Abstract
Plasma has been the focus of testing different proteomic technologies for the identification of biomarkers due to its ready accessibility. However, it is not clear if direct proteomic analysis of plasma can be used to discover new marker proteins from tumor that are associated with tumor progression. Here, we reported that such proteins can be detected in plasma in a chemical induced skin cancer mouse model. We analyzed glycoproteins from both benign papillomas and malignant carcinomas from mice using our recently developed platform, solid-phase extraction of glycopeptides (SPEG) and mass spectrometry, and identified 463 unique N-linked glycosites from 318 unique glycoproteins. These include most known extracellular proteins that have been reported to play roles in skin cancer development such as thrombospondin, cathepsins, epidermal growth factor receptor, cell adhesion molecules, cadherins, integrins, tuberin, fibulin, TGFβ receptor, etc. We further investigated whether these tumor proteins could be detected in plasma from tumor bearing mice using isotope labeling and 2D-LC-MALDI-MS/MS. Two tumor glycoproteins, Tenascin-C and Arylsulfatase B, were identified and quantified successfully in plasma from tumor bearing mice. This result indicates that analysis of tumor associated proteins in tumors and plasma by method using glycopeptide capture, isotopic labeling, and mass spectrometry can be used as a discovery tool to identify candidate tumor proteins that may be detected in plasma.
INTRODUCTION
Despite great increase in understanding of cancer at molecular level, cancer remains as the second most common cause of death in the U.S. Survival rates for many common cancer types have changed little over the past two decades 1. If cancer is detected early, prior to metastatic spread, survival rates are vastly improved 1. For this reason, improvements in ability to detect cancer early may significantly reduce mortality from cancer. Plasma has been the focus of technology developments for different proteomic technologies for the identification of biomarkers due to its ready accessibility. These include depletion of the most abundant plasma proteins 2 and extensive fractionation of proteins or peptides prior to mass spectrometric analysis 3–5. However, proteins discovered by serum profiling are often well-known, high-abundance, classical serum proteins 6, not likely to be specifically derived from cancer tissue. Useful biomarkers for cancer detection in blood are those proteins released specifically from cancer tissues (overexpression of cancer proteins), indicators of a specific response of the system to cancer cells, or leaking of organ restricted proteins to blood due to structural changes in the microenvironment surrounding cancer cells (leaking of normal proteins such as PSA) 7. The tumor proteins that are detectable in both benign and malignant tumors as well as plasma can serve as candidate proteins for early detection of cancer. Detection of these proteins in plasma is critical to evaluate proteomic technologies for the biomarker discovery in plasma.
In an attempt to identify the proteins derived from cancerous tissue that are most likely to be present in blood, we employed our recently developed glycoproteomic analysis method using solid-phase extraction of N-linked glycopeptides (SPEG) 8–10. The method has several advantages. First, most cell-surface and secreted proteins are glycosylated, and disease-associated glycoproteins (secreted by cells, shed from their surface, or otherwise released) are likely to enter the bloodstream and thus represent a rich source of potential disease markers 11. Second, the reduction in complexity achieved by focusing on the glycoprotein subproteome in both tissues and plasma translates into favorable limits of detection, thus increasing the likelihood that the same polypeptide will be detectable in both tissue and serum 8, 12, 13. Third, aberrant glycosylation is a fundamental characteristic of oncogenesis and tumor progression 14, and this method allows us to identify proteins changed in glycosylation but not necessarily changed in total protein abundance. Finally, specific mass-spectrometry-based methods and affinity reagents can be developed for the specific and sensitive detection of identified tissue proteins in plasma 15, selective isolation of a specific proteins or peptides using affinity reagents16, or the recently developed targeted approach using multiple reaction monitoring (MRM) 17–19.
The chemically induced two-stage mouse skin carcinogenesis model has been used for decades to study the genetic, molecular, and biologic basis of tumor development 20. For example, the concepts of tumor initiation and promotion were derived from this model. In this model, the backs of 8-week-old mice treated with the carcinogen 7, 12-dimethylben[a] anthracene (DMBA) followed by multiple treatments with the tumor promoter 12-o-tetradecanoylphorbol-13-acetate (TPA). Benign tumors (papillomas) develop after 8 weeks and a small percentage of these progresses to malignant invasive carcinomas after a long latency 20. The ability to quantify both benign and malignant tumor growth permits analysis of genes and environmental factors that affect tumor progression. More recently the two stage skin tumor model has been used to improve proteomic technologies for biomarker discovery using serum protein profiling 12. We have identified several serum proteins for which the abundance is increased in correlation with the chemical induction of skin cancer in mice. However, these proteins are likely not markers for the specific diagnosis of skin cancer. A major advantage of this mouse skin carcinogenesis model is that plasma samples can be taken from mice before and after tumor development. As both benign and malignant tumors and plasma samples can be obtained from the same mice, this facilitates analysis of protein changes in plasma associated with tumor development.
Here we reported a two-step strategy for detection of tumor-associated proteins in plasma: the first step was to analyze extracellular proteins from normal skin, papillomas, and carcinomas and identify tumor-associated proteins; the second step was to detect the tumor-associated proteins in plasma using tissue-targeted approach and isotope labeling 7. Using our recently developed method of solid-phase extraction of glycopeptides (SPEG) and mass spectrometry 8–10, we analyzed matched benign and cancerous tumors from four tumor-bearing mice as well as normal skin tissues from four control mice, and identified 463 unique N-linked glycosites from 318 glycoproteins. Over forty identified glycoproteins were elevated in carcinomas. Two of the tumor-associated proteins, Tenascin-C and Arylsulfatase B, were further detected and quantified in plasma from the same cancer-bearing mice using isotope labeling and 2D-LC-MALDI-MS/MS. This result indicates that direct proteomic analysis of tumors and plasma using glycopeptide capture, isotopic labeling, and mass spectrometry can be used to discover new cancer derived proteins in plasma for early cancer detection.
METHOD & MATERIALS
Materials
Hydrazide resin and Sodium periodate were from Bio-Rad (Hercules, CA) ; PNGase F was from New England Biolabs (Ipswich, MA); Sequencing grade trypsin was purchased from Promega (Madison, WI); C18 columns were from Waters (Milford, MA); α-cyano-4-hydroxycinnamic Acid (CHCA) was from Agilent (Palo Alto, CA); iTRAQ reagent and mass calibration standards were purchased from Applied Biosystems (Foster City, CA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Tissues and plasma from chemical induced mouse skin tumors
Skin tumors were induced in four NIH01a mice using the DMBA/TPA two step protocol. A single dose of DMBA (Sigma; 25mg in 200ml of acetone) was applied to the shaved backs of four 8-week old mice. Initiated treated skin cells were promoted with TPA twice a week for 15 weeks. This gave rise to papillomas that were hyperplastic, well differentiated, benign lesions consisting of keratinocytes together with stroma tissue. Papillomas appeared as early as 8 weeks after the first treatment of DMBA and continued to grow for the next several months. A small percentage of these benign papillomas (~20%) progressed to squamous cell carcinomas. All the mice were sacrificed when carcinomas appeared in all four treated mice. Four littermate mice were left untreated for normal skin tissues. Papillomas and carcinomas as well as normal skin from untreated mice were snapping frozen in liquid nitrogen. Retroorbital bleeds are collected from each treated mouse before chemical treatment and after development of chemical induced carcinomas. The only difference between the normal and cancer tissues is the chemical induced cancer. Retroorbital bleeds were performed on anesthetized mice using avertin (0.1ml per 3g weight). 0.25 ml of whole blood was collected from the retroorbital sinus into a long (9 inches) sterile glass Pasteur pipet. The whole blood was placed in a K3EDTA coated 1.5ml microcentrifuge tube and centrifuged at 4°C for 5 minutes at 3000rpm. Plasma will be collected, carefully avoiding cellular contamination. All tumor tissues and plasma were placed in cryovials and frozen in liquid nitrogen.
Peptides extraction from skin tumor tissues
Frozen tumor tissues (100 mg each) were sliced into 1~3mm3 thick and incubated in 200μl of 5mM phosphate buffer and vortexed for 2–3 min. Then the samples were sonicated for 5 min in an ice-water bath. 200μl of trifluoroethanol (TFE) was added to the sample and incubated at 60°C for 2 hours followed by sonication for 2 min. Protein disulfide bonds were reduced by 5mM tributylphosphine (TBP) with 30 min incubation at 60°C. 10mM Iodoacetamide was applied to the mixture and incubated in the dark at room temperature for another 30 min. The samples were diluted 5-fold with 50mM NH4HCO3 (pH7.8) to reduce the TFE concentration to 10% prior to the addition of Trypsin at a ratio of 1:50 (w/w, enzyme: protein). Samples were digested at 37°C overnight with gentle shaking. The precipitate was discarded by centrifuge. Silver staining was used to test the effect of tryptic digestion. 4mg of total peptides from each sample were extracted from each tumor tissue. 2mg of total peptide was used to extract N-linked glycopeptides according to the following steps.
Peptide extraction from plasma
Plasma (20μl) was added to 90ul 8M urea in 0.4M NH4HCO3, 0.1% (w/v) SDS solution (pH8.3) and 10μl 120mM TCEP in dH2O freshly prepared and incubated at 60°C for 1 hour. Proteins were alkylated by adding 10μl 160mM iodoacetamide and incubated at room temperature in the dark with shaking for another 30 min. Samples were diluted by trypsin digestion buffer (100mM NH4HCO3, pH8.3) to make the concentration of urea less than 2M. 40μl trypsin (0.5μg/μl) was adding to digest protein at 37°C overnight. SDS-PAGE and silver staining was employed to check whether trypsin digestion was complete.
Glycopeptide capture from tissue or plasma
N-glycopeptides were isolated from peptides using SPEG 5. The enriched N-linked glycopeptides were concentrated by C18 columns and dried down and resuspended in 40μl 0.4% acetic acid prior to MS analysis.
Isotope labeling of peptides
The amount of glycopeptides was determined by BCA assay (bicinchoninic acid, Bio-Rad, Hercules, CA) prior to isotope labeling. 1μg glycopeptides from plasma of the retroorbital bleeds before and after chemical-induced cancer, and tumor tissues were dried and resuspended in 20μl of 50% DMF, 40%H2O, 10% pyridine. 5μl 10mg/ml d013C0, d413C0, and d413C4 succinic anhydride solution was added to glycopeptide samples and reacted at room temperature for 1~2hrs, then following C18 clean up to remove access succinic anhydride 8.
Mass spectrometry analysis
The peptides and proteins were identified using MS/MS analysis using an LTQ ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). Glycopeptides (1μg) were injected into a peptide cartridge packed with C18 resin, and then passed through a 10 cm × 75 μm i.d. microcapillary HPLC (μLC) column packed with C18 resin. The effluent from the μLC column entered an electrospray ionization source in which peptides were ionized and passed directly into the mass spectrometer. A linear gradient of acetonitrile from 5%–32% over 100 min at flow rate of ~300 nL/min was applied. During the LC-MS mode, data was acquired between m/z of 400 and 2000. The MS/MS spectra were collected using data dependent mode. Each sample was analyzed three times to increase the number of spectra used for spectral count.
Succinic anhydride labeled peptide (5μg) was analyzed by 2-D Nano LC (Eksigent, Dublin, CA) and MALDI-TOF/TOF (Applied Biosystems, Foster City, CA). Briefly, on-line integration of 15-cm-long 300μm strong cation exchange column (SCX) with 15-cm-long 300 μm of C18-reverse phase liquid chromatograph (RPLC) was employed. 4 SCX fractions of 0, 5, 50 and 500mM KCl and 3–45% linear acetonitrile gradient (containing 0.1% TFA and acetonitrile) of RPLC for each fraction were applied before analysis by MALDI-TOF/TOF. Peptides eluted from columns were directly mixed with CHCA and spotted on a MALDI target plate with 768 spots followed by the analyzed by MS and MS/MS using ABI4800 MALDI-TOF/TOF.
Data analyses
Peptide identifications-MS/MS spectra from LTQ were searched with SEQUEST 21 against a mouse protein database (the International Protein Index mouse protein database, version 3.13). The precursor mass tolerance is set as 3.0 Da. Other parameters of database searching are modified as following: oxidized methionines (add Met with 16 Da), a (PNGase F-catalyzed) conversion of Asn to Asp (add Asn with 1 Da) and Cys modification (add cysteine with 57 Da). The output files were evaluated by INTERACT and PeptideProphet 22, 23. The criterion of PeptideProphet analysis is the probability score ≥ 0.9 so that low probability protein identifications can be filtered out.
Identifying tissue-derived peptides in plasma from MALDI-TOF/TOF (ABI 4800) was performed using GPS Explorer software (version 3.6). MS/MS spectra were searched against NCBInr database. GPS searches were carried out at a 0.2 Da precursor mass tolerance, a 0.6 Da fragment mass tolerance; trypsin as enzyme digested. In addition to the modifications for Met, Asp, and Cys that were used in LTQ MS/MS spectra analyses as described above, N-termini of peptides and Lys are modified by succinic anhydride (100 Da for d013C0, 104 Da for d413C0, and 108 Da for d413C4).
RESULTS and DISCUSSION
Strategy of the method
The objective of this study was to use N-linked glycopeptide isolation, isotopic labeling, and LC- MS to identify skin cancer related extracellular proteins and determine if these proteins could be detected in plasma from tumor bearing mice. This strategy is based on the fact that most of extracellular proteins are glycoproteins and extracellular proteins from cancer are most likely to be detected in plasma due to the fact that they are likely to be secreted by cells or shed from cell surface to enter into the blood stream.
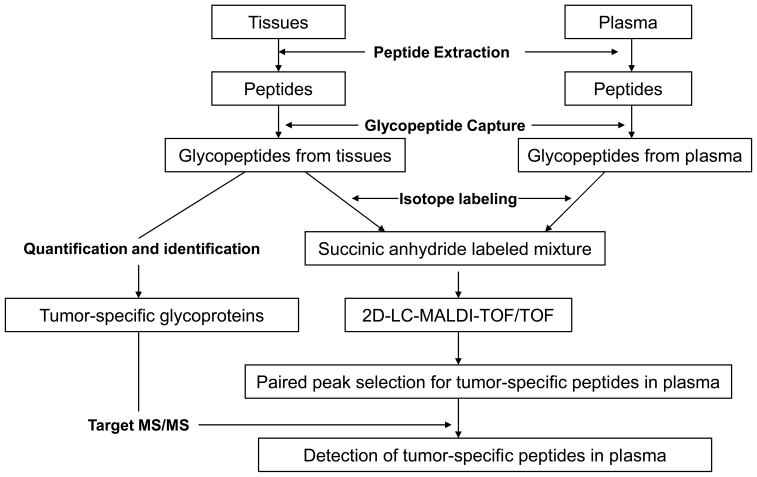
The strategy is schematically illustrated in Figure 1 and consists of four steps: 1) peptide extraction from tissue or plasma; 2) glycopeptide extraction: peptides that contain N-linked carbohydrates in extracellular proteins were isolated in their de-glycosylated form using a recently described solid-phase capture-and-release method 9, 10; 3) identification and quantification analysis of glycopeptides isolated from normal skin, papillomas, and carcinomas: isolated peptides were analyzed by LC-MS/MS and the peptides were identified and quantified using database search 21 and spectral count; 4) Detection of tissue-derived proteins in plasma. Glycopeptides from plasma samples taken from mice before and after development of skin tumors and tumor tissues were labeled with d013C0, d413C0, and d413C4 succinic anhydride respectively. The peptides containing d413C0 and d413C4 pairs indicated the tumor-derived peptides detected in plasma from tumor-bearing mice, and they were selected for MS/MS analysis for peptide identifications.
Figure 1.
Flow chart for detection of tumor-specific proteins in plasma
Identification of proteins from mouse model of skin cancer
To detect tumor-specific proteins in plasma, we first identified tumor-associated proteins from cancer (carcinomas) and benign (pipallomas) tissues. These tumor-associated proteins are likely to be secreted or shed to blood stream and fall into the detection range of current proteomic methodology.
To identify extracellular proteins from mouse skin tumors, four tissue samples each from normal skin, benign papillomas, and malignant carcinomas were collected to generate pooled normal, benign, and cancer tissues. Proteins were extracted from homogenized frozen tissues and digested to peptides. Glycopeptides were then captured using SPEG from each tissue. The N-linked glycopeptides were analyzed by LC- MS/MS by three repeated analyses for each sample. The MS/MS spectra were used to search protein database using SEQUEST 21. There were a total of 4764 peptide identifications with PeptideProphet of at least 0.9 (with error rate of 0.007) from all the tissues. 90% of these identifications (4284 identifications) contained a consensus N-linked glycosylation motif (N-X-S/T, X is any amino acid except proline). These identifications were from 463 unique glycosylation sites, representing 318 unique glycoproteins (Table 1). This indicated that the procedure was specific to N-linked glycoproteins. Therefore, we limited our subsequent analysis solely to the identified peptide sequences that contained at least one such consensus motif in order to reduce false positive rates. Since tissues are vascularized and some proteins identified from tissues are from contamination by common circulating blood proteins 13, 24. We next examined the glycoproteins identified from tissues to determine glycoproteins identified from tissues that were also identified from the normal mouse plasma 10, 25 and 59 glycoproteins were previously identified from normal mouse plasma and were not included for further study of skin cancer tissues.
Table 1.
Identified N-linked glycoproteins and glycosites.
| IPI | Protein Name | P | Identified Sequences |
|---|---|---|---|
| IPI00120245 | Integrin alpha-V | 1 | K.AN#TTQPGIVEGGQVLK.C |
| IPI00120245 | Integrin alpha-V | 1 | R.TAADATGLQPILNQFTPAN#VSR.Q |
| IPI00127447 | Lysosome membrane protein II | 1 | R.N#QSVGDPNVDLIR.T |
| IPI00127447 | Lysosome membrane protein II | 1 | T.GEDNYLN#FSK.I |
| IPI00127447 | Lysosome membrane protein II | 1 | R.TMVFPVMYLN#ESVLIDK.E |
| IPI00127447 | Lysosome membrane protein II | 1 | R.YKVPAEILAN#TSENAGF.C |
| IPI00322447 | RA175 | 1 | K.VSLTN#VSISDEGR.Y |
| IPI00322447 | RA175 | 1 | R.FQLLN#FSSSELK.V |
| IPI00118413 | Thrombospondin 1 | 1 | L.DNNVVN#GSSPAIR.T |
| IPI00118413 | Thrombospondin 1 | 1 | K.VSCPIMPCSN#ATVPDGECCPR.C |
| IPI00118413 | Thrombospondin 1 | 1 | W.PNENLVCVAN#ATYHCK.K |
| IPI00123678 | Cadherin-22 | 0.98 | R.ETAGWHN#ITVLAMEADN.H |
| IPI00154057 | Protocadherin 1 | 0.99 | N.DNAPFITAPSN#TSHR.L |
| IPI00126090 | Integrin alpha-3 | 1 | I.AMN#YSLPLR.M |
| IPI00126090 | Integrin alpha-3 | 1 | W.LECPLPDTSN#ITN#VTVK.A |
| IPI00132474 | Integrin beta-1 | 1 | R.NPCTSEQN#CTSPFSYK.N |
| IPI00132474 | Integrin beta-1 | 1 | R.KEN#SSEICSNNGECVCGQCVCR.K |
| IPI00132474 | Integrin beta-1 | 1 | K.DTCAQECSHFN#LTK.V |
| IPI00227969 | Integrin alpha-6 | 1 | K.YQTLN#CSVNVR.C |
| IPI00227969 | Integrin alpha-6 | 0.91 | R.VEQKN#NTFFDMNIF.E |
| IPI00320605 | Integrin beta-2 | 1 | K.LN#FTGPGEPDSLR.C |
| IPI00320605 | Integrin beta-2 | 0.99 | Y.LRPGQAAAFN#VTFR.R |
| IPI00415773 | Integrin alpha-M | 1 | R.TPVLN#CSVAVCK.R |
| IPI00415773 | Integrin alpha-M | 1 | V.GGPQDFN#MSVTLR.N |
| IPI00415773 | Integrin alpha-M | 1 | R.LN#YTLVGEPLR.S |
| IPI00132067 | Fibulin-2 | 1 | Y.QLPGCHGN#FSDAEEGDSER.Q |
| IPI00132067 | Fibulin-2 | 1 | K.DLDECALGTHN#CSEAETCHNIQGSFR.C |
| IPI00132067 | Fibulin-2 | 1 | K.SCVAGVMGAKEGETCGAEDN#DTCGVSLYK.A |
| IPI00223769 | CD44 antigen | 1 | R.TEAADLCQAFN#STLPTMDQMK.L |
| IPI00110810 | Prostate stem cell antigen | 1 | R.DCLNVQN#CSLDQHSCFTSR.I |
| IPI00110852 | Translocon-associated protein alpha, muscle specific isoform | 1 | K.DLNGNVFQDAVFN#QTVT.V |
| IPI00110852 | Translocon-associated protein alpha, muscle specific isoform | 1 | R.YPQDYQFYIQN#FTALPLNTVVPPQR.Q |
| IPI00112326 | Epithelial membrane protein 1 | 1 | K.N#CTGGNCDGSLSYGNEDAIK.A |
| IPI00113480 | Myeloperoxidase | 1 | R.ALMPFDSLHDDPCLLTN#R.S |
| IPI00111013 | Cathepsin D | 1 | K.YYHGELSYLN#VTR.K |
| IPI00111013 | Cathepsin D | 1 | K.N#GTSFDIHYGSGSL.S |
| IPI00128154 | Cathepsin L | 1 | R.AEFAVAN#DTGFVDIPQQEK.A |
| IPI00403938 | Tenascin C | 1 | L.EADTTQTVQN#LTVPGGLR.S |
| IPI00403938 | Tenascin C | 1 | R.EPEIGNLN#VSDVTPK.S |
| IPI00403938 | Tenascin C | 1 | R.LLQTAEHN#ISGAER.T |
| IPI00403938 | Tenascin C | 1 | N.NVEAAQN#LTVPGSLR.A |
| IPI00403938 | Tenascin C | 0.99 | N.NVETAHN#FTVPGNLR.A |
| IPI00403938 | Tenascin C | 1 | R.ESGLN#MTLPEENQPVVFNHIYNIK.L |
| IPI00403938 | Tenascin C | 1 | K.ASTEEVPSLEN#LTVT.E |
| IPI00403938 | Tenascin C | 1 | R.LN#YSLPTGQSMEVQLPK.D |
| IPI00108535 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 1 | R.FVPNSNMN#FTGQAYSGR.E |
| IPI00108535 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 1 | K.N#ITVLEPVTQPFLQVTN#TTVK.E |
| IPI00313428 | CEA-related cell adhesion molecule 2 | 1 | R.TLTLLN#VTR.N |
| IPI00122977 | Plasma protease C1 inhibitor | 1 | R.DTYVN#ASQSLYGSSPR.V |
| IPI00122977 | Plasma protease C1 inhibitor | 1 | K.VGQLQLSHN#LSFVIVVPVFPK.H |
| IPI00128689 | Collagen alpha 1(V) chain | 1 | K.VYCN#FTAGGSTCVFPDKK.S |
| IPI00130249 | GPI-anchored metastasis-associated protein homolog | 1 | A.N#VTVSLPVR.G |
| IPI00130249 | GPI-anchored metastasis-associated protein homolog | 1 | K.CQGSMPPVVNCYN#ASGR.V |
| IPI00130486 | FK506-binding protein 9 | 1 | R.YHYN#GTLLDGTLFDSSYSR.N |
| IPI00130486 | FK506-binding protein 9 | 1 | R.YHYN#GTFLDGTLFDSSHNR.M |
| IPI00132600 | Niemann-Pick C1 protein | 1 | R.LYN#VTHQFCN#ASVMDPTCVR.C |
| IPI00132600 | Niemann-Pick C1 protein | 1 | R.LIASN#ITETMR.S |
| IPI00131881 | ADAM 10 | 1 | R.IN#TTSDEKDPTNPFR.F |
| IPI00130342 | Lymphocyte antigen 6 complex locus G6C protein | 1 | K.LGLNYN#TTCCDK.D |
| IPI00130342 | Lymphocyte antigen 6 complex locus G6C protein | 1 | R.EVFN#ETNHK.L |
| IPI00133082 | CD177 antigen | 1 | K.VQGCMAQPDCNLLN#GTQAI.G |
| IPI00134549 | Lysosome-associated membrane glycoprotein 2 | 1 | A.LIVN#LTDSK.G |
| IPI00134549 | Lysosome-associated membrane glycoprotein 2 | 1 | K.VPFIFNINPATTN#FTGSCQPQSAQLR.L |
| IPI00134549 | Lysosome-associated membrane glycoprotein 2 | 1 | K.EVNVYMYLAN#GSAFN#ISNK.N |
| IPI00121430 | Collagen alpha 1(XII) chain | 1 | K.EAGN#ITTDGYEILGK.L |
| IPI00122272 | Extracellular matrix protein 1 | 1 | K.QIPGLIQN#MTVR.C |
| IPI00122272 | Extracellular matrix protein 1 | 1 | R.NVALVAGDTGN#ATGLGEQGPTR.G |
| IPI00122493 | FK506-binding protein 10 | 1 | R.YHYN#CSLLDGTR.L |
| IPI00122493 | FK506-binding protein 10 | 1 | R.YHYN#GSLMDGTLFDSSYSR.N |
| IPI00123342 | Hypoxia up-regulated 1 | 1 | R.VFGSQN#LTTVK.L |
| IPI00123342 | Hypoxia up-regulated 1 | 1 | R.LSALDNLLN#HSSIFLK.G |
| IPI00123342 | Hypoxia up-regulated 1 | 1 | K.EN#GTDAVQEEEESPAEGSK.D |
| IPI00123831 | SDR1 protein | 1 | K.ENGVFEEISN#SSGR.F |
| IPI00123831 | SDR1 protein | 1 | R.FFITNKEN#YTEL.S |
| IPI00123831 | SDR1 protein | 1 | R.ESLLPVTLQCN#LTSSSH.T |
| IPI00224728 | Cd63 antigen | 1 | K.DRVPDSCCIN#ITVGCGNDFK.E |
| IPI00462199 | Basigin | 1 | K.TSDTGEEEAITN#STEANGK.Y |
| IPI00462199 | Basigin | 1 | K.TQLTCSLN#SSGVDIVGHR.W |
| IPI00462199 | Basigin | 1 | K.SQLTISNLDVNVDPGTYVCN#ATNAQGTTR.E |
| IPI00308609 | VESICULAR INTEGRAL-MEMBRANE PROTEIN VIP36 | 1 | R.VFPYISVMVNN#GSLSYDHSK.D |
| IPI00308990 | Monocyte differentiation antigen CD14 | 1 | R.N#PSPDELPQVGN#LSLK.G |
| IPI00308785 | Prostaglandin G/H synthase 2 | 1 | R.TGFYGEN#CTTPEFLTR.I |
| IPI00308971 | Cation-independent mannose-6-phosphate receptor | 1 | K.ISTN#ITLVCKPGDLESAPVLR.A |
| IPI00308971 | Cation-independent mannose-6-phosphate receptor | 1 | R.SLLEFN#TTMGCQPSDSQHR.I |
| IPI00124836 | Beta-sarcoglycan | 1 | R.ITSN#ATSDLNIK.V |
| IPI00124836 | Beta-sarcoglycan | 0.99 | I.ILN#GTVMVSPTR.L |
| IPI00122737 | 222 kDa protein | 1 | R.QAEEAEEQANTN#LSK.F |
| IPI00122737 | 222 kDa protein | 0.98 | R.VQLLHSQN#TSLINQKK.K |
| IPI00119063 | AM2 receptor | 1 | K.LTSCATN#ASMCGDEAR.C |
| IPI00119063 | AM2 receptor | 1 | K.LNLDGSN#YTLLK.Q |
| IPI00119063 | AM2 receptor | 1 | A.VAN#DTNSCELSPCR.I |
| IPI00119063 | AM2 receptor | 1 | R.MGCQHHCVPTPSGPTCYCN#SSFQLE.A |
| IPI00119063 | AM2 receptor | 0.99 | R.GVTHLN#ISGLK.M |
| IPI00119063 | AM2 receptor | 1 | R.FN#STEYQVVTR.V |
| IPI00124265 | Latent transforming growth factor beta binding protein 4 | 1 | R.N#ATSVDSGAPGGAAPGGPGFR.A |
| IPI00124265 | Latent transforming growth factor beta binding protein 4 | 1 | R.CTPACDPGYQPTPGGGCQDVDECRN#R.S |
| IPI00129304 | Collectin sub-family member 12 | 1 | R.HTDDLTSLN#NTLVNIR.L |
| IPI00129304 | Collectin sub-family member 13 | 1 | K.ETLQN#NSFLITTVN#K.T |
| IPI00153959 | Stabilin-1 | 1 | H.ADLISN#MSQDELAR.I |
| IPI00153959 | Stabilin-1 | 1 | K.GFVDN#MTLSGPDLELH.A |
| IPI00316575 | Cathepsin K | 1 | Y.VGQDESCMYN#ATAK.A |
| IPI00126769 | Cathepsin F | 0.94 | K.VYIN#DSVELSR.N |
| IPI00121190 | Epidermal growth factor receptor | 1 | R.DIVQNVFMSN#MSMDLQSHPSSCPK.C |
| IPI00320420 | Clusterin | 1 | R.QELN#DSLQVAER.L |
| IPI00320420 | Clusterin | 0.99 | K.MLN#TSSLLEQLNDQFNWVSQLAN#LTQGEDK.Y |
| IPI00406459 | Arylsulfatase B | 1 | H.EACAPIESLN#GTR.C |
| IPI00406459 | Arylsulfatase B | 1 | R.IYAGMVSLMDEAVGN#VTK.A |
| IPI00409393 | Latent transforming growth factor beta binding protein, isoform 1L | 1 | R.YGQEQGTAPFQVSN#HTGR.I |
| IPI00409393 | Latent transforming growth factor beta binding protein, isoform 1L | 1 | Y.NLNDASLCDNVLAPN#VTK.Q |
| IPI00409393 | Latent transforming growth factor beta binding protein, isoform 1L | 0.91 | K.VCTN#GSCTNLEGSYM.C |
| IPI00108535 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 1 | R.EIIYSN#GSLLFQMITMK.D |
| IPI00117424 | Intercellular adhesion molecule 2 | 1 | K.IN#CSTNCAAPDMGGLETPTNK.I |
| IPI00122971 | N-CAM 180 of Neural cell adhesion molecule 1, 180 kDa isoform | 1 | R.DGQLLPSSN#YSNIK.I |
| IPI00406901 | Platelet/endothelial cell adhesion molecule | 1 | K.EETVLSQYQN#FSK.I |
| IPI00115976 | Integrin alpha-5 | 1 | K.VTGLSN#CTSN#YTPN.S |
| IPI00313479 | Integrin beta 4 Isoform 2 | 1 | K.TCN#CSTGSLSDTQPCLR.E |
| IPI00466371 | Integrin alpha 1 | 1 | K.DSCESNQN#ITCR.V |
| IPI00230432 | Fibulin-1 | 0.98 | H.SYN#SSLETIFIK.R |
| IPI00119756 | OX-2 membrane glycoprotein | 1 | K.GTGTGIEN#STESHFHSN#GTTSVTSILR.V |
| IPI00222589 | PTK7 protein tyrosine kinase 7 | 0.98 | R.MHIFQN#GSLVIH.D |
| IPI00314779 | TGF-beta receptor type III | 1 | R.AGVVVFN#CSLR.Q |
| IPI00112787 | Cell surface glycoprotein OX2 receptor | 1 | W.SPDGDCVTTSESHSN#GTVTVR.S |
| IPI00113528 | Transmembrane 9 superfamily protein member 3 | 1 | R.IVDVN#LTSEGK.V |
| IPI00114304 | Thrombospondin-3 | 1 | R.LGFLGN#QSQGCVPAR.T |
| IPI00119809 | Mama protein | 1 | R.ALGYEN#ATQALGR.A |
| IPI00119809 | Mama protein | 1 | K.GLN#LTEDTYKPR.L |
| IPI00469218 | Lysosomal membrane glycoprotein 1 | 1 | R.LN#MTLPDALVPTFSISN#HSLK.A |
| IPI00469218 | Lysosomal membrane glycoprotein 2 | 1 | K.N#VTVVLR.D |
| IPI00120025 | Similar to KALLIKREIN 9 | 1 | R.LTPAVQPLN#LTESRPPVGTQ.C |
| IPI00338790 | Glandular kallikrein KLK13 | 1 | K.ILN#GTN#GTSGFLPGGYTCLPH.S |
| IPI00116993 | Tuberin | 0.94 | A.PKQGLN#NSPPVK.E |
| IPI00111550 | Mucin and cadherin-like protein | 1 | R.VTN#SSEFMMNK.D |
| IPI00108041 | Stromal interaction molecule 1 | 1 | R.LAVTN#TTMTGTVLK.M |
| IPI00108328 | Methylated-DNA-- protein-cysteine methyltransferase containing protein | 0.93 | M.ETTSLLLCIGN#NSSGIRSRHR.S |
| IPI00108811 | Glucosylceramidase | 1 | R.DLGPALAN#SSHDVK.L |
| IPI00109281 | Enabled protein homolog | 1 | W.ERTNTMN#GSK.S |
| IPI00109612 | Laminin, beta 2 | 1 | L.ASGN#VSGGVCDGCQHNTAGR.H |
| IPI00109727 | Thy-1 membrane glycoprotein | 1 | K.VLTLAN#FTTK.D |
| IPI00109908 | Ig gamma-2A chain C region, membrane-bound form | 1 | R.EDYN#STLR.V |
| IPI00111014 | Elongation of very long chain fatty acids protein 4 | 1 | T.AFN#DTVEFYR.W |
| IPI00111115 | Similar to METASTASIS-ASSOCIATED GPI- ANCHORED PROTEIN | 1 | R.MNIGN#FSVPVYIR.T |
| IPI00111960 | Lysosomal alpha-glucosidase | 1 | R.GVFITN#ETGQPLIGK.V |
| IPI00112176 | Copper homeostasis protein cutC homolog | 0.94 | R.N#SSVAMGASLAHSEYSLK.V |
| IPI00113057 | Plasma kallikrein | 1 | K.LQTPLN#YTEFQKPICLPSK.A |
| IPI00113797 | Napsin A | 0.96 | W.FN#LTGQDYVIK.I |
| IPI00113824 | Basement membrane-specific heparan sulfate proteoglycan core protein | 0.98 | K.LTVPSSQN#SSFR.L |
| IPI00113824 | Basement membrane-specific heparan sulfate proteoglycan core protein | 1 | R.SLTQGSLIVGNLAPVN#GTSQGK.F |
| IPI00113824 | Basement membrane-specific heparan sulfate proteoglycan core protein | 1 | R.VAQQDSGQYICN#ATNSAGH.T |
| IPI00113853 | Desmocollin-3 | 0.99 | K.AN#FTILK.G |
| IPI00113854 | Eosinophil peroxidase | 0.99 | F.DNLHEDPCLLTN#R.S |
| IPI00114065 | Complement factor B | 1 | K.IVLDPSGSMNIYLVLDGSDSIGSSN#FTGAK.R |
| IPI00114065 | Complement factor B | 0.94 | R.SPFYN#LSDQI.S |
| IPI00114206 | Prothrombin | 1 | R.WVLTAAHCILYPPWDKN#FTENDLLVR.I |
| IPI00114206 | Prothrombin | 1 | R.ITDNMFCAGFKVN#DTK.R |
| IPI00400016 | Laminin gamma-1 chain | 1 | K.LLNN#LTSIK.I |
| IPI00400016 | Laminin gamma-1 chain | 1 | R.TLAGEN#QTALEIEELNR.K |
| IPI00400016 | Laminin gamma-1 chain | 1 | L.SYGQN#LSFSFR.V |
| IPI00400016 | Laminin gamma-1 chain | 1 | R.KYEQAKN#ISQDLEKQ.A |
| IPI00317340 | Lactotransferrin | 1 | I.PMGLLAN#QTR.S |
| IPI00317340 | Lactotransferrin | 1 | K.N#SSNFHLNQLQGLR.S |
| IPI00113539 | Fibronectin | 1 | R.DQCIVDDITYNVN#DTFHK.R |
| IPI00113539 | Fibronectin | 1 | K.LDAPTNLQFVN#ETDR.T |
| IPI00113539 | Fibronectin | 1 | R.HEEGHMLN#CTCFGQGR.G |
| IPI00119818 | Inter alpha-trypsin inhibitor, heavy chain 4 | 1 | K.AFITN#FSMIIDGVTYPGVVK.E |
| IPI00119818 | Inter alpha-trypsin inhibitor, heavy chain 5 | 1 | R.GLMLLLN#DTQHFSNNVK.G |
| IPI00114256 | Synaptophysin-like protein | 1 | K.N#QTVTATFGYPFR.L |
| IPI00114319 | Extracellular superoxide dismutase [Cu-Zn] | 1 | R.LEAYFSLEGFPAEQN#ASNR.A |
| IPI00114641 | CD98 heavy chain | 1 | K.LMNAPLYLAEWQN#ITK.N |
| IPI00114810 | Suppressor of tumorigenicity 14 | 0.99 | R.VIN#QTTCEDLMPQQITPR.M |
| IPI00114958 | HMW of Kininogen-1 | 1 | K.HSIEHFNN#NTDHSHLFTLR.K |
| IPI00114958 | HMW of Kininogen-1 | 1 | T.YTIVQTN#CSK.E |
| IPI00114958 | HMW of Kininogen-1 | 1 | K.IAN#FSQSCTLYSGDDLVEALPKPCPGCPR.D |
| IPI00115089 | Ectonucleoside triphosphate diphosphohydrolase 2 | 1 | R.LLN#LTSPEATAK.V |
| IPI00115516 | EMILIN-1 | 1 | R.FN#STLGPSEEQEK.N |
| IPI00115530 | Beta-hexosaminidase beta chain | 1 | K.TQVFGPVDPTVN#TTYA.F |
| IPI00115762 | Neural cell adhesion molecule L1 | 1 | K.EQLFFN#LSDPELR.T |
| IPI00115817 | PREDICTED: similar to ribosomal protein L21 | 0.95 | K.TGRVYN#VTQHAMGIIVNK.Q |
| IPI00115854 | TROP2 protein | 1 | R.AFN#HSDLDSELR.R |
| IPI00116105 | Corticosteroid-binding globulin | 1 | K.DLFTN#QSDFADTTK.D |
| IPI00116105 | Corticosteroid-binding globulin | 1 | R.EEDFYVN#ETSTVK.V |
| IPI00116105 | Corticosteroid-binding globulin | 1 | K.VPMMVQSGN#ISYFR.D |
| IPI00116105 | Corticosteroid-binding globulin | 1 | R.GSTQYLENLGFN#MSK.M |
| IPI00116599 | p130Cas-associated protein | 0.92 | R.RQVDEGMWPPPNNLLN#QSPK.K |
| IPI00116913 | Laminin alpha-5 chain | 1 | R.QLLAN#SSALEETILGHQGR.L |
| IPI00116913 | Laminin alpha-5 chain | 1 | H.N#FSGCISNVFVQR.L |
| IPI00116945 | Complement factor D | 1 | K.LSQN#ASLGPHVRPLPLQYEDK.E |
| IPI00117093 | Laminin beta-3 chain | 1 | R.QTACTPGDCPGELCPQDN#GTACGSHCR.G |
| IPI00117140 | Fc receptor, IgG, low affinity IIb | 1 | R.YHHYSSN#FSIPK.A |
| IPI00117735 | Myelin P0 protein | 1 | K.DGSIVIHNLDYSDN#GTFTCDVK.N |
| IPI00117831 | Ceruloplasmin | 1 | K.EYEGAVYPDN#TTDFQR.A |
| IPI00117857 | Alpha-1-antitrypsin 1–6 | 1 | K.GDTHTQILEGLQFN#LTQTSEADIHK.S |
| IPI00117932 | Paired amphipathic helix protein Sin3a | 0.96 | P.DAN#SSVLLSKTTAEK.V |
| IPI00117957 | Asporin | 1 | R.ITDIEN#GTFANIPR.V |
| IPI00118011 | mannosidase, beta A, lysosomal | 0.97 | V.AEILFNN#VTIGK.T |
| IPI00118130 | Alpha-1-acid glycoprotein 1 | 1 | R.ESQTIGDQCVYN#STHLGFQR.E |
| IPI00118130 | Alpha-1-acid glycoprotein 1 | 1 | R.QAIQTMQSEFFYLTTNLIN#DTIELR.E |
| IPI00118130 | Alpha-1-acid glycoprotein 1 | 1 | R.EN#GTFSKYEGGVETFAHLIVLR.K |
| IPI00118191 | Receptor-type tyrosine-protein phosphatase N2 | 0.98 | K.VSANIQN#MTTADVIK.A |
| IPI00118385 | Glutamate [NMDA] receptor subunit zeta 1 | 1 | K.VICTGPN#DTSPGSPR.H |
| IPI00118437 | Complement component C8 gamma chain homolog | 1 | R.EAN#LTEDQILFFPK.Y |
| IPI00119004 | Hypothetical Lipolytic enzyme, G-D-S-L containing protein | 0.91 | R.KGPGMENPVAVTIFFGAN#DSSLK.D |
| IPI00119299 | Leukemia inhibitory factor receptor | 1 | K.VVLAGSN#MTICCMSPTK.V |
| IPI00119299 | Leukemia inhibitory factor receptor | 1 | R.IEGLTN#ETYR.L |
| IPI00119299 | Leukemia inhibitory factor receptor | 1 | R.LGVQMHPGQEIHN#FTLTGR.N |
| IPI00119522 | Carboxypeptidase N, polypeptide 2 homolog | 1 | R.LQDLEITGSPVSN#LSAHIFSN#LSSLEK.L |
| IPI00119627 | Insulin receptor substrate 1 | 0.93 | K.LLPCTGDYMN#MSPVGDSN#TS.S |
| IPI00120187 | Fibromodulin | 0.97 | R.VPNNALEGLEN#LT.A |
| IPI00120751 | Proton myo-inositol transporter homolog | 1 | K.IN#GSAVIDSSCVPVNK.A |
| IPI00120769 | Solute carrier family 29 (nucleoside transporters), member 1 | 1 | R.LDVSQN#VSSDTDQSCESTK.A |
| IPI00120848 | Mimecan | 0.95 | I.SSLTDDTFCKAN#DTR.Y |
| IPI00121038 | Versican core protein | 1 | R.FEN#QTCFPLPDSR.F |
| IPI00121120 | Procollagen, type V, alpha 2 | 1 | K.EASQN#LTYICR.N |
| IPI00121312 | MFIRE1 | 1 | K.IDLTDFEKN#SSFA.Q |
| IPI00121362 | F11r protein | 1 | R.AFMN#SSFTIDPK.S |
| IPI00121418 | Retinoblastoma-associated protein | 0.97 | K.QLEN#DTRIIEVLCKEHECNIDEVKN.V |
| IPI00121550 | Sodium/potassium-transporting ATPase beta-1 chain | 1 | K.LDWLGN#CSGLNDDSYGYR.E |
| IPI00121634 | High-affinity cationic amino acid transporter-1 | 0.94 | K.FLAKINN#RTKTPVIATVTSGAIAAVM.A |
| IPI00122293 | Prolargin | 1 | R.VPVIPPRIHYLYLQNNFITELPLESFQN#ATGLR.W |
| IPI00122302 | Neutrophil elastase homolog | 0.91 | R.LGTNRPSPSVLQELN#VT.V |
| IPI00122368 | P2X4c receptor subunit | 1 | K.TSICDSDAN#CTLGSSDTHSSGIGTGR.C |
| IPI00122438 | Fibrillin-1 | 1 | K.AWGTPCELCPSVN#TSEYK.I |
| IPI00122438 | Fibrillin-1 | 1 | V.DTDECSVGNPCGN#GTCK.N |
| IPI00122438 | Fibrillin-1 | 1 | V.N#VTDYCQLVR.Y |
| IPI00122438 | Fibrillin-1 | 1 | R.NYYADN#QTCDGELLFN#MTK.K |
| IPI00122438 | Fibrillin-1 | 1 | R.N#CTDIDECR.I |
| IPI00123194 | Biglycan | 1 | R.MIEN#GSLSFLPTLR.E |
| IPI00123196 | Decorin | 1 | K.LGLSFNSITVMEN#GSLANVPHLR.E |
| IPI00123196 | Decorin | 1 | K.YIQVVYLHNNN#ISAVGQNDFCR.A |
| IPI00123223 | Murinoglobulin-1 | 1 | R.NYEVQLFHVN#ATVTEEGTGLEFSR.S |
| IPI00123223 | Murinoglobulin-1 | 1 | R.N#ASFVYTK.A |
| IPI00123824 | Amiloride-sensitive sodium channel beta-subunit | 1 | K.GEPYSPCTMN#GSDVAIK.N |
| IPI00123957 | Cd97 protein | 1 | R.DFNPATVN#YTIQK.L |
| IPI00123996 | Neuropilin-1 | 1 | K.RGPECSQN#YTAPTGVIK.S |
| IPI00124283 | Macrophage scavenger receptor types I and II | 1 | R.VLNN#ITNDLR.L |
| IPI00124640 | Osteoclast-like cell cDNA, clone:I420031M06 product:granulin | 1 | K.SDTPCDDFTRCPTN#NTCCK.L |
| IPI00124830 | Leukocyte surface antigen CD47 | 1 | I.EFTSCN#ETVVIPCIVR.N |
| IPI00125058 | Laminin alpha-3 chain | 1 | K.IESINQQLLPLGN#ISDNVDR.I |
| IPI00125058 | Laminin alpha-3 chain | 0.99 | K.TTFNLN#TTEVEPCRR.R |
| IPI00125266 | Acid ceramidase | 1 | R.SVLEN#TTSYEEAK.N |
| IPI00125293 | Eosinophil cationic protein 1 | 0.97 | R.VHITVCN#ITSR.A |
| IPI00125310 | Complement C1q subcomponent, A chain | 1 | K.VLTNQESPYQN#HTGR.F |
| IPI00125325 | Peroxisomal 2,4-dienoyl-CoA reductase | 0.96 | F.RDHGGVIVN#ITATLSMR.G |
| IPI00125514 | Ectonucleoside triphosphate diphosphohydrolase 5 | 1 | R.GYLTSFEMFN#STFK.L |
| IPI00125877 | Hypothetical protein | 1 | N.YQN#NTEVIQGIR.T |
| IPI00125877 | Hypothetical protein | 1 | R.GLTFLKN#VSSTCAASPSTDILTFTIPPSFADIFLSK.S |
| IPI00126050 | Plasma glutamate carboxypeptidase | 1 | K.EVMNLLQPLN#VTK.V |
| IPI00126186 | Macrophage mannose receptor 1 | 1 | R.TSYCN#ESFYFLCK.K |
| IPI00126194 | Alpha-2-macroglobulin | 1 | K.N#ITSVVSPLGYLSIFTTDEHGLAN#ISIDTSN#FTAPFLR.V |
| IPI00126194 | Alpha-2-macroglobulin | 1 | R.IN#VSYTGERPSSNMVIVDVK.M |
| IPI00126194 | Alpha-2-macroglobulin | 1 | Y.LN#ETQQLTEAIK.S |
| IPI00126194 | Alpha-2-macroglobulin | 1 | K.VN#LSFPSAQSLPASDTHLK.V |
| IPI00126316 | Mast cell carboxypeptidase A | 1 | R.NQN#STCIGTDLNR.N |
| IPI00126834 | Vascular cell adhesion protein 1 | 1 | K.ETTIWVSPSPILEEGSPVN#LTCSSDGIPAPK.I |
| IPI00127280 | Myeloid bactenecin | 1 | K.DCDFLEDGEERN#CTGK.F |
| IPI00127352 | AMBP protein | 1 | K.EDSCQLN#YSEGPCLGMQER.Y |
| IPI00127560 | Transthyretin | 1 | K.TLGISPFHEFADVVFTAN#DSGHR.H |
| IPI00127672 | PREDICTED: hypothetical protein LOC66967 | 1 | K.LLPAFN#TTSGLPYPR.I |
| IPI00127856 | Alpha-1-acid glycoprotein 2 | 1 | R.EYHTIDDHCVYN#STHLGIQR.E |
| IPI00127856 | Alpha-1-acid glycoprotein 2 | 1 | D.PITN#ETLSWLSDK.W |
| IPI00127933 | Androgen binding protein alpha | 1 | R.KVDLFLN#GTTEEY.V |
| IPI00128249 | Alpha-2-HS-glycoprotein | 1 | R.RPFGVVYEMEVDTLETTCHALDPTPLAN#CSVR.Q |
| IPI00128249 | Alpha-2-HS-glycoprotein | 1 | R.CPLLTPFN#DTNVVHTVNTALAAFNTQNN#GTYFK.L |
| IPI00128484 | Hemopexin | 1 | R.VAEVEN#GTKPD.S |
| IPI00128484 | Hemopexin | 1 | R.SWSTVGN#CTAALR.W |
| IPI00128484 | Hemopexin | 1 | K.SLGPNTCSSN#GSSLYFIHGPNLYCYSSIDK.L |
| IPI00128484 | Hemopexin | 1 | M.DHN#GTMLFFK.G |
| IPI00128905 | Golgi phosphoprotein 2 | 1 | K.AVLVNN#ITTGEK.L |
| IPI00128989 | Vacuolar ATP synthase subunit S1 | 1 | A.IHPPVSYN#DTAPR.I |
| IPI00129158 | Tyrosine-protein phosphatase non-receptor type substrate 1 | 1 | R.GIAN#LSNFIR.V |
| IPI00129243 | Gamma-glutamyl hydrolase | 1 | K.LPLN#FTEGAR.K |
| IPI00129243 | Gamma-glutamyl hydrolase | 0.99 | L.ALEN#LTANFHK.W |
| IPI00129250 | Leucine-rich alpha-2-glycoprotein | 1 | L.SVEFSN#LTQLPAAALQGCPGLR.E |
| IPI00129250 | Leucine-rich alpha-2-glycoprotein | 1 | K.MFSQN#DTR.C |
| IPI00129359 | zinc finger protein 68 | 0.97 | K.ELAGIGNTCN#VSTNH.I |
| IPI00129965 | PREDICTED: similar to alpha-1-B glycoprotein | 1 | K.LLFVGPQHAGN#YSCR.Y |
| IPI00129966 | PREDICTED: similar to alpha-1-B glycoprotein | 0.99 | R.VYQPGN#YSCSYQTHGECTSSTPSR.I |
| IPI00129968 | Embigin | 1 | K.DDEPLETTGDFN#TTK.M |
| IPI00130010 | Complement factor H | 1 | K.DNSCVDPPHVPN#ATIVTR.T |
| IPI00130010 | Complement factor H | 1 | K.LTEFTHN#STMDYK.C |
| IPI00130010 | Complement factor H | 1 | R.TKCIN#GTINYPTCV.- |
| IPI00130015 | Dipeptidyl-peptidase I | 1 | R.ILTN#NSQTPILSPQEVVSCSPYAQGCDGGFPYLIAGK.Y |
| IPI00130483 | KH domain RNA binding protein QKI-5A | 0.96 | R.KDMYN#DTLN#GSTEK.R |
| IPI00130627 | Legumain | 0.97 | Y.DDIANSEEN#PTPGVVINRPN#GTDVYK.G |
| IPI00130630 | Glutamate carboxypeptidase II | 1 | K.VPYNVGPGFAGN#FSTQK.V |
| IPI00130654 | Afamin | 1 | P.TKPQDVDHFN#ATQK.F |
| IPI00130654 | Afamin | 1 | L.ADLVLGELCGVNTN#R.T |
| IPI00130661 | Tripeptidyl-peptidase I | 0.97 | K.DVGSGTTN#NSQACAQFLEQYFHNSDLTEFMR.L |
| IPI00130661 | Tripeptidyl-peptidase I | 1 | K.SSSHLPPSSYFN#ASGR.A |
| IPI00131114 | Type VI collagen alpha 3 subunit | 1 | R.GPPGVN#GTQGFQGCPGQR.G |
| IPI00131114 | Type VI collagen alpha 3 subunit | 1 | R.ALN#GSALYTGSSLDFVR.N |
| IPI00131114 | Type VI collagen alpha 3 subunit | 1 | R.QLINALQIN#NTAVGHALVLPAR.R |
| IPI00131137 | 9 kDa protein | 0.96 | K.GKAN#ASEDANNPAENGDAK.T |
| IPI00131209 | Keratin intermediate filament 16a | 1 | R.KTEELNKEVASNSDLIQSN#R.S |
| IPI00131366 | Keratin, type II cytoskeletal 6B | 1 | R.VPGLN#RSGFSSVSVCR.S |
| IPI00131526 | CD209 antigen-like protein B | 1 | R.IPIFQGQN#ESIQEK.I |
| IPI00131830 | Serine protease inhibitor A3K | 1 | K.NLINDYVSN#QTQGMIK.E |
| IPI00131830 | Serine protease inhibitor A3K | 1 | K.YTGN#ASALLILPDQGR.M |
| IPI00131951 | Serpin A12 | 0.91 | L.SLGAQN#STLEEIR.E |
| IPI00133035 | NAD(P)(+)--arginine ADP-ribosyltransferase | 1 | R.LGN#FTLAYSAKPETADNQR.V |
| IPI00133035 | NAD(P)(+)--arginine ADP-ribosyltransferase | 1 | K.GTSNDLVLQSIN#STCSYYECAFLGGLK.T |
| IPI00133172 | Serpin B11 | 1 | K.N#SSECSQVGVMHPDFR.A |
| IPI00133257 | Hematopoietic progenitor cell antigen CD34 | 1 | M.VLAN#STELPSK.L |
| IPI00133751 | Microfibril-associated glycoprotein 4 | 1 | R.VDLEDFEN#NTAYAK.Y |
| IPI00133751 | Microfibril-associated glycoprotein 4 | 1 | R.FN#GSVSFFR.G |
| IPI00134191 | Solute carrier family 2, facilitated glucose transporter member 3 | 1 | K.DFLN#YTLEER.L |
| IPI00134483 | Lectin lambda | 1 | R.PGACTN#ITMGVVCK.L |
| IPI00134483 | Lectin lambda | 1 | R.VTPVCN#ASLPAQR.W |
| IPI00134547 | Zinc finger autosomal protein | 0.98 | V.ELLDPN#NSICVPREK.M |
| IPI00134652 | Type VII collagen | 1 | K.LQILN#ASSDVLR.V |
| IPI00134808 | C4b-binding protein | 1 | R.LACLN#GTVLR.G |
| IPI00134808 | C4b-binding protein | 1 | R.LVGSPFIGCTVVN#K.T |
| IPI00136642 | Antithrombin-III | 1 | K.LGACN#DTLK.Q |
| IPI00136902 | Piccolo protein | 0.97 | Y.RRQISAVQPSIIN#LSAASSLGTPVTMDSK.T |
| IPI00136925 | Immunoglobulin J chain | 1 | R.EN#ISDPTSPLR.R |
| IPI00137177 | Lysosomal protective protein | 1 | R.LDPPCTN#TTAPSNYLNNPYVR.K |
| IPI00137987 | Zinc-alpha-2-glycoprotein | 1 | K.DTTGSHTFQGMFGCEITNN#R.S |
| IPI00138342 | Liver carboxylesterase N | 1 | R.FHSELN#ISESMIPAVIEK.Y |
| IPI00139788 | Serotransferrin | 1 | K.N#STLCDLCIGPLK.C |
| IPI00153187 | Sulfatase modifying factor 1 | 1 | K.FVN#STGYLTEAEK.F |
| IPI00153202 | Angiotensin-converting enzyme 2 | 0.99 | Y.FFVTSPQN#VSDVIPR.S |
| IPI00153258 | Protein Z-dependent protease inhibitor | 1 | R.ASQQLSN#ETSSFGFNLLR.K |
| IPI00153548 | Hypothetical protein | 0.9 | C.QFGVGTFANVFLFVYN#FSPISTGSK.Q |
| IPI00169815 | Procollagen, type VI, alpha 2 | 1 | R.GTFTDCALAN#MTQQIR.Q |
| IPI00169815 | Procollagen, type VI, alpha 2 | 1 | I.GYTN#FTLEK.N |
| IPI00169815 | Procollagen, type VI, alpha 2 | 1 | R.MALLQYGSQNQQQVAFPLTYN#VTTIHEALER.A |
| IPI00169815 | Procollagen, type VI, alpha 2 | 1 | R.N#MTLFSDLVAEK.F |
| IPI00169858 | Hypothetical protein LOC435366 | 0.97 | R.HERN#QSAEKPSEYTQHGKAFALHAHSHAQ.R |
| IPI00169896 | Choline transporter-like protein 2 | 1 | K.TCNPETFPLRN#ESLQCPTAR.C |
| IPI00221418 | Hypothetical Phospholipase D/Transphosphatidylase | 1 | K.VFIVPVGN#HSNIPFSR.V |
| IPI00221426 | Glucosamine (N-acetyl)-6-sulfatase | 1 | K.YYN#YTLSINGK.A |
| IPI00221456 | Synaptic vesicle glycoprotein 2 b | 0.96 | K.KVLSMSLAIN#ASFASLSSFVQGY.G |
| IPI00221833 | Hypothetical Zinc finger, C2H2 type containing protein | 0.96 | D.WMPNN#HSVILIDDFESPQK.L |
| IPI00223446 | Laminin alpha-4 chain | 1 | R.HVTDMN#STIHLLR.T |
| IPI00223987 | Insulin-regulated membrane aminopeptidase IRAP homolog | 1 | R.MAFDLIDYLKN#ETHTAPI.T |
| IPI00224456 | Sarcalumenin | 1 | A.PLIN#VTEPPR.V |
| IPI00224456 | Sarcalumenin | 1 | K.TN#VSKFDLPNR.E |
| IPI00224584 | Calsequestrin 2 | 1 | K.IDLFKPQIGVVN#VTDADSI.W |
| IPI00224654 | Hypothetical protein | 1 | R.AYIQDFQEFSKN#ISIMLGR.C |
| IPI00225355 | Target of Nesh-SH3 variant 1 | 0.99 | K.VHIN#TTSDSILLK.F |
| IPI00226310 | Hypothetical von Willebrand factor type A domain containing protein | 1 | R.DLSVFAPN#MTEIIK.D |
| IPI00226310 | Hypothetical von Willebrand factor type A domain containing protein | 1 | K.LGN#FSELATHN#QTFLK.K |
| IPI00226310 | Hypothetical von Willebrand factor type A domain containing protein | 0.99 | L.LDMAIN#GSQEDLDHLK.A |
| IPI00226790 | GPI transamidase component PIG-T | 0.92 | L.GLAN#DTDDYFLR.Y |
| IPI00226932 | Quinoprotein alcohol dehydrogenase structure containing protein | 1 | R.FINYN#QTVSR.M |
| IPI00227834 | Inter-alpha trypsin inhibitor, heavy chain 2 | 1 | K.GAFISN#FTMTVNGMTFTSSIK.E |
| IPI00227857 | Hepatocyte growth factor activator | 1 | R.FCNIVPTEHCFLGN#GTEYR.G |
| IPI00229117 | Tenascin-N | 1 | Y.ILTYQFPN#GTVK.E |
| IPI00230289 | Excitatory amino acid transporter 2 | 1 | K.VLVAPPSEEAN#TTK.A |
| IPI00266902 | PREDICTED: similar to type V P-type ATPase isoform 3 | 0.99 | K.VCDPNSDVCN#TTR.S |
| IPI00271166 | Huntington disease gene homolog | 0.94 | R.GYSLLPSITDVTMENN#LSR.V |
| IPI00271262 | Murinoglobulin-2 | 1 | K.ELIFYYLVMAQGSIIQTGN#HTHQVEPGEAPVK.G |
| IPI00272381 | Proline 4-hydroxylase, alpha 1 | 1 | K.DMSDGFISN#LTIQR.Q |
| IPI00279010 | Lu protein | 1 | F.VFLN#SSSTVVN#CSAR.G |
| IPI00279051 | RIKEN cDNA A930025J12 | 1 | R.LFQN#CSELYK.A |
| IPI00279079 | Fibrinogen beta chain | 1 | K.GTAGNALMDGASQLVGEN#R.T |
| IPI00281188 | 140 kDa protein | 0.99 | K.VLEPPHIN#GSEGPGEV.S |
| IPI00281344 | Hypothetical Glycosyl transferase, family 8 containing protein | 0.93 | R.TGVNSGVMLMN#MTR.M |
| IPI00308213 | Ig gamma-1 chain C region, membrane-bound form | 1 | R.EEQFN#STFR.S |
| IPI00308658 | Olfactomedin-like protein 3 | 1 | K.IYVLDGTQN#DTAFVFPR.L |
| IPI00309068 | E130014G12 product:Kaiso protein | 1 | K.EDLPSN#NT.A |
| IPI00309214 | Serum amyloid P-component | 1 | K.LIPHLEKPLQN#FTLCFR.T |
| IPI00309230 | Beta-glucuronidase | 0.98 | R.ITIAIN#NTLTPH.T |
| IPI00309999 | Laminin alpha-2 chain | 1 | R.LEQMTMNIN#LTGPLPAPYK.I |
| IPI00309999 | Laminin alpha-2 chain | 1 | K.LN#ETLGNQDK.T |
| IPI00309999 | Laminin alpha-2 chain | 1 | R.ICNQN#SSNPYQR.H |
| IPI00309999 | Laminin alpha-2 chain | 1 | K.VFQAESHAAQLN#DSSAVLDGILDEAK.N |
| IPI00309999 | Laminin alpha-2 chain | 1 | K.VCN#CSTVGSLASQCNVNTGQCSCHPK.F |
| IPI00309999 | Laminin alpha-2 chain | 1 | K.ILYGLEN#TTQELK.H |
| IPI00309999 | Laminin alpha-2 chain | 1 | K.YIGGGVCIN#CTHNTA.G |
| IPI00309999 | Laminin alpha-2 chain | 1 | Y.VGGLPIN#YTTR.R |
| IPI00309999 | Laminin alpha-2 chain | 1 | L.NLASNALITTN#ATCGEK.G |
| IPI00310049 | Carboxypeptidase B2 | 1 | K.EVHFFVN#ASDVDSVK.A |
| IPI00311808 | Transmembrane glycoprotein NMB | 1 | R.DLPIVFDVLIHDPSHFLN#DSAISYK.W |
| IPI00313900 | Lumican | 1 | K.LHINYNN#LTESVGPLPK.S |
| IPI00313900 | Lumican | 1 | R.LSHNELADSGVPGNSFN#ISSLLELDLSYNK.L |
| IPI00313900 | Lumican | 1 | K.AFEN#VTDLQWLILDHNLLENSK.I |
| IPI00313900 | Lumican | 1 | K.LGSFDGLVN#LTFIYLQHNQLK.E |
| IPI00316329 | keratin complex 2, basic, gene 1 | 1 | R.MSGECTPN#VSVSVSTSHTSMSGSSSR.G |
| IPI00318012 | T-cell immunomodulatory protein | 1 | V.PCNN#ASCEEVHR.M |
| IPI00318595 | Adipocyte-derived leucine aminopeptidase | 1 | L.N#SSHPVSTPVENPAQIR.E |
| IPI00319814 | Suprabasal-specific protein suprabasin | 1 | K.EANQLLN#GSHQGQGGYGGQHGGAATT.T |
| IPI00320204 | RIKEN cDNA 2210023G05 | 1 | R.DGTSQPAICPQN#VTMNMEGLK.E |
| IPI00320675 | Complement factor I | 1 | R.WGEVDLIGN#CSQFYPDR.Y |
| IPI00320675 | Complement factor I | 1 | N.FN#VSLIYGR.T |
| IPI00321190 | Sulfated glycoprotein 1 | 1 | K.TN#SSFIQGFVDHVKEDCDR.L |
| IPI00321190 | Sulfated glycoprotein 1 | 1 | K.DN#ATQEEILHYLEK.T |
| IPI00322304 | Histidine-rich glycoprotein HRG | 1 | R.LPPLNIGEVLTLPEANFPSFSLPNCN#R.S |
| IPI00322463 | Beta-2-glycoprotein I | 1 | K.DYRPSAGN#NSLYQDTVVFK.C |
| IPI00322463 | Beta-2-glycoprotein I | 1 | K.N#ISFACNPGFFLN#GTSSSK.C |
| IPI00322575 | ATP-binding cassette transporter sub-family A member 8a | 0.95 | K.NTQNILVQN#LSGGQKRK.L |
| IPI00330747 | 5730439E10Rik protein | 0.94 | R.YLMGN#NSSEDSFLTANTVQPLAETGLQLSK.R |
| IPI00331214 | Platelet glycoprotein IV | 1 | R.QFWIFDVQNPDDVAKN#SSK.I |
| IPI00331214 | Platelet glycoprotein IV | 1 | K.DPFLSLVPYPISTTVGVFYPYN#DTVDGVYK.V |
| IPI00331214 | Platelet glycoprotein IV | 1 | K.VISNN#CTSYGVLDIGK.C |
| IPI00331214 | Platelet glycoprotein IV | 1 | K.RPYIVPILWLN#ETGTIGDEK.A |
| IPI00331214 | Platelet glycoprotein IV | 1 | K.EN#ITQDPEDH.T |
| IPI00331214 | Platelet glycoprotein IV | 1 | R.N#LSYWPSYCDMIN#GTDAASFPPFVEK.S |
| IPI00331259 | Desmoglein-1 gamma | 1 | K.LN#ATDADEPNNLNSMIAFK.I |
| IPI00331617 | Hypothetical olfactomedin-like domain containing protein | 1 | R.VDKLEEEVSKN#LTK.E |
| IPI00338565 | Mutant fibrillin-1 | 1 | R.VLPFN#VTDYCQLVR.Y |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | 1 | K.MEMPSTPQQLQN#LTEDIR.E |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | 1 | K.QADEDIQGTQNLLTSIESETAASEETLTN#ASQR.I |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | 1 | L.ATGN#VSGGVCDNCQHNTMGR.N |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | 1 | R.VN#ASTTDPN#STVEQSALTR.D |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | 1 | K.LTDTASQSN#STAGELGALQAEAESLDK.T |
| IPI00339885 | Collagen alpha 1(VI) chain | 1 | R.AALQFLQN#YTVL.A |
| IPI00339885 | Collagen alpha 1(VI) chain | 1 | L.DDGFLKN#ITAQICIDKK.C |
| IPI00340463 | PREDICTED: similar to hypothetical protein A030003A19 | 1 | K.LLNDYVSN#QTQGMIK.E |
| IPI00346978 | Spink5 protein | 0.99 | E.TNKNSASRSN#GTGSATGKDVCDQFR.S |
| IPI00346978 | Spink5 protein | 0.96 | K.GNQDPCMKFQAQMKN#GTLTCPK.G |
| IPI00348602 | Weakly similar to Zinnc finger protein GLI4 | 0.98 | R.FRN#SSNLARHR.R |
| IPI00350715 | PREDICTED: similar to protocadherin 9 | 1 | R.IDPVTGN#ITLEEKPAPTDVGLHR.L |
| IPI00355606 | PREDICTED: expressed sequence AL022779 | 0.96 | M.QN#NSVFGDLK.S |
| IPI00378430 | Ortholog of human Ras association | 0.94 | R.QETNMAN#FSYR.F |
| IPI00381122 | Weakly similar to Tiarin | 1 | K.IN#LTTNVVDVNRPLPL.A |
| IPI00403586 | Hypothetical Lipolytic enzymes | 1 | M.IVNN#HTSLDVER.A |
| IPI00405742 | Plexin B2 | 0.95 | K.QDLALSGN#LSSLYAMTQDK.V |
| IPI00406434 | Mini-agrin | 1 | K.NELMLN#SSLMR.I |
| IPI00407222 | PREDICTED: similar to human KIAA1815 protein | 0.99 | H.IPEIN#DTIR.A |
| IPI00408344 | PREDICTED: similar to solute carrier family 4 member 11 | 0.94 | R.EDSLGDEVFDTVN#SSIVSGESIR.F |
| IPI00409148 | Haptoglobin | 1 | K.NLFLN#HSETASAK.D |
| IPI00409148 | Haptoglobin | 1 | K.N#LTSPVGVQPILNEHTFCAGLTK.Y |
| IPI00409148 | Haptoglobin | 1 | K.CVVHYEN#STVPEK.K |
| IPI00409148 | Haptoglobin | 1 | K.VVLHPN#HSVVDIGLIK.L |
| IPI00410951 | Thyroxine-binding globulin homolog | 1 | K.VTTCHLPQQN#ATLYK.M |
| IPI00420489 | Von Willebrand factor | 1 | V.LEGSDEVGEANFN#K.S |
| IPI00420955 | Sortilin 1 | 1 | K.DITNLIN#NTFIR.T |
| IPI00453607 | Killer cell inhibitory receptor-like protein p91A | 1 | R.LSVLPSPVVTAGGN#MTLH.C |
| IPI00458917 | Sodium/glucose cotransporter 1 | 1 | K.VSNGN#FTAK.E |
| IPI00459432 | Kidney predominant protein | 1 | S.ADFQGRPVDDPTGAFAN#GSLTFK.V |
| IPI00460063 | Prenylcysteine oxidase | 1 | K.GELN#STLFSSRPK.D |
| IPI00461281 | NudC domain containing protein 2 | 1 | K.ENPGFDFSGAEISGN#YTK.G |
| IPI00462999 | Ahi-1 isoform III | 0.92 | D.EFVNTEN#NSSR.K |
| IPI00463311 | PREDICTED: similar to RIKEN cDNA E330026B02 | 0.99 | R.DLGMFAPN#MTR.I |
| IPI00467180 | Translocon-associated protein beta subunit | 1 | R.IAPASN#VSHTVVLRPLK.A |
| IPI00467944 | 61 kDa protein | 1 | K.VVN#VSELYGTPCTK.R |
| IPI00468097 | 340 kDa protein | 1 | R.NLQVYN#ATSNSLTVK.W |
| IPI00469000 | Zinc transporter SLC39A6 | 0.98 | R.NTNDNIQECFN#TTK.L |
| IPI00469387 | GUGU alpha | 1 | R.VLYLPAYN#CTLRPVSK.R |
| IPI00469387 | GUGU alpha | 1 | R.SPPAPPLPQRPLSPLHPLGCN#DSEVLAVAGFALQNINR.D |
| IPI00469542 | Histidine-rich calcium-binding protein | 1 | R.EVGEEN#VSEEVFR.G |
| IPI00469839 | 19 kDa protein | 0.91 | K.TRTIDVVYN#ASNNELVCTK.T |
| IPI00471081 | RIKEN cDNA 1100001H23 | 1 | K.NGDAYGYYN#DSIK.T |
| IPI00471273 | Apoptosis-related protein 3 | 1 | A.LPEICTLCPGGMHN#LSR.V |
| IPI00473625 | PREDICTED: laminin, alpha 3 | 0.98 | R.FN#ISTPAFQGCMK.N |
| IPI00473830 | Biliary glycoprotein | 1 | R.MTLSQN#NSILR.I |
| Dolichyl-diphosphooligosaccharide--protein | |||
| IPI00475154 | glycosyltransferase 63 kDa | 1 | Q.VLSGCEISVSN#ETK.E |
| IPI00475157 | Serpina1b protein | 1 | R.ELVHQSN#TSNIFFSPVSIATAFAMLSLGSK.G |
| IPI00475157 | Serpina1b protein | 1 | N.ASAVFLLPEDGK.M |
| IPI00551354 | PREDICTED: ring finger and KH domain containing 3 | 0.91 | R.N#GSGGGGGGGGGGGGGGSGGGETLDDQR.A |
| IPI00553278 | H-2D cell surface glycoprotein | 1 | R.NLLGYYN#QSAGGSHTLQQM.S |
| IPI00554833 | Eosinophil-associated ribonuclease 12 | 1 | V.GVCGN#PSGLCSDN#ISQNCHN#SSSR.V |
| IPI00606550 | Ig gamma-2B chain C region, membrane-bound form | 1 | R.EDYN#STIR.V |
| IPI00607976 | Serine (or cysteine) proteinase inhibitor, clade A, member 3A | 1 | K.FN#LTETPEADIH.Q |
| IPI00621319 | 43 kDa protein | 0.92 | K.RLFLLDLLN#ATGK.D |
| IPI00624663 | Pzp protein | 0.99 | K.ACVSLNHVN#ETVM.L |
| IPI00624761 | 44 kDa protein | 1 | R.PVDDPTGAFAN#GSLTFK.V |
| IPI00626315 | 38 kDa protein | 0.94 | P.PSSTDLLWSILN#ASALALLYKTQRDN#ASESK.D |
| IPI00627061 | MKIAA4087 protein | 0.94 | R.CNIN#GSFSEICHTR.T |
| IPI00649090 | Adult male thymus cDNA, clone:5830446P09 product:CD72 antigen | 0.96 | V.GSEQPTATWSSVN#SSALRQIPR.C |
| IPI00649281 | 52 kDa protein | 0.98 | R.YHYN#GTLLDGTAFDNSYSR.N |
| IPI00654271 | Myosin light chain, regulatory B | 0.91 | K.N#PTDAYLDAMMNEAPAPIN#FTMFL.T |
| IPI00654907 | Hypothetical protein CEACAM1/2sec | 1 | R.FHVHQPVTQPFLQVTN#TTVK.E |
P: peptide probability
N#: N-linked glycosylation site
To identify skin tumor-specific proteins, we compared the glycoproteins identified from normal skin, benign, and malignant tumors. Despite the same amount of glycopeptides from each tissue type were analyzed with the same procedures, the number of unique glycosites identified from different tissue types was different. A total of 405 glycosites were identified in cancer tissue, while 252 in benign tissue and 112 in normal skin when using PeptideProphet score of ≥0.9. The number of glycoproteins identified from papillomas and carcinoma was higher than that of normal tissue. This could be caused by the increased expression of glycoproteins in tumor tissues. A similar observation was also reported from the proteomic analysis of tryptic peptides in mouse breast cancer model 24.
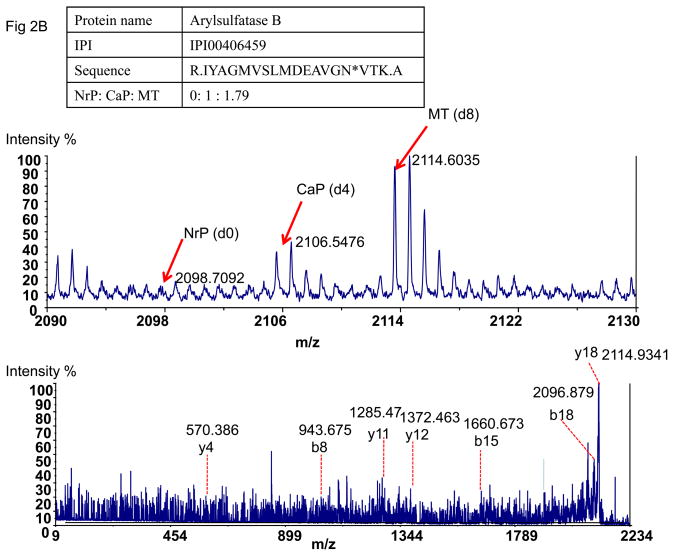
To determine the glycoprotein changes associated with cancer development, we calculated the relative protein abundance using the number of redundant MS/MS spectra from the same glycoprotein in different tissues. 26 To eliminate the spectral count due to random events, only proteins identified with at least three spectra were included for quantitation. A number of proteins identified in this study were only detected in tumor tissues (benign or cancer) but not in normal tissues (the ratio of such proteins was arbitrary assigned to 100, Table 2). Among the 111 proteins identified with spectral count ratio at least 3-fold in cancer or benign tumor tissues comparing to normal tissues, 47 proteins (Table 2) were increased at least 3 folds in cancer tissues comparing to benign tissues. Some of these have been reported to play roles in skin cancer development. These include most of known extracellular proteins such as thrombospondin, cathepsins, epidermal growth factor receptor, cell adhesion molecules, cadherins, integrins, tuberin, fibulin, TGFβ receptor, etc. Tenascin-C is an extracellular matrix glycoprotein, and plays multiple functions in cell adhesion, migration, growth and angiogenesis 27, 28. Tenascin-C has many cell surface receptors, such as intergrin, EGFR etc., which may affect genome stability associated with interference with genome safeguard functions and escape from cell cycle checkpoints 28. Tenascin-C has twenty potential N-linked glycosylation sites but only one glycosylation site (LLQTAEHN#ISGAER, Table 1) has been identified previously (Swiss-Prot Protein knowledgebase, http://us.expasy.org/sprot). In this study, eight N-linked glycosites including the previously identified site were identified in carcinomas (Table 1). They showed increased expression in carcinomas compared to papillomas (Table 2). This observation indicated that Tenascin-C might have increased its glycosylation or abundance during tumor development. In addition, 20 glycoproteins were identified in skin cancer only (Table 2) and these proteins might be used as protein markers to discriminate the malignant and benign tumors. An example of these proteins is Arylsulfatase B. In this study, Arylsulfatase B was identified three times only in cancer tissues with two unique glycosylation sites. Arylsulfatase B is lysosomal enzyme and can degrade proteoglycans in the extracellular matrix and basement membrane. In this way, preteoglycans can obstruct the cancer cell spread. Therefore, Arylsulfatase B plays a key role of accelerating cancer cell migration 29.
Table 2.
Glycoproteins upregulated in skin tumors
| IPI | Protein Name | Protein Location | Ca | Pa | Nr | Total | Ca/Nr | Pa/Nr | Ca/Pa |
|---|---|---|---|---|---|---|---|---|---|
| IPI00119063 | AM2 receptor | Transmembrane | 17 | 0 | 0 | 17 | 100.0 | 100.0 | 100.0 |
| IPI00381122 | Weakly similar to Tiarin | Cell Surface | 11 | 0 | 0 | 11 | 100.0 | 100.0 | 100.0 |
| IPI00308971 | Cation-independent mannose-6-phosphate receptor | Transmembrane | 8 | 0 | 0 | 8 | 100.0 | 100.0 | 100.0 |
| IPI00124265 | Latent transforming growth factor beta binding protein 4 | Secreted | 7 | 0 | 0 | 7 | 100.0 | 100.0 | 100.0 |
| IPI00129304 | Collectin sub-family member 12 | Transmembrane | 7 | 0 | 0 | 7 | 100.0 | 100.0 | 100.0 |
| IPI00129968 | Embigin | Transmembrane | 7 | 0 | 0 | 7 | 100.0 | 100.0 | 100.0 |
| IPI00153959 | Stabilin-1 | Transmembrane | 5 | 0 | 0 | 5 | 100.0 | 100.0 | 100.0 |
| IPI00316575 | Cathepsin K | Cell Surface | 4 | 0 | 0 | 4 | 100.0 | 100.0 | 100.0 |
| IPI00321190 | Sulfated glycoprotein 1 | Secreted | 4 | 0 | 0 | 4 | 100.0 | 100.0 | 100.0 |
| IPI00475154 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa | Transmembrane | 4 | 0 | 0 | 4 | 100.0 | 100.0 | 100.0 |
| IPI00308785 | Prostaglandin G/H synthase 2 | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00122737 | 222 kDa protein | Intracellular | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00406459 | Arylsulfatase B | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00409393 | Latent transforming growth factor beta binding protein, isoform 1L | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00119809 | Mama protein | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00111960 | Lysosomal alpha-glucosidase | Transmembrane | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00118011 | Mannosidase, beta A, lysosomal | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00121120 | Procollagen, type V, alpha 2 | Secreted | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00129158 | Tyrosine-protein phosphatase non-receptor type substrate 1 | Transmembrane | 3 | 0 | 0 | 3 | 100.0 | 100.0 | 100.0 |
| IPI00120769 | Solute carrier family 29 (nucleoside transporters), member 1 | Transmembrane | 5 | 0 | 1 | 6 | 5.0 | 0.0 | 100.0 |
| IPI00415773 | Integrin alpha-M | Transmembrane | 34 | 3 | 1 | 38 | 34.0 | 3.0 | 11.3 |
| IPI00338785 | CDNA, clone:M5C1012G13 product:laminin B1 subunit 1 | Intracellular | 9 | 1 | 3 | 13 | 3.0 | 0.3 | 9.0 |
| IPI00130627 | Legumain | Secreted | 17 | 2 | 0 | 19 | 100.0 | 100.0 | 8.5 |
| IPI00113480 | Myeloperoxidase | Secreted | 8 | 1 | 0 | 9 | 100.0 | 100.0 | 8.0 |
| IPI00113824 | Basement membrane-specific heparan sulfate proteoglycan core protein | Cell Surface | 15 | 2 | 5 | 22 | 3.0 | 0.4 | 7.5 |
| IPI00124830 | Leukocyte surface antigen CD47 | Transmembrane | 7 | 1 | 1 | 9 | 7.0 | 1.0 | 7.0 |
| IPI00320605 | Integrin beta-2 | Transmembrane | 20 | 3 | 0 | 23 | 100.0 | 100.0 | 6.7 |
| IPI00308990 | Monocyte differentiation antigen CD14 | Cell Surface | 13 | 2 | 0 | 15 | 100.0 | 100.0 | 6.5 |
| IPI00133082 | CD177 antigen | Secreted | 6 | 1 | 0 | 7 | 100.0 | 100.0 | 6.0 |
| IPI00130486 | FK506-binding protein 9 | Cell Surface | 5 | 1 | 0 | 6 | 100.0 | 100.0 | 5.0 |
| IPI00308609 | VESICULAR INTEGRAL-MEMBRANE PROTEIN VIP36 | Transmembrane | 5 | 1 | 0 | 6 | 100.0 | 100.0 | 5.0 |
| IPI00403938 | Tenascin C | Cell Surface | 133 | 29 | 0 | 162 | 100.0 | 100.0 | 4.6 |
| IPI00462199 | Basigin | Transmembrane | 13 | 3 | 0 | 16 | 100.0 | 100.0 | 4.3 |
| IPI00120245 | Integrin alpha-V | Transmembrane | 8 | 2 | 0 | 10 | 100.0 | 100.0 | 4.0 |
| IPI00120245 | Integrin alpha-V | Transmembrane | 8 | 2 | 0 | 10 | 100.0 | 100.0 | 4.0 |
| IPI00110852 | Translocon-associated protein alpha, muscle specific isoform | Cell Surface | 4 | 1 | 0 | 5 | 100.0 | 100.0 | 4.0 |
| IPI00125266 | Acid ceramidase | Secreted | 4 | 1 | 0 | 5 | 100.0 | 100.0 | 4.0 |
| IPI00121038 | Versican core protein | Cell Surface | 11 | 3 | 2 | 16 | 5.5 | 1.5 | 3.7 |
| IPI00124283 | Macrophage scavenger receptor types I and II | Transmembrane | 7 | 2 | 0 | 9 | 100.0 | 100.0 | 3.5 |
| IPI00132067 | Fibulin-2 | Secreted | 31 | 9 | 0 | 40 | 100.0 | 100.0 | 3.4 |
| IPI00223769 | CD44 antigen | Transmembrane | 10 | 3 | 0 | 13 | 100.0 | 100.0 | 3.3 |
| IPI00127447 | Lysosome membrane protein II | Transmembrane | 32 | 10 | 0 | 42 | 100.0 | 100.0 | 3.2 |
| IPI00322447 | RA175 | Transmembrane | 6 | 2 | 0 | 8 | 100.0 | 100.0 | 3.0 |
| IPI00154057 | Protocadherin 1 | Cell Surface | 3 | 1 | 0 | 4 | 100.0 | 100.0 | 3.0 |
| IPI00121312 | MFIRE1 | Secreted | 3 | 1 | 0 | 4 | 100.0 | 100.0 | 3.0 |
| IPI00124640 | Osteoclast-like cell cDNA, clone:I420031M06 product:granulin | Secreted | 3 | 1 | 0 | 4 | 100.0 | 100.0 | 3.0 |
| IPI00134483 | Lectin lambda | Cell Surface | 3 | 1 | 1 | 5 | 3.0 | 1.0 | 3.0 |
| IPI00272381 | Proline 4-hydroxylase, alpha 1 | Secreted | 17 | 6 | 0 | 23 | 100.0 | 100.0 | 2.8 |
| IPI00122493 | FK506-binding protein 10 | Secreted | 7 | 3 | 0 | 10 | 100.0 | 100.0 | 2.3 |
| IPI00123831 | SDR1 protein | Transmembrane | 11 | 5 | 0 | 16 | 100.0 | 100.0 | 2.2 |
| IPI00224728 | Cd63 antigen | Transmembrane | 8 | 4 | 0 | 12 | 100.0 | 100.0 | 2.0 |
| IPI00128689 | Collagen alpha 1(V) chain | Secreted | 6 | 3 | 0 | 9 | 100.0 | 100.0 | 2.0 |
| IPI00125877 | Hypothetical protein | Transmembrane | 6 | 3 | 0 | 9 | 100.0 | 100.0 | 2.0 |
| IPI00130015 | Dipeptidyl-peptidase I | Secreted | 4 | 2 | 0 | 6 | 100.0 | 100.0 | 2.0 |
| IPI00318012 | T-cell immunomodulatory protein | Transmembrane | 4 | 2 | 0 | 6 | 100.0 | 100.0 | 2.0 |
| IPI00123678 | Cadherin-22 | Transmembrane | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00126316 | Mast cell carboxypeptidase A | Secreted | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00130661 | Tripeptidyl-peptidase I | Cell Surface | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00131366 | Keratin, type II cytoskeletal 6B | Transmembrane | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00221418 | hypothetical Phospholipase D/Transphosphatidylase | Transmembrane | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00279051 | RIKEN cDNA A930025J12 | Transmembrane | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00554833 | Eosinophil-associated ribonuclease 12 | Secreted | 2 | 1 | 0 | 3 | 100.0 | 100.0 | 2.0 |
| IPI00127280 | Myeloid bactenecin | Secreted | 43 | 22 | 0 | 65 | 100.0 | 100.0 | 2.0 |
| IPI00118413 | Thrombospondin 1 | Secreted | 20 | 11 | 0 | 31 | 100.0 | 100.0 | 1.8 |
| IPI00127352 | AMBP protein | Secreted | 22 | 14 | 0 | 36 | 100.0 | 100.0 | 1.6 |
| IPI00132600 | Niemann-Pick C1 protein | Transmembrane | 3 | 2 | 0 | 5 | 100.0 | 100.0 | 1.5 |
| IPI00137177 | Lysosomal protective protein | Secreted | 3 | 2 | 0 | 5 | 100.0 | 100.0 | 1.5 |
| IPI00132474 | Integrin beta-1 | Transmembrane | 18 | 13 | 1 | 32 | 18.0 | 13.0 | 1.4 |
| IPI00123342 | Hypoxia up-regulated 1 | Secreted | 19 | 14 | 0 | 33 | 100.0 | 100.0 | 1.4 |
| IPI00126090 | Integrin alpha-3 | Transmembrane | 4 | 3 | 0 | 7 | 100.0 | 100.0 | 1.3 |
| IPI00131881 | ADAM 10 | Cell Surface | 4 | 3 | 0 | 7 | 100.0 | 100.0 | 1.3 |
| IPI00406434 | Mini-agrin | Secreted | 4 | 3 | 0 | 7 | 100.0 | 100.0 | 1.3 |
| IPI00410951 | Thyroxine-binding globulin homolog | Secreted | 4 | 3 | 0 | 7 | 100.0 | 100.0 | 1.3 |
| IPI00125058 | Laminin alpha-3 chain | Secreted | 9 | 7 | 1 | 17 | 9.0 | 7.0 | 1.3 |
| IPI00112326 | Epithelial membrane protein 1 | Transmembrane | 6 | 5 | 0 | 11 | 100.0 | 100.0 | 1.2 |
| IPI00128154 | Cathepsin L | Secreted | 23 | 20 | 0 | 43 | 100.0 | 100.0 | 1.2 |
| IPI00121362 | F11r protein | Transmembrane | 9 | 9 | 0 | 18 | 100.0 | 100.0 | 1.0 |
| IPI00108535 | Carcinoembryonic antigen-related cell adhesion molecule 1 | Cell Surface | 7 | 7 | 0 | 14 | 100.0 | 100.0 | 1.0 |
| IPI00407222 | PREDICTED: similar to human KIAA1815 protein | Transmembrane | 6 | 6 | 0 | 12 | 100.0 | 100.0 | 1.0 |
| IPI00128989 | Vacuolar ATP synthase subunit S1 | Transmembrane | 5 | 5 | 0 | 10 | 100.0 | 100.0 | 1.0 |
| IPI00471081 | RIKEN cDNA 1100001H23 | Cell Surface | 5 | 5 | 0 | 10 | 100.0 | 100.0 | 1.0 |
| IPI00226932 | Quinoprotein alcohol dehydrogenase structure containing protein | Secreted | 4 | 4 | 0 | 8 | 100.0 | 100.0 | 1.0 |
| IPI00127672 | PREDICTED: hypothetical protein LOC66967 | Secreted | 2 | 2 | 0 | 4 | 100.0 | 100.0 | 1.0 |
| IPI00346978 | Spink5 protein | Secreted | 2 | 2 | 0 | 4 | 100.0 | 100.0 | 1.0 |
| IPI00469387 | GUGU alpha | Secreted | 23 | 23 | 3 | 49 | 7.7 | 7.7 | 1.0 |
| IPI00134549 | Lysosome-associated membrane glycoprotein 2 | Transmembrane | 8 | 9 | 0 | 17 | 100.0 | 100.0 | 0.9 |
| IPI00121430 | Collagen alpha 1(XII) chain | Secreted | 11 | 14 | 0 | 25 | 100.0 | 100.0 | 0.8 |
| IPI00122272 | Extracellular matrix protein 1 | Secreted | 11 | 14 | 0 | 25 | 100.0 | 100.0 | 0.8 |
| IPI00227969 | Integrin alpha-6 | Transmembrane | 6 | 8 | 1 | 15 | 6.0 | 8.0 | 0.8 |
| IPI00134652 | Type VII collagen | Secreted | 5 | 7 | 0 | 12 | 100.0 | 100.0 | 0.7 |
| IPI00114256 | Synaptophysin-like protein | Transmembrane | 10 | 14 | 3 | 27 | 3.3 | 4.7 | 0.7 |
| IPI00110810 | Prostate stem cell antigen | Secreted | 9 | 13 | 0 | 22 | 100.0 | 100.0 | 0.7 |
| IPI00467180 | Translocon-associated protein beta subunit | Transmembrane | 15 | 22 | 0 | 37 | 100.0 | 100.0 | 0.7 |
| IPI00133172 | Serpin B11 | Intracellular | 2 | 3 | 0 | 5 | 100.0 | 100.0 | 0.7 |
| IPI00111013 | Cathepsin D | Secreted | 19 | 30 | 0 | 49 | 100.0 | 100.0 | 0.6 |
| IPI00117093 | Laminin beta-3 chain | Cell Surface | 3 | 6 | 0 | 9 | 100.0 | 100.0 | 0.5 |
| IPI00130342 | Lymphocyte antigen 6 complex locus G6C protein | Secreted | 2 | 4 | 0 | 6 | 100.0 | 100.0 | 0.5 |
| IPI00125293 | Eosinophil cationic protein 1 | Secreted | 1 | 2 | 0 | 3 | 100.0 | 100.0 | 0.5 |
| IPI00320204 | RIKEN cDNA 2210023G05 | Secreted | 1 | 2 | 0 | 3 | 100.0 | 100.0 | 0.5 |
| IPI00468097 | 340 kDa protein | Secreted | 4 | 8 | 1 | 13 | 4.0 | 8.0 | 0.5 |
| IPI00113853 | Desmocollin-3 | Transmembrane | 2 | 6 | 0 | 8 | 100.0 | 100.0 | 0.3 |
| IPI00319814 | Suprabasal-specific protein suprabasin | Secreted | 3 | 10 | 0 | 13 | 100.0 | 100.0 | 0.3 |
| IPI00115854 | TROP2 protein | Transmembrane | 1 | 4 | 0 | 5 | 100.0 | 100.0 | 0.3 |
| IPI00127933 | Androgen binding protein alpha | Secreted | 1 | 4 | 0 | 5 | 100.0 | 100.0 | 0.3 |
| IPI00130249 | GPI-anchored metastasis-associated protein homolog | Secreted | 10 | 60 | 0 | 70 | 100.0 | 100.0 | 0.2 |
| IPI00111014 | Elongation of very long chain fatty acids protein 4 | Transmembrane | 3 | 20 | 2 | 25 | 1.5 | 10.0 | 0.2 |
| IPI00129243 | Gamma-glutamyl hydrolase | Secreted | 1 | 8 | 0 | 9 | 100.0 | 100.0 | 0.1 |
| IPI00338790 | Glandular kallikrein KLK13 | Cell Surface | 0 | 6 | 0 | 6 | 0.0 | 100.0 | 0.0 |
| IPI00111115 | Similar to METASTASIS-ASSOCIATED GPI- ANCHORED PROTEIN | Secreted | 0 | 4 | 0 | 4 | 0.0 | 100.0 | 0.0 |
| IPI00473830 | Biliary glycoprotein | Transmembrane | 0 | 4 | 0 | 4 | 0.0 | 100.0 | 0.0 |
| IPI00153548 | Hypothetical protein | Transmembrane | 0 | 3 | 0 | 3 | 0.0 | 100.0 | 0.0 |
Ca/Nr: Ratio of spectral count of carcinomas to normal tissue
Pa/Nr: Ratio of spectral count of apillomas to normal tissue
Ca/Pa: Ratio of spectral count of carcinomas to papillomas
Here, we determined the relative abundance of glycosylated proteins using identified glycosylated peptides from the protein. However, glycosylation for individual glycosite from the same protein might be different and can be determined by quantitative analysis of each glycosite. In addition, changes in glycan structure that may be important to the disease cannot be determined by this method, and specific enrichment of glycopeptides with certain glycan structure is needed.
Detected tissue-derived protein in plasma
Since the plasma proteome is dominated by several highly abundant proteins, proteins released from specific tissues would normally be present at low abundance in plasma, and their detection might be obscured by the high abundant plasma proteins. To detect tumor-specific proteins in plasma, we used isotopic labeling to detect the isotopic peaks that consisted of the tissue-derived proteins from both plasma and tissues.
The glycopeptides from four carcinomas tissues were labeled with d413C4-succinic anhydride. The glycopeptides from plasma of the four mice before and after cancer development were labeled with d013C0 and d413C0-succinic anhydride respectively. To monitor the labeling efficiency, we spiked same amount of standard peptide from Angiotensin (0.1 μg) in the glycopeptides isolated from carcinomas and plasma samples as labeling control. Then, all the labeled peptides were combined for MS analysis. The mixture was separated by 2-D Nano-LC then analyzed by MALDI-TOF/TOF. Free Angiotensin (ms 1296.68) was not observed after labeling. Instead, 100Da, 104Da and 108Da shifted from 1296.68 were observed in equal amount in the mixed sample. This indicates the efficient and quantitative isotopic labeling using succinic anhydride.
The mixed glycopeptides from carcinomas and plasma samples contained both skin cancer related peptides and peptides from plasma. In order to detect glycopeptides associated with skin cancer in plasma, we focused our analysis on glycopeptides previously identified as cancer associated glycoproteins from skin tumors in the mixture (Table 2) and avoid the analysis of plasma proteins. To achieve this goal, the peptide peaks that contained masses from glycopeptides specifically identified from carcinomas and their isotopic pairs from plasma were selected for MS/MS analysis.
Two types of paired patterns were observed. One was that the intensity of d413C4- labeled peptides (with 8 mass unit shift for each amino group from peptides derived from cancer tissues) was much greater than d413C0-labeled peptide (with 4 mass unit shift for each amino group from peptides derived from plasma of cancer-bearing mice) and intensity of d013C0-labeled peptide (with 0 mass unit shift for each amino group from peptides derived from plasma before carcinogen induction) was lower than that of peptides from plasma of cancer-bearing mice. This pattern indicated that the peptide was from tumor-specific protein and detectable in cancer plasma at low intensity. The other pattern was that similar or lower intensity of peptides from cancer tissues than in plasma, and peptides with this pattern were derived from plasma proteins.
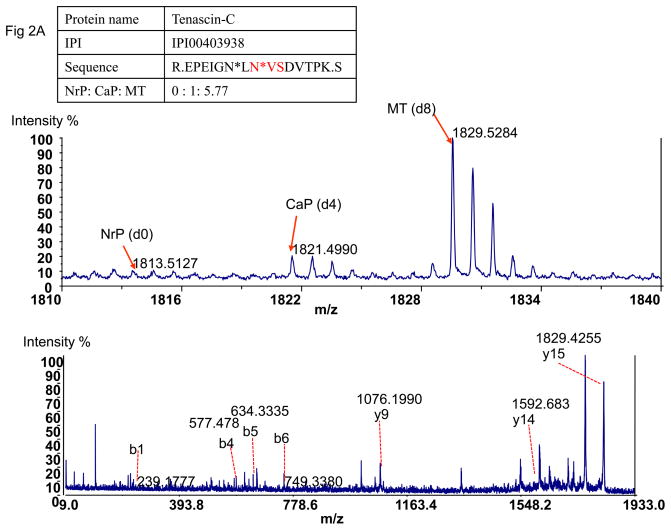
Tumor-associated glycopeptides could be detected plasma. Tenascin-C was identified in carcinomas with 133 spectra, and it was also identified in benign papillomas with 29 spectra. However, none of these glycopeptides were identified in normal tissue (Table 2). In plasma, the labeled peptide peak of Tenascin-C was found with its paired peaks with eight-Da mass difference (Fig 2A), which indicated that it was also detected in plasma after cancer development, but not in control plasma before the carcinogen treatment. Another skin tumor-specific glycoprotein, Arylsulfatase B, was also detected in plasma successfully in the similar way (Fig 2B). These data indicated that extracellular proteins associated with tumor development were identifiable in plasma from tumor-bearing mice using glycopeptide capture, isotopic labeling, and mass spectrometry.
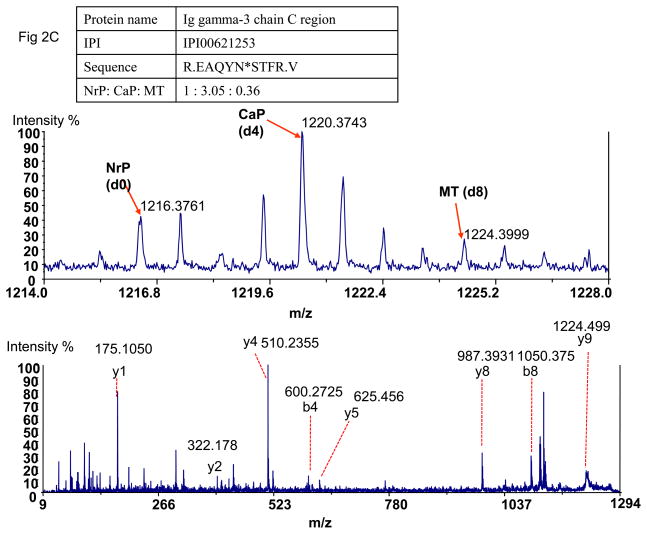
Figure 2.
Detection of tumor-specific proteins in plasma. A). The detected paired peaks of succinic anhydride labeled Tenascin-C and MS/MS spectrum of Tenascin-C. B) The paired peak of succinic anhydride labeled Arylsulfatase B and MSMS of Arylsulfatase B. C) The paired peak of succinic anhydride labeled Ig gamma-3 chain C region showed different peak pattern from Tenascin-C and MS/MS of Ig gamma-3 chain C region. NrP: Mouse plasma without carcinogen treatment; CaP: Mouse plasma from cancer-bearing mice after carcinogen treatment; MT: Mouse cancer tissues.
One of the advantages using this tissue-targeted approach is that tumor-associated proteins can be identified in plasma even they present in very low abundance. The peptides from cancer tissue are likely to be at higher abundance compared to the same peptides in plasma. These allowed us to determine their masses and peptide sequences in the mixture using isotopic peaks from tumors. Using this information, tumor-derived peptides in plasma can be identified while they are not identifiable by data-dependent MS/MS acquisition and database search. Both Tenascin-C and Arylsulfatase B are low abundant proteins. They were not identified in plasma before cancer development and their detection in plasma was associated with cancer development. Their peak intensities in cancer plasma were at least 100 folds lower than that for plasma proteins detected in the same mixture.
Proteins from plasma can also detected in tissues and plasma as isotopic pairs due to visualization of the tissue. If a glycopeptide detected in both cancer tissues and plasma was derived from plasma, the peptide peak showed similar or lower intensity in cancer tissues than that in plasma. An example of this was the identification and quantification of glycoprotein, Ig gamma-3 chain C region, in tissue and plasma. However, its paired peptide peaks were found in a different pattern from that observed with Tenascin-C (Fig 2C). The intensities of d013C0- and d413C0- labeled peptides from plasma before and after tumor induction were much higher than that from d413C4- labeled peptides from tumors. This indicated that this peptide was from a plasma-derived proteins and Ig gamma-3 could be detected from tissue due to the blood circulation in tissue.
The methodology of targeted detection of tumor proteins using glycopeptide capture, isotopic labeling, and mass spectrometry is based on the analysis of N-linked glycopeptides to study extracellular proteins from tumors and plasma, and it has shown to increase the delectability of tumor proteins by focusing the same subset of glycopeptides in both tumors and plasma 13. The tumor-associated glycopeptides could be detected in plasma on account of the several advantages of our methodologies. First, glycopeptides capture method dramatically reduces the sample complexity. Non-glycoproteins and non-glycopeptides from glycoproteins were removed from the pool of samples. For example, albumin, the most abundant serum protein, was automatically transparent to this method since it does not contain N-linked glycosylation. Second, the glycopeptides isolation method could be used to enrich extracellular proteins due to the fact that most extracellular proteins are glycosylated and likely to enter the bloodstream. Third, we used isotopic labeling method to facilitate the detection of tumor proteins within complex plasma by identifying paired peptide peaks from tumor tissues and plasma. However, the method described here is only for proteins that contain N-linked glycosylation. For proteins that do not contain N-linked glycosylation, this method will miss the detection of those proteins.
These results show our strategy for detection of tumor-specific proteins in plasma is specific and sensitive for low abundant tumor-associated proteins. Differ from the previous report of identification of prostate cancer-derived proteins in serum using xenograft-bearing mice 30, our study is more focus on tumor-associated extracellular proteins that are likely to be used in early detection.
CONCLUSIONS
In this study, we described a platform for quantitative detection of tumor-specific extracellular proteins in tumor and plasma. This suggests that it possible of detection of cancer from plasma.
The fact that tumor-specific proteins were detectable in plasma from tumor-bearing mice indicates that cancer-specific markers could be detected in plasma using targeted approaches and these proteins could be serum tumor marker candidates 7. Once such candidate proteins are identified, the homologues of the proteins can be verified in human sera using the targeted approach. ELISA assays can be developed using a pair of antibodies. However, if antibodies against the candidate proteins are not available, mass-spectrometry-based methods can be applied to detect candidate proteins in plasma. One approach is referred as a multiple reaction monitoring (MRM) 17–19. In another approach called stable isotope standards and capture by anti-peptide antibodies (SISCAPA), a specific peptide from sample and the synthetic heavy isotope labeled peptide of the candidate protein are captured by peptide antibody. The mass spectrometer is then used to detect and quantify the specific peptide with known precursor mass and fragmentation ions 31.
Acknowledgments
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, by Grants R21-CA-114852. We gratefully acknowledge the support from the Mass Spectrometry Facility at the Johns Hopkins University and the support of Trans-Proteomic Pipeline (TPP) Software tools available from Aebersold group at the Institute for Systems Biology.
References
- 1.Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Pieper R, et al. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 3.Adkins JN, et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Tirumalai RS, et al. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, et al. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 6.Coombes KR. Analysis of mass spectrometry profiles of the serum proteome. Clin Chem. 2005;51:1–2. doi: 10.1373/clinchem.2004.040832. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Chan DW. Cancer Biomarker Discovery in Plasma Using a Tissue-targeted Proteomic Approach. Cancer Epidemiol Biomarkers Prev. 2007;16:1915–1917. doi: 10.1158/1055-9965.EPI-07-0420. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protocols. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 11.Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, et al. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Schulz BL, Laroy W, Callewaert N. Clinical laboratory testing in human medicine based on the detection of glycoconjugates. Curr Mol Med. 2007;7:397–416. doi: 10.2174/156652407780831629. [DOI] [PubMed] [Google Scholar]
- 15.Pan S, et al. High throughput proteome screening for biomarker detection. Mol Cell Proteomics. 2005;4:182–190. doi: 10.1074/mcp.M400161-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Yan W, Aebersold R. Chemical probes and tandem mass spectrometry: a strategy for the quantitative analysis of proteomes and subproteomes. Curr Opin Chem Biol. 2004;8:66–75. doi: 10.1016/j.cbpa.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahl-Zeng J, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Kemp CJ. Multistep skin cancer in mice as a model to study the evolution of cancer cells. Semin Cancer Biol. 2005;15:460–473. doi: 10.1016/j.semcancer.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 24.Whiteaker JR, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 25.Zou Z, et al. Synthesis and evaluation of superparamagnetic silica particles for extraction of glycopeptides in the microtiter plate format. Anal Chem. 2008;80:1228–1234. doi: 10.1021/ac701950h. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Jones FS, Jones PL. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev Dyn. 2000;218:235–259. doi: 10.1002/(SICI)1097-0177(200006)218:2<235::AID-DVDY2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Orend G. Potential oncogenic action of tenascin-C in tumorigenesis. Int J Biochem Cell Biol. 2005;37:1066–1083. doi: 10.1016/j.biocel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh D. Human sulfatases: a structural perspective to catalysis. Cell Mol Life Sci. 2007;64:2013–2022. doi: 10.1007/s00018-007-7175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Bemd GJ, et al. Mass spectrometric identification of human prostate cancer-derived proteins in serum of xenograft-bearing mice. Mol Cell Proteomics. 2006;5:1830–1839. doi: 10.1074/mcp.M500371-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]