Abstract
Little is known about how the general lack of efficiency with which recombinant Saccharomyces cerevisiae strains utilize xylose affects the yeast metabolome. Quantitative metabolomics was therefore performed for two xylose-fermenting S. cerevisiae strains, BP000 and BP10001, both engineered to produce xylose reductase (XR), NAD+-dependent xylitol dehydrogenase and xylulose kinase, and the corresponding wild-type strain CEN.PK 113-7D, which is not able to metabolize xylose. Contrary to BP000 expressing an NADPH-preferring XR, BP10001 expresses an NADH-preferring XR. An updated protocol of liquid chromatography/tandem mass spectrometry that was validated by applying internal 13C-labeled metabolite standards was used to quantitatively determine intracellular pools of metabolites from the central carbon, energy, and redox metabolism and of eight amino acids. Metabolomic responses to different substrates, glucose (growth) or xylose (no growth), were analyzed for each strain. In BP000 and BP10001, flux through glycolysis was similarly reduced (∼27-fold) when xylose instead of glucose was metabolized. As a consequence, (i) most glycolytic metabolites were dramatically (≤120-fold) diluted and (ii) energy and anabolic reduction charges were affected due to decreased ATP/AMP ratios (3- to 4-fold) and reduced NADP+ levels (∼3-fold), respectively. Contrary to that in BP000, the catabolic reduction charge was not altered in BP10001. This was due mainly to different utilization of NADH by XRs in BP000 (44%) and BP10001 (97%). Thermodynamic analysis complemented by enzyme kinetic considerations suggested that activities of pentose phosphate pathway enzymes and the pool of fructose-6-phosphate are potential factors limiting xylose utilization. Coenzyme and ATP pools did not rate limit flux through xylose pathway enzymes.
Most of the biomass-to-ethanol processes that are currently advancing to the commercial-production scale rely on microbial strains for efficient fermentation of all sugars, that is, hexoses (mainly d-glucose) and pentoses (d-xylose, l-arabinose), contained in hydrolysates of lignocellulosic feedstocks (15, 36, 65). The yeast Saccharomyces cerevisiae is among the top candidate microorganisms to be used for mixed hexose-pentose fermentation (15). However, because neither xylose nor l-arabinose is a substrate naturally utilized by S. cerevisiae, extensive metabolic redesign was required to obtain strains producing pentose-derived ethanol in a useful yield (16, 21, 37). Despite notable achievements, even the current best-performing yeast strains fall short in specific ethanol productivity (qethanol; 0.13 to 0.45 g ethanol/g biomass/h) from xylose compared to the corresponding qethanol from glucose (1.2 g/g biomass/h) (33, 50). The low qethanol for pentose fermentation is the result of at least two limiting factors. First of all, the yield coefficient (Yethanol/sugar) for consumption of pentose substrates is usually far lower (0.30 to 0.46) than theoretically expected (Yethanol/xylose = 0.51), reflecting the formation of reduced fermentation by-products (e.g., xylitol) in substantial amounts (7, 37). While enhancements of Yethanol/xylose by a diversity of metabolic engineering strategies have received much attention in the recent past (7, 21, 22, 37), a second component of qethanol has not been well studied. Besides issues of transport (13, 17), this is the general lack of efficiency by which S. cerevisiae handles pentoses as substrates for alcoholic fermentation.
Irrespective of whether direct isomerization or multistep oxidoreductive transformations are utilized for metabolic integration of xylose and l-arabinose in engineered strains of S. cerevisiae (see Fig. S1 in the supplemental material), either pentose is eventually assimilated as d-xylulose-5-phosphate (Xyl5P) via the pentose phosphate (PP) pathway (15). Therefore, this implies a catabolic function for the PP pathway during pentose utilization. Evidence from different studies in which the yeast transcriptome and proteome were analyzed under different substrate (glucose, xylose) conditions suggests that S. cerevisiae is not well adapted for fueling lower glycolysis via the PP pathway (24, 28, 51-54). It also appeared from the “omics” data that xylose was poorly “sensed” as a substrate for alcoholic fermentation. A persuasive explanation for this lack of recognition of xylose (and probably, by extension, l-arabinose) is currently not available. Intracellular pools of a few metabolites were determined for different xylose-utilizing recombinant S. cerevisiae strains (29, 56, 57, 63, 72). However, a comprehensive quantitative description of the S. cerevisiae metabolic response to xylose utilization is currently not available, and therefore a metabolomic basis for the differences in qethanol from xylose compared to glucose remains elusive.
To address these issues, quantitative metabolomics was performed. Two previously described strains of S. cerevisiae, BP000 and BP10001, both engineered for xylose fermentation (46), and the corresponding wild-type strain (CEN.PK 113-7D) not able to utilize xylose were employed. These recombinant strains integrate xylose via the two-step oxidoreductive pathway (see Fig. S1 in the supplemental material) catalyzed by xylose reductase (CtXR; from Candida tenuis) and xylitol dehydrogenase (XDH; from Galactocandida mastotermitis). While strain BP000 expresses the gene encoding CtXR in the NADPH-preferring wild-type form, strain BP10001 produces a doubly mutated XR (XR-Dm; Lys274→Arg, Asn276→Asp) that prefers NADH over NADPH (45). The XDH used is specific for NAD+ (30). Under anaerobic conditions, neither of the strains was able to grow on xylose (46). It was shown that because of improved recycling of NADH in the XR-XDH sequence of reactions, strain BP10001 displayed 42% higher Yethanol and 52% lower Yxylitol than strain BP000 during fermentation of 20 g/liter xylose (46). Results from flux balance (FB) analysis of xylose fermentation data reported previously for BP10001 provided assertion that coenzyme usage by XR-Dm and XDH is balanced in BP10001 and that XR-Dm therefore reduces xylose in vivo preferentially by NADH (31).
MATERIALS AND METHODS
Materials.
Medium components used were as described elsewhere (46). Two recombinant S. cerevisiae strains, BP000 and BP10001 (46), and the corresponding wild-type strain CEN.PK 113-7D (64) were employed (see Table S1 in the supplemental material). Authentic standards of intracellular metabolites were obtained from Sigma-Aldrich (Schellendorf, Switzerland) in the highest purity available (see Table S2 in the supplemental material).
Fermentations.
Anaerobic cultivations were carried out using a Labfors bioreactor system (Infors-HT, Bottmingen, Switzerland) equipped with pH and pO2 sensors and an Innova 1313 acoustic online off-gas analyzer (LumaSense Technologies A/S, Ballerup, Denmark). The working volume was 2,000 ml. Biological samples for metabolite analysis were obtained from anaerobic fermentations performed in a Sixfors bioreactor system (Infors-HT). The working volume was 300 ml. Off-gas condensers were cooled to 10°C. All cultivations were performed at 30°C. A buffered (100 mM citrate, pH 5.5) mineral medium containing 20 g/liter glucose was used. The medium was supplemented with vitamins and trace elements (23), ergosterol (8 mg/liter), Tween 80 (336 mg/liter), and polypropylene glycol 2000 (1 ml/liter of a 10% [vol/vol] aqueous emulsion). Xylose was added to a final concentration of 20 g/liter when glucose was completely consumed. Stirring was at 250 rpm. N2 (oxygen concentration was <1.5 ppm) was sparged at a constant flow rate of 0.16 volumes of air per volume of medium per minute (vvm). Analysis of external metabolites by high-performance liquid chromatography (HPLC), and calculations of physiological parameters were performed as described previously (30, 46).
Determination of internal metabolites—sampling and workup.
Glucose-growing cells were harvested at mid-exponential growth. In the xylose phase, cells were withdrawn after 24 h of incubation. This time range represented steady-state conversion of xylose by BP strains and was considered to be sufficient for CEN.PK 113-7D cells to fully adapt to the new substrate (4). Typically, an aliquot of a cell suspension containing 0.4 ± 0.1 (mean ± standard deviation) mg cell dry weight (CDW) (1 ml [glucose phase], 0.25 ml [xylose phase]) was transferred to 4 ml of a 60% (vol/vol) methanolic solution containing a 10 mM ammonium acetate buffer (pH 7.5). The quenching solution was precooled to −40°C. In order to minimize oxygen transfer into the bioreactor, the outlet of the off-gas cooler was closed by a clamp before sampling. Quenched cells were separated promptly by centrifugation (8,000 rpm for 3 min at −20°C in a Sigma 3K30 tabletop centrifuge [Sigma, Osterode am Harz, Germany]) and immediately frozen in liquid N2 after decanting the supernatant. Cell pellets were treated for 3 min at 90°C by adding 1 ml of a 10 mM ammonium acetate buffer (pH 7.5) containing 75% (vol/vol) ethanol. The extraction solution was preheated to 90°C. A yeast 13C-labeled metabolite extract solution was prepared as described elsewhere (6) and was used as an internal standard. Appropriate aliquots of this solution (0°C) were added to the cell pellets or standards (−40°C) directly prior to the addition of the extraction solution. Metabolite extracts were dried in a stepwise manner, at 30°C, using reduced pressure at 0.12 mbar and 0.001 mbar in a speed vac setup described elsewhere (6). Dried samples were dissolved in 50 μl of Nanopure water before analysis by liquid chromatography/tandem mass spectrometry (LC/MS-MS).
Analysis of internal metabolites—LC/MS-MS measurements.
Metabolites were analyzed on an Agilent 1100 series high-performance liquid chromatography system (Agilent Technologies, Waldbronn, Germany) coupled to an Applied Biosystems/MDS Sciex 4000 QTRAP (AB/MDS Sciex, Concord, Ontario, Canada). Chromatographic separation at 40°C using a Waters Atlantis T3 column was performed by applying protocols reported previously (6, 12). Analyses were carried out in negative-ion mode and selected multiple-reaction-monitoring mode (MRM) (6, 34). Data acquisition, compound identification, and peak integration were done by using the Analyst software version 1.4.2 (AB/MDS Sciex). Twelve solutions, each containing all compounds to be analyzed, except Xyl5P, for which separate dilutions were made (0.2 to 10 μM), were prepared in the range of 0.2 to 40 μM. Xyl5P was produced enzymatically as described in Materials and Methods in the supplemental material. Ten mM ammonium acetate buffer (pH 7.5) was used to dilute the standards. Sedoheptulose-7-phosphate (S7P) was not available. Therefore, these pools were analyzed by its MS signals (mother ion, 298; daughter ion, 97 [34]). LC/MS-MS measurements were carried out with metabolite extracts, standards and heat-treated standards each containing the 13C-labeled internal standard and 13C-labeled metabolite extract alone.
Thermodynamic analysis.
To determine how far away from thermodynamic equilibrium metabolic reactions operate in S. cerevisiae, we used the relation ΔΔG = ΔGKeq − ΔGMAR. Herein, ΔGKeq and ΔGMAR are free energies of a reaction (ΔG) at equilibrium (Keq) and at a particular mass-action ratio (MAR), respectively. Values of free energies were calculated by applying the equation ΔG = −RTlnK, where R and T are gas constant and absolute temperature (303.15 K), respectively, while K is Keq or an MAR. Reactions analyzed are summarized in Table S4 in the supplemental material. Reactions of glucose-6-phosphate (Glc6P) dehydrogenase and 6-phosphogluconolactonase (G6PDH-lactonase), transketolase and transaldolase (TKL-TAL), as well as those of glyceraldehyde-3-phosphate (Ga3P) dehydrogenase and 3-phosphoglycerate kinase (GA3PDH-PGK), were lumped into one reaction. Independent of the strain or substrate used, the reaction, 3-phosphoglycerate (3PG) ⇆ 2-phosphoglycerate (2PG), catalyzed by phosphoglycerate mutase was assumed to be at equilibrium (14). If not mentioned otherwise, concentrations of phosphate were set to 5 mM (10) and that of CO2 was set to 28.6 mM, corresponding to the maximum solubility of CO2 in aqueous solutions at 30°C (26). A cell density of 0.42 g/ml was used to convert μmol/g CDW into mmol/liter (11). Lower and upper bounds of concentrations of pyruvate were set to 0.6 and 2.0 mM, respectively (62, 70).
RESULTS
Cultivation of BP000, BP10001, and CEN.PK 113-7D.
Cultivation conditions were chosen to elicit three different physiological states in S. cerevisiae that are henceforth abbreviated “glucose-growing,” “xylose-metabolizing,” and “xylose-resting.” To induce these states, the following experimental protocol was used. Batch fermentations were carried out under anaerobic conditions in two phases. In the first phase, each strain was individually grown in a mineral medium supplemented with 20 g/liter glucose as the sole carbon source. After glucose was completely utilized, the second phase was started by the addition of xylose to a final concentration of 20 g/liter. Representative time courses are displayed in Fig. 1, and results are summarized in Table 1. Physiological parameters obtained at mid-exponential growth on glucose were nearly identical for each strain. Neither strain applied showed any detectable growth in the xylose phase in the time range investigated (100 h for BP strains and 24 h for CEN.PK 113-7D). BP000 and BP10001 utilized xylose with constant and similar rates of 0.07 to 0.08 g/g CDW/h within the first 100 h of conversion and displayed product yields that were almost identical to those reported recently for anaerobic xylose-to-ethanol conversions (46). The specific substrate uptake rates for glucose obtained for BP000 and BP10001 were 29- and 25-fold higher than qxylose, respectively. The parent strain, CEN.PK 113-7D, showed no utilization of xylose in the time range studied.
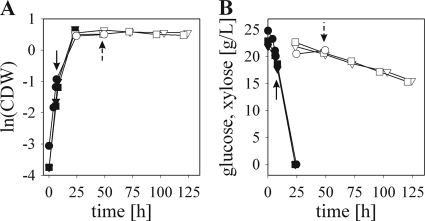
FIG. 1.
Representative time courses of anaerobic glucose and xylose fermentations performed for CEN.PK 113-7D (circles), BP000 (triangles), and BP10001 (squares). Arrows with solid and dashed stems indicate the time of quenching of cells fermenting glucose and xylose, respectively. Glucose (filled symbols) and xylose (open symbols) phases are depicted.
TABLE 1.
Summary of results obtained from anaerobic glucose and xylose fermentations
| Fermentation and strain | Parametera |
|||||||
|---|---|---|---|---|---|---|---|---|
| q (g/gCDW/h) | μ (h−1)b | Yethanol (g/g)c | Yxylitol (g/g)b | Yglycerol (g/g)b | Yacetate (g/g)b | Ybiomass (g/g)b | % C recoveryc | |
| Glucose | ||||||||
| CEN.PK 113-7D | 2.2 ± 0.3 | 0.34 | 0.42 | 0.10 | 0.023 | 0.073 | 106 | |
| BP000 | 2.0 ± 0.3 | 0.34 | 0.41 | 0.11 | 0.019 | 0.075 | 104 | |
| BP10001 | 1.9 ± 0.2 | 0.32 | 0.40 | 0.10 | 0.027 | 0.070 | 103 | |
| Xylose | ||||||||
| BP000 | 0.070 | 0.24 | 0.32 | 0.07 | 0.05 | ND | 104 | |
| BP10001 | 0.077 | 0.36 | 0.19 | 0.04 | 0.06 | ND | 102 | |
Parameters were calculated from the mid-exponential growth phase on glucose and from the steady-state phase of xylose utilization. Estimated standard errors were <15%. ND, not detected.
Values represent averages from 4 individual fermentations performed either with Sixfors (2×) or Labfors (2×) bioreactor systems.
The ethanol yield and the carbon balance were calculated based on data acquired by the Labfors bioreactor system.
LC/MS-MS analysis.
Metabolomic characterization involved the determination of 14 compounds from the central carbon metabolism (CCM), 10 from energy and redox metabolism, and 8 amino acids. Metabolites measured for each biological sample are summarized in Table S2 in the supplemental material and, together with the respective MRM quantitative data obtained, provided the basis for subsequent thermodynamic and metabolic control analysis (MCA). Of the isomeric metabolites not distinguishable by MS, fructose-6-phosphate (Fru6P) and dihydroxyacetone-phosphate (DHAP) were well separated from Glc6P and Ga3P, respectively, in LC analysis, whereas 2PG and 3PG were coeluted. Figure S2 in the supplemental material shows elution profiles of ribose-5-phosphate (Rib5P), Xyl5P, and ribulose-5-phosphate (Ru5P) and of their 13C-labeled counterparts. Only Rib5P eluted as a single peak, while Xyl5P and Ru5P were not clearly separated by LC. The elution profile of the CEN.PK 113-7D metabolite extract showed an additional peak at 9.1 min. The assignment of this signal was not further pursued due to the lack of an appropriate reference compound.
After correction for the relevant 13C-labeled standard, all compounds measured showed a linear correlation between the MS signal and the applied concentration ranges. Data for CCM metabolites are displayed in Fig. 2. Energy and redox metabolites are shown together with amino acids in Fig. 3. Values of data presented in Fig. 2 and 3 are collected in Table S3 in the supplemental material.
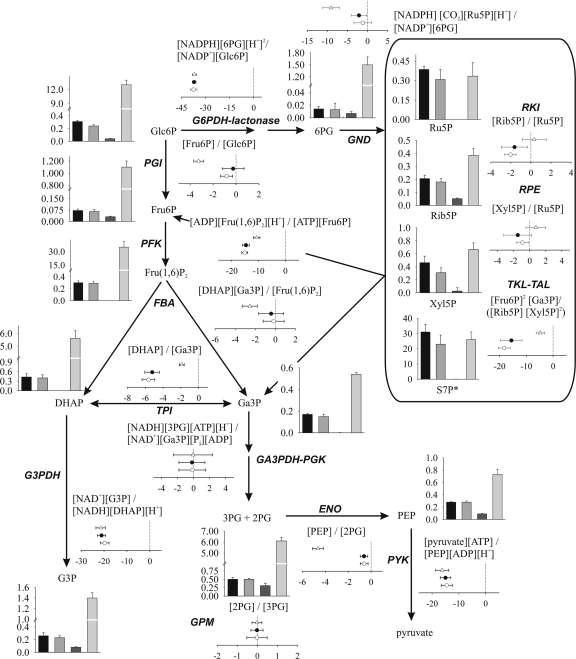
FIG. 2.
Results obtained from quantitative metabolomics and thermodynamic analysis of the central carbon metabolism. Concentrations of metabolites are given in μmol/g CDW, except for S7P (indicated by an asterisk), for which the relative MS signal per g CDW is shown. Metabolite pools obtained for BP10001, BP000, CEN.PK 113-7D on xylose, and CEN.PK 113-7D on glucose are shown as bars from left to right, respectively. Results from thermodynamic analysis are displayed as ΔΔG values (given in kJ/mol) for BP000 (open circles) and BP10001 (filled circles) on xylose and CEN.PK 113-7D on glucose (open triangles). Corresponding numbers are summarized in Tables S3 and S4 in the supplemental material.
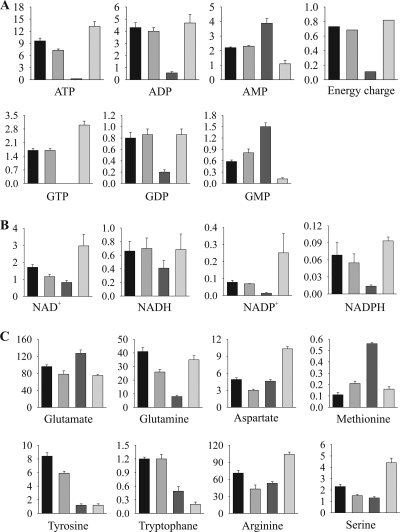
FIG. 3.
Intracellular pools of energy metabolites (A), redox metabolites (B), and amino acids (C). Concentrations of metabolites are given in μmol/g CDW. Data obtained for BP10001, BP000, and CEN.PK 113-7D on xylose and for CEN.PK 113-7D on glucose are shown as bars from left to right, respectively. Corresponding values are summarized in Table S3 in the supplemental material.
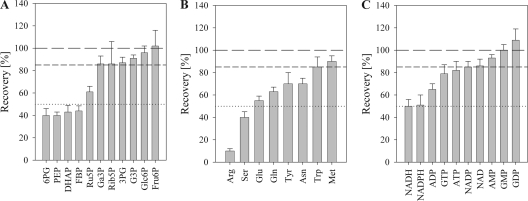
We observed large differences among the measured metabolites concerning the sensitivity to the workup procedure used (Fig. 4). Only about half (45%) of the compounds were reasonably stable and gave ≥85% recovery overall. The remaining metabolites were highly unstable, and it is important to note that in the absence of appropriate internal referencing, measurement of these metabolites would have been completely false.
FIG. 4.
Stability of measured compounds during metabolite isolation. Metabolites from the central carbon metabolism (A), amino acids (B), as well as metabolites reflecting redox and energy metabolism (C) are shown. 100%, 85%, and 50% recoveries are depicted as long dahes, short dashes, and dotted lines, respectively. FBP, fructose 1,6-bisphosphate.
Results from internal metabolite profiling.
Quantitative metabolomics was performed for glucose-growing, xylose-metabolizing, and xylose-resting cells. Samples representing these physiological states were obtained from cells (i) at mid-exponential glucose growth (all strains), (ii) after the 24-h conversion of xylose at the steady state (BP strains), and (iii) after 24 h of resting on xylose (CEN.PK 113-7D), as indicated in Fig. 1.
Intracellular metabolites determined for xylose-metabolizing BP000 and BP10001 and CEN.PK 113-7D either grown on glucose or rested on xylose are shown in Fig. 2 and 3 (numbers are given in Table S3 in the supplemental material). In addition, metabolite pools of Glc6P, Fru6P, 6-phosphogluconate (6PG), adenosine phosphates (AXPs), Xyl5P, Ru5P, and Rib5P were collected for both BP strains at mid-exponential growth on glucose. These data subsets were identical within experimental errors to corresponding metabolite pools obtained for glucose-growing CEN.PK 113-7D cells (data not shown), verifying that accumulation of metabolites was independent of pathway engineering. Values for glycolytic compounds, amino acids, AXPs, NAD(H), and NADP+ obtained for CEN.PK 113-7D grown on glucose under anaerobic conditions were in reasonable agreement with previous reports elsewhere (35, 44, 48, 61, 62). The NADPH pool of 0.09 μmol/g CDW was, in comparison to data from literature (1.2 μmol/g CDW [44]), considerably lower, and the NADPH-to-NADP+ ratio (0.36) is therefore inverse. NADPH/NADP+ ratios reported for S. cerevisiae cells, however, showed large variations, and values between ∼0.03 and 7.5 can be found in the literature (20, 55). In these works, no 13C-labeled internal standard was used (20, 44, 55), which together with different protocols used for quenching (55), extracting (20, 44, 55), or determination of NADPH concentration (44, 55) might be the reason for the observed discrepancies.
Relative changes of metabolite concentrations resulting from the switch in fermentation substrate from glucose to xylose were analyzed for xylose-metabolizing and xylose-resting cells. Data are presented as heat maps in Fig. S3 in the supplemental material. Metabolite maps of the CCM were nearly identical for BP000- and BP10001-metabolizing xylose. All compounds from the upper glycolysis, DHAP, the sum of 2PG and 3PG, and 6PG were reduced dramatically (13- to 120-fold) when xylose replaced glucose as a substrate. Intracellular concentrations of all other metabolites were, in comparison, affected only moderately (<4-fold). Among coenzymes measured, only the NAD+ pool was significantly different (∼2-fold higher) in BP10001 than in BP000. Effects on metabolite pools were enhanced in xylose-resting cells.
Levels of AMP, GMP, and Trp were higher in cells (>2-fold) cultivated on xylose, whereas pools of Tyr were selectively elevated (>5-fold) in xylose-metabolizing cells. Increased accumulation of Tyr and Trp pools in xylose-metabolizing cells was not anticipated, as these are most costly in regard to ATP molecules required for their biosynthesis (5).
Thermodynamic analysis of the central carbon metabolism.
Metabolomic data were used to calculate a thermodynamic landscape for CCM of cells growing on glucose and metabolizing xylose. ΔΔG values (ΔGKeq − ΔGMAR) obtained are displayed in Fig. 2, and respective values are summarized in Table S4 in the supplemental material. Equilibration of GA3PDH-PGK for xylose-metabolizing cells (3, 62) required phosphate concentrations of 1.2 mM (BP000) and 1.8 mM (BP10001) instead of 5 mM (glucose-growing cells). Concentrations of dissolved CO2 of ≤2 mM were necessary for xylose-metabolizing BP cells to shift ΔΔG values for the 6-phosphogluconate dehydrogenase (GND) reaction from a positive value, which is rather unlikely, toward 0.
The energy profile of glycolysis for glucose-growing cells was in excellent agreement with data reported elsewhere (3, 62). ΔΔG values for reactions from the PP pathway were close to 0 (RKI [ribose-5-phosphate ketol-isomerase], RPE [d-ribulose-5-phosphate 3-epimerase]) or slightly shifted to a negative value (TKL-TAL; −4.7 kJ/mol). Values of ΔΔG close to zero for RKI- and RPE-catalyzed reactions indicated that both enzymes operate at equilibrium, implying that the rate of RKI and RPE compared to the carbon flux through these reactions is high. In contrast, G6PDH-lactonase and GND worked far away from equilibrium, suggesting some flux control or regulatory function by these enzymes (32, 69). Results supported the proposed role of G6PDH in controlling the rate of carbon flux through the oxidative PP pathway (3, 67).
Free energy profiles of the CCM were only slightly affected when xylose instead of glucose was utilized by BP000 and BP10001. Reactions catalyzed by phosphofructokinase (PFK), pyruvate kinase (PYK), and G6PDH-lactonase were again largely displaced from equilibrium. Those for phosphoglucose isomerase (PGI) and fructose-1,6-bisphosphate aldolase (FBA) shifted closer to the equilibrium as a result of low qglycolysis. Except for the net reaction, representing combined activity of TKL and TAL (−18 kJ/mol), reactions from the PP pathway worked as in glucose-growing cells at equilibrium. The large displacement from Keq observed for TKL-TAL can be ascribed to the fast isomerization of Fru6P to Glc6P catalyzed by PGI (ΔΔG, ∼0 kJ/mol), resulting in withdrawing of 67% of the Fru6P formed by TKL-TAL.
Estimation of coenzyme usage of XR in BP000 and BP10001.
With intracellular concentrations of NAD(P)H at hand, we were able to calculate estimates of intracellular coenzyme usages of CtXR and XR-Dm. Assuming that xylose transport (i) takes place by facilitated diffusion and (ii) is not rate limiting qxylose in BP000 and BP10001 (31), the concentration of xylose was set to a value equal to the concentration in the medium (133 mM at 24 h of xylose conversion). Kinetic parameters from a full kinetic study at pH 7.0 reported elsewhere for CtXR and XR-Dm (45) and the kinetic expression accounting for the simultaneous utilization of NAD(P)H were applied to calculate steady-state rate constants for xylose reduction with either NADH (vNADH) or NADPH (vNADPH) (47). The ratio of vNADH to vNADPH reflects the coenzyme preference. Results are summarized in Table S5 in the supplemental material. Coenzyme preferences of 30 ± 5 and 0.8 ± 0.2 were obtained for XR-Dm and CtXR, respectively, implying that XR-Dm uses solely NADH, while CtXR utilizes both NADH and NADPH equally well. Data were supported by results from FB analysis (31; see also Table S5 in the supplemental material). Using a reported concentration of 508 μM for NADPH (44), CtXR displayed an almost strict dependency on NADPH (vNADPH/vNADH = 32). NADPH-dependent reduction of xylose in BP000 is not very likely, as (i) predominant production of glycerol would be expected (66) and (ii) strict utilization of NADPH by XR was not compatible with the applied genome scale network (this work and reference 31).
Metabolic control analysis based on enzyme kinetic considerations.
We used kinetic parameters and appropriate rate laws reported elsewhere for CtXR and XR-Dm (45, 47), XDH (43), and xylulose kinase from S. cerevisiae (XK) (49) to analyze to which extent measured concentrations of respective (co)substrates exerted control over the reaction rate of these enzymes. Elasticity coefficients (ɛ) were determined in accordance to reference 59. Results are summarized in Table S5 in the supplemental material. For CtXR and XR-Dm, ɛNADH and ɛNADPH were close to 0 ± 0.1, while ɛxylose was close to ∼0.45, implying some rate control by xylose but not by NAD(P)H. For the applied concentrations of xylitol (50 and 150 mM), the rate through XDH was weakly (maximal 47% of the turnover number) sensitive to both xylitol and NAD+. The reaction catalyzed by XK was saturated with ATP by 66% (BP000) and 72% (BP10001), suggesting weak if any rate control by ATP.
Reactions operating far away from equilibrium, reflected by a large negative shift of the ΔΔG value, are considered to impose some flux control (69). Corresponding enzymes are often regulated by the cell (9). For both BP strains metabolizing xylose, ΔΔG values of reactions catalyzed by PFK, PYK, and TAL-TKL were in the range of −15 to −18 kJ/mol. MCA of these reactions was performed for glucose-growing and xylose-metabolizing cells (see Table S6 in the supplemental material). Xylose-metabolizing cells showed a very low (∼3% of Vmax) saturation of the rate catalyzed by PFK. This was due mainly to small concentrations of Fru6P, reflected by a large ɛFru6P of 1.8. Compared to the large difference in qglycolysis of growing and metabolizing cells, the rate through the PYK-catalyzed reaction was similarly saturated by ADP and phosphoenolpyruvate (PEP) for glucose-growing (70% of Vmax) and xylose-metabolizing (∼50% of Vmax) cells.
In the case of TAL and TKL, a complete analysis could not be performed. The rate of erythrose-4-phosphate-plus-Xyl5P formation, catalyzed by TKL, was 0.02% of Vmax for xylose-metabolizing cells and reached a value of 0.9% of Vmax for glucose-growing cells. In comparison, formation of S7P plus Ga3P was saturated by 20% to 28% of Vmax for xylose-metabolizing cells and by 42% of Vmax for glucose-growing cells. Concentrations of Rib5P (ɛRib5P = 0.50 to 0.64 [all strains]) exerts more control on the conversion rate than those of Xyl5P (ɛXyl5P = 0.2 [glucose-growing cells], 0.25 [BP1001], 0.4 [BP000]). For xylose-metabolizing cells, TAL-catalyzed Ga3P-plus-S7P conversion displayed an ∼42% saturation by Ga3P, while Fru6P hardly saturated (8% of Vmax) the reverse conversion. Data implied that the preferred net reaction of TAL-TKL in xylose-metabolizing cells is 2Xyl5P + Rib5P → 2Fru6P + Ga3P. In glucose-growing cells, Ga3P (59% of Vmax) and Fru6P (71% of Vmax) similarly saturated either direction of the reaction of TAL.
DISCUSSION
The coenzyme usage of XR in vivo.
Balanced coenzyme recycling between XR and XDH during xylose assimilation is required to quantitatively ferment xylose to ethanol. While wild-type forms of XDH usually display a strict preference for NAD+, XRs typically utilize both NADPH and NADH. In the case of CtXR, coenzyme preferences for NADPH vary considerably between 1.5, 15, and 33 depending on the kinetic parameter, kcat, kcat/Kcoenzyme, and kcat/Ki,coenzymeKxylose, respectively, used as bases for calculation (45, 47), precluding even a rough estimation of the coenzyme usage by XR in the cell. By combining measured intracellular concentrations of NAD(P)H, reported values of kinetic parameters (45), and the appropriate kinetic model (47), we were able to calculate representative estimates of coenzyme usages in vivo for CtXR and XR-Dm. Coenzyme preferences (vNADH/vNADPH) were balanced (0.8 ± 0.2) for CtXR but strongly shifted toward NADH (30 ± 5) for XR-Dm. Kinetic estimation of coenzyme usage by CtXR and XR-Dm therefore reinforces previous conclusions (31, 46). In contrast to BP000, the conversion of xylose into xylulose in BP10001 is therefore balanced with respect to NADH and NAD+.
Metabolic response of the central carbon metabolism to low qxylose.
Assuming that ∼5% of the carbon from glucose is channeled through the PP pathway (reference 31 and references therein), the expected flux through the PP pathway (1/20 of qglucose) would be similar to qxylose, consistent with the observation that PP pathway metabolite levels were similar in growing and metabolizing cells. The carbon flux through glycolysis was 25 to 30 times higher in glucose-growing compared to xylose-metabolizing BP cells. Slow qglycolysis caused dramatic reduction of most glycolytic metabolites. Pools of compounds from the CCM therefore became rather uniformly distributed (0.2 to 0.3 μmol/g CDW). Concentrations of Ga3P and PEP were not as strongly affected (<3.6-fold). The formation of both metabolites is thermodynamically not favored. The “conservation” of PEP and Ga3P pools has been reported for an S. cerevisiae strain containing low PGI activity (8), suggesting that cellular mechanisms are tuned to maintain substantial levels of Ga3P and PEP at low qglycolysis.
The energy and redox state of xylose-metabolizing cells.
How do the energy and redox states of a nongrowing xylose-metabolizing cell differ from those of a glucose-fermenting cell? Comprehensive quantitative metabolite profiling presented in this work provided relevant evidences. Independent of the physiological state investigated, AXP and guanine (GXP) nucleotide pools were balanced by the action of adenylate kinase I and II, consistent with reports from the literature (18, 19, 60). Levels of AXPs and GXPs were similarly affected when xylose replaced glucose as a substrate, supporting previous findings that GXPs are kept under tight control in correlation with AXPs and the energy charge (EC) (11). The ECs were almost identical for both BP strains metabolizing xylose but significantly lower (0.7) than the EC obtained for glucose-growing cells (0.8). Similar values for xylose-metabolizing and glucose-growing cells were reported elsewhere (1, 63). Differences in the ECs were predominantly due to decreased and increased levels of ATP and AMP, respectively, yielding enhanced AMP-to-ATP ratios (3- to 4-fold). The AMP level and the AMP-to-ATP ratio were proposed to serve as a highly sensitive sensor of the cellular energy state (18). It has been shown that high levels of AMP (AMP/ATP) indirectly activate the SNF1 complex, which in turn, when in its active phosphorylated form, triggers the derepression of glucose-repressed genes in S. cerevisiae (38, 71). Observed changes in the AMP pool and the AMP-to-ATP ratio of xylose-metabolizing cells relative to those of glucose-growing and xylose-resting cells imply that cells utilizing xylose under anaerobic conditions are in an intermediate state, between the glucose-repressed and glucose-derepressed states.
The catabolic ([NADH]/[NADH] + [NAD+]; CRC) and the anabolic (NADPH]/([NADPH] + [NADP+]; ARC) reduction charges play essential roles in coordinating fueling and biosynthetic reactions (59). CRC and ARC were differently affected in BP000- and BP10001-metabolizing xylose. Irrespective of the substrate or strain applied, pools of NAD(P)H were held at a constant level. However, their oxidized counterparts were selectively reduced by a factor of ∼2 in BP000 (NAD+) and by a factor of ∼3 in both BP cells (NADP+) compared to BP10001 and glucose-growing cells, respectively, resulting in ∼2-fold-increased levels of respective reduction charges. The CRC level was sensitive to the coenzyme usage of the XR-XDH pair and maintained in BP10001 cells upon switching from glucose to xylose. Unlike CRC, the ARC was not dependent on whether coenzyme preferences of the XR-XDH pair were matching. Low levels of NADP+, mirroring unbalanced utilization of NADP+ and NADPH, appeared to be a specific response of both BP000 and BP10001 to xylose.
Sources limiting enzymes constituting the xylose pathway.
The net fluxes, through steps comprising xylose uptake and xylose reduction, were very similar (∼0.074 g/g CDW/h) in BP000 and BP10001. Calculated rate constants for xylose reduction were also almost identical for both XRs (∼6 s−1). The specific xylose transport rate of 0.9 g/g CDW/h, determined for S. cerevisiae at 20 g/liter xylose, clearly surpasses qxylose of BP strains, implying (some) limitation of qxylose by XR. MCA analysis revealed that the rate through CtXR and XR-Dm was sensitive to concentrations of xylose (ɛxylose, ∼0.45) but not of NAD(P)H. This result is in line with the observed 1.75-fold (0.14/0.08) increase of qxylose when 50 g/liter instead of 20 g/liter xylose was utilized by BP10001 (31).
As a consequence of undifferentiated NAD(P)H usage by CtXR, NAD+ generation relative to its utilization by XDH is slower in BP000 than in BP10001 and more xylitol can accumulate. This imbalance is further supported by the measured intracellular NAD+ pool, which is ∼2-fold smaller in BP000. The rate through XDH, however, displayed a weak sensitivity to NAD+ concentrations (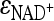 ≤ 0.34), similar for both strains. The reaction catalyzed by XK was, independent of the strain, saturated with ATP (∼70% of Vmax). Therefore, the present evidence, which we feel is the most conclusive thus far, indicates that the cofactor level per se is not a major limiting factor for qxylose. Obviously, other “players” (e.g., xylose, XR, or other sources apart from the xylose pathway [see below]) are more involved in limiting qxylose.
≤ 0.34), similar for both strains. The reaction catalyzed by XK was, independent of the strain, saturated with ATP (∼70% of Vmax). Therefore, the present evidence, which we feel is the most conclusive thus far, indicates that the cofactor level per se is not a major limiting factor for qxylose. Obviously, other “players” (e.g., xylose, XR, or other sources apart from the xylose pathway [see below]) are more involved in limiting qxylose.
Targets limiting qxylose apart from the xylose pathway.
PFK, PYK, and the net reaction represented by TKL and TAL were identified for both BP strains based on results of thermodynamic analysis as potential candidates limiting qxylose. Data from MCA revealed that reaction catalyzed by PFK displayed 33-fold-reduced Vmax for both BP strains. The 25- to 30-fold-slower qglycolysis when xylose instead of glucose was converted was accompanied by an ∼20-fold-lower rate of fructose 1,6-bisphosphate formation. Using elasticity coefficients, Fru6P pools were identified to be the primary reason for low PFK activity. The net flux through the reaction catalyzed by PYK was in the same range for growing (fast qglycolysis) and metabolizing (slow qglycolysis) cells. The availability of PFK and PYK might not be a relevant factor limiting qxylose, consistent with results from previous transcriptomics and proteomics studies (51, 53). Increasing the intracellular concentration of Fru6P instead could be of relevance when improving qxylose. Larger Fru6P pools could lead to higher levels of Glc6P and other glycolytic intermediates, which in turn can affect transcription of glycolytic and ethanologenic enzymes (39, 40). This result is interesting, as it may provide the metabolic basis to explain why xylose is utilized at a higher rate when glucose is cofermented (31).
Rates catalyzed by TAL and TKL displayed saturations of ∼42% and 30% of Vmax, with respect to their preferred substrates. This implies that flux through these reactions should be susceptible to the availability of both the level of TAL and TKL and the supply of their reactants. Results are consistent with reports from literature (2, 25, 27, 33, 42, 56, 68). To increase flux through the PP pathway in BP000 or BP10001, combined overexpression of TAL and TKL (increasing enzyme abundances) together with RPE and RKI (maintenance of substrate supply) is suggested.
Resting on xylose.
CEN.PK 113-7D cells were neither able to grow on nor utilize xylose under anaerobic conditions. Resting on xylose had dramatic consequences on the yeast metabolome. Except for pools of 3PG and PEP, which accumulated to similar extents compared to those of xylose-metabolizing BP cells, all other metabolite pools from the CCM were nearly depleted in xylose-resting cells. 3PG represents a redox sink, and accumulation therefore is thermodynamically favored (14). Maintenance of substantial 3PG pools was also observed for other resting yeast cells (10, 41, 58).
The energy state in xylose-resting cells, reflected by an EC of 0.1, was completely different from that of cells displaying an active flux through glycolysis and resembled that of starving cells (1). The high AMP/ATP ratio of 19 indicated that xylose-resting cells were in a glucose-derepressed state (71). A comparison with nitrogen- and carbon-starved yeast cells showed that the metabolic response to xylose-resting cells shared more similarities with that of nitrogen-starved cells (regarding 6PG, 3PG, ADP, Asp, Tyr, and Gln) than that of carbon-starved cells (regarding NADP+, Glu, and Trp) (4). Cells resting on xylose were no longer capable of maintaining total pools of energy and redox metabolites. Most of the AXP (75%), GXP (65%), and NADP(H) (92%) pools were metabolized as a consequence of the inability to utilize xylose. Interestingly, NAD(H) pools, in particular the NADH pool (2-fold lower than glucose-growing cells), were only moderately affected.
Conclusions.
Quantitative metabolomics presented in this work provided the basis for a comprehensive analysis of metabolomic responses in the central carbon, energy, and redox metabolism of S. cerevisiae to fermentation substrates (glucose, xylose) and growth conditions (growing, metabolizing, resting cells). Internal referencing of each metabolite measured by the corresponding 13C-labeled compound was essential for quantification. With intracellular concentrations available for NAD(P)H, we were able to calculate representative estimates for the coenzyme usage of XR in the cell. Unlike in BP000, xylose reduction in BP10001 was strictly NADH dependent, and the conversion of xylose to xylulose therefore balanced with respect to NADH. Closed coenzyme recycling between XR and XDH was reflected by the ability of BP10001 to keep the CRC constant upon switching from glucose to xylose, while in BP000 the CRC was increased 2-fold due to a selective decrease of NAD+. The utilization of xylose instead of glucose had several effects on the yeast metabolome that were similar for both BP000 and BP10001 and therefore specific to cells metabolizing xylose under anaerobic conditions: (i) most glycolytic metabolites were dramatically diluted (≤120-fold), while those of the PP pathway were hardly affected; (ii) energy and anabolic reduction charges were impaired due to the selective decrease of ATP/AMP and NADP+, respectively; (iii) the net reaction catalyzed by TKL-TAL was strongly shifted away from its equilibrium; and (iv) concentrations of Fru6P were limiting the conversion rate of PFK.
Supplementary Material
Acknowledgments
Simone Pival and Karin Longus are gratefully acknowledged for the preparation of xylulose-5-phosphate and for performing fermentations with the Labfors bioreactor system, respectively.
Financial support from the Austrian Science Fund FWF (grant J2698 to M.K.) is gratefully acknowledged.
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ball, W. J., Jr., and D. E. Atkinson. 1975. Adenylate energy charge in Saccharomyces cerevisiae during starvation. J. Bacteriol. 121:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengtsson, O., M. Jeppsson, M. Sonderegger, N. S. Parachin, U. Sauer, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2008. Identification of common traits in improved xylose-growing Saccharomyces cerevisiae for inverse metabolic engineering. Yeast 25:835-847. [DOI] [PubMed] [Google Scholar]
- 3.Berg, J. M., J. L. Tymoczko, and L. Streyer. 2003. Biochemistry, 5th ed. W. H. Freeman and Company, New York, NY.
- 4.Brauer, M. J., J. Yuan, B. D. Bennett, W. Lu, E. Kimball, D. Botstein, and J. D. Rabinowitz. 2006. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. U. S. A. 103:19302-19307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braus, G. H. 1991. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55:349-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Büscher, J. M., D. Czernik, J. C. Ewald, U. Sauer, and N. Zamboni. 2009. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal. Chem. 81:2135-2143. [DOI] [PubMed] [Google Scholar]
- 7.Chu, B. C., and H. Lee. 2007. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol. Adv. 25:425-441. [DOI] [PubMed] [Google Scholar]
- 8.Ciriacy, M., and I. Breitenbach. 1979. Physiological effects of seven different blocks in glycolysis in Saccharomyces cerevisiae. J. Bacteriol. 139:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabtree, B., E. A. Newsholme, and N. B. Reppas. 1997. Principles of regulation and control in biochemistry: a paradigmatic, flux-oriented approach, p. 117-180. In J. F. Hoffman and J. D. Jamieson (ed.), Handbook of physiology, vol. 14. Oxford University Press, New York, NY. [Google Scholar]
- 10.den Hollander, J. A., K. Ugurbil, T. R. Brown, and R. G. Shulman. 1981. Phosphorus-31 nuclear magnetic resonance studies of the effect of oxygen upon glycolysis in yeast. Biochemistry 20:5871-5880. [DOI] [PubMed] [Google Scholar]
- 11.Ditzelmüller, G., W. Wöhrer, C. P. Kubicek, and M. Röhr. 1983. Nucleotide pools of growing, synchronized and stressed cultures of Saccharomyces cerevisiae. Arch. Microbiol. 135:63-67. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrer, T., and U. Sauer. 2009. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191:2112-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gárdonyi, M., M. Jeppsson, G. Lidén, M. F. Gorwa-Grauslund, and B. Hahn-Hägerdal. 2003. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 82:818-824. [DOI] [PubMed] [Google Scholar]
- 14.Grisolia, S., and J. Carreras. 1975. Phosphoglycerate mutase from yeast, chicken breast muscle, and kidney (2,3-PGA-dependent). Methods Enzymol. 42:435-450. [DOI] [PubMed] [Google Scholar]
- 15.Hahn-Hägerdal, B., K. Karhumaa, C. Fonseca, I. Spencer-Martins, and M. F. Gorwa-Grauslund. 2007. Towards industrial pentose-fermenting yeast strains. Appl. Microbiol. Biotechnol. 74:937-953. [DOI] [PubMed] [Google Scholar]
- 16.Hahn-Hägerdal, B., K. Karhumaa, M. Jeppsson, and M. F. Gorwa-Grauslund. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147-177. [DOI] [PubMed] [Google Scholar]
- 17.Hamacher, T., J. Becker, M. Gárdonyi, B. Hahn-Hägerdal, and E. Boles. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783-2788. [DOI] [PubMed] [Google Scholar]
- 18.Hardie, D. G. 2003. Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology 144:5179-5183. [DOI] [PubMed] [Google Scholar]
- 19.Hardie, D. G., and S. A. Hawley. 2001. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23:1112-1119. [DOI] [PubMed] [Google Scholar]
- 20.Hou, J., N. F. Lages, M. Oldiges, and G. N. Vemuri. 2009. Metabolic impact of redox cofactor perturbations in Saccharomyces cerevisiae. Metab. Eng. 11:253-261. [DOI] [PubMed] [Google Scholar]
- 21.Jeffries, T. W. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320-326. [DOI] [PubMed] [Google Scholar]
- 22.Jeffries, T. W., and Y. S. Jin. 2004. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63:495-509. [DOI] [PubMed] [Google Scholar]
- 23.Jeppsson, M., O. Bengtsson, K. Franke, H. Lee, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2006. The expression of a Pichia stipitis xylose reductase mutant with higher KM for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:665-673. [DOI] [PubMed] [Google Scholar]
- 24.Jin, Y. S., J. M. Laplaza, and T. W. Jeffries. 2004. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 70:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson, B., and B. Hahn-Hägerdal. 2002. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2:277-282. [DOI] [PubMed] [Google Scholar]
- 26.Jones, R. P., and P. F. Greenfield. 1982. Effect of carbon dioxide on yeast growth and fermentation. Enzyme Microb. Technol. 4:210-223. [Google Scholar]
- 27.Karhumaa, K., B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2005. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359-368. [DOI] [PubMed] [Google Scholar]
- 28.Karhumaa, K., A. K. Påhlman, B. Hahn-Hägerdal, F. Levander, and M. F. Gorwa-Grauslund. 2009. Proteome analysis of the xylose-fermenting mutant yeast strain TMB 3400. Yeast 26:371-382. [DOI] [PubMed] [Google Scholar]
- 29.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 30.Krahulec, S., M. Klimacek, and B. Nidetzky. 2009. Engineering of a matched pair of xylose reductase and xylitol dehydrogenase for xylose fermentation by Saccharomyces cerevisiae. Biotechnol. J. 4:684-694. [DOI] [PubMed] [Google Scholar]
- 31.Krahulec, S., B. Petschacher, M. Wallner, K. Longus, M. Klimacek, and B. Nidetzky. 2010. Fermentation of mixed glucose-xylose substrates by engineered strains of Saccharomyces cerevisiae: role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb. Cell Fact. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kümmel, A., S. Panke, and M. Heinemann. 2006. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2:2006.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuyper, M., M. M. Hartog, M. J. Toirkens, M. J. Almering, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399-409. [DOI] [PubMed] [Google Scholar]
- 34.Luo, B., K. Groenke, R. Takors, C. Wandrey, and M. Oldiges. 2007. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A 1147:153-164. [DOI] [PubMed] [Google Scholar]
- 35.Luttik, M. A., Z. Vuralhan, E. Suir, G. H. Braus, J. T. Pronk, and J. M. Daran. 2008. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 10:141-153. [DOI] [PubMed] [Google Scholar]
- 36.Margeot, A., B. Hahn-Hägerdal, M. Edlund, R. Slade, and F. Monot. 2009. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 20:372-380. [DOI] [PubMed] [Google Scholar]
- 37.Matsushika, A., H. Inoue, T. Kodaki, and S. Sawayama. 2009. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 84:37-53. [DOI] [PubMed] [Google Scholar]
- 38.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 39.Meinander, N. Q., I. Boels, and B. Hahn-Hägerdal. 1999. Fermentation of xylose/glucose mixtures by metabolically engineered Saccharomyces cerevisiae strains expressing XYL1 and XYL2 from Pichia stipitis with and without overexpression of TAL1. Bioresour. Technol. 68:79-87. [Google Scholar]
- 40.Müller, S., E. Boles, M. May, and F. K. Zimmermann. 1995. Different internal metabolites trigger the induction of glycolytic gene expression in Saccharomyces cerevisiae. J. Bacteriol. 177:4517-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navon, G., R. G. Shulman, T. Yamane, T. R. Eccleshall, K. B. Lam, J. J. Baronofsky, and J. Marmur. 1979. Phosphorus-31 nuclear magnetic resonance studies of wild-type and glycolytic pathway mutants of Saccharomyces cerevisiae. Biochemistry 18:4487-4499. [DOI] [PubMed] [Google Scholar]
- 42.Ni, H., J. M. Laplaza, and T. W. Jeffries. 2007. Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on d-xylose. Appl. Environ. Microbiol. 73:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nidetzky, B., H. Helmer, M. Klimacek, R. Lunzer, and G. Mayer. 2003. Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in Escherichia coli. Chem. Biol. Interact. 143-144:533-542. [DOI] [PubMed] [Google Scholar]
- 44.Nissen, T. L., M. Anderlund, J. Nielsen, J. Villadsen, and M. C. Kielland-Brandt. 2001. Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 18:19-32. [DOI] [PubMed] [Google Scholar]
- 45.Petschacher, B., S. Leitgeb, K. L. Kavanagh, D. K. Wilson, and B. Nidetzky. 2005. The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem. J. 385:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petschacher, B., and B. Nidetzky. 2008. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb. Cell Fact. 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petschacher, B., and B. Nidetzky. 2005. Engineering Candida tenuis xylose reductase for improved utilization of NADH: antagonistic effects of multiple side chain replacements and performance of site-directed mutants under simulated in vivo conditions. Appl. Environ. Microbiol. 71:6390-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reibstein, D., J. A. den Hollander, S. J. Pilkis, and R. G. Shulman. 1986. Studies on the regulation of yeast phosphofructo-1-kinase: its role in aerobic and anaerobic glycolysis. Biochemistry 25:219-227. [DOI] [PubMed] [Google Scholar]
- 49.Richard, P., M. H. Toivari, and M. Penttilä. 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190:39-43. [DOI] [PubMed] [Google Scholar]
- 50.Runquist, D., B. Hahn-Hägerdal, and M. Bettiga. 2009. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell Fact. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salusjärvi, L. 2008. Transcriptome and proteome analysis of xylose-metabolising Saccharomyces cerevisiae. Ph.D. thesis. University of Helsinki, Helsinki, Finland.
- 52.Salusjärvi, L., M. Kankainen, R. Soliymani, J. P. Pitkänen, M. Penttilä, and L. Ruohonen. 2008. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salusjärvi, L., J. P. Pitkänen, A. Aristidou, L. Ruohonen, and M. Penttilä. 2006. Transcription analysis of recombinant Saccharomyces cerevisiae reveals novel responses to xylose. Appl. Biochem. Biotechnol. 128:237-261. [DOI] [PubMed] [Google Scholar]
- 54.Salusjärvi, L., M. Poutanen, J. P. Pitkänen, H. Koivistoinen, A. Aristidou, N. Kalkkinen, L. Ruohonen, and M. Penttilä. 2003. Proteome analysis of recombinant xylose-fermenting Saccharomyces cerevisiae. Yeast 20:295-314. [DOI] [PubMed] [Google Scholar]
- 55.Satrustegui, J., J. Bautista, and A. Machado. 1983. NADPH/NADP+ ratio: regulatory implications in yeast glyoxylic acid cycle. Mol. Cell. Biochem. 51:123-127. [DOI] [PubMed] [Google Scholar]
- 56.Senac, T., and B. Hahn-Hägerdal. 1991. Effects of increased transaldolase activity on d-xylulose and d-glucose metabolism in Saccharomyces cerevisiae cell extracts. Appl. Environ. Microbiol. 57:1701-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senac, T., and B. Hahn-Hägerdal. 1990. Intermediary metabolite concentrations in xylulose- and glucose-fermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 56:120-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shanks, J. V., and J. E. Bailey. 1988. Estimation of intracellular sugar phosphate concentrations in Saccharomyces cerevisiae using 31P nuclear magnetic resonance spectroscopy. Biotechnol. Bioeng. 32:1138-1152. [DOI] [PubMed] [Google Scholar]
- 59.Stephanopoulos, G. N., A. Aristidou, and J. Nielsen. 1998. Metabolic engineering, principles and methodologies. Academic Press, San Diego, CA.
- 60.Swedes, J. S., M. E. Dial, and C. S. McLaughlin. 1979. Regulation of protein synthesis during early limitation of Saccharomyces cerevisiae. J. Bacteriol. 138:162-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tai, S. L., P. Daran-Lapujade, M. A. Luttik, M. C. Walsh, J. A. Diderich, G. C. Krijger, W. M. van Gulik, J. T. Pronk, and J. M. Daran. 2007. Control of the glycolytic flux in Saccharomyces cerevisiae grown at low temperature: a multilevel analysis in anaerobic chemostat cultures. J. Biol. Chem. 282:10243-10251. [DOI] [PubMed] [Google Scholar]
- 62.Teusink, B., J. Passarge, C. A. Reijenga, E. Esgalhado, C. C. van der Weijden, M. Schepper, M. C. Walsh, B. M. Bakker, K. van Dam, H. V. Westerhoff, and J. L. Snoep. 2000. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267:5313-5329. [DOI] [PubMed] [Google Scholar]
- 63.Toivari, M. H., A. Aristidou, L. Ruohonen, and M. Penttilä. 2001. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab. Eng. 3:236-249. [DOI] [PubMed] [Google Scholar]
- 64.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindelov, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 65.van Maris, A. J., D. A. Abbott, E. Bellissimi, J. van den Brink, M. Kuyper, M. A. Luttik, H. W. Wisselink, W. A. Scheffers, J. P. van Dijken, and J. T. Pronk. 2006. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek 90:391-418. [DOI] [PubMed] [Google Scholar]
- 66.van Maris, A. J., A. A. Winkler, M. Kuyper, W. T. de Laat, J. P. van Dijken, and J. T. Pronk. 2007. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv. Biochem. Eng. Biotechnol. 108:179-204. [DOI] [PubMed] [Google Scholar]
- 67.Vaseghi, S., A. Baumeister, M. Rizzi, and M. Reuss. 1999. In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab. Eng. 1:128-140. [DOI] [PubMed] [Google Scholar]
- 68.Walfridsson, M., J. Hallborn, M. Penttilä, S. Keränen, and B. Hahn-Hägerdal. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61:4184-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, L., I. Birol, and V. Hatzimanikatis. 2004. Metabolic control analysis under uncertainty: framework development and case studies. Biophys. J. 87:3750-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiebe, M. G., E. Rintala, A. Tamminen, H. Simolin, L. Salusjärvi, M. Toivari, J. T. Kokkonen, J. Kiuru, R. A. Ketola, P. Jouhten, A. Huuskonen, H. Maaheimo, L. Ruohonen, and M. Penttilä. 2008. Central carbon metabolism of Saccharomyces cerevisiae in anaerobic, oxygen-limited and fully aerobic steady-state conditions and following a shift to anaerobic conditions. FEMS Yeast Res. 8:140-154. [DOI] [PubMed] [Google Scholar]
- 71.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 72.Zaldivar, J., A. Borges, B. Johansson, H. P. Smits, S. G. Villas-Bôas, J. Nielsen, and L. Olsson. 2002. Fermentation performance and intracellular metabolite patterns in laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 59:436-442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.