Abstract
Accumulation of galactose in dairy products due to partial lactose fermentation by lactic acid bacteria yields poor-quality products and precludes their consumption by individuals suffering from galactosemia. This study aimed at extending our knowledge of galactose metabolism in Lactococcus lactis, with the final goal of tailoring strains for enhanced galactose consumption. We used directed genetically engineered strains to examine galactose utilization in strain NZ9000 via the chromosomal Leloir pathway (gal genes) or the plasmid-encoded tagatose 6-phosphate (Tag6P) pathway (lac genes). Galactokinase (GalK), but not galactose permease (GalP), is essential for growth on galactose. This finding led to the discovery of an alternative route, comprising a galactose phosphotransferase system (PTS) and a phosphatase, for galactose dissimilation in NZ9000. Introduction of the Tag6P pathway in a galPMK mutant restored the ability to metabolize galactose but did not sustain growth on this sugar. The latter strain was used to prove that lacFE, encoding the lactose PTS, is necessary for galactose metabolism, thus implicating this transporter in galactose uptake. Both PTS transporters have a low affinity for galactose, while GalP displays a high affinity for the sugar. Furthermore, the GalP/Leloir route supported the highest galactose consumption rate. To further increase this rate, we overexpressed galPMKT, but this led to a substantial accumulation of α-galactose 1-phosphate and α-glucose 1-phosphate, pointing to a bottleneck at the level of α-phosphoglucomutase. Overexpression of a gene encoding α-phosphoglucomutase alone or in combination with gal genes yielded strains with galactose consumption rates enhanced up to 50% relative to that of NZ9000. Approaches to further improve galactose metabolism are discussed.
Lactococcus lactis is a lactic acid bacterium widely used in the dairy industry for the production of fermented milk products. Because of its economic importance, L. lactis has been studied extensively in the last 40 years. A small genome, a large set of genetic tools, a wealth of physiological knowledge, and a relatively simple metabolic potential render L. lactis an attractive model with which to implement metabolic engineering strategies (reviewed in references 21 and 57).
In the process of milk fermentation by L. lactis, lactose is taken up and concomitantly phosphorylated at the galactose moiety (C-6) by the lactose-specific phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTSLac), after which it is hydrolyzed to glucose and galactose 6-phosphate (Gal6P) (64). The glucose moiety enters the glycolytic pathway upon phosphorylation via glucokinase to glucose 6-phosphate (G6P), whereas Gal6P is metabolized to triose phosphates via the d-tagatose 6-phosphate (Tag6P) pathway, encompassing the steps catalyzed by galactose 6-phosphate isomerase (LacAB), Tag6P kinase (LacC), and tagatose 1,6-bisphosphate aldolase (LacD) (Fig. 1). Curiously, during the metabolism of lactose by L. lactis, part of the Gal6P is dephosphorylated and excreted into the growth medium, while the glucose moiety is readily used (2, 7, 51, 56, 60).
FIG. 1.
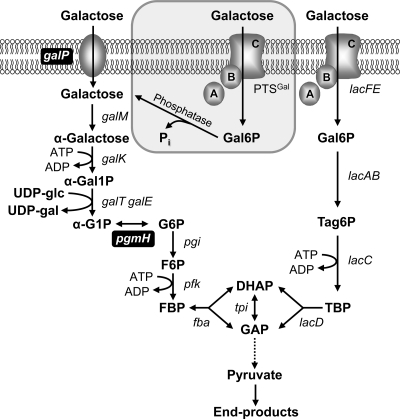
Schematic overview of the alternative routes for galactose uptake and further catabolism in L. lactis. Galactose can be imported by the non-PTS permease GalP and metabolized via the Leloir pathway (galMKTE) to α-G1P, which is converted to the glycolytic intermediate G6P by α-phosphoglucomutase (pgmH). Alternatively, galactose can be imported by PTSLac (lacFE) and further metabolized to triose phosphates by the Tag6P pathway (lacABCD). Here, we propose a new uptake route consisting of galactose translocation via the galactose PTS, followed by dephosphorylation of the internalized Gal6P to galactose, which is further metabolized via the Leloir pathway (highlighted in the gray box). galP, galactose permease; galM, galactose mutarotase; galK, galactokinase; galT, galactose 1-phosphate uridylyltransferase; galE, UDP-galactose-4-epimerase; pgmH, α-phosphoglucomutase; lacAB, galactose 6-phosphate isomerase; lacC, Tag6P kinase; lacD, tagatose 1,6-bisphosphate aldolase; lacFE, PTSLac; PTSGal, unidentified galactose PTS; Phosphatase; unidentified Gal6P-phosphatase; pgi, phosphoglucose isomerase; pfk, 6-phosphofructo-1-kinase; fba, fructose 1,6-bisphosphate aldolase; tpi, triose phosphate isomerase; α-Gal1P, α-galactose 1-phosphate; α-G1P, α-glucose 1-phosphate; UDP-gal, UDP-galactose; UDP-glc, UDP-glucose; G6P, glucose 6-phosphate; Gal6P, galactose 6-phosphate; Tag6P, tagatose 6-phosphate; TBP, tagatose 1,6-bisphosphate; FBP, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate. The dotted arrow represents the conversions of GAP to pyruvate via the glycolytic pathway. Steps essential to improve galactose consumption are shown in black boxes.
As a result of incomplete lactose utilization, some fermented dairy products contain significant residual amounts of galactose. The presence of galactose has been associated with shoddier qualities of the fermented product (6, 27, 43). In particular, galactose is a major contributor to the browning that occurs when dairy products (e.g., yogurt and mozzarella, Swiss, and cheddar cheese) are cooked or heated in the manufacture of pizzas, sauce preparation, or processed cheese. In addition, availability of residual galactose may result in production of CO2 by heterofermentative starters and, consequently, in textural defects such as the development of slits and fractures in cheeses. Therefore, the availability of starter strains with improved galactose utilization capacity is desirable to develop higher-quality dairy products. Additionally, strains with increased galactose metabolism could provide galactose-free foods for individuals and, in particular, children suffering from the rare disease galactosemia (36). To this end, a comprehensive understanding of galactose catabolism is essential.
Galactose metabolism in L. lactis was thoroughly studied in the past and has been and still is the subject of some controversy. Indeed, conflicting results regarding the type of PTS involved in galactose uptake have been published. Some authors advocated that galactose is exclusively transported via the plasmid-encoded PTSLac, whereas others proposed transport via a galactose-specific PTS (PTSGal) to the extreme of questioning the contribution of the PTSLac (17, 20, 50, 59). However, a gene encoding PTSGal has never been identified in L. lactis. Independently of the nature of the PTS, it is generally accepted that the resulting Gal6P is metabolized via the Tag6P pathway (lac operon) (Fig. 1). On the other hand, galactose translocated via the highly specific galactose permease (GalP) is metabolized via the Leloir pathway to α-glucose 1-phosphate (α-G1P) through the sequential action of galactose mutarotase (GalM), galactokinase (GalK), and galactose 1-phosphate uridylyltransferase (GalT)/UDP-galactose-4-epimerase (GalE) (gal operon). Entry in glycolysis is preceded by the α-phosphoglucomutase (α-PGM)-catalyzed isomerization of α-G1P to G6P. The use of the Leloir and/or the Tag6P pathway for galactose utilization is currently viewed as being strain dependent (9, 16, 25, 32, 33, 58), but the relative efficacy in the degradation of the sugar has not been established.
The ultimate aim of this study was to engineer L. lactis for improved galactose-fermenting capacity as a means to minimize the galactose content in dairy products. To gain insight into galactose catabolism via the Leloir (gal genes) and the Tag6P (lac genes) pathways, a series of L. lactis subsp. cremoris NZ9000 isogenic gal and lac mutants were constructed. Carbon 13 labeling experiments coupled with nuclear magnetic resonance (NMR) spectroscopy were used to investigate galactose metabolism in the gal and lac strains. The data obtained revealed a novel route for galactose dissimilation and provided clues to further enhance galactose utilization.
MATERIALS AND METHODS
Microbial strains used and growth conditions.
The strains and plasmids used in this study are listed in Table 1. For genetic manipulation, strains were routinely grown at 30°C in M17 broth (Difco, Sparks, MD) containing 0.5% (wt/vol) glucose. For physiological characterization, cultures were grown in chemically defined medium (CDM) (52) with 1% (wt/vol) galactose under anaerobic conditions in rubber-stoppered bottles (200 ml) or in a 2-liter fermentor (Biostat MD; B. Braun Biotech International, Melsungen, Germany) and at a temperature of 30°C. When appropriate, erythromycin or chloramphenicol was used at 5 μg ml−1. Expression of genes cloned downstream of the nisin-inducible PnisA promoter was induced when the optical density at 600 nm (OD600) was between 0.25 and 0.5 by addition of a supernatant (0.01%, vol/vol) of an overnight culture of the nisin producer L. lactis NZ9700 (29) or by addition of a nisin solution (1 μg liter−1 in a 50% [vol/vol] ethanol solution). In the fermentor, the medium was gassed with argon for 60 min prior to inoculation (4% inoculum from a culture grown overnight). The pH was kept at 6.5 by automated addition of 10 M NaOH, and an agitation rate of 70 rpm was used to keep the system homogeneous. Growth was monitored by measuring the OD600. Specific growth rates (μ) were calculated through linear regressions of the plots of ln(OD600) versus time during the exponential growth phase.
TABLE 1.
Lactococcal strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| L. lactis strains | ||
| MG1363 | L. lactis subsp. cremoris, plasmid-free derivative of NCDO712 | 24 |
| NZ9000 | MG1363 ΔpepN::nisRK | 31 |
| NZ9700 | Nisin-producing transconjugant with the nisin-sucrose transposon Tn5276 | 29 |
| LL302 | MG1363 RepA+ carrying a single copy of pWV01 repA in pepX | 34 |
| NZ9000ΔgalP | NZ9000 with deletion of galP | This work |
| NZ9000ΔgalPMK | NZ9000 with deletion of galPMK | This work |
| S. thermophilus ST11 | Gal-negative strain | 44 |
| Plasmids | ||
| pNZ8048 | Cmr, nisin-inducible PnisA | 19 |
| pNG8048e | Cmr Emr PnisA, pNZ8048 derivative containing erythromycin resistance gene | Lab coll.a |
| pORI280 | Emr LacZ+ RepA−ori+ | 35 |
| pVE6007 | Cmrori(Ts), derivative of pWV01 | 42 |
| pMG820 | 23.7-kb derivative of pLP712 containing lacFEGABCD | 41 |
| pGalP | pNG8048e with galP cloned in the NcoI/XbaI site, Cmr | This work |
| pGalPMKT | pNG8048e with galPMKT cloned in the NcoI/XbaI site, Cmr | This work |
| pPgmA | pNZ8048 with pgmA of S. thermophilus cloned in the NcoI/XbaI site | This work |
| pGalPpgmA | pGalP with pgmA of S. thermophilus cloned in the XbaI site | This work |
| pGalPMKTpgmA | pGalPMKT with pgmA of S. thermophilus cloned in the XbaI site | This work |
| pLacFE | pNZ8048 with lactococcal pMG820 lacFE cloned in the RcaI/XbaI site | This work |
| pLacABCD | pNZ8048 with lactococcal pMG820 lacABCD cloned in the PtsI/SpeI site | This work |
| pLacABCDFE | pNZ8048 with lactococcal pMG820 lacABCDFE cloned in the SpeI/SpeI site | This work |
| pLacABCDFEG | pLacABCDFE with lactococcal pMG820 lacG cloned in the SacI site | This work |
Lab coll., laboratory collection.
General DNA manipulations.
General DNA techniques were performed essentially as described before (55). Plasmid DNA was isolated by the method of Birnboim and Doly (8). Restriction enzymes, T4 DNA ligase, Expand polymerase, and Taq DNA polymerase were obtained from Roche Diagnostics GmbH (Mannheim, Germany) and were used according to the supplier's instructions. PCR amplifications were performed in an Eppendorf thermal cycler (Eppendorf, Hamburg, Germany) with L. lactis MG1363 chromosomal DNA as the template, unless described otherwise, using appropriate conditions. The primers used in this study are described in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Enzyme |
|---|---|---|
| GalA1-fw | CGGTCTCCCATGAAAGAGGGAAAAATGAAACAACG | Eco31I |
| GalA-rev | CTAGTCTAGATTATTTCAAACGTTCTTC | XbaI |
| GalT-rev | CTAGTCTAGATTATTGATTCACAAAATC | XbaI |
| PgmA-fw1 | CATGCCATGGTAGTTGTGATACAATGTAAGCG | NcoI |
| PgmA-fw2 | GCTCTAGATAGTTGTGATACAATGTAAGCG | XbaI |
| PgmA-rev | GCTCTAGATTGGTGTAGCAGCGAAAG | XbaI |
| GalA-KO1 | GCTCTAGACTTTCGGGAGAAACCGTGG | XbaI |
| GalA-KO2 | CGGGATCCCCCTCTTTCATGGGAATCC | BamHI |
| GalA-KO3 | CGGGATCCCCTTTGTAGTCCCAGCGG | BamHI |
| GalA-KO4 | CGGAATTCGAATGCTATCTTCTCCACC | EcoRI |
| GalAMK-KO5 | CGGGATCCGATGATTACGAAGTCACTGG | BamHI |
| GalAMK-KO6 | CGGAATTCAATCGCCAGAAGTTGGTCC | EcoRI |
| LacFE-fw | CCTGATCATGAACAGAGAAGAGATGAC | RcaI |
| LacFE-rev | TGCTCTAGATTAATCAAACTGTTGTTG | XbaI |
| LacAD-fw | AAACTGCAGATGGCTATTGTTGTTGGTGC | PstI |
| LacAD-rev | GGACTAGTCTATACTTTATCAGTCCATGGAC | SpeI |
| LacAE-fw | ACATGCATGCATGGCTATTGTTGTTGGTGC | SphI |
| LacAE-rev | GGACTAGTTTAATCAAACTGTTGTTGAACAAATG | SpeI |
| LacG-fw | CGAGCTCAATATATCAAATTGACACGTGACGG | SacI |
| LacG-rev | CGAGCTCTTACTCTATCACTTGAGTTTCTGC | SacI |
Construction of overexpression vectors.
Cloning of genes involved in the Leloir pathway was performed as follows: the PCR products obtained with primer pairs GalA1-fw/GalA-rev and GalA1-fw/GalT-rev were cloned as 1.39-kb and 6.60-kb Eco31I/XbaI restriction fragments in NcoI/XbaI-restricted pNG8048e, resulting in plasmids pGalP (galP cloned downstream of the nisin-inducible promoter PnisA) and pGalPMKT (galPMKT downstream of PnisA), respectively. The plasmids were introduced by electrotransformation (26) into L. lactis NZ9000.
To clone the pgmA gene (1.72 kb) of Streptococcus thermophilus ST11, primers were designed on the sequence of S. thermophilus LY03 (accession number IMDST01, culture collection at VU, Brussels, Belgium). The PCR products obtained with primer pair PgmA-fw1/PgmA-rev or PgmA-fw2/PgmA-rev were cloned as NcoI/XbaI or XbaI restriction fragments in NcoI/XbaI-restricted pNZ8048 to obtain pPgmA and in XbaI-restricted pGalP and pGalPMKT to obtain pGalPpgmA and pGalPMKTpgmA, respectively. Variants of the plasmids that carried pgmA downstream and in the same orientation as the gal gene(s) were selected in L. lactis NZ9000. Plasmid constructs were checked by EcoRI restriction enzyme analysis.
The Tag6P pathway genes lacFE, lacABCD, and lacABCDFE were amplified by PCR using pMG820 DNA as the template and primer pairs LacFE-fw/LacFE-rev, LacAD-fw/LacAD-rev, and LacAE-fw/LacAE-rev (Table 2). The overexpression plasmids pLacFE, pLacABCD, and pLacABCDFE were constructed by cloning the 2.04-kb RcaI/XbaI, 2.89-kb PtsI/SpeI, and 4.95-kb SphI/SpeI restriction fragments into NcoI/XbaI-, PtsI/SpeI-, and SphI/SpeI-restricted pNZ8048, respectively. lacG was amplified using primer pair LacG-fw and LacG-rev, restricted with SacI, and cloned into SacI-restricted pLacABCDFE. The lacG orientation in pLacABCDFEG was confirmed by restriction enzyme analysis. The resulting constructs were transformed (26) into L. lactis NZ9000ΔgalPMK.
Construction of galP- and galPMK-deletion strains.
The PCR products obtained with primer pairs GalA-KO1/GalA-KO2 and GalA-KO3/GalA-KO4 were cloned together as XbaI/BamHI and BamHI/EcoRI restriction fragments in XbaI/EcoRI-restricted pORI280 (35), resulting in pORI280-galP′. This plasmid was obtained using L. lactis LL302 (34) as the cloning host. PCR products obtained with primer pairs GalA-KO1/GalA-KO2 and GalAMK-KO5/GalAMK-KO6 were cloned as XbaI/BamHI and BamHI/EcoRI restriction fragments in XbaI/EcoRI-restricted pORI280, resulting in pORI280-galPMK′, which was obtained and maintained in L. lactis LL302. Introducing pORI280-galP′ or pORI280-galPMK′ together with helper plasmid pVE6007 in L. lactis NZ9000, followed by a two-step homologous recombination event (35), yielded strains NZ9000ΔgalP and NZ9000ΔgalPMK, respectively. The chromosomal structures of both strains were confirmed by PCR analysis and Southern blotting using enhanced chemiluminescence (ECL) detection (Amersham Pharmacia Biotech) with a PCR fragment obtained with primer pair GalA-KO1/GalA-KO2 as the probe.
Enzyme assays.
For enzymatic assays, cells were disrupted using 0.5 g glass beads (diameter, 50 to 105 μm; Fischer Scientific BV, Den Bosch, Netherlands) and a Mini-BeadBeater-8 apparatus (Biospec Products, Inc., Bartlesville, OK) with two 1-min pulses separated by 1 min of cooling down on ice. Cell debris was pelleted, and activities were assayed at 30°C. Protein concentrations were determined by the method of Bradford (11).
(i) α-Phosphoglucomutase.
Cell cultures grown to an OD600 of 0.25 were treated with nisin (0.01% [vol/vol] supernatant of NZ9700), as described above. When the cultures reached an OD600 of 1 (approximately 4 h after induction), the cells were centrifuged (2,500 × g, 7 min), washed twice with potassium phosphate (KPi) buffer (10 mM, pH 7.2), and resuspended in the same buffer. α-PGM specific activity in cell extracts was assayed as described by Qian et al. (54). The 1-ml assay mixture contained 50 mM triethanolamine-HCl (TEA; pH 7.2), 5 mM MgCl2, 0.5 mM NADP+, 50 μM glucose-1,6-bisphosphate, and 1.75 U glucose-6-phosphate dehydrogenase. Reactions were started by the addition of 1.5 mM α-G1P. One unit of enzyme is defined as the amount of enzyme catalyzing the conversion of 1 μmol of substrate (NADP+) per min assayed at 340 nm.
Activities involved in galactose catabolism in L. lactis strain NZ9000ΔgalP.
Cells were grown in galactose (0.5%, wt/vol)-CDM, harvested in the mid-exponential phase of growth, centrifuged (2,500 × g, 7 min), washed twice with TEA buffer (50 mM, pH 7.2), and resuspended in the same buffer, after which cell extracts were prepared. All the reaction mixtures contained 50 mM TEA buffer, pH 7.2, 5 mM MgCl2, and cell extract (about 2 mg total protein) in a final volume of 0.55 ml. The reactions were initiated by the addition of 5 mM sugar substrate (Gal6P, Gal1P, or galactose). After 1 h of incubation at 30°C, 50 μl of 2H2O was added and the reaction products were analyzed by 31P NMR spectroscopy. Alternatively, the reactions were monitored online, and 31P spectra were acquired every 2.5 min. ATP and the phosphatase inhibitor sodium fluoride were added to the reaction mixtures to final concentrations of 2.5 mM and 10 mM, respectively. α-Glucose 1,6-bisphosphate (50 μM) was added when appropriate.
31P NMR spectra were recorded using a selective probe head (31P-SEX) at 30°C on an Avance II 500-MHz spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany) by using standard Bruker pulse programs. Spectra were referenced to the resonance of external 85% H3PO4, designated 0 ppm.
Galactose PTS assay.
L. lactis strain NZ9000ΔgalP was grown to an optical density at 600 nm of 2.2 in galactose-CDM, and PTS activity was determined as described by LeBlanc et al. (32). Briefly, 20 ml of late-exponential-phase cells was harvested, washed twice with KPi buffer (100 mM, pH 7.2), and suspended in 5 ml of the same buffer supplemented with MgCl2 (5 mM). The cells in this suspension were permeabilized with 250 μl of toluene-acetone (1:9), and 50 μl of permeabilized cells was used in the assays. The 1-ml assay mixture contained 100 mM KPi (pH 7.2), 5 mM MgCl2, 15 mM NADH, 10 U lactate dehydrogenase, 10 mM NaF, and 20 mM galactose. Reactions were started by the addition of 5 mM PEP. The oxidation of NADH was measured in a PEP-dependent manner.
In vivo NMR spectroscopy.
Carbon-13 spectra were acquired at 125.77 MHz on an Avance II 500-MHz spectrometer (Bruker BioSpin GmbH). All in vivo experiments were run using a quadruple nucleus probe head at 30°C, as described before (46). Cells were harvested in the mid-logarithmic phase of growth, washed twice with 5 mM KPi buffer (pH 6.5), and resuspended in 50 mM KPi (pH 6.5) to a protein concentration of approximately 15 mg protein ml−1. In vivo NMR experiments were performed using the online system described earlier, which consists of a minibioreactor coupled to NMR detection with a circulating system that allows noninvasive studies of metabolism under controlled conditions of pH, gas atmosphere, and temperature (46). Galactose specifically labeled with 13C on carbon 1 (20 mM) was added to the cell suspension at time zero. The time courses of galactose consumption, product formation, and changes in the pools of intracellular metabolites were monitored in vivo. When the substrate was exhausted and no changes in the resonances of intracellular metabolites were observed, an NMR sample extract was prepared as described previously (46, 48). The lactate in the NMR sample extract was quantified by 1H NMR in an AMX300 spectrometer (Bruker BioSpin GmbH). The concentrations of the other metabolites were determined in fully relaxed 13C spectra of the NMR sample extracts as described previously (48). Due to the fast pulsing conditions used for acquiring in vivo 13C spectra, correction factors were determined to convert peak intensities into concentrations (45, 46). The quantitative kinetic data for intracellular metabolites were calculated as described elsewhere (47, 48). The lower limit for in vivo NMR detection of intracellular metabolites under these conditions was 3 to 4 mM. Intracellular metabolite concentrations were calculated using a value of 2.9 μl (mg of protein)−1 for the intracellular volume of L. lactis (53). Although the results of individual experiments are illustrated in each figure, each experiment was repeated at least twice and the results were highly reproducible. The values reported are averages of two experiments, and the accuracy varied from ±2% (extracellular products) to ±10% in the case of intracellular metabolites with concentrations below 5 mM and varied from ±5% to ±10% for the maximal galactose consumption rate.
Analysis of [1-13C]galactose fermentation products by 1H NMR.
L. lactis NZ9000ΔgalPMK and derivatives carrying pMG820 or each of the lac constructs were grown in M17 broth with 1% glucose and an initial pH of 6.5. When appropriate, chloramphenicol (5 μg ml−1) was added. Nisin (Sigma, St. Louis, MO) at a concentration of 1 μg l−1 was added when the OD600 of the cultures reached approximately 0.4. After 2 h of induction, cells were harvested, washed twice (50 mM KPi, pH 6.5), resuspended at a concentration of approximately 5 mg protein ml−1 in KPi buffer (50 mM, pH 6.5), and placed in tubes with a rubber stopper. Argon was bubbled through the suspension for 10 min to ensure anaerobiosis. [1-13C]galactose (20 mM) was added, and the supernatant was recovered following 1 h of incubation at 30°C with mild agitation to keep the system homogeneous. The 13C-labeling pattern of the end products formed from 13C-enriched galactose was determined by 1H NMR analysis. 1H NMR spectra were acquired in a Bruker AMX300 spectrometer using a 5-mm inverse detection probe head, 32,000 data points, a 90° flip angle, and a repetition delay of 42.7 s. The water resonance was suppressed with a presaturation pulse.
Transcriptome analysis.
The levels of transcripts in NZ9000ΔgalP grown on galactose were compared by transcriptome analysis using full-genome amplicon-based L. lactis MG1363 DNA microarrays, and the levels of mRNA in NZ9000ΔgalP grown on glucose or the levels of mRNA in NZ9000 grown on galactose were determined as described elsewhere (30). Cells were grown in CDM in rubber-stoppered bottles as described above and harvested at an OD600 of 0.25. The experiments were performed essentially as described elsewhere (62) with the modifications introduced previously (51).
Chemicals.
[1-13C]galactose (99% enrichment) was obtained from Euriso-top (Gif-Sur-Yvette, France). Formic acid (sodium salt) was purchased from Merck Sharp & Dohme (Whitehouse Station, NJ). Nisin was obtained from Sigma-Aldrich (St. Louis, MO). All other chemicals were reagent grade.
RESULTS
L. lactis NZ9000 can import galactose via more than one transport system.
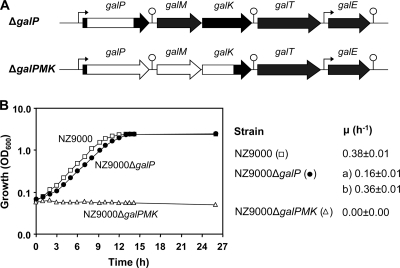
L. lactis can use the Leloir pathway, the Tag6P pathway, or both pathways for galactose metabolism in a strain-dependent manner (58). Presumably, L. lactis subsp. cremoris NZ9000 (MG1363pepN::nisRK) internalizes galactose by a secondary carrier symporter (GalP) and further metabolizes the sugar exclusively via the Leloir pathway (25), as this strain lacks the plasmid-linked genes encoding the Tag6P pathway. To assess the potential of the Tag6P route for galactose catabolism, a strategy was devised that consisted of introduction of plasmid pMG820 (41) with genes lacABCDFEG, encoding the lactose PTS (lacFE), the Tag6P pathway enzymes (lacABCD), and a β-phosphogalactosidase (lacG) (64), in mutants in which galactose utilization via the Leloir pathway was prevented. Grossiord et al. (25) previously reported blockage of the Leloir pathway by inactivation of the galactose permease gene. In our study, galP was deleted in strain NZ9000 using a double-crossover recombination method. The extent of the deletion is shown in Fig. 2 A. Unexpectedly, L. lactis NZ9000ΔgalP was still able to grow in a medium with galactose as the sole carbon source (Fig. 2B); growth was biphasic and characterized by an initial growth rate that was 2.3-fold lower than a second growth rate, which was similar to that of parent strain NZ9000 (0.38 ± 0.01 h−1). To exclude the possibility of residual GalP activity due to partial deletion only (Fig. 2A), a new out-of-frame deletion was made in which only the first four amino acids of the original protein were left. The behavior of the resulting strain was in all aspects similar to that of our original galP-deletion strain. Subsequently, a mutant strain of L. lactis NZ9000 was made in which, apart from galP, the downstream genes of the operon, galM (galactose mutarotase) and galK (galactose kinase), were also deleted (Fig. 2A). Deletion of galPMK resulted in total loss of the capacity to grow in a medium with galactose as the sole source of carbon (Fig. 2B). These data imply that L. lactis NZ9000 has an additional transport system with specificity for galactose.
FIG. 2.
Lactococcal strains used in this study and their respective growth profiles on galactose. (A) Schematic overview of the gal operon and the genetic makeup of the galP and galPMK deletions in strain NZ9000. Hooked arrow, putative promoter; lollipop, putative terminator structure; white empty areas, deleted sequence. (B) Growth of L. lactis NZ9000 and derivatives in CDM supplemented with 55 mM galactose at 30°C in rubber-stoppered bottles without pH control (initial pH 6.5). The growth rate (μ) for each strain is also shown.
L. lactis NZ9000 can catabolize galactose via the Tag6P pathway.
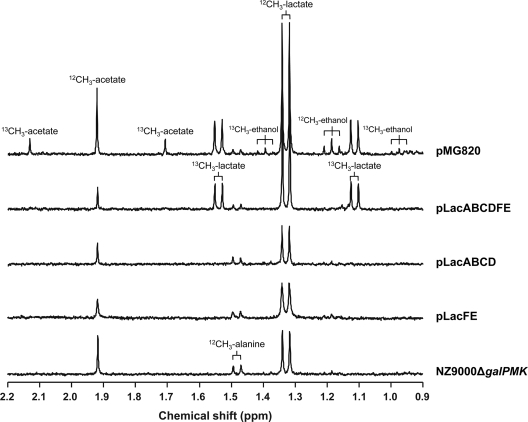
Introduction of pMG820 in L. lactis NZ9000ΔgalPMK rendered a strain that could grow in CDM containing lactose (or glucose, both at 1% [wt/vol]) but not when galactose (1% wt/vol) was the sole carbon source (data not shown). However, resting cells of lactose-grown cultures were able to convert [1-13C]galactose (20 mM) to a mixture of fermentation end products, including [3-13C]lactate, [2-13C]acetate, and [2-13C]ethanol, as determined by 1H NMR spectroscopy (Fig. 3). To determine which of the genes within the lac operon were essential for galactose metabolism, the following combinations of genes were cloned into pNZ8048 under the control of the nisin promoter: lacABCD, lacFE, lacABCDFE, and lacABCDFEG. The various constructs were introduced into L. lactis NZ9000ΔgalPMK. Expression of the lac genes in the different constructs was confirmed by SDS-PAGE of cell extracts obtained from nisin-induced (1 μg liter−1) glucose-grown cultures (data not shown). Also, strain NZ9000ΔgalPMK(pLacABCDFEG) was able to grow on lactose (1%, wt/vol)-CDM when the inducer nisin (1 μg liter−1) was added at time 0 h (inoculation). Strain NZ9000ΔgalPMK(pLacABCDFE) showed moderate growth under the same conditions (Table 3), thus showing functional expression of the lac genes. Like NZ9000ΔgalPMK(pMG820), none of the resulting strains was able to grow in CDM with galactose (1%, wt/vol) as the carbon source, even though nisin was added at time 0 h. To determine the galactose-fermenting capacity of NZ9000ΔgalPMK and derivatives harboring the lac constructs, resting cell suspensions were incubated with [1-13C]galactose (20 mM), and the end products in the supernatants were examined by 1H NMR (Fig. 3). Strains NZ9000ΔgalPMK(pLacABCD) and NZ9000ΔgalPMK(pLacFE) and the negative control, NZ9000ΔgalPMK, were unable to metabolize galactose, as indicated by the absence of labeled end products in the supernatants. Lactate labeled on carbon 3 was detected in the supernatants of NZ9000ΔgalPMK(pLacABCDFE) and NZ9000ΔgalPMK(pLacABCDFEG), showing that, under the conditions tested (glucose-grown cells), both the Tag6P enzymes (lacABCD) and PTSLac (lacFE) are required for galactose consumption in L. lactis NZ9000ΔgalPMK.
FIG. 3.
1H NMR spectra of end products formed from the metabolism of [1-13C]galactose (20 mM) by cell suspensions of NZ9000ΔgalPMK and derivatives carrying different lac operon constructs (pLacFE, pLacABCD, pLacABCDFE) or pMG820 at an initial pH of 6.5 and under anaerobic conditions. Cell suspensions were prepared as described in Materials and Methods. Expression of the lac genes under the control of PnisA was induced by the addition of 1 μg liter−1 nisin. 1H NMR spectra were acquired after a 10-fold dilution in 2H2O in a Bruker AMX300 spectrometer with a 5-mm inverse detection probe head and processed with 0.1-Hz line broadening. Isotopomers of lactate, acetate, and ethanol are indicated. The end-product profile of NZ9000(pLacABCDFEG) was identical to that of NZ9000(pLacABCDFE) and was omitted for the sake of simplicity. pLacFE, PTSLac; pLacABCD, Tag6P pathway; pLacABCDFE, PTSLac and Tag6P pathway.
TABLE 3.
Maximal OD600s obtained for L. lactis NZ9000ΔgalPMK derivatives during growth on CDM with lactosea
| Strain | OD600 | Growth rate |
|---|---|---|
| NZ9000(pNZ8048) | 0.40 | −/+ |
| NZ9000ΔgalPMK | 0.05 | − |
| NZ9000ΔgalPMK(pLacABCD) | 0.05 | − |
| NZ9000ΔgalPMK(pLacFE) | 0.05 | − |
| NZ9000ΔgalPMK(pLacABCDFE) | 0.70 | + |
| NZ9000ΔgalPMK(pLacABCDFEG) | 2.2 | ++ |
| NZ9000ΔgalPMK(pMG820) | 3.7 | +++ |
Lactose was present at 0.5% (wt/vol). The OD600 was evaluated every 12 h for a period of 48 h. Precultures were grown in CDM with glucose (1%, wt/vol).
Consumption rate and glycolytic dynamics depend on the route for galactose utilization.
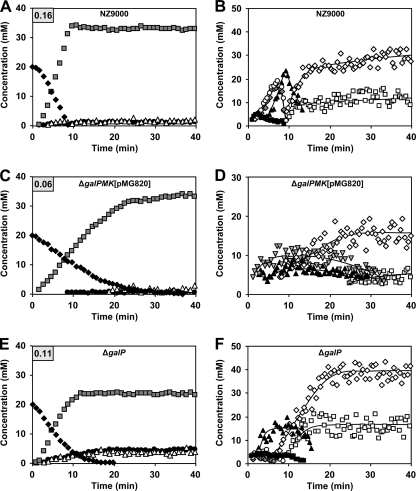
The metabolism of [1-13C]galactose (20 mM) in resting cells of strains NZ9000 and NZ9000ΔgalP grown on galactose and strain NZ9000ΔgalPMK(pMG820) grown on lactose was studied by in vivo 13C NMR (Fig. 4). The kinetics of galactose consumption in NZ9000 was characterized by an initial lag phase followed by quasilinear galactose consumption with a maximal rate of 0.16 ± 0.01 μmol min−1 (mg of protein)−1 (Fig. 4A). During the quasilinear consumption period, α-Gal1P and α-G1P, the phosphorylated intermediates of the Leloir pathway, accumulated to maximal concentrations of 16.7 ± 0.6 and 18.7 ± 0.7 mM, respectively (Fig. 4B). Buildup of fructose 1,6-bisphosphate (FBP; maximal concentration, 23.0 ± 1.5 mM), the predominant metabolite during glucose metabolism, was slightly delayed. At the onset of galactose depletion, 3-phosphoglycerate (3-PGA) and PEP pools build up to 28.8 ± 2.6 and 11.8 ± 2.3 mM, respectively. Curiously, the Tag6P pathway intermediate Gal6P accumulated (maximal concentration, 5.0 ± 0.3 mM) immediately after galactose addition, decreasing to undetectable levels upon galactose exhaustion.
FIG. 4.
Metabolism of galactose in suspensions of nongrowing cells of L. lactis NZ9000 and its derivatives. Kinetics of [1-13C]galactose (20 mM) consumption and end-product formation (A, C, and E) as well as pools of intracellular metabolites (B, D, and F) in resting cells of L. lactis strains NZ9000 (A and B), NZ9000ΔgalPMK(pMG820) (C and D), and NZ9000ΔgalPMK (E and F) under anaerobic conditions at 30°C with the pH controlled at 6.5, as monitored by in vivo 13C NMR. Maximal galactose consumption rates (μmol min−1 mg protein−1) are boxed in the upper-left corners (A, C, and E). Symbols: closed diamond, galactose; gray square, lactate; open triangle, acetate; closed circle, 2,3-butanediol; closed triangle, fructose 1,6-bisphosphate; open circle, α-glucose 1-phosphate; gray diamond, α-galactose 1-phosphate; open diamond; 3-phosphoglycerate; open square, phosphoenolpyruvate; closed square, galactose 6-phosphate; gray inverted triangle, tagatose 1,6-bisphosphate.
Strain NZ9000ΔgalPMK(pMG820) presumably assimilates galactose via the plasmid-encoded PTSLac and Tag6P pathway activities. In this strain, the kinetics of galactose consumption was biphasic: quasilinear consumption with a maximal rate of 0.06 ± 0.01 μmol min−1 (mg of protein)−1 was followed by a second phase at a rate approximately 3-fold lower (Fig. 4C). A similar profile for substrate consumption had previously been observed in an L. lactis strain having cellobiose-specific PTS (PTSCel) as the single glucose transporter (13), and it resulted from a strong preference for the β-glucose anomer. In contrast, the second phase of galactose consumption is not associated with PTSLac anomeric specificity, as the ratio between α- and β-galactose utilization rates remains constant throughout the experiment (data not shown). Therefore, in strain NZ9000ΔgalPMK(pMG820), the slowdown in galactose consumption 17 min after its addition (galactose concentration, <4.5 mM) is probably a consequence of a low affinity for galactose transport via PTSLac, which is in agreement with previously reported Km values in the mM range (32). Using a mathematical model with Michaelis-Menten formalism (13), a Km value of approximately 8.4 mM was estimated for galactose uptake in NZ9000ΔgalPMK(pMG820). The same model predicted a Km of galactose uptake by strain NZ9000 of approximately 0.4 mM, a value close to that reported for galactose uptake via a permease (Km, 0.13 mM) (59). Accumulation of the Tag6P pathway intermediate tagatose 1,6-bisphoshate (maximal concentration, about 10 mM) occurred within seconds after addition of galactose (Fig. 4D), but Gal6P was not detected in vivo by 13C NMR. In the spectra acquired during the metabolism of [1-13C]galactose, a resonance due to FBP labeled in C-6 (δ, 65.1 ppm) appeared soon after (about 1.5 min) the C-1 peak of tagatose 1,6-bisphoshate (δ, 66.4 ppm) became detectable. Tagatose 1,6-bisphoshate labeled in C-6 (δ, 63.4 ppm) was not detected. The observed labeling pattern is consistent with scrambling of the 13C label at the level of triose phosphates and backflux through aldolase (46). Indeed, the FBP pool showed a profile identical to that of tagatose 1,6-bisphoshate, but the maximal concentration was somewhat lower (6.5 ± 1.1 mM). Accumulation of 3-PGA, a metabolite generally associated with substrate starvation, occurred while galactose was still abundant; likewise, PEP was also detected before galactose exhaustion.
The profile of galactose consumption in strain NZ9000ΔgalP resembled that in NZ9000ΔgalPMK(pMG820), as it also displayed biphasic kinetics, but the rate of the first phase was nearly 2-fold higher [0.11 ± 0.01 μmol min−1 (mg of protein)−1] and that of the second phase was considerably shorter (Fig. 4E). The observed behavior (biphasic kinetics) strongly indicates the presence of a transporter with a low affinity for galactose in this strain. The estimated Km value for galactose uptake is about 6.3 mM. Inactivation of galP apparently caused no loss of expression of the downstream genes galMKTE in the gal operon (galPMKTE), as accumulation of α-Gal1P and α-G1P during the metabolism of galactose denoted functional GalKTE enzyme activities (Fig. 4F). Indeed, a whole-genome transcription analysis revealed no significant differences in the expression levels of galMKTE genes in NZ9000ΔgalP and NZ9000 cells grown on galactose. In contrast, in the galP mutant the expression levels of galMKT and galE were about 50-fold and 8-fold higher, respectively, during growth on galactose compared to growth on glucose. FBP was detected only about 2.5 min after galactose addition, reached a steady concentration of 15.9 ± 1.5 mM during the rapid phase, and decreased to undetectable levels before galactose depletion. As was the case in NZ9000, Gal6P accumulated immediately (concentration, about 4 mM) after galactose addition, decreasing to undetectable levels, however, during the transition from the first to the second phase of galactose consumption. Concomitantly, 3-PGA and PEP pools rose to 38.1 ± 2.8 and 11.0 ± 2.2 mM, respectively.
Initial steps for galactose utilization in strain NZ9000ΔgalP.
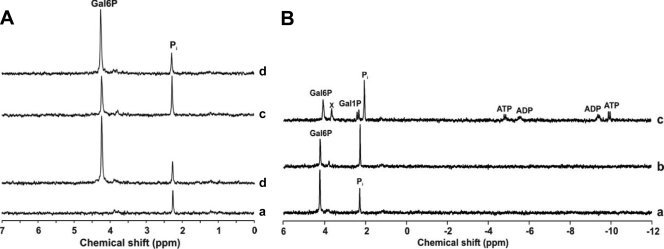
The in vivo 13C NMR data obtained during the metabolism of galactose in strain NZ9000ΔgalP showed accumulation of α-Gal1P, which originates from the ATP-dependent phosphorylation of galactose by GalK. However, mining of the L. lactis MG1363 genome revealed no clear candidate gene for a galactose permease alternative to galP. Additionally, detection of Gal6P immediately after the addition of galactose to cell suspensions of NZ9000ΔgalP (and also NZ9000) is indicative of a galactose PTS activity. Indeed, galactose translocation by a PTS other than the PTSLac has previously been suggested in L. lactis (50, 59). Therefore, we hypothesized that transport of galactose in strain NZ9000ΔgalP is mediated via a PTS, leading to Gal6P, which is subsequently (in)directly converted to α-Gal1P and further metabolized via the Leloir pathway. Specific galactose PTS activity was assayed using permeabilized cells of galactose-grown NZ9000 ΔgalP: a value of 50 ± 2 nmol min−1 (mg of protein)−1 was determined. The step(s) involved in the conversion of Gal6P to α-Gal1P was investigated by incubating cell-free NZ9000ΔgalP extracts with appropriate phosphorylated substrates coupled to 31P NMR analysis. A considerable (2-fold) decrease in the resonance due to Gal6P (δ, 4.25 ppm) accompanied by an increase in Pi (δ, 2.28 ppm) was observed upon incubation of Gal6P with cell extract (Fig. 5 A). When the reaction was performed in the presence of the phosphatase inhibitor NaF, no changes in the Gal6P and Pi resonances were detected. The data show that Gal6P phosphatase activity is present in L. lactis. Direct conversion of Gal6P into α-Gal1P was not observed, ruling out the presence of a phosphogalactomutase activity. The paucity of phosphohexomutase genes in the genome sequence of L. lactis MG1363 (65) and the lack of specificity for phosphogalactose by the lactococcal α-phosphoglucomutase (45) further support the absence of such a phosphogalactomutase activity.
FIG. 5.
31P NMR spectra showing galactose 6-phosphate processing in cell extracts of strain NZ9000ΔgalP. The reaction mixtures contained 50 mM TEA buffer, pH 7.2, 5 mM MgCl2, and cell extract (about 2 mg total protein). (A) Traces a and b represent spectra of the reaction mixture before and after addition of Gal6P (5 mM), respectively. Traces c and d represent spectra of the reaction mixture after 60 min incubation at 30°C in the absence and in the presence of the phosphatase inhibitor NaF (10 mM), respectively. (B) Conversion of Gal6P in cell extracts of NZ9000ΔgalP monitored online at 30°C by 31P NMR. Traces a and b represent spectra acquired 1.5 and 80 min after addition of Gal6P, respectively. ATP (2.5 mM) was added to the reaction mixture at time 81 min. Trace c represents a spectrum acquired at time 86 min, 5 min after addition of ATP. The shifts in Pi and Gal6P resonances are due to pH variation during the assay (online monitoring without pH control). The resonance labeled x is due to an unidentified compound.
A resonance due to α-Gal1P (δ, 2.55 ppm) was readily detected upon addition of ATP (2.5 mM) to a reaction mixture in which dephosphorylation of Gal6P by the cell extract had occurred (Fig. 5B). The results show that conversion of Gal6P to α-Gal1P entails sequential dephosphorylation and phosphorylation steps, which are catalyzed by a yet unidentified phosphatase and GalK, respectively. The requirement for a functional galactokinase is clear from the inability of strain NZ9000ΔgalPMK to metabolize galactose (Fig. 3). At this point, it should be noted that the inability of strain NZ9000ΔgalPMK(pLacABCD) to metabolize galactose is not in contradiction to the presence of an alternative galactose PTS in the genome. Rather, this result implies that expression of such a transporter needs to be induced by galactose. In agreement, suspensions of glucose-grown NZ9000ΔgalPMK cells incubated with [1-13C]galactose failed to accumulate [1-13C]Gal6P (data not shown), as opposed to NZ9000 and NZ9000ΔgalP cells grown on galactose (Fig. 4). Also in line with this view is the observed growth behavior of strain NZ9000ΔgalP in galactose-CDM (Fig. 2), with the initial low growth rate suggesting the need to express new activities for full growth on galactose.
Searching for alternative galactose PTS in L. lactis using transcriptomics.
Our data strongly indicate that strain NZ9000ΔgalP transports galactose via an unknown PTS transporter(s) displaying low affinity for the sugar. Previously, a transcriptome analysis approach proved useful to identify PTSCel as an additional glucose transporter in L. lactis (51). Assuming that the galactose PTS would be expressed at a higher level in the galP mutant when it was grown on galactose, using full-genome L. lactis DNA microarrays, we compared the mRNA levels of galactose-grown NZ9000ΔgalP cells with those of glucose-grown NZ9000ΔgalP or galactose-grown NZ9000. Genes encoding PTS proteins and their respective expression levels under the conditions studied are shown in Table 4. On galactose-grown cells, loss of GalP did not significantly alter the expression of genes encoding PTS components, the exception being llmg_pseudo_54. L. lactis MG1363 llmg_pseudo_54, yidB in strain IL1403, is annotated as a putative cellobiose-specific PTS IIC component (10). Curiously, when galactose- and glucose-grown NZ9000ΔgalP cells are compared, llmg_pseudo_54 showed the second-highest fold overexpression, after ptcA. However, the frameshift mutation in llmg_pseudo_54 (nucleotide 422 is missing) was confirmed by (re)sequencing llmg_pseudo_54 from strains MG1363, NZ9000, and NZ9000ΔgalP (grown on glucose or galactose), and thus, this gene fragment does not seem to produce a functional protein in either strain. Identification of the galactose PTS is hindered by the number of PTS-encoding genes (55% of the total) upregulated in galactose-grown NZ9000ΔgalP compared to the number in glucose-grown cells.
TABLE 4.
Transcription of PTS genes in galactose-grown NZ9000ΔgalP compared to that in parent strain NZ9000 or glucose-grown NZ9000ΔgalP
| Gene | Familya | Product | Substrate (reference) | Up- or downregulationc |
|
|---|---|---|---|---|---|
| ΔgalP vs NZ9000 | Gal vs Glc | ||||
| llmg_0022 (mtlA) | Fru | IIBC | Mannitol (23) | 1.23 | 1.06 |
| llmg_0024 (mtlF) | Fru | IIA | −1.01 | −1.03 | |
| llmg_0187 (celB) | Lac | IIC | Cellobiose/lactose (28) | 1.00 | 1.09 |
| llmg_0437 (ptcB) | Lac | IIB | Cellobiose/glucose (13) | 1.14 | 1.28 |
| llmg_0438 (ptcA) | Lac | IIA | 1.12 | 5.06 | |
| llmg_0440 (ptcC) | Lac | IIC | 1.02 | 1.32d | |
| llmg_0453 (yedE) | Glc | IIA | Trehalose (4) | −1.19 | 2.17 |
| llmg_0454 (yedF) | Glc | IIBC | −1.15 | 2.76 | |
| llmg_0727 (ptnD) | Man | IID | Mannose/glucose (13) | 1.19 | 2.92 |
| llmg_0728 (ptnC) | Man | IIC | 1.10 | 2.03 | |
| llmg_0729 (ptnAB) | Man | IIAB | −1.10 | 2.11 | |
| llmg_0865 | Fru | IIA | NDb | 1.28 | 1.25 |
| llmg_0866 | Gat | IIB | 1.03 | 2.33 | |
| llmg_0867 | Asc | IIC | −1.07 | 1.02 | |
| llmg_0963 | Lac | IIC | ND | 1.14 | −1.01 |
| llmg_1045 (ptbA) | Glc | IIABC | Salicin (28) | −1.23 | 1.33 |
| llmg_1244 | Lac | IIC | ND | 1.16 | −1.21 |
| llmg_1426 (yleD) | Glc | IIABC | Sucrose (39, 65) | 1.20 | 1.67 |
| llmg_1568 (fruA) | Glc | IIABC | Fructose (5) | 1.08 | 3.51 |
| llmg_pseudo54 (yidB) | Lac | IIC | ND | 1.31d | 4.16 |
PTS families according to the transport classification database (www.tcdb.org): Fru, fructose-mannitol family; Lac, lactose-N,N′-diacetylchitobiose-β-glucoside family; Glc, glucose-glucoside family; Man, mannose-fructose-sorbose family; Gat, galactitol family; Asc, ascorbate family.
ND, not determined.
Positive values indicate upregulation, and negative values indicate downregulation. Genes with both significantly altered expression (P < 0.001) and an expression ratio of ≥|±1.6| are shown in boldface.
Significantly altered expression.
Engineering strategies to improve the capacity for galactose utilization.
From our results on the characterization of the galactose dissimilation routes, engineering the Leloir pathway appeared to be the most promising strategy to increase galactose consumption in L. lactis NZ9000 since (i) the strain possessing the GalP/Leloir pathway showed the highest rate of galactose utilization and (ii) the PTSLac and the galactose PTS in L. lactis NZ9000ΔgalP display an apparent low affinity for galactose. Furthermore, improving the capacity to use galactose is likely to require increased α-PGM activity, as this enzyme was shown to be a metabolic bottleneck in galactose metabolism (45). Because the native α-PGM, an enzyme essential for growth, may be tied to unknown physiological control elements in L. lactis, we chose to bypass it by engineering the expression of S. thermophilus pgmA, together with the lactococcal gal operon genes galPMKT.
The genes galPMKT were cloned in pNZ8048, downstream of the nisin-inducible promoter, and the resulting vector, pGalPMKT, was introduced into L. lactis NZ9000. The S. thermophilus pgmA was cloned downstream of galT in pGalPMKT. Nisin-inducible expression of the Leloir genes (galPMKT) alone or together with pgmA in NZ9000 was examined by SDS-PAGE. Bands due to GalP (50.5 kDa) and GalK (43.7 kDa), but not GalM (37.6 kDa) or GalT (56.3 kDa), were detected in NZ9000(pGalPMKT). Besides GalP and GalK, an additional band due to α-PGM was readily detected in the crude extracts of NZ9000(pGalPMKTpgmA) (data not shown). In addition, a 3-fold increased α-PGM activity was present in the crude extracts of induced NZ9000(pGalPMKTpgmA) compared to that observed in NZ9000(pNZ8048) (Table 5), demonstrating the functional overexpression of pgmA.
TABLE 5.
α-Phosphoglucomutase activity in L. lactis cells grown to mid-exponential phase in CDM containing galactosea
| Strain | α-Phosphoglucomutase activity (μmol min−1 mg protein−1) | Fold overexpressionb |
|---|---|---|
| NZ9000(pNZ8048) | 0.33 | 1.0 |
| NZ9000(pPgmA) | 1.26 | 3.8 |
| NZ9000(pGalP) | 0.43 | 1.3 |
| NZ9000(pGalPpgmA) | 1.15 | 3.4 |
| NZ9000(pGalPMKT) | 0.43 | 1.3 |
| NZ9000(pGalPMKTpgmA) | 0.85 | 2.6 |
Galactose was present at 1% (wt/vol). Induction was performed by adding a supernatant (0.01%, vol/vol) of a full-grown culture of the nisin producer L. lactis NZ9700 when an OD600 of 0.25 was reached.
Relative to that in the empty vector pNZ8048 control strain. The results are averages of two independent cultivations, and the error is below 10%.
To examine the effect of overproducing the genes in the gal operon alone or in combination with α-PGM, in vivo NMR coupled to 13C labeling was used to follow the metabolism of galactose in suspensions of nongrowing cells (Fig. 6). All strains were grown in CDM containing galactose and were induced with nisin (0.01% [vol/vol] supernatant of NZ9700) at an OD600 of 0.5.
FIG. 6.
Galactose consumption and time courses for α-Gal1P, α-G1P, and FBP pools in resting cells of L. lactis strains engineered to improve galactose utilization. [1-13C]galactose (20 mM) was supplied at time 0 min to nongrowing suspensions of strains NZ9000(pGalPMKT) (A), NZ9000(pGalPMKTpgmA) (B), control strain NZ9000(pNZ8048) (C), and NZ9000(pGalPpgmA) (D); and its metabolism was monitored by in vivo 13C NMR under anaerobic conditions at pH 6.5 and 30°C. Maximal galactose consumption rates (μmol min−1 mg protein−1) are boxed in the upper-left corners. Symbols: closed diamond, galactose; closed triangle, fructose 1,6-bisphosphate; open circle, α-glucose 1-phosphate; gray diamond, α-galactose 1-phosphate.
Nisin-induced expression of galPMKT did not improve galactose consumption in strain NZ9000 (compare Fig. 6A with 6C). Instead, it caused increases of about 5 and 15 mM in the maximal concentrations of the Leloir pathway intermediates α-Gal1P and α-G1P, respectively. These data are in line with the fact that wild-type α-PGM activity is insufficient to support a high flux via the Leloir pathway (45). Indeed, coexpression of pgmA together with the gal operon genes reduced the α-Gal1P and α-G1P pools by approximately 3-fold and led to a 38% increase in the galactose consumption rate (Fig. 6B). Previously, a 6-fold increase in the native α-PGM activity resulted in a 25% greater galactose consumption rate (45). As expected, nisin-induced expression of the streptococcal pgmA per se also had a positive effect on the galactose consumption rate (an increase of about 19% when the α-PGM activity was 3.8 times higher; Table 5). Altogether these results prompted a pull-and-push strategy that consisted of cloning the first gene needed for galactose utilization, the galactose permease, and pgmA, the gateway to glycolysis, with the obvious advantage being that only two genes needed to be cloned. Under the conditions studied, the resulting strain, NZ9000(pGalPpgmA), presented the highest galactose consumption rate, which was increased 50% compared to that for control strain NZ9000(pNZ8048). Furthermore, α-Gal1P and α-G1P levels were even lower than those in strain NZ9000(pGalPMKTpgmA) (compare Fig. 6B with 6D). Curiously, increasing the galactose consumption rate led to an FBP profile more similar to that observed during the metabolism of glucose (45). Our data show that to improve galactose utilization in L. lactis subsp. cremoris, at least α-PGM and galactose uptake activities must be above the wild-type levels.
DISCUSSION
In the 1970s and early 1980s, the efforts of several laboratories to elucidate galactose metabolism in L. lactis led to the publication of conflicting results regarding the uptake step and the relative contributions/efficiencies of the Leloir and Tag6P pathways. In this work, we used directed genetically engineered strains to examine galactose utilization via the chromosomal Leloir pathway or the plasmid-encoded Tag6P pathway in L. lactis subsp. cremoris NZ9000. This strain lacks a lactose plasmid (Tag6P pathway) but possesses the gal operon (galPMKTE), which encodes a galactose permease (galP) and the enzymes of the Leloir pathway. Assessment of Tag6P pathway activity in strain NZ9000 requires inactivation of the Leloir route. To this end, galP- and galPMK-deletion strains were constructed, but growth on galactose was abolished only when a gal operon fragment covering galP, galM, and galK was removed. Thus, GalM and/or GalK is crucial for growth of strain NZ9000 on galactose, whereas GalP is not. GalM is not present in all bacteria known to metabolize galactose via the Leloir pathway (1), most likely because the anomerization reaction catalyzed by the mutarotase (β-Gal ↔ α-Gal) can occur spontaneously in solution. On the other hand, loss of GalK abolished galactose fermentation in L. lactis MG1363 (25) and severely impaired growth of the phylogenetically related organisms Streptococcus mutans and Streptococcus salivarius (1, 14), denoting the key role of the kinase in organisms relying on the Leloir pathway for galactose utilization. In fact, a low level of expression of galK and, thus, insufficient galactokinase activity could explain the inability of a previously described lactococcal galP-disruption mutant (MG1363 galP::ery) to grow on galactose (25). Interestingly, this galP-disruption mutant displayed galactose uptake activity (25), which corroborates the occurrence of an additional transporter(s) with specificity for galactose in L. lactis MG1363 and derivatives.
Detection of galactose phosphorylated at position C-6 (Gal6P) both in the galP mutant and in NZ9000 by in vivo NMR strongly indicated galactose PTS activity, since the formation of Gal6P is usually associated with translocation of galactose or lactose via PTS transporters in bacteria. To the best of our knowledge, production of Gal6P through galactokinase phosphorylation on C-6 or a phosphogalactomutase activity (interconversion of α-Gal1P and Gal6P) has never been reported. In agreement, we failed to detect either activity in cell extracts of strain NZ9000ΔgalP by 31P NMR analysis. Galactose PTS activity was, however, readily measured in permeabilized cells of galactose-grown NZ9000ΔgalP. Thus, strain NZ9000 possesses a PTS for galactose uptake, in addition to the secondary carrier GalP (Fig. 1). The presence of a galactose PTS in strains ML3 and C2, which, like strain NZ9000, derive from the dairy strain L. lactis NCDO712, has been suggested (50, 59). In an attempt to identify the additional galactose PTS, we used a transcriptomics approach and compared mRNA levels of galactose-grown and glucose-grown NZ9000ΔgalP. In L. lactis MG1363, 12 operons (20 open reading frames) potentially encode PTS-type transporters (65). Of these, genes in seven operons showed significant increases in their expression levels during growth on galactose compared to the levels during growth on glucose. Upregulation of genes encoding the mannose/glucose PTS, fructose PTS, and putative trehalose PTS domains is more likely related to relief of glucose-mediated catabolite repression than to induction by galactose (5, 13, 66). In fact, genes encoding the putative trehalose PTS (llmg_0453 and llmg_0454) showed significantly altered expression levels in an L. lactis ccpA mutant (40, 66). Transcriptional regulation of the lactococcal fru operon (encoding the fructose PTS and a 1-phosphofructokinase) by CcpA has been reported (5), and disruption of the fructose PTS affected only the growth of L. lactis on fructose; thus, a role of the transporter on galactose metabolism is unlikely. The mannose/glucose PTS is characterized by a broad substrate specificity being able to transport mannose, glucose, fructose, N-acetylglucosamine, and 2-deoxyglucose (15, 22, 37), but not galactose (15). Unexpectedly, loss of GalP did not induce significantly altered expression of genes encoding PTS components, except for a modest, but significant, increase in the level of llmg_pseudo_54 (Table 4). This gene, which encodes a protein with homology to enzymes IIC of the PTS lactose-N,N′-diacetylchitobiose-β-glucoside family in L. lactis strains IL1403 and SK11, was the second most upregulated in galactose-grown cells; ptcA, which encodes a IIACel domain involved in transport processes by two distinct Lac-family EII integral membrane domains, PtcC and CelB (13, 28), showed the highest expression increase. In view of these results, llmg_pseudo_54 and ptcA appeared to be promising candidates for the galactose PTS domains IIC and IIA. As such, ptcB emerged as the most likely IIB domain to complete the sugar-specific PTS complex. However, the frameshift mutation in MG1363 llmg_pseudo_54 was confirmed in strains NZ9000 and NZ9000ΔgalP by sequencing. Consequently, the identity of the gene encoding the galactose PTS remains elusive. Considering our expression data, firm identification of the transporter in question most likely requires inactivation in the galP mutant of each PTS encoded in the genome, a task beyond the scope of this work.
In the absence of the Tag6P pathway, metabolism of Gal6P, the galactose PTS product, requires phosphatase or phosphohexomutase activities. In this work, evidence for a dephosphorylation step is presented (Fig. 1), since incubation of NZ9000ΔgalP cell extracts with Gal6P resulted in accumulation of phosphate at the expense of Gal6P. Complete inhibition of Gal6P hydrolysis by a phosphatase inhibitor (sodium fluoride) further supports this hypothesis. The occurrence in L. lactis of phosphatases with specificity for Gal6P has been reported (61), and excretion of galactose to the medium during growth of L. lactis on lactose is usually rationalized as resulting from dephosphorylation of Gal6P (7, 51). The genome sequence of L. lactis MG1363 possesses at least 18 genes whose products potentially exhibit phosphatase activity (65). Among those, 11 genes showed differential expression, but only 5 (llmg_0264, llmg_1288, llmg_1517, llmg_1854, and llmg_2075) were upregulated when NZ9000ΔgalP cells grown on galactose were compared with NZ9000ΔgalP cells grown on glucose. Specific functions have been attributed only to llmg_0264 (fbp), which encodes the lactococcal fructose 1,6-bisphosphatase (38). The gene llmg_1288 (hisK) is not a strong candidate, since it encodes a histidinol phosphatase (HisK), and all of its neighboring genes (9 in total), also involved in histidine metabolism, are upregulated (data not shown). llmg_2075 is annotated as an ADP-ribose pyrophosphatase, while llmg_1157 is annotated as a membrane-associated putative phosphatidic acid phosphatase but shares 98% homology with a histidine protein kinase-like kinase (49). llmg_1854 is annotated as an alkaline phosphatase. These enzymes are characterized by a wide substrate specificity and alkaline pH optimum (pH 8 to 10). Among the genes determined to be upregulated, llmg_1854 emerges as the best candidate for encoding a phosphatase with Gal6P activity. However, if this is the case, the enzyme is most likely unspecific, as typical of alkaline phosphatases. In fact, a lactococcal phosphatase that exhibited affinity for several hexose 6-phosphates, including Gal6P, has been partially purified, but this enzyme showed a pH optimum of 6.0, and its activity decreased sharply above pH 6.5 (61). The presence of several unspecific phosphatases catalyzing the hydrolysis of Gal6P cannot be ruled out. Most likely, identification of the phosphatase(s) involved requires its purification, which is a task outside the scope of the present study.
An L. lactis NZ9000 strain devoid of Leloir pathway activity (NZ9000ΔgalPMK) can metabolize galactose via the Tag6P pathway upon introduction of the lactose plasmid pMG820 (Fig. 4), but the sugar does not sustain growth. It seems unlikely that this feature is related to a low level of expression of the metabolic genes in NZ9000ΔgalPMK(pMG820), since the lac operon inducer, Tag6P, is formed during metabolism of galactose via this route (63) and the strain can grow on lactose (Table 3). Likewise, L. lactis subsp. cremoris FD1 actively metabolizes galactose but does not grow on the sugar (7). This behavior was explained by a nil fructose 1,6-bisphosphatase activity, impeding the use of galactose as a gluconeogenic sugar and, consequently, the production of phosphorylated precursors (G6P and fructose 6-phosphate) for biosynthesis purposes. The activity of fructose 1,6-bisphosphatase in L. lactis MG1363-related strains is very low (<15 μU) (38, 47) and might be insufficient to sustain growth of the Tag6P-dependent strain on galactose. This hypothesis is further supported by the elevated expression of fructose 1,6-bisphosphatase during growth on galactose (data not shown). Previously, low fructose 1,6-bisphosphatase activity was shown to be limiting for growth of L. lactis on fructose (38), a substrate that also requires gluconeogenic conversion of FBP for biosynthetic purposes (generation of fructose 6-phosphate and G6P).
The pattern of galactose degradation in strain NZ9000ΔgalPMK(pMG820) revived the question as to whether the PTSLac is able to take up galactose (16, 17, 18, 20, 32, 50, 59, 60). Introduction of plasmids carrying different combinations of the lac genes in NZ9000ΔgalPMK showed the necessity of lacFE for utilization of galactose (Fig. 3), indicating uptake of the sugar via PTSLac.
Internalization of galactose in L. lactis NZ9000 occurs via a secondary carrier (GalP) or a phosphotransfer-driven group translocator (galactose PTS). Our data show that GalP is a high-affinity uptake system (Km in the μM range), whereas the galactose PTS is a low-affinity transporter (Km in the mM range), which is in accordance with the findings described in previous reports (32, 59). The plasmid-encoded PTSLac also has a low affinity for galactose. Indeed, in strains devoid of GalP, the galactose consumption rate slowed markedly at concentrations of galactose in the medium of about 6 to 7 mM. The strain equipped with GalP (NZ9000) showed the highest rate of galactose consumption, while the lowest was observed in the strain mainly importing galactose via PTSLac [NZ9000ΔgalPMK(pMG820)] (Fig. 4). The data pinpoint GalP as the transporter displaying the highest capacity to take up galactose. In summary, the GalP/Leloir pathway consumes galactose faster and is fully active with galactose at concentrations down to about 1.5 mM. Given these properties, the GalP/Leloir route appeared to be the best target for manipulations aiming at enhanced galactose consumption in L. lactis. Overexpression of the gal genes (galPMKT) in strain NZ9000 did not lead to an enhanced galactose consumption rate. A considerable increase in the maximal concentrations of the phosphorylated Leloir intermediates α-Gal1P and α-G1P pointed to an obstruction at the level of α-PGM. This result was not completely unexpected, as we have recently shown that the step catalyzed by α-PGM is a major bottleneck in the utilization of galactose (45). In line with this observation, the α-PGM step was also critical for the improvement of the galactose uptake capacity in Saccharomyces cerevisiae (12). In the present study, overproduction of a heterologous α-PGM (from S. thermophilus) alone or in combination with gal gene products, namely, GalP or GalPMKT, resulted in higher galactose consumption rates. The best performance, corresponding to a 50% increase relative to wild-type levels, was obtained with the NZ9000 strain that overexpressed the genes encoding GalP and α-PGM. However, the galactose consumption rate in this engineered strain is 2-fold lower than the rate of glucose utilization in strain NZ9000 (13). Although a full explanation cannot be put forward at this stage, it would not be surprising if regulatory events such as CcpA-mediated regulation of catabolic genes are involved. CcpA-mediated activation of the las operon (pfk, pyk, and ldh) is not in effect during growth on galactose (40, 66), which could in part explain the decreased glycolytic flux. In view of these findings, we postulate that further improvement of galactose utilization may require engineering of regulatory genes, such as ccpA or even specific sugar regulators. Future approaches, combining increased expression of catabolic genes, namely, galactose permease and α-PGM, with engineering of the catabolite control network could create L. lactis strains preferring galactose over glucose and lactose, a desirable trait considering that the concentration of lactose in milk fermentations is normally higher than that of galactose (3). Thus, galactose scavengers would be ideal starters in the manufacture of galactose-free dairy products.
Acknowledgments
This work was supported by Fundação para a Ciência e a Tecnologia, Portugal (FCT) projects POCTI/BIO/48333/2002, PTDC/EEA-ACR/69530/2006, and PTDC/SAU-MII/100964/2008 and by contract QLK1-CT-2000-01376 of the Commission of the European Communities. The NMR spectrometers are part of the National NMR Network (REDE/1517/RMN/2005), supported by the Programa Operacional Ciência e Inovação (POCTI) 2010 and FCT.
We thank Luis Fonseca for determination of kinetic parameters related to galactose consumption and Eddy Smid (NIZO Food Research) for kindly providing pure tagatose 1,6-bisphosphate.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Abranches, J., Y. Y. Chen, and R. A. Burne. 2004. Galactose metabolism by Streptococcus mutans. Appl. Environ. Microbiol. 70:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleksandrzak-Piekarczyk, T., J. Kok, P. Renault, and J. Bardowski. 2005. Alternative lactose catabolic pathway in Lactococcus lactis IL1403. Appl. Environ. Microbiol. 71:6060-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, A. 1982. Effect of fermentation on lactose, glucose, and galactose content in milk and suitability of fermented milk products for lactose intolerant individuals. J. Dairy Sci. 65:346-352. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, U., F. Levander, and P. Radstrom. 2001. Trehalose-6-phosphate phosphorylase is part of a novel metabolic pathway for trehalose utilization in Lactococcus lactis. J. Biol. Chem. 276:42707-42713. [DOI] [PubMed] [Google Scholar]
- 5.Barriere, C., M. Veiga-da-Cunha, N. Pons, E. Guedon, S. A. van Hijum, J. Kok, O. P. Kuipers, D. S. Ehrlich, and P. Renault. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC Gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskaran, D., and S. Sivakumar. 2003. Galactose concentration in pizza cheese prepared by three different culture techniques. Int. J. Dairy Technol. 56:229-232. [Google Scholar]
- 7.Benthin, S., J. Nielsen, and J. Villadsen. 1994. Galactose expulsion during lactose metabolism in Lactococcus lactis subsp. cremoris FD1 due to dephosphorylation of intracellular galactose 6-phosphate. Appl. Environ. Microbiol. 60:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bissett, D. L., and R. L. Anderson. 1974. Lactose and d-galactose metabolism in group N streptococci: presence of enzymes for both the d-galactose 1-phosphate and d-tagatose 6-phosphate pathways. J. Bacteriol. 117:318-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolotin, A., S. Mauger, K. Malarme, S. D. Ehrlich, and A. Sorokin. 1999. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Van Leeuwenhoek 76:27-76. [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Bro, C., S. Knudsen, B. Regenberg, L. Olsson, and J. Nielsen. 2005. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl. Environ. Microbiol. 71:6465-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro, R., A. R. Neves, L. L. Fonseca, W. A. Pool, J. Kok, O. P. Kuipers, and H. Santos. 2009. Characterization of the individual glucose uptake systems of Lactococcus lactis: mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 71:795-806. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y. Y., M. J. Betzenhauser, J. A. Snyder, and R. A. Burne. 2002. Pathways for lactose/galactose catabolism by Streptococcus salivarius. FEMS Microbiol. Lett. 209:75-79. [DOI] [PubMed] [Google Scholar]
- 15.Cochu, A., C. Vadeboncoeur, S. Moineau, and M. Frenette. 2003. Genetic and biochemical characterization of the phosphoenolpyruvate:glucose/mannose phosphotransferase system of Streptococcus thermophilus. Appl. Environ. Microbiol. 69:5423-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cords, B. R., and L. L. McKay. 1974. Characterization of lactose-fermenting revertants from lactose-negative Streptococcus lactis C2 mutants. J. Bacteriol. 119:830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crow, V. L., G. P. Davey, L. E. Pearce, and T. D. Thomas. 1983. Plasmid linkage of the d-tagatose 6-phosphate pathway in Streptococcus lactis: effect on lactose and galactose metabolism. J. Bacteriol. 153:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demko, G. M., S. J. Blanton, and R. E. Benoit. 1972. Heterofermentative carbohydrate metabolism of lactose-impaired mutants of Streptococcus lactis. J. Bacteriol. 112:1335-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Vos, W. M., and E. E. Vaughan. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217-237. [DOI] [PubMed] [Google Scholar]
- 21.de Vos, W. M., and J. Hugenholtz. 2004. Engineering metabolic highways in lactococci and other lactic acid bacteria. Trends Biotechnol. 22:72-79. [DOI] [PubMed] [Google Scholar]
- 22.Erni, B., B. Zanolari, and H. P. Kocher. 1987. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J. Biol. Chem. 262:5238-5247. [PubMed] [Google Scholar]
- 23.Gaspar, P., A. R. Neves, A. Ramos, M. J. Gasson, C. A. Shearman, and H. Santos. 2004. Engineering Lactococcus lactis for production of mannitol: high yields from food-grade strains deficient in lactate dehydrogenase and the mannitol transport system. Appl. Environ. Microbiol. 70:1466-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossiord, B. P., E. J. Luesink, E. E. Vaughan, A. Arnaud, and W. M. de Vos. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J. Bacteriol. 185:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 27.Hutkins, R., S. M. Halambeck, and H. A. Morris. 1986. Use of galactose-fermenting Streptococcus thermophilus in the manufacture of Swiss, mozzarella, and short-method cheddar cheese. J. Dairy Sci. 69:1-8.3700799 [Google Scholar]
- 28.Kowalczyk, M., M. Cocaign-Bousquet, P. Loubiere, and J. Bardowski. 2008. Identification and functional characterisation of cellobiose and lactose transport systems in Lactococcus lactis IL1403. Arch. Microbiol. 189:187-196. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers, O. P., A. de Jong, R. J. Baerends, S. A. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 31.Kuipers, O. P., P. G. G. A. De Ruyter, M. Kleerebezem, and W. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 32.LeBlanc, D. J., V. L. Crow, L. N. Lee, and C. F. Garon. 1979. Influence of the lactose plasmid on the metabolism of galactose by Streptococcus lactis. J. Bacteriol. 137:878-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, R., T. Molskness, W. E. Sandine, and P. R. Elliker. 1973. Carbohydrate metabolism in lactic streptococci: fate of galactose supplied in free or disaccharide form. Appl. Microbiol. 26:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 35.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 36.Leslie, N. D. 2003. Insights into the pathogenesis of galactosemia. Annu. Rev. Nutr. 23:59-80. [DOI] [PubMed] [Google Scholar]
- 37.Liberman, E. S., and A. S. Bleiweis. 1984. Transport of glucose and mannose by a common phosphoenolpyruvate-dependent phosphotransferase system in Streptococcus mutans GS5. Infect. Immun. 43:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Looijesteijn, P. J., I. C. Boels, M. Kleerebezem, and J. Hugenholtz. 1999. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremoris by the sugar source. Appl. Environ. Microbiol. 65:5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luesink, E. J., J. D. Marugg, O. P. Kuipers, and W. M. de Vos. 1999. Characterization of the divergent sacBK and sacAR operons, involved in sucrose utilization by Lactococcus lactis. J. Bacteriol. 181:1924-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luesink, E. J., R. E. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 41.Maeda, S., and M. J. Gasson. 1986. Cloning, expression and location of the Streptococcus lactis gene for phospho-beta-d-galactosidase. J. Gen. Microbiol. 132:331-340. [DOI] [PubMed] [Google Scholar]
- 42.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel, V., and F. G. Martley. 2001. Streptococcus thermophilus in cheddar cheese-production and fate of galactose. J. Dairy Res. 68:317-325. [DOI] [PubMed] [Google Scholar]
- 44.Mollet, B., J. Knol, B. Poolman, O. Marciset, and M. Delley. 1993. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J. Bacteriol. 175:4315-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neves, A. R., W. A. Pool, R. Castro, A. Mingote, F. Santos, J. Kok, O. P. Kuipers, and H. Santos. 2006. The α-phosphoglucomutase of Lactococcus lactis is unrelated to the α-d-phosphohexomutase superfamily and encoded by the essential gene pgmH. J. Biol. Chem. 281:36864-36873. [DOI] [PubMed] [Google Scholar]
- 46.Neves, A. R., A. Ramos, M. C. Nunes, M. Kleerebezem, J. Hugenholtz, W. M. de Vos, J. Almeida, and H. Santos. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200-212. [DOI] [PubMed] [Google Scholar]
- 47.Neves, A. R., A. Ramos, C. Shearman, M. J. Gasson, J. S. Almeida, and H. Santos. 2000. Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo 13C-NMR. Eur. J. Biochem. 267:3859-3868. [DOI] [PubMed] [Google Scholar]
- 48.Neves, A. R., R. Ventura, N. Mansour, C. Shearman, M. J. Gasson, C. Maycock, A. Ramos, and H. Santos. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD(+) and NADH pools determined in vivo by 13C-NMR. J. Biol. Chem. 277:28088-28098. [DOI] [PubMed] [Google Scholar]
- 49.O'Connell-Motherway, M., G. F. Fitzgerald, and S. D. van. 1997. Cloning and sequence analysis of putative histidine protein kinases isolated from Lactococcus lactis MG1363. Appl. Environ. Microbiol. 63:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, Y. H., and L. L. McKay. 1982. Distinct galactose phosphoenolpyruvate-dependent phosphotransferase system in Streptococcus lactis. J. Bacteriol. 149:420-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pool, W. A., A. R. Neves, J. Kok, H. Santos, and O. P. Kuipers. 2006. Natural sweetening of food products by engineering Lactococcus lactis for glucose production. Metab. Eng. 8:456-464. [DOI] [PubMed] [Google Scholar]
- 52.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poolman, B., E. J. Smid, H. Veldkamp, and W. N. Konings. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 169:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian, N., G. A. Stanley, B. Hahn-Hagerdal, and P. Radstrom. 1994. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J. Bacteriol. 176:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 56.Solem, C., B. Koebmann, F. Yang, and P. R. Jensen. 2007. The las enzymes control pyruvate metabolism in Lactococcus lactis during growth on maltose. J. Bacteriol. 189:6727-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steidler, L., and P. Rottiers. 2006. Therapeutic drug delivery by genetically modified Lactococcus lactis. Ann. N. Y. Acad. Sci. 1072:176-186. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, T. D., K. W. Turner, and V. L. Crow. 1980. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J. Bacteriol. 144:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, J. 1980. Galactose transport systems in Streptococcus lactis. J. Bacteriol. 144:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, J. 1978. In vivo regulation of glycolysis and characterization of sugar: phosphotransferase systems in Streptococcus lactis. J. Bacteriol. 136:465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J., and B. M. Chassy. 1983. Intracellular hexose-6-phosphate:phosphohydrolase from Streptococcus lactis: purification, properties, and function. J. Bacteriol. 156:70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Hijum, S. A., A. de Jong, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Rooijen, R. J., K. J. Dechering, C. Niek, J. Wilmink, and W. M. de Vos. 1993. Lysines 72, 80 and 213 and aspartic acid 210 of the Lactococcus lactis LacR repressor are involved in the response to the inducer tagatose-6-phosphate leading to induction of lac operon expression. Protein Eng. 6:201-206. [DOI] [PubMed] [Google Scholar]
- 64.van Rooijen, R. J., S. van Schalkwijk, and W. M. de Vos. 1991. Molecular cloning, characterization, and nucleotide sequence of the tagatose 6-phosphate pathway gene cluster of the lactose operon of Lactococcus lactis. J. Biol. Chem. 266:7176-7181. [PubMed] [Google Scholar]
- 65.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. P. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]