand incision in repair of alkylated DNA. Nature. 1977 Oct 27;269(5631):829–832. doi: 10.1038/269829a0. [DOI] [PubMed] [Google Scholar]
- Lawley P. D., Orr D. J. Specific excision of methylation products from DNA of Escherichia coli treated with N-methyl-N'-nitro-N-nitrosoguanidine. Chem Biol Interact. 1970 Aug;2(2):154–157. doi: 10.1016/0009-2797(70)90047-5. [DOI] [PubMed] [Google Scholar]
- Ljungquist S., Lindahl T. Relation between Escherichia coli endonucleases specific for apurinic sites in DNA and exonuclease III. Nucleic Acids Res. 1977 Aug;4(8):2871–2879. doi: 10.1093/nar/4.8.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Pegg A. E. Enzymatic release of 7-methylguanine from methylated DNA by rodent liver extracts. Proc Natl Acad Sci U S A. 1981 Feb;78(2):861–865. doi: 10.1073/pnas.78.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbaugh D. W., Linn S. Characterization of the action of Escherichia coli DNA polymerase I at incisions produced by repair endodeoxyribonucleases. J Biol Chem. 1982 Jan 10;257(1):575–583. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Physical association of pyrimidine dimer DNA glycosylase and apurinic/apyrimidinic DNA endonuclease essential for repair of ultraviolet-damaged DNA. Proc Natl Acad Sci U S A. 1981 May;78(5):2742–2746. doi: 10.1073/pnas.78.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. R., Boiteux S., Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988 Jul 11;16(13):5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre J., Laval J. Micrococcus luteus endonucleases for apurinic/apyrimidinic sites in deoxyribonucleic acid. 1. Purification and general properties. Biochemistry. 1980 Oct 28;19(22):5018–5024. doi: 10.1021/bi00563a013. [DOI] [PubMed] [Google Scholar]
- Radany E. H., Friedberg E. C. A pyrimidine dimer-DNA glycosylase activity associated with the v gene product of bacterophage T4. Nature. 1980 Jul 10;286(5769):182–185. doi: 10.1038/286182a0. [DOI] [PubMed] [Google Scholar]
- Radany E. H., Naumovski L., Love J. D., Gutekunst K. A., Hall D. H., Friedberg E. C. Physical mapping and complete nucleotide sequence of the denV gene of bacteriophage T4. J Virol. 1984 Dec;52(3):846–856. doi: 10.1128/jvi.52.3.846-856.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recinos A., 3rd, Lloyd R. S. Site-directed mutagenesis of the T4 endonuclease V gene: role of lysine-130. Biochemistry. 1988 Mar 22;27(6):1832–1838. doi: 10.1021/bi00406a006. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Saporito S. M., Smith-White B. J., Cunningham R. P. Nucleotide sequence of the xth gene of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4542–4547. doi: 10.1128/jb.170.10.4542-4547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump D. G., Lloyd R. S. Site-directed mutagenesis of the T4 endonuclease V gene: role of tyrosine-129 and -131 in pyrimidine dimer-specific binding. Biochemistry. 1988 Mar 22;27(6):1839–1843. doi: 10.1021/bi00406a007. [DOI] [PubMed] [Google Scholar]
- Valerie K., Henderson E. E., deRiel J. K. Identification, physical map location and sequence of the denV gene from bacteriophage T4. Nucleic Acids Res. 1984 Nov 12;12(21):8085–8096. doi: 10.1093/nar/12.21.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verly W. G., Paquette Y. An endonuclease for depurinated DNA in Escherichia coli B. Can J Biochem. 1972 Feb;50(2):217–224. doi: 10.1139/o72-029. [DOI] [PubMed] [Google Scholar]
- Weiss B., Cunningham R. P. Genetic mapping of nth, a gene affecting endonuclease III (thymine glycol-DNA glycosylase) in Escherichia coli K-12. J Bacteriol. 1985 May;162(2):607–610. doi: 10.1128/jb.162.2.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Grossman L. Phosphodiesterases involved in DNA repair. Adv Enzymol Relat Areas Mol Biol. 1987;60:1–34. doi: 10.1002/9780470123065.ch1. [DOI] [PubMed] [Google Scholar]




