Abstract
A significant fraction (46/108, 43%) of swine isolates of Campylobacter coli but none of 81 isolates of C. coli from turkeys had genomic DNA that was resistant to digestion by MboI, suggesting methylation of adenines at GATC sites. No consistent association was noted between antimicrobial resistance and MboI resistance. Seven swine-associated multilocus sequence typing-based sequence types (STs) were detected among multiple isolates with MboI-resistant DNA. The data suggest host-associated DNA modification system(s) specific for adenine at GATC sites in C. coli from swine.
Campylobacter coli commonly colonizes swine but has also been isolated from turkeys (7, 12-15, 23, 26, 29, 31). Applications of several DNA-based typing schemes have revealed that genotypes of C. coli isolates from swine are largely different than those from other meat animals (7, 13, 19). Characterization of antimicrobial susceptibility profiles in C. coli isolates derived from different animals has shown noticeable differences in the prevalence of resistance to several antibiotics (23). In addition, we have found that C. coli isolates from turkeys were more prone to acquire resistance to erythromycin via natural transformation than their counterparts from swine (8). Such findings suggest that C. coli isolates from different meat animals harbor genetic attributes that confer on them distinct phenotypes and host-associated adaptations. Other host-associated phenotypes, however, remain unidentified. To date, only one C. coli strain (RM2228, a multidrug-resistant strain from a chicken) has had its genome completely sequenced and described in detail (5), and limited information is available on genome-wide comparisons of C. coli isolates from different animal hosts (11).
DNA modification at specific sites is a common feature of bacterial genomes and is frequently associated with restriction-modification systems (30). A type of DNA modification that has been extensively identified in various bacterial genomes involves methylation of either adenines or cytosines at GATC sites, rendering the genomic DNA of the organism resistant to enzymes such as MboI and Sau3AI, respectively (18, 22). Cytosine methylation at GATC sites is frequently encountered in bacteria, and in several species it has been found to be characteristic of specific clonal groups (e.g., see references 2, 24, 28, and 32). On the other hand, methyltransferases specific for adenine at GATC sites are encountered not only as part of type II restriction-modification systems but also as solitary enzymes that are evolutionarily related to the Dam methylase of Escherichia coli (16, 20). In E. coli and other bacteria, such solitary adenine DNA methyltransferases with specificity for GATC sites have key functions in DNA mismatch repair mechanisms and in a variety of regulatory processes (3, 17, 21).
Surprisingly limited information is available on the presence of either adenine or cytosine modification at GATC sites in Campylobacter spp. A survey of 12 Campylobacter strains identified three (one each of C. jejuni, C. coli, and C. upsaliensis) that harbored methylated adenines at GATC sites; none of the isolates harbored methylated cytosines at these sites (4). However, only two strains of C. coli were included in the survey. In this study, we investigated the prevalence of adenine or cytosine methylation at GATC sites among C. coli isolates from turkeys (n = 81) and swine (n = 108). The isolates were derived from several different conventional turkey and swine farms in eastern North Carolina at various time points between 2003 and 2005 (31).
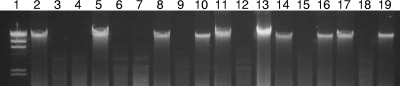
DNA from all 189 isolates was susceptible to Sau3AI, suggesting the absence of cytosine methylation at GATC sites. Furthermore, DNA from all 81 turkey isolates was readily digestible with MboI (data not shown), suggesting the absence of adenine methylation at the GATC sites of these turkey-derived C. coli isolates. In contrast, 46 of the 108 (43%) swine-derived isolates had DNA that was resistant to digestion by MboI (Fig. 1), suggesting adenine methylation at GATC sites. DNA from 18 randomly chosen isolates with MboI-resistant DNA was digested with DpnI, which is specific for GATC sites at which the adenine is methylated (10). The DNA of all 18 isolates was fully susceptible to DpnI digestion (data not shown), confirming that the adenines at GATC sites were methylated.
FIG. 1.
MboI resistance among swine-derived C. coli. Genomic DNA was extracted with a Qiagen DNeasy kit and digested with Sau3AI and MboI. Lanes (after lane 1, the sets of three lanes for the isolates contain DNA that is undigested, digested with Sau3AI, and digested with MboI, respectively): 1, molecular size markers (Roche, Indianapolis, IN); 2 to 4, DNA of the turkey-derived isolate 6017; 5 to 7, DNA of the turkey-derived isolate 6067; 8 to 10, DNA of the swine-derived isolate 6458; 11 to 13, DNA of the swine-derived isolate 6461; 14 to 16, DNA of the swine-derived isolate 6550; 17 to 19, DNA of the swine-derived isolate 7995.
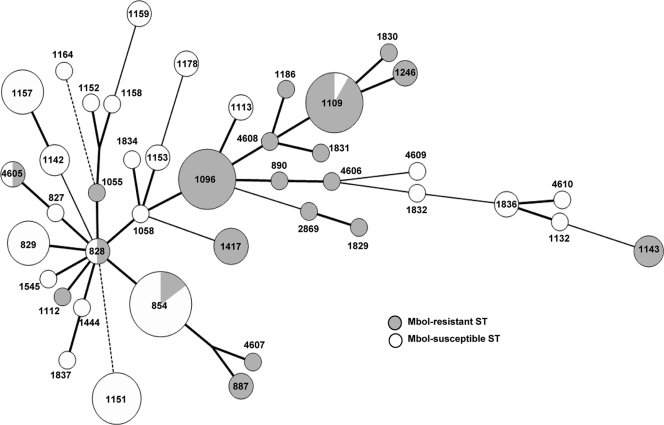
Multilocus sequence typing (MLST) of swine-derived C. coli isolates (47 and 60 with DNA that was MboI resistant and MboI susceptible, respectively) identified 42 sequence types (STs), 20 of which were encountered in one or more of the isolates with MboI-resistant DNA. Most MboI-resistant strains tended to be closely related, being placed in common branches in a minimum spanning tree depicting putative relationships among the STs (Fig. 2). Seven STs (854, 887, 1096, 1109, 1143, 1246, and 1417) were identified among multiple isolates with MboI-resistant DNA (Table 1). All 11 isolates of ST-1096 and most (10/11) isolates of ST-1109 were MboI resistant. In contrast, only two of the 13 isolates of ST-854 were resistant to MboI (Table 1). Several prevalent STs were also detected for which none of the isolates had MboI-resistant DNA (e.g., all six isolates of ST-829, all eight of ST-1151, and all six of ST-1157) (Fig. 2).
FIG. 2.
STs of swine-derived C. coli. Thick bold lines indicate one locus difference, thin solid lines indicate 2 locus differences, and dashed lines indicate 3 or more locus differences. The size of the circle indicates the number of isolates with that ST, with the smallest circles indicating unique STs. STs were determined as described previously (19). ST-4605 to ST-4610 were new STs.
TABLE 1.
STs encountered in swine-derived C. coli isolates with MboI-resistant DNA
| ST | No. of isolates | No. of isolates with MboI-resistant DNA |
|---|---|---|
| 828 | 2 | 1 |
| 854 | 13 | 2 |
| 887 | 2 | 2 |
| 1096 | 11 | 11 |
| 1109 | 11 | 10 |
| 1143 | 3 | 3 |
| 1246 | 2 | 2 |
| 1417 | 4 | 4 |
| 4605 | 2 | 1 |
| Othersa | 11 | 11 |
STs found only in one MboI-resistant isolate were 890, 1055, 1112, 1186, 1829, 1830, 1831, 2869, 4606, 4607, and 4608.
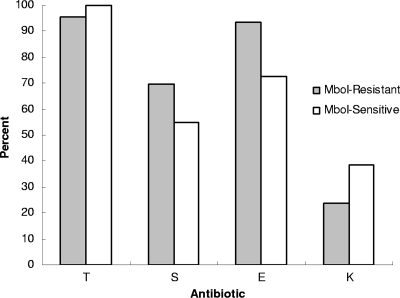
No consistent trends in antimicrobial resistance were detected between MboI-resistant and MboI-susceptible swine-derived isolates. With the exception of resistance to erythromycin, no significant differences were noted in resistance to the other antibiotics (tetracycline, streptomycin, kanamycin, and ciprofloxacin) (Fig. 3). The antimicrobial susceptibility profiles observed most frequently among both MboI-resistant and MboI-susceptible isolates were TSE (resistance to tetracycline, streptomycin, and erythromycin) and TE (resistance to tetracycline and erythromycin) (Table 2), but the TSE profile was more prevalent among MboI-resistant isolates (20/46, 43%) than among MboI-susceptible ones (16/62, 26%) (P = 0.05931).
FIG. 3.
Antibiotic resistance prevalence among Mbo-resistant and Mbo-sensitive swine-derived Campylobacter coli isolates. Erythromycin resistance prevalence was higher among MboI-resistant than among MboI-susceptible isolates (P < 0.05); differences in prevalence in resistance to the other antibiotics were not significant. T, S, E, and K indicate resistance to tetracycline, streptomycin, erythromycin, and kanamycin, respectively. None of the isolates were resistant to ciprofloxacin (data not shown). Resistance thresholds were as described previously (15).
TABLE 2.
Prevalence of antimicrobial resistance profiles in MboI-resistant versus MboI-susceptible C. coli isolates from swine
| AMR profilea | No. (%) of isolates with: |
|
|---|---|---|
| MboI-resistant DNA (n = 46) | MboI-susceptible DNA (n = 62) | |
| Pan sensitive | 0 (0) | 0 (0) |
| T | 0 (0) | 4 (6) |
| SE | 2 (4) | 0 (0) |
| TE | 12 (26) | 13 (21) |
| TK | 1 (2) | 2 (3) |
| TS | 1 (2) | 5 (8) |
| TEK | 1 (2) | 9 (15) |
| TSE | 20 (43) | 16 (26) |
| TSK | 1 (2) | 6 (10) |
| TSEK | 8 (17) | 7 (11) |
AMR, antimicrobial resistance; T, tetracycline; S, streptomycin; E, erythromycin; K, kanamycin. “AMR profile” refers to resistance phenotypes. Thus, “T” indicates isolates with resistance only to tetracycline, “TE” indicates isolates with resistance to both tetracycline and erythromycin (but susceptible to the other antibiotics in the panel), etc. “Pan sensitive” indicates isolates that were susceptible to all antibiotics in the panel.
The DNA methylation system detected here is the first to show host animal-associated differences in prevalence in Campylobacter species. Several STs of isolates with this methylation (e.g., ST-854 and ST-1096) appear to be prevalent among C. coli isolates from swine, having been detected among several isolates in this study and others (9, 13, 19, 25, 27). These findings suggest that the DNA methylation described here may be relatively common among C. coli isolates from swine but rare or absent among C. coli isolates from turkeys, although characterization of DNA methylation in C. coli isolates from other animal hosts is needed. The detection of isolates with a common ST but different DNA resistance to MboI suggests that the gene responsible for the methylation may be transferred among closely related isolates via horizontal gene transfer. Alternatively, the gene may have been inactivated or lost in those with MboI-susceptible DNA.
Further studies are needed to identify and characterize the gene mediating the genomic DNA methylation and to determine whether such a gene is accompanied by a cognate restriction endonuclease gene. The responsible gene (or restriction modification system) may have the potential to affect DNA transfer via natural transformation, as has been described in other systems (1, 6). Such involvement, along with possible roles in host adaptation and other adaptations, will need to be investigated, ideally through the use of isogenic mutants.
Acknowledgments
This research was partially supported by USDA-NRI Competitive Grant 2008-35201-04664 and USDA-ARS CRIS project 5325-42000-045. S. Wilson was partially supported with National Science Foundation grant HRD 0102892 to St. Augustine's College.
We thank all members of our laboratory for encouragement and support.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Ando, T., Q. Xu, M. Torres, K. Kusugami, D. A. Israel, and M. J. Blaser. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37:1052-1065. [DOI] [PubMed] [Google Scholar]
- 2.Burrus, V., C. Bontemps, B. Decaris, and G. Guedon. 2001. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol. 67:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadesús, J., and D. Low. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edmonds, P., B. M. Hall, W. R. Edwards, and K. M. Hartline. 1992. Presence of methylated adenine in GATC sequences in chromosomal DNAs from Campylobacter species. J. Bacteriol. 174:8156-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groot, M. N., F. Nieboer, and T. Abee. 2008. Enhanced transformation efficiency of recalcitrant Bacillus cereus and Bacillus weihenstephanensis isolates upon in vitro methylation of plasmid DNA. Appl. Environ. Microbiol. 74:7817-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopkins, K. L., M. Desai, J. A. Frost, J. Stanley, and J. M. J. Logan. 2004. Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping, and phage typing. J. Clin. Microbiol. 42:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, J.-S., D. K. Carver, and S. Kathariou. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korczak, B. M., M. Zurfluh, S. Emler, J. Kuhn-Oertli, and P. Kuhnert. 2009. Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J. Clin. Microbiol. 47:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacks, S., and B. Greenberg. 1975. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem. 250:4060-4066. [PubMed] [Google Scholar]
- 11.Lang, P., T. Lefebure, W. Wang, P. Pavinski Bitar, R. J. Meinersmann, K. Kaya, and M. J. Stanhope. 2010. Expanded MLST genotyping and comparative genomic hybridization of Campylobacter coli isolates from multiple hosts. Appl. Environ. Microbiol. 76:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, B. C., N. Reimers, H. J. Barnes, C. D'Lima, D. Carver, and S. Kathariou. 2005. Strain persistence and fluctuation of multiple-antibiotic resistant Campylobacter coli colonizing turkeys over successive production cycles. Foodborne Pathof. Dis. 2:103-110. [DOI] [PubMed] [Google Scholar]
- 13.Litrup, E., M. Torpdahl, and E. M. Nielsen. 2007. Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 103:210-218. [DOI] [PubMed] [Google Scholar]
- 14.Logue, C. M., J. S. Sherwood, L. M. Elijah, P. A. Olah, and M. R. Dockter. 2003. The incidence of Campylobacter spp. on processed turkeys from processing plants in the midwestern United States. J. Appl. Microbiol. 95:234-241. [DOI] [PubMed] [Google Scholar]
- 15.Luangtongkum, T., T. Y. Morishita, A. J. Ison, S. Huang, P. F. McDermott, and Q. Zhang. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mannarelli, B. M., T. S. Balganesh, B. Greenberg, S. S. Springhorn, and S. A. Lacks. 1985. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 82:4468-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marinus, M. G., and J. Casadesús. 2009. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33:488-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClelland, M., M. Nelson, and E. Raschke. 1994. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22:3640-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletzky, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 21.Reisenauer, A., L. S. Kahng, S. McCollum, and L. Shapiro. 1999. Bacteria DNA methylation: a cell cycle regulator? J. Bacteriol. 181:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234-D236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saenz, Y., M. Zarazaga, M. Lantero, M. J. Gastanares, F. Baquero, and C. Torres. 2000. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob. Agents Chemother. 44:267-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeber, S., C. Kessler, and F. Gotz. 1990. Cloning, expression and characterization of the Sau3AI restriction and modification genes in Staphylococcus carnosus TM300. Gene 94:37-43. [DOI] [PubMed] [Google Scholar]
- 25.Sheppard, S. K., J. F. Dallas, M. MacRae, N. D. McCarthy, E. L. Sproston, F. J. Gormley, N. J. Strachan, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, K., N. Reimers, H. J. Barnes, B. C. Lee, R. Siletzky, and S. Kathariou. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J. Food Prot. 67:1463-1468. [DOI] [PubMed] [Google Scholar]
- 27.Thakur, S., W. E. Morrow, J. A. Funk, P. B. Bahnson, and W. A. Gebreyes. 2006. Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl. Environ. Microbiol. 72:5666-5669. (Erratum, 74:342.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twomey, D. P., L. L. McKay, and D. J. O'Sullivan. 1998. Molecular characterization of the Lactococcus lactis LlaKR2I restriction-modification system and effect of an IS982 element positioned between the restriction and modification genes. J. Bacteriol. 180:5844-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wesley, I. V., W. T. Muraoka, D. W. Trampel, and H. S. Hurd. 2005. Effect of preslaughter events on prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys. Appl. Environ. Microbiol. 71:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, G. G., and N. E. Murray. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585-627. [DOI] [PubMed] [Google Scholar]
- 31.Wright, S. L., D. K. Carver, R. M. Siletzky, S. Romine, W. E. Morrow, and S. Kathariou. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J. Food Prot. 71:1791-1796. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, W., and S. Kathariou. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl. Environ. Microbiol. 63:3085-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]