Abstract
The oxazolidinone antibiotic linezolid targets the peptidyl transferase center (PTC) on the bacterial ribosome. Thirteen single and four double 23S rRNA mutations were introduced into a Mycobacterium smegmatis strain with a single rRNA operon. Converting bacterial base identity by single mutations at positions 2032, 2453, and 2499 to human cytosolic base identity did not confer significantly reduced susceptibility to linezolid. The largest decrease in linezolid susceptibility for any of the introduced single mutations was observed with the G2576U mutation at a position that is 7.9 Å from linezolid. Smaller decreases were observed with the A2503G, U2504G, and G2505A mutations at nucleotides proximal to linezolid, showing that the degree of resistance conferred is not simply inversely proportional to the nucleotide-drug distance. The double mutations G2032A-C2499A, G2032A-U2504G, C2055A-U2504G, and C2055A-A2572U had remarkable synergistic effects on linezolid resistance relative to the effects of the corresponding single mutations. This study emphasizes that effects of rRNA mutations at the PTC are organism dependent. Moreover, the data show a nonpredictable cross-resistance pattern between linezolid, chloramphenicol, clindamycin, and valnemulin. The data underscore the significance of mutations at distal nucleotides, either alone or in combination with other mutated nucleotides, in contributing to linezolid resistance.
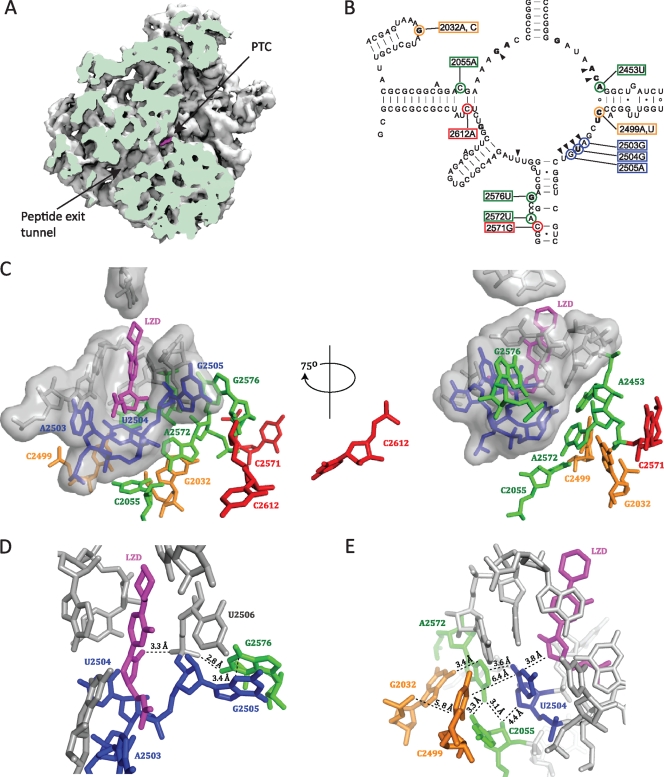
The bacterial ribosome is an important target for many clinically relevant antibiotics, including linezolid, which is effective in the treatment of infections with various multidrug-resistant Gram-positive cocci, especially methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci (26). Several lines of evidence have established that oxazolidinones bind to the peptidyl transferase center (PTC) on the large ribosomal subunit. The evidence includes competitive binding studies with chloramphenicol and lincomycin (25), the clustering of single rRNA resistance mutations in the peptidyl transferase loop (22), in vivo cross-linking experiments (4, 23), and crystal structures of linezolid-50S subunit complexes (archaeal 50S complex [20] and bacterial 50S complex [49]). These structures show that linezolid binds to the A-site portion of the PTC (Fig. 1 A) overlapping the site of the aminoacyl moiety of A-site-bound tRNA (20, 49). The linezolid binding pocket is lined by eight 23S rRNA nucleotides (marked by wedges in Fig. 1B) that are highly conserved among the three domains of life (49), and the drug is not in direct contact with any ribosomal proteins. Despite this high conservation, indirect evidence suggests that linezolid affects bacterial and mitochondrial ribosomes but not cytosolic ribosomes (23, 34). It has been proposed that the specificity may be determined by nucleotides a bit further away from the binding site acting through the nearby conserved nucleotides (23).
FIG. 1.
The positions of mutated 23S rRNA nucleotides relative to the linezolid binding site. (A) A cut view of the D. radiodurans 50S subunit showing linezolid (magenta) bound at the PTC and the peptide exit tunnel (PDB accession number 3DLL [49]). The image was made with the VMD software program (19) and contributed by Jacob Pøhlsgaard. (B) Secondary structure of the M. smegmatis 23S rRNA peptidyl transferase loop. The positions of mutated nucleotides are highlighted with colored circles, and the corresponding E. coli numbering is boxed in colored rectangles. The coloring of mutations indicates first-layer (blue), second-layer (green), third-layer (orange), and outer-layer (red) nucleotides with respect to linezolid. Wedges point to first-layer nucleotides that form the linezolid binding pocket. Nucleotides in bold type are those where a mutation(s) is identified to confer linezolid resistance in at least one organism (1, 14, 22, 30, 33, 43, 52, 53). (C to E) Close-up views of the linezolid binding site made with the PyMOL software program (7), where nucleotides are colored as described above. (C) The locations of mutated nucleotides with respect to bound linezolid. First-layer nucleotides are shown in surface representation. (D) The position of G2576 with respect to linezolid. (E) The positions of the nucleotides involved in the double mutations with respect to linezolid.
The linezolid resistance mechanisms characterized thus far involve alteration at or around the PTC by modification or mutation. Methylation of the 23S rRNA nucleotide A2503 (Escherichia coli 23S rRNA numbering is used throughout) at the C8 position by the Cfr methyltransferase confers a PhLOPSA resistance phenotype (10, 21). This is combined resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics (31) that bind to overlapping positions at the PTC (5, 13, 45, 46, 49). In contrast, lower linezolid susceptibility was found in Streptococcus pneumoniae by abolished methylation of G2445 in 23S rRNA (9). Hence, both gain and loss of methylations in the PTC area can affect linezolid binding. In addition, it has been shown that pseudouridylation of 23S rRNA nucleotide U2504 leads to a 16-fold decrease in linezolid susceptibility (47).
A number of 23S rRNA mutations in the peptidyl transferase loop have conferred linezolid resistance in laboratory selection experiments in several organisms (Fig. 1B, bold nucleotides). The mutations obtained depend on the specific organism used, and the mutations are both at nucleotides directly at the linezolid binding pocket (positions 2061, 2451, 2452, and 2504) and at nucleotides further away from the binding pocket (positions 2032, 2062, 2447, 2453, 2499, 2500, 2576, and 2608) (reviewed in reference 49). The G2576U mutation is thus far the most prevalent in clinical linezolid-resistant isolates (see reference 8 and references therein). Mutants with deletions in the ribosomal protein L4 have also been shown by complementation experiments to confer reduced susceptibility to linezolid in S. pneumoniae (50). In addition, a number of other mutations in 23S rRNA and the ribosomal proteins L3 (28, 29) and L4 (9, 51) have been associated with linezolid resistance but without substantial documentation of the cause-effect relationship. The 23S rRNA mutations reported sometimes correspond to those found in laboratory selection experiments, but in others cases there is no genetic evidence relating the specific 23S rRNA mutations to the linezolid resistance phenotype (3, 9, 27, 41).
In the present work, a series of single and double 23S rRNA mutations was engineered in an M. smegmatis strain carrying a single rRNA operon. The study was designed to answer the following questions. (i) Is selectivity of linezolid action associated with single nucleotide changes around the drug binding site? (ii) Are linezolid resistance data from RNA mutations in other organisms valid for M. smegmatis, and can those reported in the literature without genetic proof be validated in M. smegmatis? (iii) Do double mutations provide a higher level of resistance than the corresponding single mutations? (iv) Finally, what are the cross-resistance patterns from RNA mutations in the PTC area? The resistance data underscore the complexity of mutational effects in the PTC area.
MATERIALS AND METHODS
Strains and plasmids.
The M. smegmatis strains and plasmids used in this investigation are summarized in Table 1. The strains all possess a single rRNA operon encoded on a plasmid integrated at a chromosomal attB locus. The integrated plasmid carries a gentamicin resistance cassette in the parent strain (SZ558) and a hygromycin B resistance cassette in the wild-type and mutant strains (16). As a note, our rrnA and rrnB designations are reversed relative to the ones in the annotated M. smegmatis mc2 155 genome. The E. coli strain DH5α was grown in LB medium. M. smegmatis strains were grown in LB Tween (LB supplemented with 0.05% Tween 80) to minimize cell clumping. For plasmid selection and maintenance, LB agar plates were supplemented with streptomycin (50 μg ml−1), gentamicin (15 μg ml−1), or hygromycin B (100 μg ml−1).
TABLE 1.
Strains and plasmids
| Strain/plasmid | Selection marker(s)a | Source/reference |
|---|---|---|
| M. smegmatis strains | ||
| SMR5 ΔrrnA ΔrrnB attB::pMIG-sacB-rrnB (parent) | Strr Genr | 16 |
| SMR5 ΔrrnB rrnA 2055A | Strr | 30 |
| SMR5 ΔrrnA ΔrrnB attB::pMIH-rrnB (wild type) | Strr HygBr | This study |
| SMR5 ΔrrnA ΔrrnB attB::pMIH-rrnBmutation | Strr HygBr | This study |
| Plasmids | ||
| pMIHΔint-rrnB | HygBr | 16 |
| pMIHΔint-rrnBmutation | HygBr | This study |
Str, streptomycin; Gen, gentamicin; HygB, hygromycin B.
Construction of pMIH-rrnB mutant plasmids and transformation into M. smegmatis.
Single mutations were introduced onto fragments of M. smegmatis rRNA genes by overlap extension PCR (42) using pMIHΔint-rrnB as a template. The G2032A, G2032C, and A2058G mutations were introduced into a 2.1-kb M. smegmatis rRNA gene fragment, where the primers OPF3 and OPR3 were used in combination with mutagenic primers (Table 2). The OPF3 and OPR3 primers were also used to amplify a 2.1-kb fragment containing the C2055A mutation from the SMR5 ΔrrnB rrnA 2055A strain (Table 1) (30) using colony PCR. These mutant fragments were subsequently cloned into the NcoI and SpeI sites of pMIHΔint-rrnB (16). The other single mutations were introduced either into 2.6-kb fragments using the OPF4 and OPR4/OPR5 primers (A2453U, C2499A, C2499U, U2504G, A2572U, and G2576U) or 2.0-kb fragments using the OPF5 and OPR7 primers (A2503G, G2505A, C2571G, and C2612A) used in combination with mutagenic primers (Table 2), where both fragments contain the 3′ end of the 23S rRNA gene, the 5S rRNA gene, and pMIHΔint-rrnB plasmid sequence, including the MunI site. These fragments were subsequently cloned into the SpeI and MunI sites of pMIHΔint-rrnB. Double mutations were introduced by ligation of either NcoI-SpeI or SpeI-MunI fragments containing one mutation into the corresponding cut pMIHΔint-rrnB plasmids containing the second mutation. The ligated plasmids were transformed into E. coli DH5α, followed by sequencing of plasmid DNA with the primer SP1F or SP2F to verify the presence of the mutations.
TABLE 2.
DNA oligonucleotide primers
| Name | Sequence (5′ to 3′)c | Positionsa |
|---|---|---|
| OPF3 | GAGTGATCTACCCATGGCCAG | 798-818 |
| OPF4 | GACCAAGGGTTCCTGGGCCAG | 1421-1441 |
| OPF5 | GGTTTGTGTAGGATAGGTGGG | 2331-2351 |
| OPR3 | GTGCCCCTGGTGGGACAACTG | 2919-2939 |
| OPR4 | CGAGTCGCGACGTACGTTCG | Noneb |
| OPR5 | TCGAGCTCGGTACCTACGTAC | Noneb |
| OPR7 | GGTCGAGAAGTAACAGGG | Noneb |
| SP1F | CCTGCACGAATGGCGTAACG | 2188-2207 |
| SP2F | GAGACGGACATGTCGAGCAG | 2578-2597 |
| G2032AF | CTACGAGTAAAAATGCTCGTTACG | 2248-2271 |
| G2032AR | AACGAGCATTTTTACTCGTAGTG | 2246-2268 |
| G2032CF | CTACGAGTAAACATGCTCGTTACG | 2248-2271 |
| G2032CR | AACGAGCATGTTTACTCGTAGTG | 2246-2268 |
| A2058GF | GCAGGACGAGAAGACCCCGGGAC | 2276-2298 |
| A2058GR | CCGGGGTCTTCTCGTCCTGCCGC | 2273-2295 |
| A2453UF | CGGGGATAACTGGCTGATCTTCC | 2670-2692 |
| A2453UR | AAGATCAGCCAGTTATCCCCGGG | 2668-2690 |
| C2499AF | GTTTGGCACATCGATGTCGGCT | 2717-2738 |
| C2499AR | CGACATCGATGTGCCAAACCATC | 2713-2735 |
| C2499UF | GTTTGGCACTTCGATGTCGGCT | 2717-2738 |
| C2499UR | CGACATCGAAGTGCCAAACCATC | 2713-2735 |
| A2503GF | TGGCACCTCGGTGTCGGCTCGTC | 2720-2742 |
| A2503GR | CGAGCCGACACCGAGGTGCCAAAC | 2717-2740 |
| U2504GF | CACCTCGAGGTCGGCTCGTCG | 2723-2743 |
| U2504GR | GAGCCGACCTCGAGGTGCCAAAC | 2717-2739 |
| G2505AF | GGCACCTCGATATCGGCTCGTCGC | 2721-2744 |
| G2505AR | CGAGCCGATATCGAGGTGCCAAAC | 2717-2740 |
| C2571GF | ATTAAAGCGGGACGCGAGCTGGG | 2788-2810 |
| C2571GR | CAGCTCGCGTCCCGCTTTAATGGG | 2785-2808 |
| A2572UF | TAAAGCGGCTCGCGAGCTGGGT | 2790-2811 |
| A2572UR | CAGCTCGCGAGCCGCTTTAATGG | 2786-2808 |
| G2576UF | AGCGGCACGCTAGCTGGGTTTAG | 2793-2815 |
| G2576UR | AACCCAGCTAGCGTGCCGCTTTA | 2790-2812 |
| C2612AF | CAGTTCGGTCTATATCCGCCGC | 2828-2849 |
| C2612AR | GCGGCGGATATAGACCGAACTGTC | 2826-2849 |
Positions of M. smegmatis 23S rRNA gene complementarity are given.
Primers are complementary to pMIHΔint-rrnB at sites downstream of rrnB and the MunI site.
Mutated nucleotides are underlined in the sequences of primers used for site-directed mutagenesis.
Transformation of plasmids into competent M. smegmatis cells was performed essentially as described previously (43) except that cultures were grown in LB Tween medium and the cells were plated on LB hygromycin B plus 5% sucrose plates. The transformation plates were incubated at 37°C for 3 to 5 days or until single colonies were visible. Single colonies were reisolated on LB hygromycin B plus 5% sucrose plates and screened for loss of the pMIG-sacB-rrnB plasmid by testing for gentamicin sensitivity by patching cells on LB gentamicin plates. Colony PCR with the primers OPF4 and OPR3 was used to amplify fragments containing the mutations, followed by sequencing of the fragments with the primers SP1F and/or SP2F to ensure that the mutations had not reverted during the plasmid exchange process.
Growth rate measurements.
Cells from freshly streaked agar plates were used to inoculate 5 ml LB Tween medium, and the cultures were incubated at 37°C for 1 to 3 days. The cultures were diluted into 5 ml LB Tween and grown at 37°C prior to dilution into 25 or 50 ml LB Tween. The growth rates in the exponential growth phase were determined by growing strains in 25 or 50 ml LB Tween at 37°C with constant shaking. The optical density at 600 nm (OD600) values were recorded every hour for 7 to 12 h. The data were fit to the exponential function, OD = OD0eμt, where μ is the growth rate and OD0 is the optical density at time zero. The doubling times, td, were calculated from the growth rates, where td = (ln2/μ), and represent the average of data from 3 to 7 independent experiments. In some strains, cell clumping and/or very slow growth prevented determination of exact growth rates.
Drug susceptibility testing.
The drug susceptibility assay was carried out in the 96-well microtiter plate format, where plates were read manually. Cells from freshly streaked agar plates were used to inoculate 5 ml LB Tween medium, and the cultures were incubated at 37°C until growth was clearly visible. The cultures were diluted to an OD600 of 0.02, and 100 μl was mixed with 100 μl of antibiotic solutions in a series of 2-fold dilution steps. The tested concentration range of each drug was as follows: linezolid, 0.125 to 64 μg ml−1; and chloramphenicol, clindamycin, and valnemulin, 1 to 512 μg ml−1. The MIC is defined as the drug concentration at which the growth of the cultures is completely inhibited by visual inspection after an incubation at 37°C corresponding to 24 generation times as measured under the conditions described in the previous section. Each microtiter plate included the parent and/or wild-type strains as internal controls.
RESULTS
The model system and procedure for rRNA mutant isolation.
Multiple rRNA operons and heterogeneous ribosome populations complicate the study of rRNA mutations in many bacteria. Model systems containing a single rRNA operon are therefore advantageous. Genetically engineered derivatives of the Gram-positive bacterium M. smegmatis with a single functional rRNA operon are available. These strains are good models of pathogenic bacteria, such as Mycobacterium tuberculosis, and are suitable for investigating mutations related to ribosomal drug resistance (15, 17, 18, 30, 44). The recently developed M. smegmatis strain, SMR5 ΔrrnA ΔrrnB attB::pMIG-sacB-rrnB, has a deletion of its two naturally occurring rrn operons and an insertion of the rrnB operon into a chromosomal attB locus (16). In this study, mutations were introduced onto rRNA gene fragments by PCR and cloned into the integration-proficient plasmid pMIHΔint-rrnB (16). The wild-type and mutant pMIHΔint-rrnB plasmids were then transformed into M. smegmatis SMR5 ΔrrnA ΔrrnB attB::pMIG-sacB-rrnB. Plasmid exchange at the attB locus was achieved by selection with hygromycin B and sucrose (16). Since expression of the sacB gene encoding the enzyme levansucrase from Bacillus subtilis is lethal in M. smegmatis in the presence of sucrose, it can be used as a marker for positive selection of gene replacement events (35, 36). In a previously used M. smegmatis model system, the mutations were introduced by homologous recombination followed by selection with the antibiotic the mutation was thought to confer resistance to (30). Thus, the strength of the model system implemented here is the ability to introduce mutations that don't necessarily confer antibiotic resistance.
Single 23S rRNA mutations associated with domain specificity in the PTC area.
Most antibiotics are specific for ribosomes from one or two domains of life, although a few universal antibiotics inhibit ribosomes from all three domains. Indeed, the clinical utility of ribosomal antibiotics hinges upon their selectivity for bacterial versus mammalian ribosomes. Linezolid binds to the peptidyl transferase center of bacterial ribosomes but apparently not or much less to eukaryotic cytosolic ribosomes (see reference 23 and references therein). The conservation of the eight nucleotides that surround a bound linezolid molecule (here called first-layer nucleotides) (Fig. 1C) raises the question of the basis of linezolid selectivity for bacterial ribosomes. One possible explanation for the preference of linezolid for bacterial ribosomes is that nucleotides outside the drug binding pocket (here called second-, third-, and outer-layer nucleotides) influence its conformation and thereby affect drug binding and susceptibility. The identities of the second-layer nucleotide 2453 and third-layer nucleotides 2032 and 2499 differ between bacterial and human cytosolic rRNA (23). The G2032C, A2453U, and C2499U single mutations have been introduced into M. smegmatis 23S rRNA to convert their identity to that found in human cytosolic 28S rRNA to investigate their effect on linezolid susceptibility. The positions of the nucleotides relative to linezolid are illustrated in Fig. 1C. The A2058G mutation, which confers macrolide resistance (38, 48), was included as a control mutation not expected to affect linezolid susceptibility. The linezolid susceptibilities of the mutant strains were assayed by measurement of MICs and are shown in Table 3. There are only 2-fold increases in linezolid MICs for the G2032C and A2453U mutants, whereas MICs for the C2499U and A2058G mutants are the same as those for the wild-type strain (Table 3). It is thus clear that these mutations do not lead to significantly reduced linezolid susceptibility.
TABLE 3.
Antibiotic susceptibilities and growth rates of M. smegmatis mutant strains with single and double 23S rRNA mutationsa
| M. smegmatis straind | LZD MIC (μg ml−1) | LZD-nucleotide proximitye | Doubling time (h) | MIC (μg ml−1) of: |
||
|---|---|---|---|---|---|---|
| CHL | CLI | VAL | ||||
| SZ558 (parent) | 2 | NM | 3.3 ± 0.1 | 32 | 16 | 32 |
| Wild type | 2 | NM | 3.5 ± 0.1 | 32 | 16-32 | 32 |
| A2058G | 2 | C | 3.5 ± 0.3 | 32 | >1,028 | 64 |
| G2032C | 4 | 3 | 3.6 ± 0.6b | 32 | 128 | 128 |
| A2453U | 4 | 2 | 3.9 ± 0.1 | 32 | 32 | 64 |
| C2499U | 2 | 3 | 4.1 ± 0.2 | 32 | 16 | 32 |
| A2503G* | 8 | 1 | 4.6 ± 0.1 | 128 | 8 | 32 |
| G2505A* | 16 | 1 | 6.4 ± 0.3 | 128 | 16 | 32 |
| C2571G | 4 | X | 4.0 ± 0.1 | 32 | 16-32 | 32 |
| G2576U | 64 | 2 | 9.1 ± 0.2 | 256 | 64 | 32 |
| C2612A | 4 | X | 4.2 ± 0.3 | 64 | 16 | 32 |
| C2499A↓ | 2 | 3 | 3.8 ± 0.1 | 32 | 16 | 64 |
| G2032A-C2499A | 8 | 3-3 | 4.4 ± 0.1 | 64 | 16 | 128 |
| G2032A↕ | 2 | 3 | 3.7 ± 0.3 | 32 | 32 | 128 |
| G2032A-U2504G | 64 | 3-1 | >10c | 256 | 16 | 128 |
| U2504G*↕ | 8 | 1 | 5.8 ± 0.1 | 512 | 16-32 | 64 |
| C2055A-U2504G | 32 | 2-1 | >10c | 128 | 16-32 | 64 |
| C2055A↕ | 0.5 | 2 | 3.8 ± 0.2 | 32 | 32 | 64 |
| C2055A-A2572U | 64 | 2-2 | 10.2 ± 0.6 | 64 | 512 | 128 |
| A2572U↑ | 4 | 2 | 3.6 ± 0.3 | 8 | 16 | 128 |
MICs that differ from the unmutated strain sensitivity by 4-fold or more are indicated in bold type.
The large standard deviation in the measured growth rates is due to cell clumping.
Exact growth rate measurements could not be obtained due to cell clumping.
Asterisks indicate nucleotide positions that are interacting directly with linezolid. Arrows point to the corresponding double mutants.
The positions of the mutated nucleotides relative to that of the bound linezolid drug are indicated, where 1, 2, and 3 represent the first, second, and third nucleotide layers around linezolid and X represents nucleotides outside of these three layers (see Fig. 1); C, control mutation; NM, no mutation. LZD, linezolid; CHL, chloramphenicol; CLI, clindamycin; VAL, valnemulin.
Cross-resistance was investigated by determining MICs for chloramphenicol, clindamycin, and valnemulin for the mutant strains (Table 3). These antibiotics have binding sites partially overlapping with that of linezolid, as is known from crystal structures of 50S subunit-antibiotic complexes (20, 45, 46, 49). Although the streptogramin A binding site overlaps that of linezolid (13, 30, 49), the parent and wild-type strains were not sensitive to the streptogramin A antibiotic virginiamycin M1 (data not shown). The MIC values for virginiamycin and quinupristin-dalfopristin (Synercid), each containing streptogramin A and B components, were also determined. However, the streptogramin MICs of the mutant strains differed from that of the wild-type strain by no more than 2-fold (data not shown). The G2032C mutation confers considerable resistance to valnemulin and clindamycin (Table 3). As expected, the control A2058G mutation is highly resistant to clindamycin (39) (Table 3). There are only 2-fold increases in valnemulin MICs for the A2058G and A2453U mutants, and none of the four mutants (Table 3) exhibits chloramphenicol resistance.
Single 23S rRNA mutations in the PTC area associated with antibiotic resistance.
While mutations at 12 23S rRNA nucleotide positions have been shown to confer linezolid resistance (indicated by bold text in Fig. 1B), other reports of linezolid resistance are observational in nature, without genetic proof of a causal relationship between a particular 23S rRNA mutation and the linezolid resistance phenotype. We introduced the four single 23S rRNA mutations A2503G, G2505A, C2571G, and C2612A (Fig. 1C), which have previously been associated with linezolid resistance without real genetic proof of resistance. The A2503G mutation has been detected together with G2576U in S. aureus and together with C2571G and A1743U in S. pneumoniae strains with decreased linezolid susceptibilities (3, 9, 27). In addition, as mentioned in the introduction, an m8A modification at A2503 confers resistance to PhLOPSA drugs, including linezolid (10, 31). Furthermore, the A2503U mutation has been recently associated with decreased susceptibility to tiamulin, valnemulin, chloramphenicol, florfenicol, and lincomycin in Mycoplasma gallisepticum (24). The G2505A, C2571G, and C2612A mutations have been detected in linezolid-resistant isolates of enterococci or streptococci (3, 41), but it is unclear if the mutations are the cause of resistance. Although G2576 is positioned 7.9 Å from linezolid (Fig. 1D), the G2576U mutation is the most commonly detected 23S rRNA mutation in linezolid-resistant clinical isolates (see reference 8 and references therein) and has therefore been introduced for comparison.
The MIC values show that all mutations confer reduced susceptibility to linezolid, albeit to different extents. The largest effect is observed with the G2576U mutant, with a 32-fold increase in the linezolid MIC compared to that for the wild-type strain (Table 3). Moderate 4- and 8-fold increases in linezolid MICs are observed with the first-layer A2503G and G2505A mutants, respectively (Table 3). Only 2-fold increases in the linezolid MIC are observed with the outer-layer C2571G and C2612A mutants (Table 3). According to the common secondary structure (Fig. 1B), both of these mutations should break Watson-Crick base pairs, although 2612 is not base paired in the structure of the Deinococcus radiodurans 50S-linezolid complex (49). Both nucleotides are far from linezolid and are apparently not providing significant resistance as single mutations in M. smegmatis. We observe cross-resistance to chloramphenicol with the A2503G, G2505A, and G2576U mutations. The A2503G and G2505A mutations were previously shown to confer chloramphenicol resistance in Thermus thermophilus (11). The A2503G mutant exhibits a 2- to 4-fold decrease and the G2576U mutant a 2- to 4-fold increase in clindamycin MIC values relative to those for the wild-type strain. No cross-resistance to valnemulin was observed in these five mutant strains.
The single mutations G2032A, C2055A, C2499A, U2504G, and A2572U, which are either associated with or shown to confer resistance to linezolid or other PTC antibiotics (30, 37, 40), were also introduced to compare their effects with those of the other mutations in M. smegmatis. Note that although the G2032 and C2499 mutants are described above, the base changes are different. The linezolid MICs of the G2032A and C2499A mutants are identical to that of the wild-type strain (Table 3). Four- and 2-fold increases in valnemulin MICs are observed for the G2032A and C2499A mutants, respectively, but there is no cross-resistance to either chloramphenicol or clindamycin. The other single mutations have modest effects on linezolid MIC values, with a 4-fold increase for the U2504G mutant and a 2-fold increase and decrease for the A2572U and C2055A mutants, respectively (Table 3). Changes in chloramphenicol MICs are observed for the U2504G (8-fold increase) and A2572U (4-fold decrease) mutants. Only modest (2- or 4-fold) increases in valnemulin MICs are observed for the mutants, and there is no cross-resistance to clindamycin. The differences in linezolid, chloramphenicol, and clindamycin MIC values for the C2055A, U2504G, and A2572U mutant strains relative to those for the wild-type strain generally correspond to those observed previously with similar but not identical strains, whereas the valnemulin susceptibilities are apparently lower than those in an earlier study (30).
Double 23S rRNA mutations associated with antibiotic resistance.
It is conceivable that some single mutations that cannot confer resistance on their own can contribute to resistance in combination with other mutations. This possibility has been explored with a selected set of double 23S rRNA mutations together with each of the single mutations. Four double mutation combinations, G2032A-C2499A, G2032A-U2504G, C2055A-U2504G, and C2055A-A2572U, which were previously observed in tiamulin-resistant Brachyspira hyodysenteriae isolates (40), were introduced. The positions and distances relative to linezolid and to the nucleotides at the binding site are illustrated in Fig. 1E. All four double mutants exhibit markedly increased linezolid MICs compared to the wild-type strain and the corresponding single mutant strains (Table 3). There are thus synergistic effects on linezolid resistance, since the double mutations confer higher linezolid MICs than the additive effects of the two single mutations. The G2032A-U2504G mutant exhibits an 8-fold increase in the linezolid MIC with respect to that for the U2504G mutant, indicating that the G2032A single mutation, which does not confer linezolid resistance, can contribute significantly to resistance in combination with the U2504G mutation (Table 3). The 4-fold linezolid MIC increase in the G2032A-C2499A mutant shows that although the two single mutations do not confer linezolid resistance alone, they confer resistance when present together (Table 3). Moreover, the C2055A-U2504 mutant exhibits a 4-fold increase in the linezolid MIC relative to that of the U2504G mutant, and the C2055A-A2572U mutant exhibits a 16-fold increase relative to that of the A2572U mutant. These data demonstrate that even though the C2055A single mutation confers hypersensitivity to linezolid, it can contribute significantly to linezolid resistance when in combination with either the U2504G or A2572U mutation (Table 3). There is also a synergistic effect on clindamycin resistance with the C2055A-A2572U mutant, where there is a 16- to 32-fold MIC increase relative to results for the wild type and the C2055A and A2572 mutant strains (Table 3). There are small increases and decreases in the chloramphenicol and valnemulin MICs for the double mutant relative to their corresponding single mutant strains, but no synergistic effects are observed for these antibiotics.
Effect of the 23S rRNA mutations on growth rate.
A mutation preventing antibiotic binding is beneficial only if it is not causing too much harm otherwise. The single and double 23S rRNA mutations summarized in Table 3 were successfully introduced into M. smegmatis and are thus not lethal in this organism. Generation times of the mutant strains were measured to determine the effect of the introduced mutations and are shown in Table 3. The wild-type control strain with the pMIHΔint-rrnB plasmid and a generation time of 3.5 h has a slightly slower growth rate than the parent strain with the pMIG-sacB-rrnB plasmid and a generation time of 3.3 h. The single G2032A, A2058G, and A2572U 23S rRNA mutations do not cause significant changes in growth relative to that of the wild-type strain. The C2055A, A2453U, and C2499A mutations lead to small decreases in the growth rate (3.8- to 3.9-h doubling times). Moderate decreases in the growth rate are observed with the C2499U, A2503G, C2571G, and C2612A mutations (4.0- to 4.6-h doubling times). The largest decreases are observed with the U2504G (5.8-h doubling time), G2505A (6.4-h doubling time), and G2576U (9.1-h doubling time) mutations. A precise determination of the G2032C mutant growth rate was precluded by extensive cell clumping in liquid culture. The growth rates of the C2055A, U2504G, and A2572U mutants are consistent with those in a previous M. smegmatis study (30) and also with recent data for E. coli, where the Ψ2504G mutation leads to slow growth (37). The strains with double mutations all have significant decreases in the growth rate. The G2032A-C2499A and C2055A-A2572U mutants have doubling times of 4.4 and 10.2 h, respectively. Although precise growth rates could not be obtained for the G2032A-U2504G and C2055A-U2504G mutants due to cell clumping, we estimate that these strains have doubling times that are longer than 10 h.
DISCUSSION
The 23S rRNA nucleotides G2061, A2451, C2452, A2503, U2504, G2505, U2506, and U2585 interact directly with bound linezolid and are all universally conserved and packed in an RNA-only cluster at the PTC (20, 49) (Fig. 1C). Since there are quantitative differences in binding of linezolid to ribosomes from the three domains of life, specificity is likely influenced by nucleotides at a distance that cause subtle conformational changes, as has also been discussed elsewhere (6, 12, 40). This also applies to other antibiotics that bind to the PTC, such as chloramphenicol, clindamycin, and valnemulin. In an attempt to locally mimic the eukaryotic ribosomal structure around the linezolid binding site, the three 23S rRNA changes G2032C, A2453U, and C2499U were introduced at positions located on the opposite side of U2504 relative to linezolid (Fig. 1C) (6, 12, 40). Only very moderate decreases in linezolid susceptibility were observed for G2032C and A2453U (Table 3) even though elevated MICs have been reported for G2032C and -U mutations in E. coli (52). There was no MIC increase for C2499U (Table 3), in contrast to findings of a study of Halobacterium halobium, where the same mutation caused a moderate decrease in linezolid susceptibility (22). Thus, the single mutations at positions 2032, 2453, and 2499 that differ between bacterial and eukaryotic cytosolic ribosomes are not individually able to confer species specificity regarding linezolid binding. None of these mutations can significantly reduce linezolid susceptibility in M. smegmatis.
Since linezolid is an important and relatively new drug for treatment of resistant pathogenic bacteria, there has been a strong interest in determining resistance mechanisms and investigating cross-resistance. The studies have been performed in various organisms and with various degrees of verification or genetic evidence of the involved mutations. These mutations have therefore been introduced in M. smegmatis, together with mutations associated with tiamulin resistance, to compare and verify their effects and investigate resistance “from nucleotides at a distance” (Fig. 1C). Starting directly at the linezolid binding pocket (blue nucleotides in Fig. 1C), the A2503G, U2504G, and G2505A mutations confer significant, although only up to an 8-fold increase in, linezolid resistance (Table 3). U2504 interacts directly with linezolid (Fig. 1E), and U2504G confers reduced susceptibility to linezolid. In our previous work on tiamulin resistance, mutations were found nearby and at U2504, and we suggested that the mutations affected antibiotic binding indirectly through an effect on U2504 (40). The contributions from the individual mutations were not proven at that time but have been verified in our recent paper (30) and herein. All the mutations introduced at second-layer nucleotides (green nucleotides in Fig. 1C) exhibit linezolid susceptibility changes not greater than 4-fold except for G2576U, which shows a 32-fold increase in the linezolid MIC (Table 3). The G2576U data are not surprising, since this mutation has been observed in clinical isolates and laboratory strains of staphylococci, enterococci, and S. pneumoniae (3, 8, 9, 32, 33, 41). However, the G2576U mutation results in 3-fold-slower growth of M. smegmatis. This is consistent with a previous study, where only the G2447U mutation was isolated by selection in the presence of linezolid (43). Either the G2576U mutation is not so detrimental in other bacteria or perhaps compensatory mutations, which we are unaware of, are rapidly acquired for a better cost-benefit ratio. According to the D. radiodurans 50S-linezolid structure (49), the closest distance from linezolid to G2576 is 7.9 Å (Fig. 1C), but a G2576U mutation will presumably reduce the degree of stacking with G2505 and disrupt a hydrogen bond with the U2506 backbone that abuts the bound linezolid (Fig. 1D), thereby decreasing linezolid binding. This is in accordance with data from Haloarcula marismortui with the PTC antibiotic anisomycin suggesting that mutations outside the antibiotic binding site can confer resistance by inducing increased disorder in the drug binding pocket (2). Moreover, a recent study of the structural basis of cross-resistance for PTC antibiotics advocates the idea of remote mutations affecting the antibiotic binding sites (6). There are no or minor changes observed in linezolid MICs for single mutations in the third or outer layer (orange and red nucleotides, Fig. 1C), but it cannot be ruled out that they can have a larger effect in other bacteria. In summary, the RNA mutations conferring linezolid resistance in other organisms also do so in M. smegmatis, although the resistance levels are small except that for G2576U. The A2503G, G2505A, C2571G, and C2612A mutations described in the literature without genetic proof confer some resistance in M. smegmatis.
There have been few attempts to evaluate the effect of double rRNA mutations. Antibiotic resistance occurring stepwise could very likely proceed through accumulation of several mutations working additively or even synergistically. In our previous study with tiamulin, the double mutations G2032A-U2504G, G2032A-C2499A, C2055A-U2504G, and C2055A-A2572U were found (40), and some of them were also in combination with a mutation in r-protein L3. Since the linezolid and tiamulin binding sites overlap, we hypothesized that these double mutations are likely to confer linezolid resistance. Cross-resistance between linezolid and tiamulin has been observed previously in S. aureus (33). All of the double mutations yield 4- to 32-fold increases in the linezolid MIC, whereas only the single mutation U2504G confers a significant increase. There are thus highly synergistic effects, especially for the C2055A-A2572U mutant, with a combination of a hypersensitive mutation and a mutation conferring a 2-fold increase in the linezolid MIC that are both placed in the second layer relative to bound linezolid (Fig. 1E). It is worthwhile to note that the resistance effect for these two single mutations does not concur with the recent suggestion that A2055 and U2572 determine the specificity of so-called A-site antibiotics through their influence on U2504 (12).
All of the nucleotides changed in the double mutations are clustered on the opposite side of U2504 relative to the bound linezolid drug (Fig. 1E), and while two of the double mutations involve nucleotides that are located next to each other (C2055A-A2572U and U2504G-C2055A), the other two involve nucleotide pairs that are too distant to interact directly (G2032A-C2499A and G2032A-U2504G). The closest distance between G2032 and C2499 is 5.8 Å, and this double mutation presumably disturbs interactions of A2572 and C2055, which are close to G2032 and C2499, respectively (Fig. 1E), which in turn affects U2504. Since the A2572 nucleotide is sandwiched between G2032 and U2504, the G2032A-U2504G double mutation probably involves direct perturbations of A2572 and G2504 in the binding cavity. We propose that the single mutations can be accommodated individually in the structure without severely disrupting the antibiotic binding cavity but not together. Thus, the four pairs of mutations presumably confer high linezolid resistance by changing the conformation of nucleotide 2504 and its interactions. It has been established from chemical probing experiments that mutant ribosomes with the single mutations C2055A, A2572U, and U2504G exhibit reduced CMCT [1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulphonate] modifications at the otherwise accessible nucleobases U2506 and U2585 compared to wild-type ribosomes (30). Other structural changes are also observed, including enhanced background bands at position G2576 due to structural hindrance of reverse transcriptase in the A2572U mutant ribosomes (30). These alterations suggest a reduction in flexibility at the PTC in ribosomes with single mutations. We propose that the synergistic effect of the double mutations is caused by further increasing the rigidity of the PTC, possibly by stabilizing a conformation that is nonproductive for linezolid binding.
One issue is how much resistance a mutation can provide, and another is how well the mutation is tolerated. Since the PTC contains many conserved nucleotides, it is to be expected that mutations might cause slower growth. Surprisingly few of the mutants studied here severely inhibit growth, except the double mutants. Three of the four double mutants (Table 3) grow very slowly, which implies that this functionally important region accepts subtle single changes but not too many changes together. It is questionable that such deleterious double mutations will ever appear in a natural setting.
Although cross-resistance between PTC antibiotics from 23S rRNA mutations is common, there is not a simple relationship between overlapping binding sites and cross-resistance. Our data clearly illustrate the complexity of cross-resistance related to rRNA mutations. For example, the third-layer G2032C mutation confers resistance to valnemulin and clindamycin but not to linezolid and chloramphenicol. Furthermore, the G2032A mutation yields the same valnemulin resistance as G2032C but shows only a very low increase in the clindamycin MIC and no resistance to linezolid and chloramphenicol. Small increases in linezolid resistance have been observed with plasmid expression of 23S rRNA with mutations at G2032 in E. coli, indicating that the mutational effects at this position on linezolid susceptibility are species specific (52). The A2503G and the G2505A mutations confer some resistance to linezolid and cross-resistance to chloramphenicol but no resistance to clindamycin and valnemulin. The G2576U mutation is the only single mutation providing high resistance to both linezolid and chloramphenicol, as well as a 2- to 4-fold increase in clindamycin resistance. The U2504G mutation provides a 4-fold-reduced sensitivity for linezolid and high resistance to chloramphenicol, but when combined with C2055A (not providing significant resistance), the synergistic effect yields a much higher linezolid resistance (16-fold relative to the wild-type level) but much lower resistance to chloramphenicol than U2504G alone. The C2055A-A2572U double mutant also has its own specific cross-resistance pattern, showing a synergistic effect for linezolid and clindamycin. The linezolid hypersensitivity with C2055A is converted to high linezolid resistance in the double mutant, as with chloramphenicol, albeit to a lesser extent. The strongest valnemulin effects are 4-fold increases conferred by single mutations, where there is no significant cross-resistance to linezolid. There is thus a correlation between linezolid and chloramphenicol resistance for the single mutations, but for the double mutations this simple connection does not hold. On the contrary, there does not seem to be a correlation between linezolid, valnemulin, and clindamycin resistance.
In summary, mutation of individual nonconserved nucleotides outside the linezolid binding site does not account for the species specificity of PTC antibiotics. This study illustrates that mutations at some distance from an antibiotic binding site can confer resistance and the same rRNA mutation can have significantly different effects in different bacteria. Moreover, the data show that rRNA double mutations can have synergistic effects on antibiotic resistance and that cross-resistance between PTC antibiotics is not easily predictable.
Acknowledgments
We thank Tatjana Kristensen for excellent technical assistance and Jacob Pøhlsgaard for contributing Fig. 1A. Valnemulin was kindly provided by Novartis Animal Health.
K.S.L. and B.V. acknowledge support from the Danish Medical Research Council and the Danish National Research Foundation.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Beringer, M., C. Bruell, L. Xiong, P. Pfister, P. Bieling, V. I. Katunin, A. S. Mankin, E. C. Böttger, and M. V. Rodnina. 2005. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 280:36065-36072. [DOI] [PubMed] [Google Scholar]
- 2.Blaha, G., G. Gürel, S. J. Schroeder, P. B. Moore, and T. A. Steitz. 2008. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 379:505-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carsenti-Dellamonica, H., M. Galimand, F. Vandenbos, C. Pradier, P. M. Roger, B. Dunais, M. Sabah, G. Mancini, and P. Dellamonica. 2005. In vitro selection of mutants of Streptococcus pneumoniae resistant to macrolides and linezolid: relationship with susceptibility to penicillin G or macrolides. J. Antimicrob. Chemother. 56:633-642. [DOI] [PubMed] [Google Scholar]
- 4.Colca, J. R., W. G. McDonald, D. J. Waldon, L. M. Thomasco, R. C. Gadwood, E. T. Lund, G. S. Cavey, W. R. Mathews, L. D. Adams, E. T. Cecil, J. D. Pearson, J. H. Bock, J. E. Mott, D. L. Shinabarger, L. Xiong, and A. S. Mankin. 2003. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278:21972-21979. [DOI] [PubMed] [Google Scholar]
- 5.Davidovich, C., A. Bashan, T. Auerbach-Nevo, R. D. Yaggie, R. R. Gontarek, and A. Yonath. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc. Natl. Acad. Sci. U. S. A. 104:4291-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidovich, C., A. Bashan, and A. Yonath. 2008. Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc. Natl. Acad. Sci. U. S. A. 105:20665-20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 8.Farrell, D. J., R. E. Mendes, J. E. Ross, and R. N. Jones. 2009. Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn. Microbiol. Infect. Dis. 65:392-403. [DOI] [PubMed] [Google Scholar]
- 9.Feng, J., A. Lupien, H. Gingras, J. Wasserscheid, K. Dewar, D. Légaré, and M. Ouellette. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 19:1214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giessing, A. M., S. S. Jensen, A. Rasmussen, L. H. Hansen, A. Gondela, K. Long, B. Vester, and F. Kirpekar. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory, S. T., J. F. Carr, D. Rodriguez-Correa, and A. E. Dahlberg. 2005. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 187:4804-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gürel, G., G. Blaha, P. B. Moore, and T. A. Steitz. 2009. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms, J. M., F. Schlünzen, P. Fucini, H. Bartels, and A. Yonath. 2004. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillemann, D., S. Rusch-Gerdes, and E. Richter. 2008. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob. Agents Chemother. 52:800-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbie, S. N., C. Bruell, S. Kalapala, S. Akshay, S. Schmidt, P. Pfister, and E. C. Böttger. 2006. A genetic model to investigate drug-target interactions at the ribosomal decoding site. Biochimie 88:1033-1043. [DOI] [PubMed] [Google Scholar]
- 16.Hobbie, S. N., S. K. Kalapala, S. Akshay, C. Bruell, S. Schmidt, S. Dabow, A. Vasella, P. Sander, and E. C. Böttger. 2007. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 35:6086-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbie, S. N., P. Pfister, C. Bruell, P. Sander, B. François, E. Westhof, and E. C. Böttger. 2006. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 50:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbie, S. N., P. Pfister, C. Brüll, E. Westhof, and E. C. Böttger. 2005. Analysis of the contribution of individual substituents in 4,6-aminoglycoside-ribosome interaction. Antimicrob. Agents Chemother. 49:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38, 27-28. [DOI] [PubMed] [Google Scholar]
- 20.Ippolito, J. A., Z. F. Kanyo, D. Wang, F. J. Franceschi, P. B. Moore, T. A. Steitz, and E. M. Duffy. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51:3353-3356. [DOI] [PubMed] [Google Scholar]
- 21.Kehrenberg, C., S. Schwarz, L. Jacobsen, L. H. Hansen, and B. Vester. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064-1073. [DOI] [PubMed] [Google Scholar]
- 22.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 23.Leach, K. L., S. M. Swaney, J. R. Colca, W. G. McDonald, J. R. Blinn, L. M. Thomasco, R. C. Gadwood, D. Shinabarger, L. Xiong, and A. S. Mankin. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol. Cell 26:393-402. [DOI] [PubMed] [Google Scholar]
- 24.Li, B. B., J. Z. Shen, X. Y. Cao, Y. Wang, L. Dai, S. Y. Huang, and C. M. Wu. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol. Lett. 308:144-149. [DOI] [PubMed] [Google Scholar]
- 25.Lin, A. H., R. W. Murray, T. J. Vidmar, and K. R. Marotti. 1997. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livermore, D. M. 2003. Linezolid in vitro: mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 51(Suppl. 2):ii9-ii16. [DOI] [PubMed] [Google Scholar]
- 27.Livermore, D. M., M. Warner, S. Mushtaq, S. North, and N. Woodford. 2007. In vitro activity of the oxazolidinone RWJ-416457 against linezolid-resistant and -susceptible staphylococci and enterococci. Antimicrob. Agents Chemother. 51:1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locke, J. B., M. Hilgers, and K. J. Shaw. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53:5275-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locke, J. B., M. Hilgers, and K. J. Shaw. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long, K. S., J. Poehlsgaard, L. H. Hansen, S. N. Hobbie, E. C. Böttger, and B. Vester. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol. Microbiol. 71:1218-1227. [DOI] [PubMed] [Google Scholar]
- 31.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, K., C. J. Dunsmore, C. W. Fishwick, and I. Chopra. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 52:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagiec, E. E., L. Wu, S. M. Swaney, J. G. Chosay, D. E. Ross, J. K. Brieland, and K. L. Leach. 2005. Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob. Agents Chemother. 49:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J. Bacteriol. 178:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 37.Persaud, C., Y. Lu, A. Vila-Sanjurjo, J. L. Campbell, J. Finley, and M. O'Connor. 2010. Mutagenesis of the modified bases, m(5)U1939 and psi2504, in Escherichia coli 23S rRNA. Biochem. Biophys. Res. Commun. 392:223-227. [DOI] [PubMed] [Google Scholar]
- 38.Pfister, P., S. Jenni, J. Poehlsgaard, A. Thomas, S. Douthwaite, N. Ban, and E. C. Böttger. 2004. The structural basis of macrolide-ribosome binding assessed using mutagenesis of 23S rRNA positions 2058 and 2059. J. Mol. Biol. 342:1569-1581. [DOI] [PubMed] [Google Scholar]
- 39.Poehlsgaard, J., P. Pfister, E. C. Böttger, and S. Douthwaite. 2005. Molecular mechanisms by which rRNA mutations confer resistance to clindamycin. Antimicrob. Agents Chemother. 49:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pringle, M., J. Poehlsgaard, B. Vester, and K. S. Long. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295-1306. [DOI] [PubMed] [Google Scholar]
- 41.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sander, P., L. Belova, Y. G. Kidan, P. Pfister, A. S. Mankin, and E. C. Böttger. 2002. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol. Microbiol. 46:1295-1304. [DOI] [PubMed] [Google Scholar]
- 44.Sander, P., T. Prammananan, and E. C. Böttger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 45.Schlünzen, F., E. Pyetan, P. Fucini, A. Yonath, and J. M. Harms. 2004. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 54:1287-1294. [DOI] [PubMed] [Google Scholar]
- 46.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 47.Toh, S. M., and A. S. Mankin. 2008. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J. Mol. Biol. 380:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vester, B., and R. A. Garrett. 1987. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie 69:891-900. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, D. N., F. Schluenzen, J. M. Harms, A. L. Starosta, S. R. Connell, and P. Fucini. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U. S. A. 105:13339-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolter, N., A. M. Smith, D. J. Farrell, W. Schaffner, M. Moore, C. G. Whitney, J. H. Jorgensen, and K. P. Klugman. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, A., S. P. Reddy, D. S. Smyth, M. E. Aguero-Rosenfeld, G. Sakoulas, and D. A. Robinson. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents Chemother. 54:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, J., A. Golshani, H. Aoki, J. Remme, J. Chosay, D. L. Shinabarger, and M. C. Ganoza. 2005. Protected nucleotide G2608 in 23S rRNA confers resistance to oxazolidinones in E. coli. Biochem. Biophys. Res. Commun. 328:471-476. [DOI] [PubMed] [Google Scholar]