Abstract
Previous studies showed an increased prevalence of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) thumb subdomain polymorphisms Pro272, Arg277, and Thr286 in patients failing therapy with nucleoside analogue combinations. Interestingly, wild-type HIV-1BH10 RT contains Pro272, Arg277, and Thr286. Here, we demonstrate that in the presence of zidovudine, HIV-1BH10 RT mutations P272A/R277K/T286A produce a significant reduction of the viral replication capacity in peripheral blood mononuclear cells in both the absence and presence of M41L/T215Y. In studies carried out with recombinant enzymes, we show that RT thumb subdomain mutations decrease primer-unblocking activity on RNA/DNA complexes, but not on DNA/DNA template-primers. These effects were observed with primers terminated with thymidine analogues (i.e., zidovudine and stavudine) and carbovir (the relevant derivative of abacavir) and were more pronounced when mutations were introduced in the wild-type HIV-1BH10 RT sequence context. RT thumb subdomain mutations increased by 2-fold the apparent dissociation equilibrium constant (Kd) for RNA/DNA without affecting the Kd for DNA/DNA substrates. RNase H assays carried out with RNA/DNA complexes did not reveal an increase in the reaction rate or in secondary cleavage events that could account for the decreased excision activity. The interaction of Arg277 with the phosphate backbone of the RNA template in HIV-1 RT bound to RNA/DNA and the location of Thr286 close to the RNA strand are consistent with thumb polymorphisms playing a role in decreasing nucleoside RT inhibitor excision activity on RNA/DNA template-primers by affecting interactions with the template-primer duplex without involvement of the RNase H activity of the enzyme.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is a major target for antiretroviral drug development (3, 61). HIV-1 RT catalyzes the conversion of the viral single-stranded genomic RNA into a double-stranded DNA that integrates into the host genome. HIV-1 RT is a heterodimer composed of 66- and 51-kDa subunits, with subdomains termed fingers, thumb, palm, and connection in both subunits and an RNase H domain in the large subunit only (23, 26, 29).
Approved antiretroviral drugs targeting the HIV-1 RT include nucleoside and nucleotide inhibitors (zidovudine, lamivudine, stavudine, didanosine, zalcitabine, abacavir, emtricitabine, and tenofovir) and nonnucleoside inhibitors (nevirapine, delavirdine, efavirenz, and etravirine) (39). Nucleoside RT inhibitors (NRTIs) mimic natural nucleosides. Inside the cell, nucleoside (and nucleotide) analogues need to be phosphorylated to their active triphosphate forms to act as competitive inhibitors of HIV-1 RT. Since NRTIs lack a 3′-OH group, their incorporation results in chain termination (12, 38, 62). Despite the efficacy of current antiretroviral therapies, the selection and emergence of drug-resistant HIV-1 strains are major factors contributing to treatment failure.
HIV-1 RT mutations conferring resistance to nucleoside (or nucleotide) inhibitors act either by (i) improving discrimination against the RT inhibitors (15, 54, 59) or (ii) by increasing the RT's ability to remove 3′-terminal chain terminator inhibitors from blocked DNA primers, through phosphorolysis mediated by ATP or pyrophosphate (PPi) (2, 41). The nucleotide excision or primer-unblocking mechanism appears to be most relevant for the acquisition of resistance to thymidine analogues, such as zidovudine (AZT) (3′-azido-3′-deoxythymidine) and stavudine (d4T) (2′,3′-didehydro-2′,3′-dideoxythymidine), through the accumulation of the so-called thymidine analogue resistance mutations (TAMs) (i.e., M41L, D67N, K70R, L210W, T215F or T215Y, and K219E or K219Q) (for recent reviews, see references 38 and 68). TAMs increase the rate of nucleotide excision. Although primer-unblocking efficiencies are influenced by the nucleotide sequence context (44), available data indicate that thymidine analogues and tenofovir are the best substrates of the reaction, while cytidine analogues are removed very inefficiently (7, 25, 31, 33, 41, 42, 46, 55, 63, 72). Despite being excisable, there is no agreement on the unblocking efficiencies for primers terminated with carbovir monophosphate (CBVMP) (46, 56, 72) or dideoxyadenosine monophosphate (ddAMP) (25, 43, 46). CBVMP and ddAMP are active metabolites of abacavir and didanosine, respectively.
Computational analysis of the HIV genetic variability in the RT-coding region revealed an association between the presence of TAMs and mutations at codons 35, 39, 43, 122, 203, 207, 208, 214, 218, 223, and 228 in patients failing therapy with RT inhibitors (9, 11, 65). Some of those mutations appear to increase viral fitness in the presence of TAMs, as demonstrated for K43E (24), Q207D (30), and L214F (52). However, very often, genotypic analysis is restricted to RT residues 1 to 240, while growing evidence suggests that potential antiretroviral therapy-related mutations in the thumb-connection subdomains (codons 241 to 424) and RNase H domains (codons 425 to 560) can likewise contribute to resistance to RT inhibitors. Thus, recent reports have suggested that mutations in the connection subdomain (e.g., E312Q, G335C/D, N348I, A360I/V, V365I, and A376S) and in the RNase H domain of the RT (e.g., Q509L, H539N, or D549N) can significantly increase zidovudine resistance by altering the balance between excision and template RNA degradation (13, 47, 48, 73). A reduction of the specific RNase H activity of the viral RT stabilizes the RNA/DNA duplex, giving RT more time to excise the AZT from the terminated primer (8, 14, 18).
The analysis of HIV-1 subtype B genomes of isolates obtained from untreated patients reveals three major polymorphisms within the RT thumb subdomain at positions 272, 277, and 286 (Stanford University HIV Drug Resistance Database [http://hivdb.stanford.edu]). Ala and Pro were found at position 272 in 47.4% and 44.0% of the sequenced isolates, respectively. At position 277, 59.0% of the sequences contained Lys, while 40.9% contained Arg. The variability at position 286 was lower, with Thr and Ala found in 69.1% and 29.0% of the sequences, respectively. Previous cross-sectional studies carried out with a large database containing HIV-1 subtype B pol sequences from patients treated in Spanish hospitals revealed an increased prevalence of Pro272, Arg277, and Thr286 in isolates from individuals failing therapy with abacavir/d4T and other nucleoside analogue combinations (20).
The p66 thumb subdomain plays a role in DNA polymerization by making important interactions with the minor groove of the template-primer through α-helices H (residues 255 to 268) and I (residues 278 to 286) (5, 16), while the p51 thumb subdomain contributes to the stabilization of the p66/p51 heterodimer (69). In addition, interactions between the p51 thumb subdomain and the RNase H domain of p66 appear to be essential for the conversion of the RT heterodimer from an inactive to an active form (45). Amino acid substitutions G262A and W266A within α-helix H are known to decrease RT processivity and frameshift fidelity (4) while impairing the RNase H-mediated removal of the polypurine tract during reverse transcription (51). These effects suggest that mutations in the RT thumb subdomain could alter the balance between excision and template RNA degradation in a way that could be similar to that reported for resistance-associated connection subdomain and RNase H domain mutations (13, 18, 47, 48, 73). Here, we show the effects of thumb subdomain polymorphisms on viral replication capacity and provide insights into the mechanism by which Pro272, Arg277, and Thr286 confer a selective advantage for viral replication in the presence of excisable nucleotide analogues.
(A preliminary report of this work has been presented at the International HIV & Hepatitis Virus Drug Resistance Workshop & Curative Strategies, at Dubrovnik, Croatia, 8 to 12 June 2010, abstract 66 [5a].)
MATERIALS AND METHODS
Reverse transcriptases.
In this study, the HIV-1BH10 RT has been arbitrarily designated the wild-type (WT) RT, and thumb subdomain polymorphisms have been considered mutations introduced in the sequence context of the BH10 enzyme (P272A, R277K, and T286A). Expression and purification of WT HIV-1BH10 RT, MAK_SSSY RT (HIV-1BH10 RT containing a Ser-Ser insertion between codons 69 and 70 and containing mutations M41L, A62V, T69S, K70R, and T215Y) and an RNase H-deficient RT (HIV-1 group O RT with mutations V75I/E478Q) were performed with modified versions of plasmid p66RTB, as previously described (1, 6, 10, 34, 36). Other mutant RTs were obtained by using the QuikChange site-directed mutagenesis kit (Stratagene), following the manufacturer's instructions. The double mutant M41L/T215Y was obtained with previously described mutagenic primers (10, 35). To introduce the P272A and R277K mutations, we used primers 5′-CAAGTCAGATTTACGCAGGGATTAAAGTAAAGCAATTATGTAAAC-3′ and 5′-GTTTACATAATTGCTTTACTTTAATCCCTGCGTAAATCTGACTTG-3′. To introduce the T286A mutation, the mutagenic primers were 5′-CTCCTTAGAGGAGCCAAAGCACTAACAGA-3′ and 5′-TCTGTTAGTGCTTTGGCTCCTCTAAGGAG-3′. The introduced mutations were confirmed by DNA sequencing. RT p66 subunits carrying a His6 tag at their C terminus were coexpressed with HIV-1 protease in Escherichia coli XL1 Blue to obtain p66/p51 heterodimers, which were later purified by ionic exchange followed by affinity chromatography (6, 34). RT concentrations were determined by active site titration as previously described (27).
Nucleotides and template-primers.
Stock solutions (100 mM) of deoxynucleoside triphosphates (dNTPs), dideoxy-ATP (ddATP), and ATP were obtained from GE Healthcare. AZT triphosphate (AZTTP), and carbovir triphosphate (CBVTP) were purchased from Moravek Biochemicals (Brea, CA). Stavudine triphosphate (d4TTP) was obtained from Sierra Bioresearch (Tucson, AZ). Before use, nucleoside triphosphates were treated with inorganic pyrophosphatase (Roche) to remove traces of PPi (33). DNA oligonucleotides 21P (5′-ATACTTTAACCATATGTATCC-3′), 25PGA (5′-TGGTAGGGCTATACATTCTTGCAGG-3′), 31T (5′-TTTTTTTTTAGGATACATATGGTTAAAGTAT-3′), D38 (5′-GGGTCCTTTCTTACCTGCAAGAATGTATAGCCCT ACCA-3′), D38C (5′-GGGTCCTTTATTCCCTGCAAGAATGTATAGCCCTACCA-3′), D38T (5′-GGGTCCTTTCAATCCTGCAAGAATGTATAGCCC TACCA-3′), and PR26 (5′-CCTGTTCGGGCGCCACTGCTAGAGAT-3′) and RNA oligonucleotides 31rna (5′-GGGUCCUUUCUUACCUGCAAGAAUGUAUAGC-3′), 31Trna (5′-UUUUUUUUUAGGAUACAUAUGGUUAAAGUAU-3′), D38rna (5′-GGGUCCUUUCUUACCUGCAAGAAUGUAUAGCCCUACCA-3′), D38Crna (5′-GGGUCCUUUAUUCCCUGCAAGAAUGUAUAGCCCUACCA-3′), and T35rna (5′-AGAAUGGAAAAUCUCUAGCAGUGGCGCCCGAACAG-3′) were obtained from Invitrogen. Oligonucleotides were labeled at their 5′ termini with [γ-32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (Promega), and then annealed to their corresponding templates or primers depending on the experiment.
Chain terminator excision assays.
RT-catalyzed DNA rescue reactions were performed with D38/25PGA, D38C/25PGA, and D38T/25PGA DNA duplexes, as previously described (32, 35). Briefly, the phosphorylated template-primer (75 nM) was preincubated at 37°C for 10 min in the presence of the corresponding RT at an active enzyme concentration of 60 nM in 50 mM HEPES buffer (pH 7.0) containing 15 mM NaCl, 15 mM magnesium acetate, 130 mM potassium acetate, 1 mM dithiothreitol, and 5% (wt/vol) polyethylene glycol 6000. Reactions were initiated by adding an equal amount of preincubation buffer containing the NRTI in its triphosphorylated form at a final concentration of 25 μM. After the samples were incubated at 37°C for 30 min, rescue reactions were initiated by adding a mixture of all dNTPs in the presence of sodium PPi (200 μM) or ATP (3.2 mM) depending on the assay. In these assays, all dNTPs except dATP were supplied at a final concentration of 100 μM. Since the next complementary dNTP (dATP in our assay conditions) has an inhibitory effect on the rescue reaction, time course experiments of the unblocking and extension reactions were carried out with 1 μM dATP. Rescue reactions were also performed with the RNA/DNA template-primers D38rna/25PGA and D38Crna/25PGA. These reactions were carried out under the same conditions as for the DNA duplexes, but the dNTP and inhibitor concentrations were 2-fold higher. Excision and extension reactions were carried out after preincubating the corresponding RT with template-primer and triphosphorylated NRTI for 10 min at 37°C. In experiments designed to assess the inhibitory effect of dATP, extension reaction mixtures were incubated for 15 to 40 min in the presence of different concentrations of dATP. In all cases, incubation times were within the linear range of the corresponding time course experiment. The reactions were stopped by adding an equal amount of sample loading buffer (10 mM EDTA in 90% formamide containing 3 mg/ml xylene cyanol FF and 3 mg/ml bromophenol blue). Products were resolved on a denaturing 20% (wt/vol) polyacrylamide-8 M urea gel, and primer rescue was quantified by phosphorimaging with a BAS 1500 scanner (Fuji) using the program Tina version 2.09 (Raytest Isotopenmessgerate GmbH, Staubenhardt, Germany).
Pre-steady-state kinetics of the ATP-dependent excision reaction.
For these experiments, primer 25PGA was blocked at its 3′ end with d4TTP or CBVTP, using terminal deoxynucleotidyltransferase as previously described (33). Then, the free nucleotides were eliminated by repeated passage through a Quick Spin minicolumn (Roche), until the A260 remained unchanged. Blocked primers were labeled at their 5′ terminus with [γ-32P]ATP and T4 polynucleotide kinase and annealed to template D38rna or D38Crna as described above, in order to obtain the template-primers D38rna/25PGAd4T and D38Crna/25PGACBV. Single-turnover conditions were used to study the excision of d4T monophosphate (d4TMP) and CBVMP. Solutions containing 250 nM RT and 30 nM D38rna/25PGAd4T or D38Crna/25PGACBV in RT buffer (15 mM HEPES [pH 7.0], 4 mM NaCl, 4 mM magnesium acetate, 130 mM KCH3COO, 1 mM dithiothreitol, and 5% [wt/vol] polyethylene glycol) were mixed with equal volumes of RT buffer containing 6.4 mM ATP and 36 mM MgCl2. Excision reactions were carried out for 0 to 60 min at 37°C. Aliquots were removed after defined incubation periods, quenched with sample loading buffer, and analyzed by denaturing polyacrylamide gel electrophoresis as described above. The formation of product (concentration of product [P]) over time was fitted to a single exponential decay: [P] = A × e−kobs × t, where kobs is the apparent kinetic constant of the excision reaction. A linear regression was used to fit the data when the excision rates were low. In those cases, we assumed that the amount of active RT was identical to the template-primer concentration in the assay, since reactions were carried out with a large excess of RT relative to the RNA/DNA duplex.
Determination of dissociation equilibrium constants (Kd) for WT and mutant RTs and DNA/DNA and RNA/DNA template-primers.
RTs were preincubated with increasing concentrations of the 5′-32P-labeled 25/38-mer DNA-DNA (2 to 60 nM) for 10 min at 37°C in 10 μl of a buffer containing 100 mM HEPES (pH 7.0), 30 mM NaCl, 30 mM magnesium acetate, 130 mM KCH3COO, 1 mM dithiothreitol, and 5% (wt/vol) polyethylene glycol. Reactions were initiated by the addition of 10 μl of 1 mM dTTP to 10 μl of the solution indicated above, and incubated at 37°C. In these experiments, the active RT concentration was around 3 nM. Aliquots of 4 μl were then removed at 10, 20, 30, and 40 s, quenched with sample loading buffer, and analyzed by denaturing polyacrylamide gel electrophoresis as described above. The burst amplitudes (RT bound to template-primer at time zero) were plotted as a function of the template-primer concentration, and the data were fitted to a quadratic equation to obtain the equilibrium dissociation constant for RT binding to template-primer (37).
RNase H assays.
RNase H activity was evaluated with 31Trna/21P and D38rna/25PGA complexes as previously described (1). Assays were carried out at 37°C in 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, and 50 nM 32P-labeled RNA/DNA template-primer, in the presence of 100 nM RT (active site concentration). Aliquots were removed at various times, quenched with equal volumes of gel loading dye, and analyzed by denaturing polyacrylamide gel electrophoresis as described above. In addition, the RNase H activity of WT and mutant RTs was determined with an AZT-terminated RNA/DNA substrate (i.e., T35rna/PR26) (8). Briefly, the 32P-labeled T35rna/PR26 complex (80 nM) was blocked at the 3′ end of the primer with AZTTP in the presence of a previously characterized RNase H-deficient RT (HIV-1 group O RT containing mutations V75I/E478Q [1]). Reaction mixtures were incubated for 60 min at 37°C in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, and 60 μM AZTTP in the presence of 30 nM RT. After heat inactivation of the enzyme, the free AZTTP was eliminated with a Quick Spin minicolumn (Roche) as described above. T35rna/PR26AZT RNase H cleavage reactions were carried out at 37°C in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, and 10 mM MgCl2, containing 0.3 mM ATP, 20 nM 32P-labeled RNA/DNA template-primer and the corresponding RT at a 200 nM active site concentration.
Recombinant virus and drug susceptibility tests.
These recombinant virus and drug susceptibility assays were performed as described previously (32, 36). Briefly, full-length RT coding sequence DNA was amplified by PCR from the expression plasmids carrying the appropriate RT, using primers IN3 and IN5 (36). The PCR products were then cotransfected in MT-4 cells with an RT-deleted HXB2-D clone previously linearized with BstEII (28). Culture supernatants were harvested when the HIV-1 p24 antigen concentration surpassed 20 ng/ml. Progeny virus was propagated and titrated in MT-4 cells. The nucleotide sequence of the RT-coding region of the progeny virus was PCR amplified with the above-mentioned primers and checked for possible reversions or undesired mutations. The MT-4 cells and the deleted HXB2-D clone were obtained from the AIDS Reagent Program (Medical Research Council). RT inhibitors were obtained from the NIH AIDS Research and Reference Reagent Program. HIV-1 drug susceptibility data were obtained after infecting 30,000 MT-4 cells with 100 50% tissue culture infective doses (TCID50) of virus at a multiplicity of infection of 0.003 by exposing the HIV-1-infected cultures to various concentrations of each drug (5-fold dilutions). After the MT-4 cells were allowed to proliferate for 5 days, the number of viable cells was quantified by a tetrazolium-based colorimetric method [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method] as described elsewhere (50).
Replication capacity assays.
Viral replication kinetics of mutant viruses was assayed by infecting 5 × 106 peripheral blood mononuclear cells (PBMCs) (mixed from two healthy donors), previously stimulated with 3 μg/ml of phytohemagglutinin (Sigma-Aldrich) and 10 U/ml of interleukin 2 with 500 TCID50 of each viral stock (multiplicity of infection of 0.0001) (52). After incubation for 2 h at 37°C, cells were washed twice with phosphate-buffered saline and resuspended in RPMI 1640 medium supplemented with 20% fetal bovine serum and interleukin 2 (10 U/ml) at a final concentration of 1 × 106 cells/ml. Zidovudine (AZT) at different concentrations was added to the cultures when indicated. Viral replication was quantified by measuring HIV-1 p24Gag antigen production in the culture supernatant for 10 days. Growth kinetics was analyzed by fitting a linear model to the log-transformed p24Gag data during the exponential growth phase by maximum likelihood methods.
RESULTS
Thumb subdomain polymorphisms and their effects on viral replication.
Our previous statistical analysis carried out with HIV-1 pol sequences obtained from patients treated in Spain showed that several RT thumb subdomain polymorphisms (i.e., Pro272, Arg277, and Thr286) were selected during therapy with nucleoside drug combinations such as d4T/abacavir, d4T/ddI, ddI/abacavir, d4T/lamivudine, and d4T/tenofovir (20). The largest levels of statistical significance were observed with the first two combinations. In addition, correlated pairs of mutations at positions 272 and 277 and positions 272 and 286 were observed in HIV sequences from patients failing therapy with d4T and ddI. Despite the statistical correlation, the effects of thumb mutations P272A/R277K/T286A on nucleoside (or nucleotide) RT inhibitor susceptibility were not significant in phenotypic assays (Table 1). All viruses tested were found to be susceptible to d4T, abacavir, ddI, tenofovir, lamivudine, and nevirapine. Statistically significant differences between the 50% inhibitory concentrations (IC50s) obtained with WT and mutant HIV-1 were observed only in the case of AZT (P < 0.01 by Student's t test). Thus, recombinant HIV-1 containing RT mutations M41L/T215Y showed a decrease in susceptibility to AZT of about 3-fold. Interestingly, in the presence of TAMs (M41L/T215Y), the addition of the thumb subdomain mutations P272A, R277K, and T286A produced a slight decrease of the IC50 for the inhibitor.
TABLE 1.
Susceptibility of HIV-1 constructs to RT inhibitors
| RT | IC50 (nM)a |
||||||
|---|---|---|---|---|---|---|---|
| AZT | d4T | Abacavir | ddI | Tenofovir | Lamivudine | Nevirapine | |
| WT | 5.1 ± 2.6 | 315.6 ± 44.9 | 1,280.8 ± 242.4 | 2,210.4 ± 363.6 | 181.0 ± 38.6 | 1,014.1 ± 39.3 | 44.0 ± 15.7 |
| P272A/R277K/T286A | 6.1 ± 2.3 (1.2) | 289.3 ± 50.9 (0.9) | 1,169.8 ± 112.1 (0.9) | 2,499.8 ± 894.9 (1.1) | 221.1 ± 28.5 (1.2) | 1,519.2 ± 486.0 (1.5) | 23.1 ± 4.1 (0.5) |
| M41L/T215Y | 14.5 ± 0.8 (2.8) | 399.2 ± 54.3 (1.3) | 1,526.1 ± 236.7 (1.2) | 1,888.8 ± 604.0 (0.9) | 264.4 ± 38.5 (1.5) | 1,988.7 ± 628.1 (1.9) | 22.8 ± 14.4 (0.5) |
| M41L/T215Y/P272A/R277K/ T286A | 10.7 ± 6.5 (2.1) | 378.6 ± 90.3 (1.2) | 1,768.2 ± 188.1 (1.4) | 2,562.4 ± 710.3 (1.2) | 261.4 ± 78.3 (1.4) | 1,558.8 ± 854.2 (1.5) | 29.1 ± 24.2 (0.7) |
The IC50 values shown are averages ± standard deviations of at least three tests, with each one performed six times. The fold increase in IC50 relative to the wild-type HXB2 virus control carrying the RT sequence of BH10 is shown in parentheses.
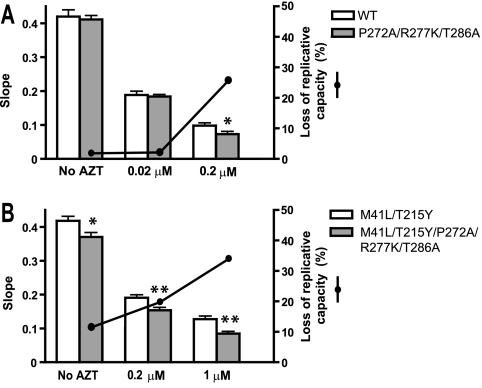
Viral replication kinetics assays carried out in PBMCs showed that mutations P272A/R277K/T286A had a negative effect on viral fitness (Fig. 1). Growth kinetics curves were analyzed by fitting a linear model to the log-transformed p24Gag data during the exponential growth phase (see Fig. S1 in the supplemental material). The slope of p24 antigen production provides a reliable estimate of the viral replication capacity. As shown in Fig. 1, the addition of thumb subdomain mutations to HIV variants containing TAMs decreased viral replication capacity by 11 to 34%. These effects were more significant at higher concentrations of AZT. A similar trend was observed when mutations were introduced in a WT recombinant virus, although the negative effects were observed only in experiments carried out in the presence of 0.2 μM AZT. These results together with the observed associations between thumb subdomain polymorphisms and failure to therapies containing combinations of excisable NRTIs suggested a link between excision and the selection of HIV-1 variants containing Pro272/Arg277/Thr286.
FIG. 1.
Replication kinetics assay in the absence and presence of AZT. The slope of p24 antigen production of each virus after infection of PBMCs (mixed from two donors) is shown by the bars. Comparisons of WT versus mutant P272A/R277K/T286A virus and mutant M41L/T215Y virus versus M41L/T215Y/P272A/R277K/T286A virus are shown in panels A and B, respectively. The significance of the difference between slopes was calculated using the GraphPrism v. 4 software (*, P < 0.05; **, P < 0.01). Solid black circles represent the percentage loss of replication capacity of the recombinant HIV-1 variants containing the P272A, R277K, and T286A mutations in the absence (A) or presence (B) of M41L/T215Y mutations under different assay conditions.
Effects of mutations on the ATP-mediated excision of NRTIs from chain-terminated DNA/DNA and RNA/DNA template-primers.
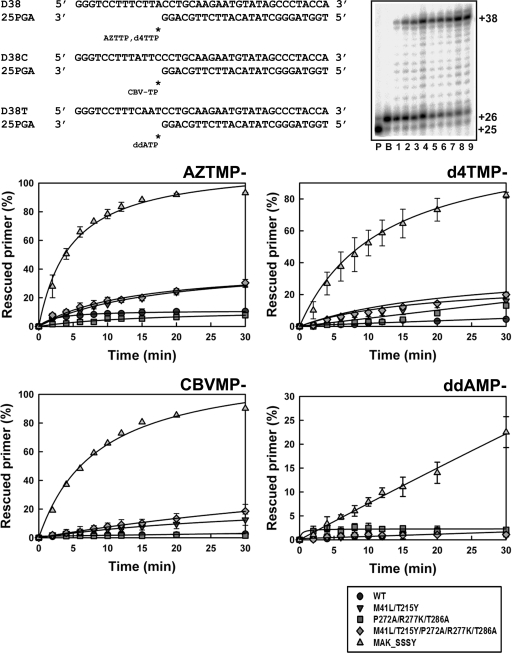
The ability of RTs to unblock NRTI-terminated primers was initially assessed with DNA/DNA template-primers (Fig. 2). An excision-proficient RT (i.e., MAK_SSSY [M41L/A62V/T69SSS/K70R/T215Y]) was used as a reference. In the presence of 3.2 mM ATP, MAK_SSSY was able to excise AZT monophosphate (AZTMP), d4TMP, CBVMP, and ddAMP, although ddAMP was removed at a very low rate. The ATP-dependent phosphorolytic activities of WT and mutant RTs (M41L/T215Y, M41L/T215Y/P272A/R277K/T286A, and P272A/R277K/T286A) were rather small in comparison with the activity shown by the MAK_SSSY RT. Although M41L/T215Y and M41L/T215Y/P272A/R277K/T286A RTs showed some activity with AZTMP-, d4TMP- and CBVMP-terminated primers, thumb subdomain mutations had a minor effect on excision. As previously observed with other RT variants, all enzymes tested showed similar excision efficiencies in the presence of PPi (200 μM) (data not shown).
FIG. 2.
ATP-mediated excision of AZTMP, d4TMP, CBVMP, and ddAMP from DNA/DNA template-primers by WT and mutant RTs. Reactions were carried out with 38/25-mer DNA/DNA heteropolymeric complexes (sequences shown at the top of the figure). First, the inhibitor was incorporated at position +1 (indicated with an asterisk) of the 25-nucleotide primer (lane P) to generate a 26-nucleotide product (lane B). Excision of the inhibitor and further primer extension in the presence of 3.2 mM ATP and a mixture of dNTPs lead to the formation of a fully extended 38-nucleotide product. A representative time course experiment of a primer rescue reaction is shown in lanes 1 to 9, which correspond to aliquots removed 2, 4, 6, 8, 10, 12, 15, 20, and 30 min after the addition of 3.2 mM ATP (gel in the top right corner of the figure). Graphs of time course experiments of primer rescue reactions initiated from inhibitor-terminated primers are given below. All dNTPs in the assays were supplied at 100 μM, except for dATP whose concentration was 1 μM. Template-primer and active RT concentrations in these assays were 30 and 24 nM, respectively. The values (averages ± standard deviations [error bars]) were obtained from three independent experiments. The MAK_SSSY RT is an excision-proficient enzyme that contains a Ser-Ser insertion between codons 69 and 70 and contains mutations M41L, A62V, T69S, K70R, and T215Y.
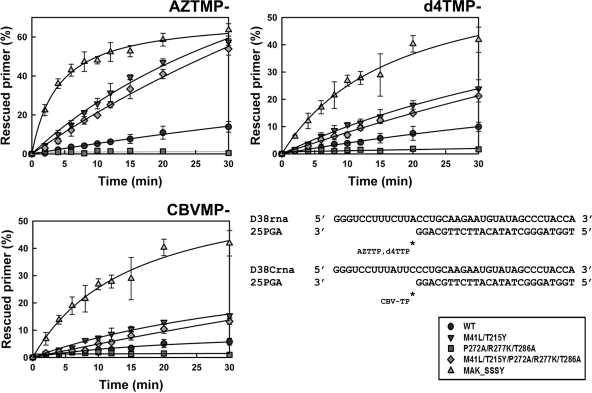
As in the case of DNA/DNA template-primers, the efficiency of the ATP-dependent phosphorolytic activity on RNA/DNA substrates in reactions catalyzed by MAK_SSSY RT followed the order AZTMP > d4TMP > CBVMP (Fig. 3). Control experiments carried out with d4T-terminated duplexes showed that the efficiencies of the unblocking and extension reactions catalyzed by all RTs were similar in the presence of 30 nM RT and 10 nM D38rna/25PGA (Fig. 3) to the efficiencies obtained in the presence of 24 nM RT and 30 nM D38rna/25PGA (data not shown). The ATP-mediated excision activity of M41L/T215Y and M41L/T215Y/P272A/R277K/T286A RTs was higher in reactions carried out with blocked RNA/DNA template-primers than with DNA/DNA substrates. Although both enzymes had remarkable excision activity, mutations P272A, R277K, and T286A produced a small reduction in the corresponding excision rates. These differences could not be attributed to their susceptibility to inhibition by the next complementary dNTP (see Table S1 in the supplemental material). Thus, in reactions catalyzed by M41L/T215Y and M41L/T215Y/P272A/R277K/T286A RTs, ATP-dependent rescue of primers terminated with d4TMP was inhibited by the next complementary dNTP at concentrations of around 4 μM. These values were similar to those reported for other RTs and obtained in reactions carried out with different DNA/DNA template-primers (10, 36, 42).
FIG. 3.
ATP-mediated excision of AZTMP, d4TMP, and CBVMP from RNA/DNA template-primers by WT and mutant RTs. Time course experiments of excision reactions were carried out in the presence of 3.2 mM ATP. Template-primers used are indicated below. All dNTPs in the assays were supplied at 200 μM, except for dATP whose concentration was 2 μM. Template-primer and active RT concentrations in these assays were 10 and 30 nM, respectively. The values (averages ± standard deviations [error bars]) were obtained from three independent experiments.
Interestingly, the introduction of P272A/R277K/T286A in an otherwise WT sequence context rendered an enzyme lacking ATP-dependent phosphorolytic activity in our assay conditions. These effects were observed in the presence of RNA/DNA duplexes containing primers terminated with AZTMP, d4TMP, or CBVMP. The presence of Pro272, Arg277, and Thr286 (as in the WT BH10 RT) conferred detectable excision activity, particularly on primers terminated with AZT and d4T, annealed to RNA templates (Fig. 3).
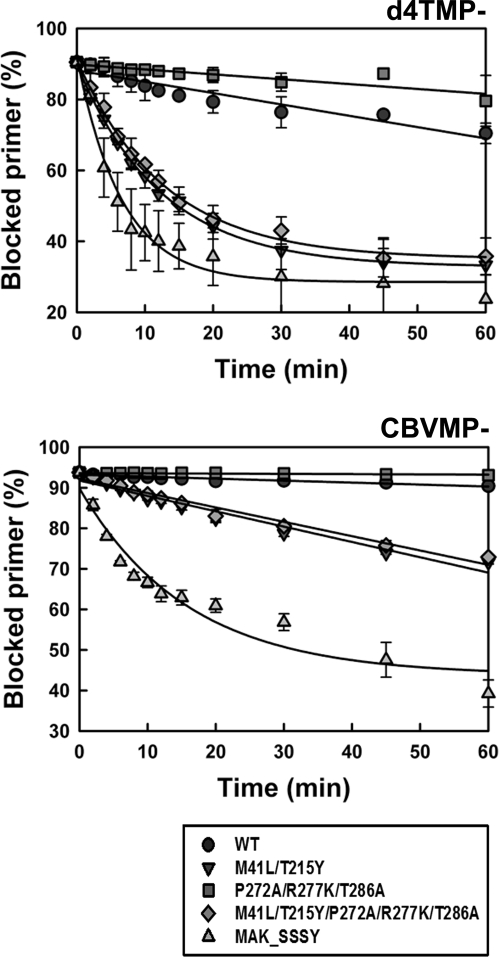
The d4TMP excision rates for WT and mutant P272A/R277K/T286A RT were determined under single-turnover conditions by using a d4TMP-terminated primer previously annealed to a 38-nucleotide RNA template in the presence of 3.2 mM ATP but without dNTPs. The WT RT showed a d4TMP excision rate (kobs) of 0.0032 min−1, which was about 2.5 times higher than the value obtained for mutant P272A/R277K/T286A RT (Fig. 4). Both enzymes showed negligible activity on CBVMP-terminated RNA/DNA substrates. The differences between M41L/T215Y and M41L/T215Y/P272A/R277K/T286A RTs were not significant in these assays. Both enzymes showed remarkable d4TMP excision activity on RNA/DNA templates (kobs around 0.08 min−1), although they were less efficient in removing CBVMP (kobs around 0.0036 min−1).
FIG. 4.
Kinetics of the ATP-dependent excision of d4TMP and CBVMP from RNA/DNA template-primers. Time course experiments for the excision reaction of d4TMP- or CBVMP-terminated primers (26-mers) annealed to their corresponding 38-nucleotide RNA templates (30 nM) were determined in the presence of 3.2 mM ATP. The excision reaction was catalyzed by WT and mutant RTs (250 nM). The calculated kobs values for the d4TMP excision reaction were 0.0032 ± 0.0003 min−1 for WT RT, 0.0014 ± 0.0002 min−1 for mutant P272A/R277K/T286A RT, 0.0810 ± 0.0025 min−1 for mutant M41L/T215Y RT, 0.0782 ± 0.0044 min−1 for mutant M41L/T215Y/P272A/R277K/T286A RT, and 0.153 ± 0.018 min−1 for the MAK_SSSY RT. For the excision of CBVMP, the kobs values for mutants M41L/T215Y, M41L/T215Y/P272A/R277K/T286A, and MAK_SSSY RTs were 0.0038 ± 0.0003 min−1, 0.0035 ± 0.0002 min−1, and 0.0629 ± 0.0138 min−1, respectively.
RNase H activity of WT and mutant RTs.
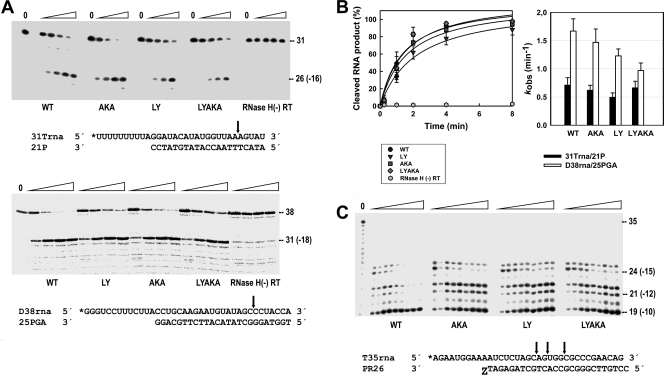
Thumb subdomain polymorphisms did not have a major impact on the RNase H activity as measured with the 31Trna/21P complex (Fig. 5A). Cleavage patterns monitored on the 5′-end-labeled RNA template revealed similar kinetics for reactions catalyzed by RTs with or without thumb subdomain mutations. No secondary cleavages were detected with the 31Trna/21P template-primer, under our assay conditions. Similar results were obtained with RNA/DNA template-primers used in rescue reactions (i.e., D38rna/25PGA) (Fig. 5A), although in this case the primary cleavage at position −18 occurred with higher efficiency. An increased frequency of secondary RNase H cleavages could diminish RNA/DNA duplex length and decrease the efficiency of NRTI excision. RNase H secondary cleavages occurring during the excision reaction were monitored with an AZT-terminated RNA/DNA complex in the presence of ATP. Catalytic rate constants for the cleavage of D38rna and 31Trna were in the range of 0.97 to 1.67 min−1 and 0.49 to 0.71 min−1, respectively (Fig. 5B). Differences between WT and P272A/R277K/T286A RTs and between M41L/T215Y and M41L/T215Y/P272A/R277K/T286A RTs were not significant in these assays (P > 0.1 by Student's t test). As shown in Fig. 5C and in Fig. S2 in the supplemental material, in an excision-competent mode, mutant RTs with P272A/R277K/T286A, M41L/T215Y, and M41L/T215Y/P272A/R277K/T286A had similar kinetics of formation of the −10 cleavage product. However, the WT enzyme showed 1.6-fold increased activity. Similar band patterns were observed for all tested RTs when the reactions were carried out in the absence of ATP (data not shown). As demonstrated for several RT connection and RNase H domain mutations (8, 48, 53), increased RNase H secondary cleavage efficiency correlates with lower levels of excision activity. Therefore, the higher ATP-mediated excision activity of WT RT relative to mutant P272A/R277K/T286A cannot be explained by invoking the effects of thumb subdomain mutations on polymerase-independent cleavages.
FIG. 5.
RNase H activity of WT and mutant RTs. (A) [32P]RNA/DNA substrates (50 nM) were cleaved at 37°C in the presence of the corresponding RT at 100 nM (active enzyme concentration). The template-primer sequences are shown below. For 31Trna/21P, the time points were obtained after incubating the samples for 1, 2, 4, and 8 min. For D38rna/25PGA, the time points were obtained after incubating the samples for 10, 20, 30, and 40 s. The time is shown above the gel by the height of the triangle above four lanes for each sample. Mutant RTs are abbreviated as follows: AKA, P272A/R277K/T286A; LY, M41L/T215Y; LYAKA, M41L/T215Y/P272A/R277K/T286A. An RNase H-deficient RT [RNase H(-) RT] (HIV-1 group O RT with mutations V75I/E478Q [1]) was included as a control. (B) Time courses of RNase H cleavage reactions carried out with 31Trna/21P (left), and apparent rate constants for the cleavage of templates 31Trna and D38rna, as obtained from 3 or 4 independent experiments (right). (C) Representative autoradiogram of the RNase H cleavage activity of WT and mutant RTs during ATP-mediated AZTMP excision. Assays were performed with the template-primer shown below (at 20 nM) in the presence of 200 nM RT. The time points in the experiments were 10, 20, 30, 45, 60, 90, 120, 150, and 180 min and are given by the height of the triangle above nine lanes for each sample. respectively. Cleavage sites are indicated by the black arrows. The labeled 5′ ends of the templates are marked with asterisks.
Effects of thumb subdomain polymorphisms on template-primer binding.
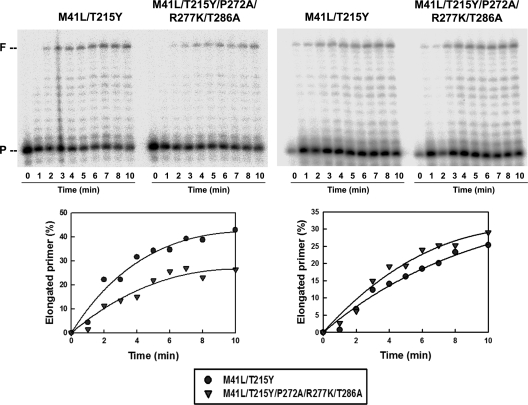
The effects of thumb subdomain mutations on substrate binding were determined by measuring the equilibrium dissociation constants (Kd) for WT and mutant RTs and DNA/DNA and RNA/DNA template-primers. Both template-primers were substrates of the excision reactions shown in Fig. 2 and 3. However, the RNA/DNA template-primer used (i.e., 31rna/25PGA) was a derivative of D38rna/25PGA lacking 7 nucleotides at the 3′ end of the RNA. These nucleotides were eliminated, since the corresponding RNase H primary cleavage (at position +18) occurs early during the ATP-mediated excision reaction (Fig. 5). As shown in Table 2, thumb subdomain polymorphisms had a minor impact on the Kd for DNA/DNA complexes (38/25-mer), with values around 2 to 3 nM. However, WT and mutant M41L/T215Y showed about 2-fold-higher affinity for RNA/DNA than their homologous counterparts having the thumb subdomain mutations P272A, R277K, and T286A. Primer extension assays carried out with 31rna/25PGA were consistent with the Kd measurements (Fig. 6). At low concentrations of template-primer (i.e., 1 nM), the amount of fully extended product was about 2-fold higher for mutant M41L/T215Y RT than for M41L/T215Y/P272A/R277K/T286A RT, while at higher concentrations (i.e., 10 nM), differences in the efficiency of the primer extension reaction were very small. On the other hand, thumb subdomain mutations had a relatively minor influence on RNase H cleavage patterns and kinetics in reactions carried out in the presence of low concentrations of RNA/DNA duplexes (i.e., 1.5 nM D38rna/25PGA) (see Fig. S3 in the supplemental material). Interestingly, the results of the primer extension assays were in agreement with fitness assays showing that in the absence of antiretroviral drugs, HIV clones containing the M41L/T215Y RT had a higher replication capacity than their homologous HIV-1 variants containing RT mutations M41L/T215Y/P272A/R277K/T286A (Fig. 1).
TABLE 2.
Dissociation equilibrium constants for WT and mutant HIV-1 RTs and DNA/DNA and RNA/DNA template-primersa
| RT | Apparent Kd (nM) |
|
|---|---|---|
| DNA/DNA | RNA/DNA | |
| WT | 2.19 ± 0.34 | 1.23 ± 0.33 |
| P272A/R277K/T286A | 1.75 ± 0.44 | 2.76 ± 0.45 |
| M41L/T215Y | 3.13 ± 0.04 | 1.14 ± 0.30 |
| M41L/T215Y/P272A/R277K/T286A | 2.77 ± 0.31 | 2.09 ± 0.46 |
The Kd values for DNA/DNA binding were obtained with the template-primer D38/25PGA. The RNA/DNA template-primer used in these experiments was D31rna/25PGA (sequences given below).
D31rna 5′ GGGUCCUUUCUUACCUGCAAGAAUGUAUAGC 3′
25PGA 3′ GGACGTTCTTACATATCGGGATGGT 5′
The values reported are averages ± standard deviations obtained from three independent experiments.
FIG. 6.
Extension of an unblocked DNA primer 25PGA by mutant RTs M41L/T215Y and M41L/T215Y/P272A/R277K/T286A in the presence of an RNA template (31rna). Reactions were carried out with template-primer concentrations of 1 nM (left panel) and 10 nM (right panel) in 50 mM HEPES buffer (pH 7.0) containing 15 mM NaCl, 15 mM magnesium acetate, 130 mM potassium acetate, 1 mM dithiothreitol, 5% (wt/vol) polyethylene glycol 6000, and 200 μM each dNTP except dATP, which was supplied at 2 μM. The RT concentration used in these assays was 3 nM (active site concentration). P, primer; F, full-length product.
DISCUSSION
The HIV-1 RT thumb subdomain residues 255 to 286 form a characteristic helix-turn-helix secondary structure (termed the helix clamp) that exhibits sequence homology with other nucleic acid polymerases (22). In p66, the two α-helices (H and I) interact with the template-primer, while in p51 they play a structural role by contributing to the stabilization of the RT heterodimer (23, 29, 40, 69). Our previous statistical analysis carried out with HIV-1 pol sequences obtained from patients treated in Spain showed that thumb subdomain polymorphisms were selected during therapy with NRTIs (20). In most cases, polymorphisms were associated with TAMs and the most significant associations were detected with samples from patients failing therapy with combinations of excisable NRTIs (e.g., abacavir/d4T, d4T/ddI, etc.). In those studies, the frequencies of Pro272, Arg277, and Thr286 in the naïve population were 45.0%, 25.7%, and 54.5%, respectively. However, those numbers increase up to 82.6%, 63.0%, and 91.3%, respectively, among isolates from patients failing therapy with abacavir/d4T (20).
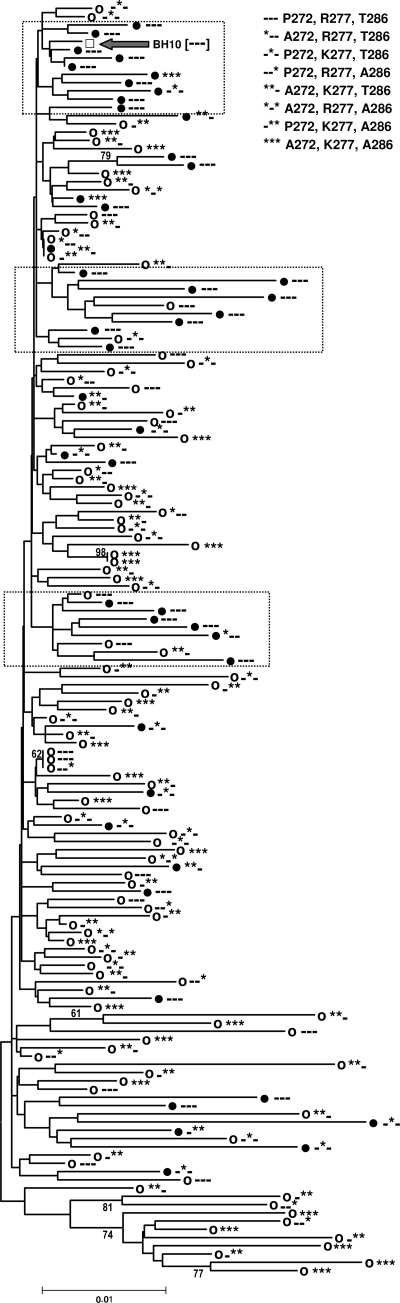
Phenotypic assays carried out with mutant RTs bearing thumb subdomain mutations like P272A, R277K, and T286A failed to detect differences in their susceptibility to NRTIs. This result was not unexpected, assuming their accessory role in resistance and the fact that assays were carried out in MT-4 cells. These cells contain high concentrations of dNTP that are known to counteract nucleotide excision mechanisms of resistance, particularly for abacavir, d4T, ddI, and tenofovir (64, 67). However, in phenotypic assays, mutant HIV-1 carrying the M41L/T215Y/P272A/R277K/T286A RT was found to be slightly more susceptible to AZT than its homologous counterpart containing the double mutant M41L/T215Y polymerase. Moreover, viral replication capacity assays carried out with PBMCs underlined these differences and showed that HIV-1 clones bearing Pro272, Arg277, and Thr286 in their RT-coding region had an increased growth rate in the presence of AZT. A phylogenetic tree obtained with the HIV-1 pol sequences derived from our previously reported cross-sectional study (20) showed the clustering of isolates obtained from patients failing therapy with abacavir and d4T bearing Pro272, Arg277, and Thr286 in their RT-coding region (Fig. 7). Despite the lack of sequence information from the connection and RNase H domains of the RT, these data argue in favor of the selection of HIV variants containing specific combinations of RT thumb polymorphisms after extensive treatment with excisable NRTIs.
FIG. 7.
Evolutionary relationships of HIV-1 protease and RT-coding sequences of isolates from naïve patients and individuals failing therapy with abacavir and d4T. Sequences (GenBank accession numbers HM460345 to HM460497) from isolates obtained from patients failing treatment with abacavir and d4T and from untreated patients (20) are represented by solid black circles and open circles, respectively. The WT BH10 sequence, encoding Pro272, Arg277 and Thr286 in the viral RT, is represented by the open square. Clusters of isolates from patients failing therapy with abacavir/d4T are boxed. The evolutionary history was inferred by the neighbor-joining method (57) by using p-distances in the substitution model. The bootstrap consensus tree was inferred from 2,000 replicates (19). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (2,000 replicates) are shown next to the branches (19). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. A total of 778 positions were considered in the final data set (all positions containing gaps and missing data were eliminated prior to analysis). Phylogenetic trees were obtained by using MEGA4 software (66).
Biochemical analyses designed to elucidate the mechanism by which thumb subdomain mutations impair viral fitness in the presence of RT inhibitors support a relevant role for ATP-mediated excision, particularly for thymidine analogues. Although thumb subdomain mutations did not affect the efficiency of the rescue reactions carried out with chain-terminated DNA/DNA template-primers, we detected significant differences in rescue reactions carried out with RNA/DNA complexes, particularly when mutations were introduced in a WT sequence context. The effects were larger with AZT- and d4T-terminated primers than with primers blocked with CBVMP. The rates of excision reactions carried out in the absence of dNTPs revealed a direct effect of the mutations in the kinetics of thymidine analogue removal in the absence of TAMs.
Our results suggest that nucleoside analogue resistance mechanisms similar to those reported for several connection subdomain and RNase H domain mutations could operate in the case of thumb polymorphisms. Thus, several mutations such as N348I (14, 18, 21, 53, 73), A360V (18, 47) and Q509L (8) were shown to increase chain-terminated primer rescue with RNA/DNA complexes, but not with DNA/DNA template-primers. However, these mutations as well as others found in the connection and RNase H domains of the RT (e.g., RNase H primer grip mutations such as G335C/D, V365I, A376S, etc.) (13, 14) had an impact on RNase H activity either by decreasing its specific activity or by altering the RNase H secondary cleavage kinetics. These effects were demonstrated in the presence of TAMs. We tested the effects of thumb subdomain polymorphisms on the kinetics of accumulation of RNase H cleavage fragments, using three different RNA/DNA complexes and different assay conditions. However, none of the experiments revealed significant differences between RTs containing or lacking thumb subdomain mutations. The G333D mutation in the connection subdomain has also been found to promote resistance to AZT in the presence of TAMs without affecting RNase H activity (74), but unlike in the case of the thumb mutations, an increased excision was reported for both RNA/DNA and DNA/DNA substrates.
In agreement with the results of rescue assays, we found that the apparent Kd for DNA/DNA was not influenced by the presence of thumb polymorphisms. However, in the case of RNA/DNA substrates, WT and M41L/T215Y RTs showed higher affinity for the template-primer than their homologous enzymes having thumb subdomain mutations. These differences could explain the lower efficiency of rescue reactions catalyzed by enzymes containing mutations P272A/R277K/T286A. It should be noted that when the amino acid substitutions M41L and T215Y were present, the negative effect of P272A/R277K/T286A on the efficiency of the rescue reaction was barely detectable at template-primer concentrations around 10 nM but become more significant at lower RNA/DNA concentrations (i.e., at 1 nM, as shown in Fig. 6). A reduction in template-primer affinity could influence nucleotide excision by decreasing the availability of NRTI in an excision-competent conformation but could also decrease the efficiency of the primer extension reaction after removal of the chain-terminating inhibitor. In the absence of M41L and T215Y, thumb subdomain polymorphisms could also affect the orientation of the blocked primer, thereby influencing excision by altering the geometry required for the attack of the PPi donor on the terminal phosphodiester bond.
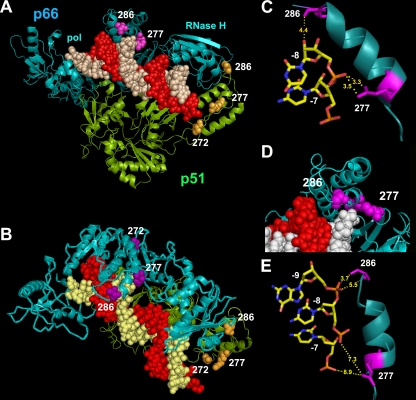
Our results raise the question of how thumb subdomain mutations could affect NRTI excision. The analysis of the crystal structure of HIV-1 RT complexed with an RNA/DNA substrate (60) revealed that Arg277 and Thr286 of the p66 subunit are close to the RNA template strand (Fig. 8). The distances between an oxygen substituent of the phosphate group linking template nucleotides −7 and −8 and the amino groups of Arg277 are around 3.3 Å, while the side chain of Thr286 points toward the RNA template. A distance of 4.4 Å is observed between the hydroxyl group of Thr286 and the 2′OH of the ribose at position −8 of the template. On the other hand, Pro272 is away from the template-primer and could play a role in facilitating the appropriate folding of the thumb subdomain. However, single amino acid substitutions at this position were shown to affect template-primer binding and RT processivity (70). In p51, the relevant thumb subdomain polymorphisms do not appear to be involved in interactions with the template-primer, although structural information is still limited. Thr286 could play a role in stabilizing the RT heterodimer (40).
FIG. 8.
Structure of HIV-1 RT in complex with RNA/DNA. The RT subunits are represented as ribbon diagrams (cyan for p66 and green for p51). The DNA primer is shown in red, while the RNA template is shown in white (in panel A) or yellow (in panel B). The side chains of relevant residues of the thumb subdomains of p66 and p51 are shown in magenta and orange, respectively, using a Corey-Pauling-Koltun (CPK) model. Structures were drawn with the PyMOL molecular viewer (http://www.pymol.org). Atom coordinates were taken from PDB file 1HYS (60). Side and top views are shown in panels A and B, respectively. (C) Relevant distances between the side chains of Arg277 and Thr286 and the RNA template. (D) Locations of positions 277 and 286 in the p66 subunit of a binary complex of HIV-1 RT and a DNA/DNA duplex (PDB file 2HMI) (16). The template is shown in white, and the primer is shown in red. (E) Relevant interatomic distances between the side chains of Arg277 and Thr286 and the template DNA backbone.
Compared with the HIV-1 RT structure containing RNA/DNA, the analysis of crystal structures containing DNA/DNA duplexes (16, 23) revealed that there were less extensive contacts between the RT template grip residues (i.e., α-helix I [residues 278 to 286]) and the DNA/DNA template-primer. Furthermore, in these two structures, the side chain of Arg277 points away from the DNA/DNA substrate, although Thr286 appears in a similar position as in the structure containing RNA/DNA, but closer to the sugar-phosphate backbone at positions −8 and −9 of the template (interatomic distances of 3.7 to 5.4 Å). Similar conformations were observed in the structures of HIV-1 RT with pre- and posttranslocation AZTMP-terminated DNA/DNA (58). Taken together, structural data are consistent with our findings regarding the loss of affinity for RNA/DNA complexes of mutant RTs containing R277K and other amino acid substitutions in the polymerase thumb subdomain. Interestingly, another mutation in α-helix I of the thumb subdomain (i.e., R284K) has been found to be associated with the accumulation of TAMs in treated patients (9, 71). Arg284 lies close to the RNA template, and as in the case of the thumb polymorphisms described in our study, could affect NRTI excision by altering interactions with the template-primer. Despite the compelling evidence in favor of excision as the molecular mechanism facilitating the selection of thumb polymorphisms under treatment with NRTIs, we cannot exclude the potential contribution of polymorphisms to RT heterodimer stability in vivo, either by changing interactions between p66 and p51 (40, 45, 69) or by altering the susceptibility of RT subunits to cleavage by the viral protease (17, 49).
In summary, our studies provide strong evidence for a mechanism that involves RNase H-independent contributions to increases in the efficiency of NRTI excision, and in turn, a selective advantage to these drugs; as well as for the effects of thumb subdomain polymorphisms Pro272, Arg277, and Thr286 on RNA/DNA template-primer affinity that may result in a selective advantage both in the presence or in the absence of RT inhibitors. Although the contribution of thumb subdomain polymorphisms to drug resistance may not be significant, their effects on viral fitness can be relevant for the selection of NRTI-resistant HIV variants. In this context, accessory mutations in the connection and RNase H domains of the RT could also modulate the effects of thumb subdomain polymorphisms, arguing in favor of the inclusion of C-terminal portions of RT in clinical genotypic and phenotypic assays. Further studies on the role of accessory mutations should be helpful for designing prediction algorithms for the viral replication capacity.
Supplementary Material
Acknowledgments
Funding for this work was provided by grants from the Spanish Ministry of Science and Innovation (BIO2007/60319), Fundación para la Investigación y Prevención del SIDA en España (FIPSE) (grant 36771/08), Fondo de Investigación Sanitaria (through the “Red Temática de Investigación Cooperativa en SIDA” RD06/0006), and an institutional grant of Fundación Ramón Areces. Work at the Fundació irsiCaixa was supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under the “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)” project grant agreement 223131 and the Spanish Ministry of Science and Innovation through grant PI07/0098 (to M.A.M.).
Footnotes
Published ahead of print on 23 August 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Álvarez, M., T. Matamoros, and L. Menéndez-Arias. 2009. Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 392:872-884. [DOI] [PubMed] [Google Scholar]
- 2.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 3.Basavapathruni, A., and K. S. Anderson. 2007. Reverse transcription of the HIV-1 pandemic. FASEB J. 21:3795-3808. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek, K., W. A. Beard, J. R. Casas-Finet, H.-R. Kim, T. A. Darden, S. H. Wilson, and T. A. Kunkel. 1995. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem. 270:19516-19523. [DOI] [PubMed] [Google Scholar]
- 5.Bebenek, K., W. A. Beard, T. A. Darden, L. Li, R. Prasad, B. A. Luxon, D. G. Gorenstein, S. H. Wilson, and T. A. Kunkel. 1997. A minor groove binding track in reverse transcriptase. Nat. Struct. Biol. 4:194-197. [DOI] [PubMed] [Google Scholar]
- 5a.Betancour, G., M. C. Puertas, M. Nevot, C. Garriga, M. A. Martínez, J. Martinez-Picado, and L. Menéndez-Arias. 2010. Mechanisms of nucleoside analogue resistance associated with the selection of HIV-1 reverse transcriptase thumb subdomain polymorphisms. Antivir. Ther. 15(Suppl. 2):A80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boretto, J., S. Longhi, J.-M. Navarro, B. Selmi, J. Sire, and B. Canard. 2001. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 292:139-147. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brehm, J. H., J. W. Mellors, and N. Sluis-Cremer. 2008. Mechanism by which a glutamine to leucine substitution at residue 509 in the ribonuclease H domain of HIV-1 reverse transcriptase confers zidovudine resistance. Biochemistry 47:14020-14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cane, P. A., H. Green, E. Fearnhill, and D. Dunn, on behalf of the UK Collaborative Group on HIV Drug Resistance. 2007. Identification of accessory mutations associated with high-level resistance in HIV-1 reverse transcriptase. AIDS 21:447-455. [DOI] [PubMed] [Google Scholar]
- 10.Cases-González, C. E., S. Franco, M. A. Martínez, and L. Menéndez-Arias. 2007. Mutational patterns associated with the 69 insertion complex in multi-drug-resistant HIV-1 reverse transcriptase that confer increased excision activity and high-level resistance to zidovudine. J. Mol. Biol. 365:298-309. [DOI] [PubMed] [Google Scholar]
- 11.Ceccherini-Silberstein, F., V. Svicher, T. Sing, A. Artese, M. M. Santoro, F. Forbici, A. Bertoli, S. Alcaro, G. Palamara, A. d'Arminio Monforte, J. Balzarini, A. Antinori, T. Lengauer, and C. Perno. 2007. Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J. Virol. 81:11507-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Clercq, E., and J. Neyts. 2009. Antiviral agents acting as DNA or RNA chain terminators. Handb. Exp. Pharmacol. 189:53-84. [DOI] [PubMed] [Google Scholar]
- 13.Delviks-Frankenberry, K. A., G. N. Nikolenko, R. Barr, and V. K. Pathak. 2007. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J. Virol. 81:6837-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delviks-Frankenberry, K. A., G. N. Nikolenko, P. L. Boyer, S. H. Hughes, J. M. Coffin, A. Jere, and V. K. Pathak. 2008. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc. Natl. Acad. Sci. U. S. A. 105:10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deval, J., J.-M. Navarro, B. Selmi, J. Courcambeck, J. Boretto, P. Halfon, S. Garrido-Urbani, J. Sire, and B. Canard. 2004. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J. Biol. Chem. 279:25489-25496. [DOI] [PubMed] [Google Scholar]
- 16.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, Jr., A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structural and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284:1095-1111. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, L. L., M. J. McWilliams, K. Das, E. Arnold, and S. H. Hughes. 2009. Mutations in the thumb allow human immunodeficiency virus type 1 reverse transcriptase to be cleaved by protease in virions. J. Virol. 83:12336-12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehteshami, M., G. L. Beilhartz, B. J. Scarth, E. P. Tchesnokov, S. McCormick, B. Wynhoven, P. R. Harrigan, and M. Götte. 2008. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 283:22222-22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 20.Garriga, C., M. J. Pérez-Elías, R. Delgado, L. Ruiz, L. Pérez-Álvarez, T. Pumarola, A. López-Lirola, J. González-García, and L. Menéndez-Arias, on behalf of the Spanish Group for the Study of Antiretroviral Drug Resistance. 2009. HIV-1 reverse transcriptase thumb subdomain polymorphisms associated with virological failure to nucleoside drug combinations. J. Antimicrob. Chemother. 64:251-258. [DOI] [PubMed] [Google Scholar]
- 21.Hachiya, A., E. N. Kodama, S. G. Sarafianos, M. M. Schuckmann, Y. Sakagami, M. Matsuoka, M. Takiguchi, H. Gatanaga, and S. Oka. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermann, T., T. Meier, M. Götte, and H. Heumann. 1994. The ‘helix clamp’ in HIV-1 reverse transcriptase: a new nucleic acid binding motif common in nucleic acid polymerases. Nucleic Acids Res. 22:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 24.Huigen, M. C., P. M. van Ham, L. de Graaf, R. M. Kagan, C. A. Boucher, and M. Nijhuis. 2008. Identification of a novel resistance (E40F) and compensatory (K43E) substitution in HIV-1 reverse transcriptase. Retrovirology 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isel, C., C. Ehresmann, P. Walter, B. Ehresmann, and R. Marquet. 2001. The emergence of different resistance mechanisms toward nucleoside inhibitors is explained by the properties of the wild type HIV-1 reverse transcriptase. J. Biol. Chem. 276:48725-48732. [DOI] [PubMed] [Google Scholar]
- 26.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. U. S. A. 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 28.Kellam, P., and B. A. Larder. 1994. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 38:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 30.Lu, L., J. Whitcomb, and D. R. Kuritzkes. 2005. Effect of the Q207D mutation in HIV type 1 reverse transcriptase on zidovudine susceptibility and replicative fitness. J. Acquir. Immune Defic. Syndr. 40:20-23. [DOI] [PubMed] [Google Scholar]
- 31.Marchand, B., K. L. White, J. K. Ly, N. A. Margot, R. Wang, M. McDermott, M. D. Miller, and M. Götte. 2007. Effects of the translocation status of human immunodeficiency virus type 1 reverse transcriptase on the efficiency of excision of tenofovir. Antimicrob. Agents Chemother. 51:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mas, A., M. Parera, C. Briones, V. Soriano, M. A. Martínez, E. Domingo, and L. Menéndez-Arias. 2000. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 19:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mas, A., B. M. Vázquez-Álvarez, E. Domingo, and L. Menéndez-Arias. 2002. Multidrug-resistant HIV-1 reverse transcriptase: involvement of ribonucleotide-dependent phosphorolysis in cross-resistance to nucleoside analogue inhibitors. J. Mol. Biol. 323:181-197. [DOI] [PubMed] [Google Scholar]
- 34.Matamoros, T., J. Deval, C. Guerreiro, L. Mulard, B. Canard, and L. Menéndez-Arias. 2005. Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by α-phosphate-modified thymidine analogues. J. Mol. Biol. 349:451-463. [DOI] [PubMed] [Google Scholar]
- 35.Matamoros, T., S. Franco, B. M. Vázquez-Álvarez, A. Mas, M. A. Martínez, and L. Menéndez-Arias. 2004. Molecular determinants of multi-nucleoside analogue resistance in HIV-1 reverse transcriptases containing a dipeptide insertion in the fingers subdomain: effect of mutations D67N and T215Y on removal of thymidine nucleotide analogues from blocked DNA primers. J. Biol. Chem. 279:24569-24577. [DOI] [PubMed] [Google Scholar]
- 36.Matamoros, T., M. Nevot, M. A. Martínez, and L. Menéndez-Arias. 2009. Thymidine analogue resistance suppression by V75I of HIV-1 reverse transcriptase. Effects of substituting valine 75 on stavudine excision and discrimination. J. Biol. Chem. 284:32792-32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menéndez-Arias, L. 1998. Studies on the effects of truncating α-helix E′ of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry 37:16636-16644. [DOI] [PubMed] [Google Scholar]
- 38.Menéndez-Arias, L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124-146. [DOI] [PubMed] [Google Scholar]
- 39.Menéndez-Arias, L. 2010. Molecular basis of human immunodeficiency drug resistance: an update. Antiviral Res. 85:210-231. [DOI] [PubMed] [Google Scholar]
- 40.Menéndez-Arias, L., A. Abraha, M. E. Quiñones-Mateu, A. Mas, M.-J. Camarasa, and E. J. Arts. 2001. Functional characterization of chimeric reverse transcriptases with polypeptide subunits of highly divergent HIV-1 group M and O strains. J. Biol. Chem. 276:27470-27479. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer, P. R., A. J. Smith, S. E. Matsuura, and W. A. Scott. 2004. Effects of primer-template sequence on ATP-dependent removal of chain-terminating nucleotide analogues by HIV-1 reverse transcriptase. J. Biol. Chem. 279:45389-45398. [DOI] [PubMed] [Google Scholar]
- 45.Morris, M. C., C. Berducou, J. Mery, F. Heitz, and G. Divita. 1999. The thumb domain of the p51-subunit is essential for activation of HIV reverse transcriptase. Biochemistry 38:15097-15103. [DOI] [PubMed] [Google Scholar]
- 46.Naeger, L. K., N. A. Margot, and M. D. Miller. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikolenko, G. N., K. A. Delviks-Frankenberry, S. Palmer, F. Maldarelli, M. J. Fivash, Jr., J. M. Coffin, and V. K. Pathak. 2007. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc. Natl. Acad. Sci. U. S. A. 104:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolenko, G. N., S. Palmer, F. Maldarelli, J. W. Mellors, J. M. Coffin, and V. K. Pathak. 2005. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc. Natl. Acad. Sci. U. S. A. 102:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivares, I., A. Mulky, P. I. Boross, J. Tözsér, J. C. Kappes, C. López-Galíndez, and L. Menéndez-Arias. 2007. HIV-1 protease dimer interface mutations that compensate for viral reverse transcriptase instability in infectious virions. J. Mol. Biol. 372:369-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 51.Powell, M. D., W. A. Beard, K. Bebenek, K. J. Howard, S. F. J. Le Grice, T. A. Darden, T. A. Kunkel, S. H. Wilson, and J. G. Levin. 1999. Residues in the αH and αI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J. Biol. Chem. 274:19885-19893. [DOI] [PubMed] [Google Scholar]
- 52.Puertas, M. C., M. J. Buzón, A. Artese, S. Alcaro, L. Menéndez-Arias, C. F. Perno, B. Clotet, F. Ceccherini-Silberstein, and J. Martinez-Picado. 2009. Effect of the human immunodeficiency virus type 1 reverse transcriptase polymorphism Leu-214 on replication capacity and drug susceptibility. J. Virol. 83:7434-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radzio, J., S.-H. Yap, G. Tachedjian, and N. Sluis-Cremer. 2010. N348I in reverse transcriptase provides a genetic pathway for HIV-1 to select thymidine analogue mutations and mutations antagonistic to thymidine analogue mutations. AIDS 24:659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray, A. S., A. Basavapathruni, and K. S. Anderson. 2002. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J. Biol. Chem. 277:40479-40490. [DOI] [PubMed] [Google Scholar]
- 55.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 56.Ray, A. S., E. Murakami, C. N. Peterson, J. Shi, R. F. Schinazi, and K. S. Anderson. 2002. Interactions of enantiomers of 2′,3′-didehydro-2′,3′-dideoxy-fluorocytidine with wild type and M184V mutant HIV-1 reverse transcriptase. Antiviral Res. 56:189-205. [DOI] [PubMed] [Google Scholar]
- 57.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 58.Sarafianos, S. G., A. D. Clark, Jr., K. Das, S. Tuske, J. J. Birktoft, P. Ilankumaran, A. R. Ramesha, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 21:6614-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc. Natl. Acad. Sci. U. S. A. 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarafianos, S. G., B. Marchand, K. Das, D. M. Himmel, M. A. Parniak, S. H. Hughes, and E. Arnold. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schinazi, R. F., B. Hernandez-Santiago, and S. J. Hurwitz. 2006. Pharmacology of current and promising nucleosides for the treatment of human immunodeficiency viruses. Antiviral Res. 71:322-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sluis-Cremer, N., C.-W. Sheen, S. Zelina, P. S. A. Torres, U. M. Parikh, and J. W. Mellors. 2007. Molecular mechanism by which the K70E mutation in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 51:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith, A. J., and W. A. Scott. 2006. The influence of natural substrates and inhibitors on the nucleotide-dependent excision activity of HIV-1 reverse transcriptase in the infected cell. Curr. Pharm. Des. 12:1827-1841. [DOI] [PubMed] [Google Scholar]
- 65.Svicher, V., T. Sing, M. M. Santoro, F. Forbici, F. Rodríguez-Barrios, A. Bertoli, N. Beerenwinkel, M. C. Bellocchi, F. Gago, A. d'Arminio Monforte, A. Antinori, T. Lengauer, F. Ceccherini-Silberstein, and C. F. Perno. 2006. Involvement of novel human immunodeficiency virus type 1 reverse transcriptase mutations in the regulation of resistance to nucleoside inhibitors. J. Virol. 80:7186-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetic Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 67.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 68.Vivet-Boudou, V., J. Didierjean, C. Isel, and R. Marquet. 2006. Nucleoside and nucleotide inhibitors of HIV-1 replication. Cell. Mol. Life Sci. 63:163-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, J., S. J. Smerdon, J. Jäger, L. A. Kohlstaedt, P. A. Rice, J. M. Friedman, and T. A. Steitz. 1994. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. U. S. A. 91:7242-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Y.-X., H.-J. Zhang, J. Xu, B.-J. Zheng, and Y.-M. Wen. 2008. Mutational analysis of the “turn” of helix clamp motif of HIV-1 reverse transcriptase. Biochem. Biophys. Res. Commun. 377:915-920. [DOI] [PubMed] [Google Scholar]
- 71.Waters, J. M., W. O'Neal, K. L. White, C. Wakeford, E. B. Lansdon, J. Harris, E. S. Svarovskaia, M. D. Miller, and K. Borroto-Esoda. 2009. Mutations in the thumb-connection and RNase H domain of HIV type-1 reverse transcriptase of antiretroviral treatment-experienced patients. Antivir. Ther. 14:231-239. [PubMed] [Google Scholar]
- 72.White, K. L., J. M. Chen, J. Y. Feng, N. A. Margot, J. K. Ly, A. S. Ray, H. L. MacArthur, M. J. McDermott, S. Swaminathan, and M. D. Miller. 2006. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir. Ther. 11:155-163. [DOI] [PubMed] [Google Scholar]
- 73.Yap, S.-H., C.-W. Sheen, J. Fahey, M. Zanin, D. Tyssen, V. D. Lima, B. Wynhoven, M. Kuiper, N. Sluis-Cremer, P. R. Harrigan, and G. Tachedjian. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zelina, S., C.-W. Sheen, J. Radzio, J. W. Mellors, and N. Sluis-Cremer. 2008. Mechanisms by which the G333D mutation in human immunodeficiency virus type 1 reverse transcriptase facilitates dual resistance to zidovudine and lamivudine. Antimicrob. Agents Chemother. 52:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.