Abstract
With the advent of molecular biological techniques, especially next-generation sequencing and metagenomics, the number of microbial biogeography studies is rapidly increasing. However, these studies involve the synthesis of data generated by different laboratories using different protocols, chemicals, etc., all with inherent biases. The aim of this study was to assess inter- as well as intralaboratory variations in microbial community composition when standardized protocols are applied to a single soil sample. Aliquots from a homogenized soil sample from a rice field in Italy were sent to five participating laboratories. DNA was extracted by two investigators per laboratory using an identical protocol. All DNA samples were sent to one laboratory to perform DNA quantification, quantitative PCR (QPCR), and microarray and denaturing gradient gel electrophoresis (DGGE) analyses of methanotrophic communities. Yields, as well as purity of DNA, were significantly different between laboratories but in some cases also between investigators within the same laboratory. The differences in yield and quality of the extracted DNA were reflected in QPCR, microarray, and DGGE analysis results. Diversity indices (Shannon-Wiener, evenness, and richness) differed significantly between laboratories. The observed differences have implications for every project in which microbial communities are compared in different habitats, even if assessed within the same laboratory. To be able to make sensible comparisons leading to valid conclusions, intralaboratory variation should be assessed. Standardization of DNA extraction protocols and possible use of internal standards in interlaboratory comparisons may help in rendering a “quantifiable” bias.
Microorganisms comprise a major part of total biomass and biodiversity (21, 41-43, 49). They play a critical role in biogeochemical processes and ecosystem functioning (16). However, knowledge of ecology and functioning of environmental microbial communities is still far from complete, mainly due to our inability to grow the majority of environmental microbes under laboratory conditions. The introduction of culture-independent DNA- and RNA-based techniques has led to a revolution in environmental microbiology, yielding a wealth of information on community compositions in an ever-growing range of habitats. Phylogenetic as well as functional microarrays (51) and metagenomic techniques (41, 47) enable in-depth analyses and comparison of whole microbial communities in a high-throughput manner.
The collective goal of all environmental microbial ecology studies is 2-fold: (i) to obtain an overall understanding of microbial community composition, dynamics, and functioning and (ii) to identify regulating mechanisms. Reaching these goals will necessitate the integrated analyses of data generated in different laboratories and from different habitats. The first step in most if not all environmental microbial community studies is the extraction of total DNA from environmental samples in a way that reflects the in situ community composition as closely as possible. Numerous methods, protocols, and commercial kits have been developed to improve and optimize quantity and quality of extracted community DNA from a wide range of natural environments (4, 8, 28, 37, 39). However, up-to-date bias-free extraction methods are not available, especially not for complex and highly variable matrices, like soil. Beside the challenge of lysing all cells, the incomplete removal of compounds interfering with downstream processing render the development of a bias-free protocol a “mission impossible.” Assessments of the bias introduced by DNA extraction with different methods and kits on microbial community profiling revealed that a perfect protocol fitting all types of environments is not feasible (10, 17, 20, 45). However, in light of the global biodiversity debate, assessment of local and global patterns of microbial diversity and their controlling factors (19, 26) necessitates the comparison of data collected in multiple habitats and processed in different laboratories.
In contrast to other scientific disciplines, intercalibration of protocols is not common practice in environmental microbiology. Interlaboratory comparisons (ring analyses) have been applied commonly in food control, veterinary, forensic, and soil studies to evaluate, for example, Salmonella diagnostic accuracy (25), virus isolation (18), enzyme-linked immmunosorbent assay methods (2), mitochondrial DNA sequencing (30), soil microbial biomass C (3), and quantitative PCR (QPCR) (11). Ring analyses assessing the reproducibility of DNA extraction and subsequent community analyses between different laboratories have not been carried out so far in environmental microbial ecology.
A microbial functional guild that has been investigated intensively using molecular techniques is represented by the methanotrophs (aerobic methane-oxidizing bacteria [MOB]), which can be found in a wide variety of environments (27). The unique contribution of these bacteria to the global methane cycle has rendered the diversity and ecology of MOB hot topics for decades (9, 14, 34, 46, 48). By using methane as single source of carbon and energy, these microbes represent the only biological sink of the greenhouse gas methane under aerobic conditions (13). Aerobic MOB belong to the Gamma- and Alphaproteobacteria and the Verrucomicrobia (13, 34) and have the following features that enable linking function and identity. Assimilating methane facilitates the application of stable isotope probing of diagnostic lipids and of RNA/DNA (6, 29, 33). Besides this, the key gene in methane oxidation (for methanemonooxygenase subunit A, pmoA) reflects the phylogeny of these bacteria, facilitating a direct link between methane consumption and taxonomy. These features have made this group of microbes a model group for studies in environmental microbial ecology. Combined with the broad distribution and high environmental relevance, this group is highly suited to perform a ring analysis on reproducibility of DNA extraction and subsequent community profiling.
In the present study, five independent laboratories from Norway, Finland, Netherlands, Germany, and Austria extracted DNA from the same rice field soil sample, using identical protocols and performed by two different investigators per laboratory. Subsequently, the extracted DNA was sent to one laboratory, where DNA quantification, QPCR, microarray, and denaturing gradient gel electrophoresis (DGGE) analyses were performed by one and the same person. The impacts of inter- as well as intralaboratory variations of DNA extraction are discussed, and recommendations for comparative studies are presented.
MATERIALS AND METHODS
Soil.
Soil samples were collected in rice fields in Vercelli, Italy, on 30 March 2009 before flooding. The characteristics of this rice field soil have been described previously (24). Soil was frozen and freeze-dried in the laboratory of the Max-Planck Institute for Terrestrial Microbiology (Lab D; Marburg, Germany). After homogenizing by mortar and pestle and sieving through a 2-mm mesh, equal 10-g portions were sent to four additional laboratories (Lab A [Finland], Lab B [Netherlands], Lab C [Austria], and Lab E [Norway]) for DNA extraction.
DNA extraction.
DNA was extracted using a modification of the method described by Yeates and Gillings (50), based on the FastDNA spin kit for soil (MP Biomedicals, LLC, Solon, OH). In every laboratory two investigators (investigators 1 and 2) processed four replicate soil samples in parallel on the same day, using identical chemicals and machinery.
Soil (0.3 g) and 780 μl lysis buffer (200 mM NaPO4 [pH 7.0], 1% cetyltrimethylammonium bromide, 1.5 M NaCl, 2% polyvinylpyrrolidone K30, and 5 mg/ml lysozyme [added right before use]) was added into a multimix FastPrep tube and incubated at 37°C for 30 min. MT buffer (122 μl) was added, and tubes were shaken in the FastPrep instrument (MP Biomedicals, LLC, Solon, OH) for 30 s at 5.5 m s−1. Subsequently, samples were centrifuged for 15 min at 10,000 rpm, and 700 μl of supernatant was collected. The pellet was reextracted by adding lysis buffer (500 μl) and 50 μl MT buffer to the FastPrep tubes, which were shaken in the FastPrep instrument for 30 s at 5.5 m s−1 again followed by the transfer of the second 700 μl of supernatant into separate Eppendorf tubes. At this step, two 700-μl aliquots of supernatant were obtained from each sample. Five microliters of a 10-mg ml−1 freshly made proteinase K solution was added to each tube. Tubes were incubated at 65°C for 30 min. Samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by a chloroform-isoamyl alcohol (24:1) extraction. A 125-μl volume of 7.5 M potassium acetate was added, and samples were incubated on ice for 5 min and then centrifuged at 10,000 rpm for 10 min. Supernatants (two 700-μl aliquots per soil sample) were transferred to new tubes, 700 μl of binding matrix was added, and tubes were mixed for 5 min on a rotator. Binding matrix, with bound DNA, was pelleted by 1 min of centrifugation at 10,000 rpm. The supernatant was discarded, and the pellet was resuspended in 500 μl wash buffer. The resulting suspension was added into a Spinfilter and centrifuged for 1 min at 10,000 rpm. The eluate was discarded, and the pellet was washed again in 500 μl wash buffer. After discarding the second eluate, the Spinfilter was centrifuged for another 10 s to dry the pellet. The filter was placed in a new tube, and 50 μl of Tris-EDTA (TE; pH 8.0) was added. The filter was incubated at room temperature for 1 min and centrifuged for 1 min. The filter was reeluted in the same way with 50 μl of TE (pH 8.0). The eluate collected in the catch tube contained the purified DNA. The DNA was subsequently lyophilized at 40°C maximally and shipped within 24 h to the laboratory of the Netherlands Institute of Ecology (Nieuwersluis, Netherlands) for further processing. Upon arrival, all lyophilized DNA samples were dissolved in equal amounts (100 μl) of water, after which the analyses described below were executed.
DNA quantification.
The DNA concentration was assessed using two independent methods, NanoDrop and PicoGreen. The NanoDrop spectrophotometer (ND-1000; Nanodrop Technology, Wilmington, DE) gives the DNA concentration as well as purity indices (A260/A280, A260/A230, and A320). The PicoGreen (Molecular Probes, Eugene, OR) assay kit uses an ultrasensitive fluorescent nucleic acid stain for quantitating double-stranded DNA in solution. A standard curve was constructed by a dilution series of 0, 0.002, 0.001, 0.01, 0.1, and 2 ng/μl of standard lambda phage DNA (100 ng/μl). For each sample, duplicate 50-μl aliquots of the DNA extracts supplemented with 50 μl of 1× PicoGreen solution were transferred to a 96-well microtiter plate. The plate was shaken for 5 min; fluorescence of DNA extracts was measured at 520 nm after excitation at 480 nm by using a microplate reader (BioTek). The DNA concentration was calculated from the standard curve. During the whole procedure, all samples were protected from light.
Diagnostic pmoA microarray analyses.
Microarray analysis was carried out using a modification of the method described in reference 44, as described below.
Target preparation.
The PCR amplification was based on a two-step seminested protocol. The first-step PCR mixture comprised 25 μl of 2× Premix F (Epicentre Biotechnologies, Madison, WI), 25 pmol of primers A-189f and A-682r (21a) each, 1 unit of Taq polymerase (Invitrogen), and 50 ng of genomic DNA as template, in a total volume of 50 μl. PCR was performed using a touchdown protocol with an initial incubation of 5 min at 94°C, then 25 cycles of 1 min at 94°C, 1 min at the annealing temperature, and 1 min at 72°C, followed by a final incubation of 10 min at 72°C. The annealing temperature was lowered from 62°C to 52°C over the first 11 cycles, after which it was maintained for a further 14 cycles at 52°C. Five microliters of 1/100-diluted PCR product from the first step was used as template in a subsequent nested amplification with primers A-189f and T7-661r. The second-step PCR was performed with a total 25 cycles, with an initial incubation of 5 min at 94°C, then 1 min at 94°C, 1 min at the annealing temperature, and 1 min at 72°C, followed by a final incubation of 10 min at 72°C. The annealing temperature was lowered from 62°C to 52°C over the first 11 cycles, after which it was maintained for a further 14 cycles at 52°C.
IVT.
In vitro transcription (IVT) was carried out under RNase-free conditions using the following procedure: 7 μl purified PCR product (50 ng μl−1; Qiagen kit), 4 μl of 5× T7 RNA polymerase buffer, 2 μl dithiothreitol (100 mM), 0.5 μl RNasin (40 U μl−1; Promega, Madison, WI), 1 μl each of ATP, CTP, and GTP (10 mM), 0.5 μl of UTP (10 mM), 1 μl T7 RNA polymerase (40 U ml−1; Invitrogen), and 1 μl Cy3-UTP (5 mM; GE Healthcare, Uppsala, Sweden) were added into a 1.5-ml tube and incubated at 37°C for 4 h. RNA was purified immediately by using the RNeasy minikit (Qiagen, Valencia, CA). Eight microliters of diethyl pyrocarbonate (DEPC)-treated water, 350 μl RLT, and 250 μl ethanol were added to the IVT mixture, followed by thorough mixing. Samples were transferred to an RNeasy minitube. Subsequently, 500 μl of RPE was added, followed by centrifugation at 10,000 rpm for 15 s. Another 500 μl of RPE was added, followed by 2 min of centrifugation at 10,000 rpm. The purified RNA was eluted into 50 μl DEPC-treated water and subsequently fragmented by incubation with 9.5 mM ZnCl2 and 24 mM Tris-Cl (pH 7.4) at 60°C for 30 min. Fragmentation was stopped by the addition of 12 mM EDTA (pH 8.0) to the reaction mixture and putting it on ice. RNasin (1 μl of a 40-U/μl solution) was added to the fragmented target.
Hybridization.
Microarray slides from the same production batch, spotted with the same batch of probes, were used throughout the analyses in this study to minimize effects that could be introduced by spotting. Hybridization was carried out in a Belly Dancer hybridization water bath (Stovall Life Sciences, Greensboro, NC) that was preheated to 55°C for at least 1 h. For each hybridization mixture, 62 μl DEPC-treated water, 1 μl 10% SDS, 30 μl 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2 μl 50× Denhardt's reagent, and 5 μl target RNA were added into a 1.5-ml tube and incubated at 65°C for 1 min. The preheated hybridization mixture was applied onto assembled slides with a HybriWell (Grace BioLabs, Bend, OR). In the hybridization chamber, the slides were incubated overnight at 55°C at maximum bending and the lowest rpm setting of the Belly Dancer shaking plateau. In the washing procedure following the hybridization, the slides were washed by shaking at room temperature for 5 min in 2× SSC, 0.1% (wt/vol) SDS, twice for 5 min in 0.2× SSC, and finally for 5 min in 0.1× SSC. Slides were dried individually using pressurized air.
Scanning and data analysis.
Hybridized slides were scanned at 10-μm resolution with a GenePix 4000 laser scanner (Axon, Foster City, CA) at a wavelength of 532 nm. Fluorescent images were analyzed with the GenePix software (Axon). Microsoft Excel was used for statistical analyses and presentation of results.
QPCR.
Three methanotrophic subgroups (Table 1) were quantified by pmoA-based quantitative PCR based on assays described elsewhere (22). The type Ia and II assays were carried out as described previously (7). For the type Ib assay, DNA standards were prepared by dilution of a known amount of PCR product amplified from a reference clone by using the 189-Mc468 primer set (22). A 25-μl reaction mixture contained 12.5 μl 2× SYBR green mix (AB Gene, Epsom, United Kingdom), 2.5 μl of diluted DNA template, and 0.8 mM (each) primers. The samples were diluted accurately to 1 ng/μl. The thermal cycle started with an initial denaturation at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 20 s, annealing at 64°C for 20 s, and extension at 72°C for 45 s. Fluorescence was recorded at 84°C, and DNA melting curve analysis was performed at temperatures ranging from 70°C to 99°C. All three assays were performed with a RotorGene 6000 thermal cycling system (Corbett Research, Eight Mile Plains, Concorde, QLD, Australia), and samples were added to aliquots of the master mixture by using a CAS-1200 liquid handling system (Corbett Robotics, Eight Mile Plains, Concorde, QLD, Australia). Every sample was analyzed in duplicate. Quantification analysis was performed by using the RotorGene software. Cell numbers were calculated assuming PCR product lengths of 412 bp, 279 bp, and 423 bp for type Ia, type Ib, and type II methanotrophs, respectively.
TABLE 1.
Explanation of MOB group indicators used in the microarray and QPCR analysesa
| Group indicator | Genera detected | Phylogenetic affiliation |
|---|---|---|
| Type Ia | Methylobacter, Methylomonas, Methylomicrobium, Methylosarcina | Gammaproteobacteria |
| Type Ib | Methylococcus, Methylocaldum | Gammaproteobacteria |
| Type II | Methylocystis, Methylosinus | Alphaproteobacteria |
Shown are the genera covered by the group indicators (types) and the respective phylogenetic affiliations of these genera.
16S rDNA gene PCR-DGGE.
DGGE analysis was performed as described previously (5), with the following exception. The PCR mixture contained 25 μl of 2× Premix F (Epicentre Biotechnologies), 25 pmol of each primer, 1 unit of Taq polymerase (Invitrogen), and 50 ng of genomic DNA as template. One microliter of the first-step PCR product was added to the second step. The gels were loaded with 2 μl of loading buffer and 23 μl of PCR product.
Statistical analyses.
The effects of laboratory and investigator on DNA extraction method, DNA concentration, purity, and QPCR results were analyzed using a nested analysis of variance design, where the investigator was nested within laboratory. Post hoc comparisons between laboratories were analyzed using Student's t test with a Bonferroni correction for the number of comparisons. All data were checked for normality and homogeneity of variances and log transformed, if necessary. The analysis was carried out using the Statistica software package, version 8.0 (Statsoft Inc., Tulsa, OK).
Analyses of microarray data.
The normalized signal intensities derived from the scanned microarrays formed the matrix that was used for nonmetric multidimensional scaling (MDS) analyses. The inputs of MDS analyses were Bray-Curtis similarity matrices generated using the log(x + 1)-transformed signal intensity values, to even out the contributions of very rare and very dominant probe signals. The MDS analyses result in a two-dimensional plot, where the distance between samples indicates the similarity of the samples relative to other samples in the plot. The accuracy of the two-dimensional representation is indicated by the “stress” value (Kruskall's stress formula). Stress values of <0.1 indicate a good ordination with no prospect of misleading interpretation. Stress values of <0.2 still give a good two-dimensional representation where not too much reliance should be put on the detail. In this case other methods of representation should be used in parallel, such as clustering analyses. The ANOSIM procedure was used to test whether microarray community profiles obtained in different laboratories differed. All MDS analyses were performed using the Primer-E software (Plymouth Marine Laboratory, Plymouth, United Kingdom). Theoretical aspects of the cluster and MDS analyses used have been described elsewhere (12). Due to repetition of single-sample DNA extraction in different countries, the effect of country on positive microarray probe was tested with a linear mixed effect model that was fit for observations taken on related individuals. Analysis used the R program add-on package nlme (38).
RESULTS
DNA concentration and quality.
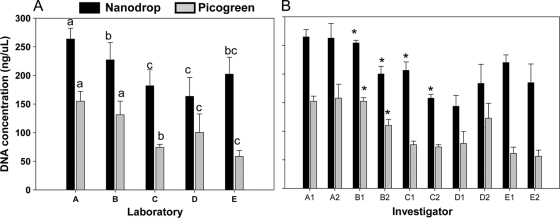
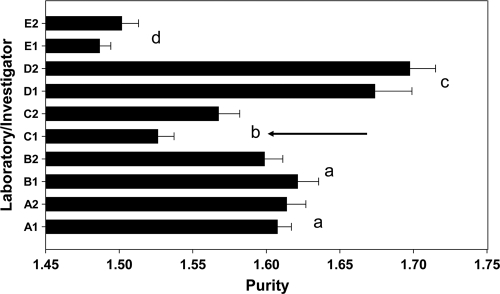
DNA concentration was analyzed using two different methods, the NanoDrop and PicoGreen assays (Fig. 1). It is obvious that the DNA concentrations measured with the NanoDrop assay were on average double the amounts measured with PicoGreen. Besides this, for both assays there were significant effects of laboratory as well as for investigator (Fig. 1A; see also Table S1 in the supplemental material). Laboratories A and B extracted more DNA than the other labs, while in three laboratories there were significant differences between investigators (Fig. 1B; see also Table S1 in the supplemental material). Differences between laboratories were also observed in the purity of the DNA (i.e., the A260/A280) (Fig. 2; see also Table S1). However, yield and purity only correlated for the PicoGreen method and not for DNA concentrations assessed by the NanoDrop method (see Table S4 in the supplemental material). An investigator effect on DNA purity was only observed in laboratory C (Fig. 2).
FIG. 1.
DNA concentrations (means ± 1 standard deviation) as analyzed with NanoDrop or PicoGreen, showing the comparisons between laboratories (A) and between investigators in the various laboratories (B). Different letters in panel A indicate significant differences between countries (P < 0.05, unequal honestly significant differences test). In panel B, the asterisk indicates a significant difference between investigators within one laboratory (as assessed using Students's t test; P < 0.01).
FIG. 2.
Purity of extracted DNA, measured as the A260/A280 ratio (means ± 1 standard deviastion). Different letters indicate significant differences between laboratories (unequal n honestly significant differences test, P < 0.05). The arrow indicates a significant difference between investigators within that laboratory.
Quantitative PCR.
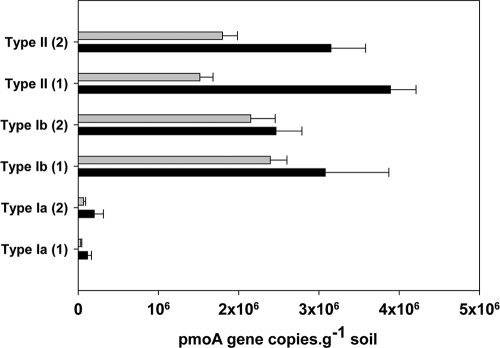
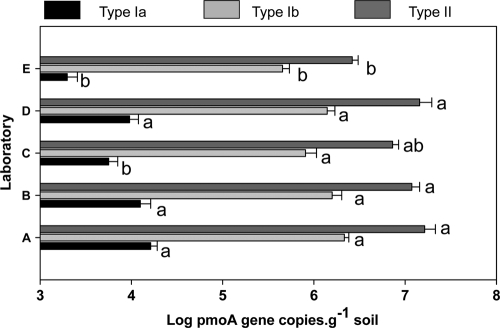
To test the effect of different DNA concentration measurements on QPCR results, both methods were compared for the samples processed in laboratory C (Fig. 3). QPCR assays targeted three groups of MOB, type Ia, type Ib, and type II. In all assays, numbers of MOB were significantly higher when initial target input amounts were based on the NanoDrop than the PicoGreen method (Fig. 3; see also Table S2 in the supplemental material). Except for type II pmoA copy numbers, there were also significant differences between the investigators (see Table S4). In Fig. 4, the QPCR data for all laboratories are presented for three different groups of MOB. Again, there were significant differences between laboratories, which were largest for type Ia, for which the numbers from laboratories E and C were lower than the rest (Fig. 4; see also Table S3 in the supplemental material). For all QPCR assays, there was no investigator effect (see Table S3). Copy numbers in all assays correlated strongly with the DNA purity (i.e., the A260/A280 ratio) and DNA concentration based on the PicoGreen assay (see Table S4).
FIG. 3.
Effect of DNA quantitation method on pmoA gene copy number (means ± 1 standard deviation) in rice soil assessed using three different assays targeting subgroups of methane-oxidizing bacteria, executed in one laboratory only. The numbers 1 and 2 in parentheses indicate the results of the two different investigators from the Austrian lab.
FIG. 4.
pmoA gene copy numbers (means ± 1 standard deviation) in rice soil of three different subgroups of methane-oxidizing bacteria as assessed in five different laboratories. Different letters per subgroup of MOB indicate significant differences between laboratories (unequal n honestly significant differences test, P < 0.05; n = 8, except for Lab E [n = 6]).
pmoA microarray analysis.
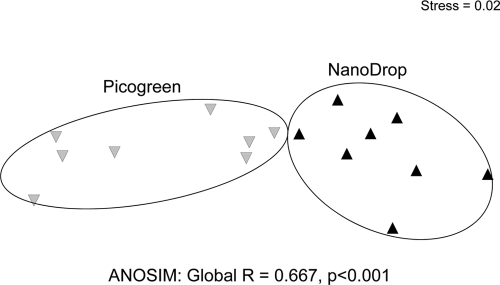
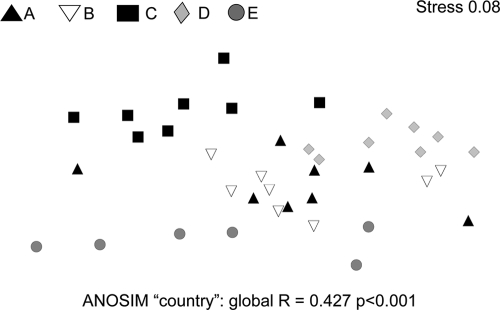
The diversity of the MOB was assessed using a pmoA diagnostic microarray. In analogy to the QPCR analysis, microarray analysis was carried out with samples from laboratory C and an initial target concentration based on the NanoDrop as well as PicoGreen DNA concentration assays. The nonmetric multidimensional scaling shown in Fig. 5 displays the dissimilarities between methanotrophic community patterns derived from DNA batches extracted in one laboratory. What is graphically obvious was also confirmed by ANOSIM: community profiles based on different DNA concentrations differed significantly from each other (Fig. 5). Microarray community profiles generated with DNA from different laboratories showed obvious differences (Fig. 6). A global ANOSIM for the laboratory effect was highly significant (P < 0.001), while there was no effect of investigator. Linear mixed effect model analyses indicated that only the profiles from laboratories A and B did not differ from each other (see Table S5 in the supplemental material). Overall, 56 probes showed positive signals. DNA from laboratory D gave higher signal intensities for eight probes than results from laboratories A and B. On the other hand, DNA from laboratories C and E gave 15 and 14 significantly reduced probe intensities, respectively. For all laboratories and investigators, 59% of potentially positive probes were not detected or showed a reduced intensity at least once. Most of these changes happened within species-specific type II probes (67% out of 21 probes were affected). However, group-specific probes for type II methanotrophs were consistently positive, while type Ia- and type Ib-specific probes differed between laboratories (see Table S5).
FIG. 5.
Nonmetric multidimensional scaling plot using log-transformed Bray-Curtis dissimilarity matrices based on signal intensity values of pmoA microarray analyses, performed on the basis of the NanoDrop or PicoGreen DNA quantitation method. Distances between symbols represent relative dissimilarity between MOB communities. Analyses of similarity (ANOSIM) resulted in a significant difference between MOB community structure when based on different DNA concentration measurements (n = 8).
FIG. 6.
Nonmetric multidimensional scaling plot using log-transformed Bray-Curtis dissimilarity matrices based on signal intensity values of the pmoA microarray analyses on DNA extracted in five different laboratories. Distances between symbols represent relative dissimilarity between MOB communities. Analyses of similarity (ANOSIM) resulted in a significant difference between MOB community structure analyzed in the different laboratories. Only samples from laboratory A and B did not differ from each other (n = 8, except for laboratory E [n = 6]).
DGGE analysis.
The DGGE patterns were rather similar in terms of the number of bands (i.e., operational taxonomic units) detected (see Fig. S1 in the supplemental material). However, the intensities of the bands differed. Since equal amounts of extracted DNA were added to the PCR mixture and equal volumes of PCR product were loaded on the gels, the intensity differences could be related to the amounts of targets in the extracts. There are differences between laboratories as well as between investigators. DNA from laboratory A, also representing the largest amount of extracted DNA, gave the highest band intensities for type I as well as for type II. Laboratories with the highest investigator differences (B and C) observed in the pmoA-based analyses also displayed the highest variability by 16S-based PCR-DGGE.
DISCUSSION
Source of variation.
The data presented in this paper clearly demonstrate that even the application of identical protocols to a single soil sample leads to significantly different community composition. It is well known that different DNA extraction protocols differ in their efficiency of liberating and purifying DNA from various environmental matrices with consequences for microbial community composition (32). This study adds another component of variation, the laboratory or the researcher carrying out the analyses. Applying the same protocol to the same sample, different laboratories with different investigators ended up with different MOB community profiles. Different methods of assessing the community composition as well as different markers (the 16S rRNA gene and pmoA) confirmed the conclusion that the origin of the differences can only reside in the DNA extraction procedure. The dependency of the outcome of microbial diversity analyses on the laboratory and even on an individual researcher has serious implications for any project or meta-analysis where results from different laboratories have to be compared.
Since all analyses following the DNA extraction were carried out in the same laboratory, using the same equipment and chemicals and executed by the same person, the source of variation between laboratories came from the extraction procedure. One explanation may be that not all laboratories used the same bead-beating machine. This may have caused the rather low DNA yield and purity in laboratory E, which used a different bead beater than the other laboratories involved. The bead beating has been found to increase the yield of soil DNA (23); however, so far there are no reports presenting data on DNA extraction efficiency as a function of the type of bead beater used.
However, laboratories using the same bead-beating machine (i.e., Labs B, C, and D) also arrived at different results. Here, the source of variation may reside in the chemicals (buffers, enzymes, solvents, etc.) used in the extraction procedures. However, the intralaboratory comparisons between two investigators using exactly the same equipment as well as chemicals also yielded significant differences in some cases. The latter can only be caused by variation in parts of the protocol that are subject to handling variation by the investigator, which can only be the pipetting routine. In the protocol, there are phase separation steps where the DNA is in the upper phase of the extract. The upper layer has to be removed without taking anything from the lower layers containing the contaminants. The efficiency of the removal of the upper phase will determine the DNA yield in a 1:1 ratio. This step requires pipetting practice and may very well differ between investigators. Nevertheless, the effect between laboratories is still greater than that between investigators. Hence, the observed variation is a mix of local differences in equipment or chemicals and also the technical skills of the person carrying out the analyses.
In addition to the phase separation step, the quality of phenol used in this part of the protocol may affect the results. Phenol (pKa 9.95) is a sensitive and reactive organic compound, having a hydroxyl group in a benzole backbone. It has a crucial role in the phase separation step by removing proteins, polyphenols, and polysaccharides from the aquatic phase, in which the DNA resides. If the phenol is not in optimal condition, the quality of the extracted DNA will be lower. In our study all laboratories used the same mixing ratios of phenol, chloroform, and isoamyl alcohol in the phase separation step except for laboratory D, which used water-saturated phenol of pH 4.0 that contained 8-hydroxyquinoline as a stabilizer. The other laboratories used Tris-HCl-buffered phenol of pH 7.0 to 8.0 and without stabilizer. The difference in pH itself is not crucial, but the storage at low pH in the presence of the stabilizer preserves the phenol better, possibly leading to a higher purity of DNA and higher values for microarray probes in the case of laboratory D.
A critical factor investigated in this study was the method of DNA concentration measurement. The popular NanoDrop method is very fast and needs only small aliquots. However, it measures DNA as well as RNA, and it is affected by the impurities in the extract. This method gave double the amount of DNA as the PicoGreen method, which only detects double-stranded DNA. The suggested overestimation by the NanoDrop method may add to the variability of final results: when starting to dilute the DNA in order to add a fixed amount to the PCR, the extract is diluted more than it would be when using the concentration measured using PicoGreen. The former method may lower the influence of residual PCR-inhibiting compounds. This may contribute to interlaboratory differences when the DNA concentration is measured with different assays. It is difficult to say which assays perform better. There are reports that above concentrations of 10 ng μl−1 both assays perform similarly, while below this concentration PicoGreen performs better (1, 15). The reproducibility is in the same range for both assays, as indicated by the coefficient of variation (see Fig. S2 in the supplemental material) and the positive correlation (see Table S4).
Consequences for MOB community structure assessment.
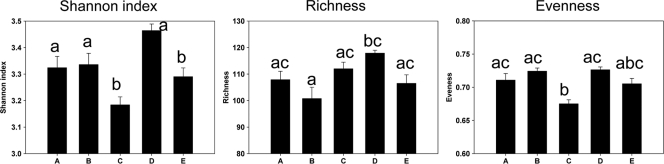
To the best of our knowledge, there are no combined inter- and intralaboratory studies on methanotrophic community structure against which to compare our results. However, it is obvious that quantitative comparisons between MOB communities have to be treated with caution, even if done in one laboratory but by different investigators. DNA extraction yields and purity may be significantly different, as are, e.g., the subsequent QPCR results. Diversity measures considering quantitative data may also be compromised. The Shannon diversity index based on the richness and evenness (microarray analyses) shows that there are significant differences between laboratories (Fig. 7). These differences already occur at the very beginning of every molecular investigation, at the DNA extraction procedure. The purity of DNA (the A260/A280 ratio) correlated positively with the Shannon and eveness indices as well as MOB abundance as assessed by QPCR, indicating that yield as well as purity influence the downstream results. In addition, higher single-probe intensities in the case of the eight probes of laboratory D could be connected to the higher purity of the DNA (see Table S5 in the supplemental material). Comparing diversity and/or abundance between habitats can be almost impossible without some information on the bias introduced at the extraction stage. A strategy overcoming these problems may be to use internal controls during DNA extraction, as previously described (31, 35, 40). However, the relative abundance as measured by QPCR stayed rather constant between laboratories. Hence, the ratios between numbers of types Ia, Ib, and II were not significantly affected by laboratory or investigator and were also not correlated to DNA quantity or quality (see Table S4). Therefore, statements such as “in this habitat there is more type II than in another habitat” or “this habitat is more diverse than another” cannot be made when analyses have been carried out in different laboratories or by different investigators, without a proper assessment of laboratory or investigator bias. What is valid is to test the effects of environmental change (assessed in different laboratories) on normalized relative abundances of MOB, which were not affected by DNA purity or yield. In a recent study it was shown that denitrification gene abundance was correlated with activity and environmental parameters only when relative abundances were used in the analyses (36). This may be caused by the fact that relative abundance levels as measured with independent assays are not influenced by biases in DNA extraction. Also, changes in community structure as assessed in one laboratory are valid results, because the same bias will be introduced at every time point, assuming that DNA yield, purity, and the investigator are identical.
FIG. 7.
Shannon index, species richness (i.e., positive probes), and evenness (means ± 1 standard deviation) of MOB communities assessed by pmoA microarray analyses, on DNA extracted in five different laboratories. Different letters indicate statistically significant differences (unequal honestly significant differences test, P < 0.05; n = 8, except for laboratory E [n = 6]).
Conclusions.
The quality and quantity of extracted DNA from a single soil sample by using identical protocols can differ between laboratories and investigators, resulting in significant bias in downstream molecular analyses. Comparison of results from different experiments necessitates assessment of this bias. To minimize the bias, we recommend performing all DNA extractions in one laboratory using identical chemicals and machinery. In a large-scale project with multiple research groups involved, all downstream processing steps should be executed using PCR ingredients purchased from the same company, and preferably from the same production batch. However, in many projects these criteria are difficult to meet. An alternative strategy would be to develop an internal control system which gives information on inter- and intralaboratory biases. Internal standards should not only help validate the quality of downstream processing but also, preferably, the DNA extraction step. In this way a correction factor might be developed, which would allow a reproducible interpretation of molecular data. For post hoc meta-analyses not meeting these requirements, however, only robust algorithms should be used, in order to avoid overinterpretation of differences.
Supplementary Material
Acknowledgments
This study was part of the METHECO Collaborative Research Project of the ESF EUROCORES-Eurodiversity program (ERAS-CT-2003-98049, 6th EU Framework Program) and was financially supported by grants from the Netherlands Organization for Scientific Research (grant no. 855.01.108).
We thank the METHECO team (Gunnar Börjesson, Xinmei Feng, Janet Andert, Werner Liesack, Pravin Shrestha, Steffen Kolb, Daniela Degelman, Anne Saari, Pertti Martikainen, Genevieve Grundmann, Colin Murrell, Deepak Kumaresan, and Guy Abell).
This publication is publication no. 4888 of the Netherlands Institute of Ecology (NIOO-KNAW).
Footnotes
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Aranda, R., S. M. Dineen, R. L. Craig, R. A. Guerrieri, and J. M. Robertson. 2009. Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 104 concentration range. Anal. Biochem. 387:122-127. [DOI] [PubMed] [Google Scholar]
- 2.Batten, C. A., K. Bachanek-Bankowska, A. Bin-Tarif, L. Kgosana, A. J. Swain, M. Corteyn, K. Darpel, P. S. Mellor, H. G. Elliott, and C. A. L. Oura. 2008. Bluetongue virus: European Community inter-laboratory comparison tests to evaluate ELISA and RT-PCR detection methods. Vet. Microbiol. 129:80-88. [DOI] [PubMed] [Google Scholar]
- 3.Beck, T., R. G. Joergensen, E. Kandeler, F. Makeschin, E. Nuss, H. R. Oberholzer, and S. Scheu. 1997. An inter-laboratory comparison of ten different ways of measuring soil microbial biomass C. Soil Biol. Biochem. 29:1023-1032. [Google Scholar]
- 4.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 5.Bodelier, P. L., M. Meima-Franke, G. Zwart, and H. J. Laanbroek. 2005. New DGGE strategies for the analyses of methanotrophic microbial communities using different combinations of existing 16S rRNA-based primers. FEMS Microbiol. Ecol. 52:163-174. [DOI] [PubMed] [Google Scholar]
- 6.Bodelier, P. L. E., M. J. B. Gillisen, K. Hordijk, J. S. S. Damste, W. I. C. Rijpstra, J. A. J. Geenevasen, and P. F. Dunfield. 2009. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 3:606-617. [DOI] [PubMed] [Google Scholar]
- 7.Bodelier, P. L. E., M. Kamst, M. Meima-Franke, N. Stralis-Pavese, and L. Bodrossy. 2009. Whole-community genome amplification (WCGA) leads to compositional bias in methane-oxidizing communities as assessed by pmoA-based microarray and QPCR. Environ. Microbiol. Rep. 1:434-441. [DOI] [PubMed] [Google Scholar]
- 8.Burgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell, S. L., J. R. Laidler, E. A. Brewer, J. O. Eberly, S. C. Sandborgh, and F. S. Colwell. 2008. Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ. Sci. Technol. 42:6791-6799. [DOI] [PubMed] [Google Scholar]
- 10.Carrigg, C., O. Rice, S. Kavanagh, G. Collins, and V. O'Flaherty. 2007. DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 77:955-964. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, C. M., K. T. Van, W. Lin, and R. M. Ruby. 2009. Interlaboratory validation of a real-time PCR 24-hour rapid method for detection of Salmonella in foods. J. Food Prot. 72:945-951. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. PRIMER-E, Plymouth, United Kingdom.
- 13.Conrad, R. 2007. Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96:1-63. [Google Scholar]
- 14.Dumont, M. G., and J. C. Murrell. 2005. Community-level analysis: key genes of aerobic methane oxidation. Environ. Microbiol. 397:413-427. [DOI] [PubMed] [Google Scholar]
- 15.English, C. A., S. Merson, and J. T. Keer. 2006. Use of elemental analysis to determine comparative performance of established DNA quantification methods. Anal. Chem. 78:4630-4633. [DOI] [PubMed] [Google Scholar]
- 16.Falkowski, P. G., T. Fenchel, and E. F. Delong. 2008. The microbial engines that drive Earth's biogeochemical cycles. Science 320:1034-1039. [DOI] [PubMed] [Google Scholar]
- 17.Feinstein, L. M., W. J. Sul, and C. B. Blackwood. 2009. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75:5428-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferris, N. P., D. P. King, S. M. Reid, G. H. Hutchings, A. E. Shaw, D. J. Paton, N. Goris, B. Haas, B. Hoffmann, E. Brocchi, M. Bugnetti, A. Dekker, and K. De Clercq. 2006. Foot-and-mouth disease virus: a first inter-laboratory comparison trial to evaluate virus isolation and RT-PCR detection methods. Vet. Microbiol. 117:130-140. [DOI] [PubMed] [Google Scholar]
- 19.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frostegard, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 21a.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 22.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krsek, M., and E. M. H. Wellington. 1999. Comparison of different methods for the isolation and purification of total community DNA from soil. J. Microbiol. Methods 39:1-16. [DOI] [PubMed] [Google Scholar]
- 24.Kruger, M., P. Frenzel, and R. Conrad. 2001. Microbial processes influencing methane emission from rice fields. Global Change Biol. 7:49-63. [Google Scholar]
- 25.Malorny, B., J. Hoorfar, M. Hugas, A. Heuvelink, P. Fach, L. Ellerbroek, C. Bunge, C. Dorn, and R. Helmuth. 2003. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 89:241-249. [DOI] [PubMed] [Google Scholar]
- 26.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A.-L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 27.McDonald, I. R., L. Bodrossy, Y. Chen, and J. C. Murrell. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty, S. R., P. L. E. Bodelier, V. Floris, and R. Conrad. 2006. Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl. Environ. Microbiol. 72:1346-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montesino, M., A. Salas, M. Crespillo, C. Albarrán, A. Alonso, V. Álvarez-Iglesias, J. A. Cano, M. Carvalho, D. Corach, C. Cruz, A. Di Lonardo, R. Espinheira, M. J. Farfán, S. Filippini, J. García-Hirschfeld, A. Hernández, G. Lima, C. M. López-Cubría, M. López-Soto, S. Pagano, M. Paredes, M. F. Pinheiro, A. M. Rodríguez-Monge, A. Sala, S. Sóñora, D. R. Sumita, M. C. Vide, M. R. Whittle, A. Zurita, and L. Prieto. 2007. Analysis of body fluid mixtures by mtDNA sequencing: an inter-laboratory study of the GEP-ISFG Working Group. Forensic Sci. Int. 168:42-56. [DOI] [PubMed] [Google Scholar]
- 31.Mumy, K. L., and R. H. Findlay. 2004. Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. J. Microbiol. Methods 57:259-268. [DOI] [PubMed] [Google Scholar]
- 32.Ning, J., J. Liebich, M. Kastner, J. Z. Zhou, A. Schaffer, and P. Burauel. 2009. Different influences of DNA purity indices and quantity on PCR-based DGGE and functional gene microarray in soil microbial community study. Appl. Microbiol. Biotechnol. 82:983-993. [DOI] [PubMed] [Google Scholar]
- 33.Noll, M., P. Frenzel, and R. Conrad. 2008. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol. Ecol. 65:125-132. [DOI] [PubMed] [Google Scholar]
- 34.Op den Camp, H. J. M., T. Islam, M. B. Stott, H. R. Harhangi, A. Hynes, S. Schouten, M. S. M. Jetten, N. K. Birkeland, A. Pol, and P. F. Dunfield. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293-306. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, D. G., and I. Dahllof. 2005. Improvements for comparative analysis of changes in diversity of microbial communities using internal standards in PCR-DGGE. FEMS Microbiol. Ecol. 53:339-348. [DOI] [PubMed] [Google Scholar]
- 36.Philippot, L., J. Cuhel, N. P. A. Saby, D. Cheneby, A. Chronakova, D. Bru, D. Arrouays, F. Martin-Laurent, and M. Simek. 2009. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ. Microbiol. 11:1518-1526. [DOI] [PubMed] [Google Scholar]
- 37.Picard, C., C. Ponsonnet, E. Paget, X. Nesme, and P. Simonet. 1992. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl. Environ. Microbiol. 58:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinheiro, J., D. Bates, S. DebRoy, D. Sarker, and the R Core team. 2009. nlme: linear and nonlinear mixed effects models. R package, version 3.1-93. Vienna University, Vienna, Austria.
- 39.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramette, A. 2009. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl. Environ. Microbiol. 75:2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roesch, L. F., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Y. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. Plos Biol. 5:398-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sogin, M. L., H. G. Morrison, J. A. Huber, D. Mark Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stralis-Pavese, N., A. Sessitsch, A. Weilharter, T. Reichenauer, J. Riesing, J. Csontos, J. C. Murrell, and L. Bodrossy. 2004. Optimization of diagnostic microarray for application in analysing landfill methanotroph communities under different plant covers. Environ. Microbiol. 6:347-363. [DOI] [PubMed] [Google Scholar]
- 45.Thakuria, D., O. Schmidt, M. Mac Siúrtáin, D. Egan, and F. M. Doohan. 2008. Importance of DNA quality in comparative soil microbial community structure analyses. Soil Biol. Biochem. 40:1390-1403. [Google Scholar]
- 46.Trotsenko, Y. A., and J. C. Murrell. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63:183-229. [DOI] [PubMed] [Google Scholar]
- 47.von Mering, C., P. Hugenholtz, J. Raes, S. G. Tringe, T. Doerks, L. J. Jensen, N. Ward, and P. Bork. 2007. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315:1126-1130. [DOI] [PubMed] [Google Scholar]
- 48.White, J. R., R. D. Shannon, J. F. Weltzin, J. Pastor, and S. D. Bridgham. 2008. Effects of soil warming and drying on methane cycling in a northern peatland mesocosm study. J. Geophys. Res. Biogeosci. 113.
- 49.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]
- 51.Yergeau, E., S. A. Schoondermark-Stolk, E. L. Brodie, S. Dejean, T. Z. DeSantis, O. Goncalves, Y. M. Piceno, G. L. Andersen, and G. A. Kowalchuk. 2009. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 3:340-351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.