Abstract
Bilateral asymmetry in Caenorhabditis elegans arises in part from cell lineages that differ on the left and right sides of the animal. The unpaired MI neuron descends from the right side of an otherwise left-right symmetric cell lineage that generates the MI neuron on the right and the e3D epithelial cell on the left. We isolated mutations in three genes that caused left-right symmetry in this normally asymmetric cell lineage by transforming MI into an e3D-like cell. These genes encode the proneural bHLH proteins NGN-1 and HLH-2 and the Otx homeodomain protein CEH-36. We identified the precise precursor cells in which ceh-36 and ngn-1 act, and showed that CEH-36 protein is asymmetrically expressed and is present in an MI progenitor cell on the right but not in its bilateral counterpart. This asymmetric CEH-36 expression promotes asymmetric ngn-1 and hlh-2 expression, which in turn induces asymmetric MI neurogenesis. Our results indicate that this left-right asymmetry is specified within the two sister cells that first separate the left and right branches of the cell lineage. We conclude that the components of an evolutionarily conserved Otx/bHLH pathway act sequentially through multiple rounds of cell division on the right to relay an initial apparently cryptic asymmetry to the presumptive post-mitotic MI neuron, thereby creating an anatomical bilateral asymmetry in the C. elegans nervous system.
Keywords: Left-right asymmetry, C. elegans, Cell lineage
INTRODUCTION
Left-right asymmetry is a widespread feature of animal anatomy. Studies over the past decade have advanced our understanding of mechanisms that establish left-right asymmetry in visceral organs of vertebrate embryos (for reviews, see Levin, 2005; Shiratori and Hamada, 2006). In addition to visceral organs, the nervous systems of both vertebrates and invertebrates display anatomical asymmetry (Concha and Wilson, 2001; Toga and Thompson, 2003), and in some cases such neuroanatomical asymmetries are important for nervous system function (Barth et al., 2005; Pascual et al., 2004). The identification of the molecular mechanisms by which neuroanatomical asymmetry is established is important to understand both neural development and neural function. Our current knowledge of the molecular and cellular mechanisms that specify anatomical asymmetry within the nervous system is limited.
The nervous system of Caenorhabditis elegans displays bilateral asymmetry (Hobert et al., 2002). Because the complete cell lineage of C. elegans has been described and the somatic cell lineage is essentially invariant from animal to animal (Kimble and Hirsh, 1979; Sulston and Horvitz, 1977; Sulston et al., 1983), the developmental origins of all cells, including all left-right paired and unpaired cells, are known. Much of the left-right symmetry arises from pairs of left-right analogous blastomeres, which through bilaterally symmetric cell lineages give rise to sets of left-right paired cells. Left-right asymmetry and threefold symmetry can be generated by disrupting the bilateral symmetry in such cell lineages.
The C. elegans pharynx contains single unpaired neurons as well as sets of cells arranged with threefold symmetry (Albertson and Thomson, 1976). For example, the MI motoneuron is a single unpaired neuron located in the pharynx. The MI cell body sends a single unilateral process circumferentially around the pharyngeal nerve ring. Numerous epithelial cells in the pharynx display three-fold radial symmetry. For example, the three e3 epithelial cells are located on the ventral left (VL), ventral right (VR) and dorsal (D) regions of the pharynx. e3VL and e3VR are generated from a left-right symmetric cell lineage, whereas the e3D epithelial cell and the MI neuron are generated as lineally homologous descendants from a left-right asymmetric cell lineage. At the 50-cell stage of embryogenesis, the blastomere ABaraap divides and generates two multipotent daughter cells, ABaraapa and ABaraapp, which are a pair of left-right analogous blastomeres that give rise to six identical sets of left-right paired cells of diverse cell types, including the M2, M3 and NSM pharyngeal neurons, the m1 and m2 pharyngeal muscles, and the mc2 pharyngeal marginal cells (Fig. 1A; see Fig. S1 in the supplementary material for positions of these blastomeres). The anteriormost descendants of ABaraapa and ABaraapp differ in their cell fates: whereas ABaraapaaaa differentiates into the e3D epithelial cell, its lineally homologous cell ABaraappaaa becomes the MI neuron. When this left-right asymmetry is specified and how information concerning the asymmetry is transduced to generate MI on the right and e3D on the left are unknown.
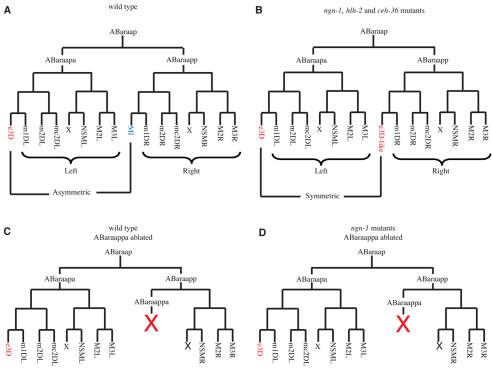
Fig. 1.
The MI neuron is generated from a left-right asymmetric cell lineage. (A) In the wild type, left-right asymmetry in the ABaraap cell lineage is seen in the different cell fates of the MI neuron and the e3D epithelial cell. (B) In ngn-1, hlh-2 and ceh-36 mutants, left-right asymmetry in the cell lineage is lost as a result of the cell-fate transformation of the presumptive MI neuron into an e3D-like cell. (C,D) The predicted ABaraap cell lineages of (C) the wild type and (D) ngn-1(n1921) mutants after laser ablation of the grandmother cell of the MI neuron, ABaraappa. X in red indicates the cell killing of ABaraappa by laser microsurgery.
Here, we report that ngn-1, a bHLH neurogenin gene, hlh-2, the C. elegans ortholog of E2A/Daughterless, and ceh-36,a C. elegans homolog of the mammalian homeodomain gene Otx, are required for establishing left-right asymmetry in the ABaraap cell lineage. Loss-of-function mutations in these genes transform MI into an e3D-like cell, resulting in left-right symmetry in this normally asymmetric cell lineage. We show that ceh-36 promotes expression of ngn-1 and hlh-2 and identify the precise precursor cells in which ceh-36 and ngn-1 act: the bilateral asymmetry is specified by the asymmetric expression of ceh-36 between ABaraapa and ABaraapp, the pair of left-right sister cells that generate the left and right branches of the cell lineage, respectively, and a CEH-36/NGN-1/HLH-2 transcriptional cascade acts through three rounds of cell division on the right side to generate MI and establish this aspect of neuronal bilateral asymmetry.
MATERIALS AND METHODS
C. elegans strains
C. elegans strains were cultured at 20°C as described previously (Brenner, 1974). N2 (Bristol) was the wild-type strain.
Identification of MI and e3D cell fate reporters
The reporter sams-5::gfp is expressed in a single pharyngeal neuron of wild-type animals (The Genome BC C. elegans Gene Expression Consortium). We identified this cell as MI based on the morphology and position of its nucleus (Albertson and Thomson, 1976). The reporter D2096.6::gfp has previously been shown to be expressed in pharyngeal epithelial cells, pharyngeal muscle cells and pharyngeal marginal cells (Gaudet and Mango, 2002). We generated a variant, D2096.6::pes-10::gfp, that retains expression in the e3 pharyngeal epithelial cells, including e3D, but lacks expression in the pharyngeal muscle cells or pharyngeal marginal cells.
Isolation of ngn-1(n1921), ngn-1(n5020), ngn-1(n5052) and hlh-2(n5053)
The wild-type strain was mutagenized with ethyl methanesulfonate (EMS) and F2 progeny were observed using Nomarski optics (Brenner, 1974). ngn-1(n5020), ngn-1(n5052) and hlh-2(n5053)/+ were isolated from screens looking for animals in which MI was missing and an extra e3D-like cell was present. ngn-1(n1921) was recovered from screens looking for animals in which one or more extra neuronal cells were present in the anterior pharynx. The extra neuronal cells in the anterior pharynx of ngn-1(n1921) mutants result from a dislocation of the M2 neurons, which are normally located in the posterior pharynx (R.E.E. and H.R.H., unpublished).
Isolation of ceh-36(n5333), ceh-36(n5339) and ceh-36(n5340)
Wild-type animals carrying the D2096.6::pes-10::gfp reporter were mutagenized with EMS and F3 progeny were observed using a fluorescence-equipped dissecting microscope. ceh-36(n5333), ceh-36(n5339) and ceh-36(n5340) were isolated as animals that contained an extra cell expressing D2096.6::pes-10::gfp.
Isolation of hlh-2(n5287Δ)
Genomic DNA pools from EMS-mutagenized animals were screened by PCR for deletion alleles of hlh-2, essentially as described (Jansen et al., 1997; Liu et al., 1999). hlh-2(n5287Δ) was isolated from a frozen stock and backcrossed to the wild type four times. The hlh-2(n5287Δ) allele removes sequence between nucleotide 18388 of cosmid M05B5 and nucleotide 2042 of cosmid C01H6 and inserts the 15 bp sequence GAGCAATGGCGGCAG at that site.
RNA interference (RNAi)
We performed RNAi of ngn-1 or hlh-2 by growing eri-1(mg366) or wild-type animals, respectively, on HT115(DE3) E. coli harboring a ngn-1 RNAi construct or a hlh-2 RNAi construct, respectively. The presence or absence of MI in the progeny of the animals grown on these bacteria was determined using Nomarski optics.
Cell ablation
Wild-type or ngn-1(n1921) mutant embryos carrying the D2096.6::pes-10::gfp reporter were directly observed from the 28-cell stage. Laser microsurgery of ABaraappa was performed as described (Avery and Horvitz, 1987). The operated embryos were recovered and grown at 20°C. After hatching, the number of e3D-like cells was determined using the D2096.6::pes-10::gfp reporter.
Mosaic analyses
We observed animals of genotype ngn-1(n1921); nEx1638[ngn-1(+), sur-5::gfp, unc-119::gfp] and of genotype ceh-36(n5333) lin-15AB(n765); nEx1703[ceh-36(+), sur-5::gfp, unc-119::gfp, lin-15AB(+)] using Nomarski optics equipped with epifluorescence. Fisher's tests were used for statistical analyses of the association between the fate specification of MI and the presence of the array in ABaraappaaa. To determine the cell division at which the array was lost in the Class IV animals, we determined the presence or absence of the array as judged by GFP fluorescence in the following cells: e1D, e1VL, e1VR, e2VR, e3D, e3VR, I2R, I5, M2L, M2R, M3L, M3R, mc2DL, mc2DR, MCR, NSML and NSMR. Based on the cell lineages that give rise to these cells (Sulston et al., 1983), we determined the cell division at which the array was lost.
Expression analyses of ngn-1::gfp, hlh-2::gfp and ceh-36::gfp
Gravid hermaphrodites carrying the ngn-1::gfp, hlh-2::gfp and ceh-36::gfp reporters were dissected to obtain early-stage embryos. The cell divisions of these embryos were directly observed using Nomarski optics and epifluorescence.
Plasmid constructions and germline transformation experiments
A description of the plasmid constructs and concentrations used is available from the authors. Germline transformation experiments were performed as described (Mello et al., 1991).
Physical interaction between His-NGN-1 and HLH-2
We purified His-NGN-1 proteins and HLH-2 proteins from E. coli. His-NGN-1 (1 μM) was mixed with HLH-2 protein or MBP (maltose-binding protein) (1 μM) in the presence of Ni-NTA agarose (Qiagen), and the protein mixtures were washed three times. We eluted proteins bound to the Ni-NTA with 200 mM imidazole solution and assessed the amount of HLH-2 or MBP proteins in the eluates by western blot analysis using anti-HLH-2 antibody (provided by Michael Krause) or anti-MBP antibody (New England BioLabs).
RESULTS
ngn-1 and hlh-2 mutants lack the MI neuron and contain an extra e3D-like epithelial cell
To identify genes required for establishing left-right asymmetry in the cell lineage that gives rise to MI on the right and e3D on the left (Fig. 1A), we performed genetic screens for mutations that generate left-right symmetry in this normally asymmetric cell lineage. Such mutants might lack MI and contain an extra e3D-like cell, or alternatively lack e3D and contain an extra MI-like neuron. MI and e3D can be identified using Nomarski optics based on their sizes, morphologies and the positions of their nuclei (Fig. 2A). We performed screens using Nomarski optics and recovered four mutations (n1921, n5020, n5052 and n5053) that cause the apparent absence of MI and presence of an extra e3D-like cell (Fig. 2B,C). As described below, n1921, n5020 and n5052 confer recessive phenotypes, fail to complement, and are alleles of the gene ngn-1. n5053 is an allele of the gene hlh-2 (see below) and semi-dominantly causes the absence of MI and the presence of an extra e3D-like cell. Strains homozygous for the n5053 mutation displayed embryonic lethality.
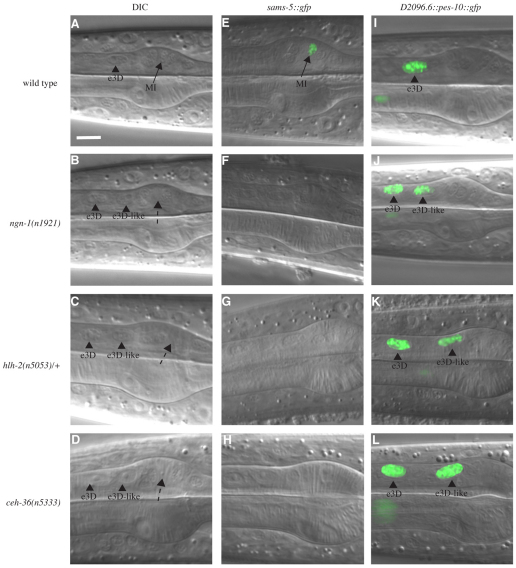
Fig. 2.
ngn-1(n1921), hlh-2(n5053) and ceh-36(n5333) transform the MI neuron into an e3D-like epithelial cell. (A) Nomarski image of a region of the anterior wild-type pharynx. The wild-type pharynx contains MI (arrow) and e3D (arrowhead). The nucleus of the MI neuron is small and granular, whereas that of the e3D epithelial cell is larger, oval in shape and has a distinct nucleolus. (B-D) Nomarski image of a region of anterior (B) ngn-1(n1921), (C) hlh-2(n5053) and (D) ceh-36(n5333) pharynges. The pharynx in these mutants lacks MI (dotted arrow) and contains two e3D-like cells (arrowheads). (E) sams-5::gfp reporter expression in the wild type. The sams-5::gfp reporter is expressed in MI (arrow). (F-H) sams-5::gfp reporter expression in (F) ngn-1(n1921), (G) hlh-2(n5053) and (H) ceh-36(n5333) mutants. The sams-5::gfp reporter failed to be expressed in the pharynx of these mutants. (I) D2096.6::pes-10::gfp reporter expression in the wild type. The D2096.6::pes-10::gfp reporter was expressed in e3D (arrowhead). (J-L) D2096.6::pes-10::gfp reporter expression in (J) ngn-1(n1921), (K) hlh-2(n5053) and (L) ceh-36(n5333) mutants. The D2096.6::pes-10::gfp reporter was expressed in e3D and the extra e3D-like cell (arrowheads). Scale bar: 5 μm.
ngn-1 and hlh-2 mutants lack a neuronal nucleus in the region of the pharynx where MI is normally located in wild-type animals. To confirm that the missing neuron in these mutants is MI, we tested whether these mutant animals express a gfp cell-fate reporter that is normally expressed in MI in wild-type animals. The reporter sams-5::gfp was expressed in MI of wild-type animals (Table 1A; Fig. 2E). We introduced sams-5::gfp into ngn-1 and hlh-2 mutants and found that sams-5::gfp was not expressed in these mutants (Table 1A; Fig. 2F,G), indicating that MI is absent in these mutants.
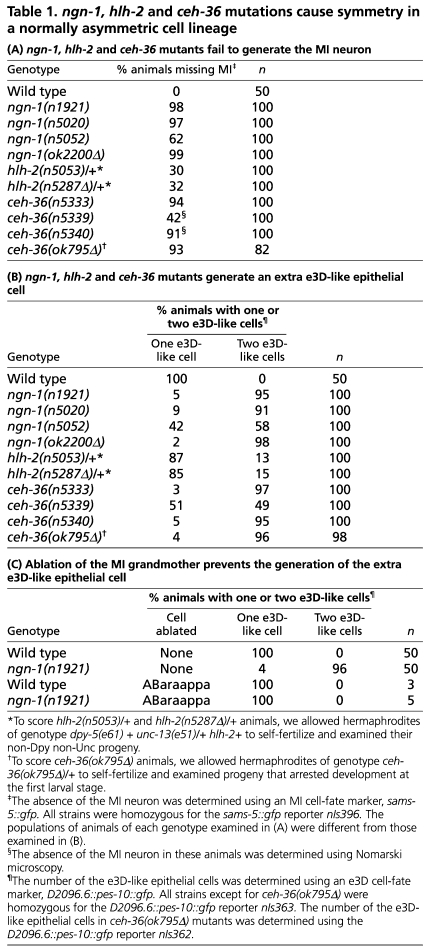
Table 1.
ngn-1, hlh-2 and ceh-36 mutations cause symmetry in a normally asymmetric cell lineage
ngn-1 and hlh-2 mutants contain an extra nucleus that resembles that of e3D (Fig. 2B,C). To determine whether this extra cell adopts the e3D fate, we tested whether these extra cells express a gfp cell-fate marker that is expressed in e3D of wild-type animals. The reporter D2096.6::pes-10::gfp was expressed in the e3 epithelial cells, including e3D, of wild-type animals (Fig. 2I; Table 1B). We introduced D2096.6::pes-10::gfp into ngn-1 and hlh-2 mutants and found that D2096.6::pes-10::gfp was also expressed in the extra cell with a nucleus morphologically similar to that of e3D (Fig. 2J,K; Table 1B), indicating that these mutants generate an extra e3D-like cell.
ngn-1 and hlh-2 mutations generate symmetry in a normally left-right asymmetric cell lineage
Using Nomarski optics, we found that the absence of MI and the presence of an extra e3D-like cell in ngn-1 and hlh-2 mutants were perfectly correlated: when MI was absent, the extra e3D-like cell was always present; when MI was present, the extra e3D-like cell was always absent (n=600, data not shown). We observed no animals that contained or lacked both MI and the extra e3D-like cell, suggesting that these mutations transform MI into an e3D-like cell, thereby generating symmetry in a normally left-right asymmetric cell lineage (Fig. 1B).
We performed laser microsurgery to determine whether the extra e3D-like cell in ngn-1(n1921) mutants is generated from the cell lineage that normally gives rise to MI. If ABaraappaaa, which normally becomes MI in wild-type animals, instead becomes an e3D-like cell in ngn-1 mutants, then elimination of a precursor cell of ABaraappaaa from ngn-1(n1921) mutant embryos by laser ablation should result in the absence of the extra e3D-like cell (Fig. 1C,D). We observed developing ngn-1(n1921) embryos and identified ABaraappa, the grandmother cell of the presumptive MI neuron. Elimination of ABaraappa by laser ablation prevented the generation of the extra e3D-like cell in ngn-1(n1921) animals (Table 1C). By contrast, elimination of the MI grandmother cell in wild-type embryos did not change the number of the e3D cells compared with that in unoperated wild-type embryos (Fig. 1C; Table 1C). We concluded that the extra e3D-like cell in ngn-1(n1921) mutants is generated from the cell lineage that normally gives rise to MI and that MI is transformed into an e3D-like cell in ngn-1 and hlh-2 mutants, resulting in the loss of left-right asymmetry in the cell lineage (Fig. 1B).
ngn-1 encodes a bHLH protein of the Neurogenin subfamily
We mapped n1921 to a 153 kb region of LG IV that includes the ngn-1 locus. We generated a 15.1 kb ngn-1 genomic clone that contains 12.4 kb 5′ upstream and 1.5 kb 3′ downstream sequence of the predicted ngn-1-coding region and found that this ngn-1 clone rescued the MI transformation of n1921 (see Table S1A in the supplementary material). We also observed that ngn-1(RNAi) caused MI transformation (see Table S2 in the supplementary material).
ngn-1 is predicted to encode a 184 amino acid protein similar to members of the neurogenin bHLH subfamily (see Fig. S2 in the supplementary material). We determined the DNA sequence of ngn-1 in our mutants and identified a mutation in each case (Fig. 3A, see Fig. S3 for details concerning the mutation found in n5052). We also characterized an ngn-1 deletion allele, ngn-1(ok2200Δ), which lacks the second and third exons of ngn-1, thus likely defining a null allele of ngn-1 (Fig. 3A). ngn-1(ok2200Δ) mutants exhibited MI transformation (Table 1A,B).
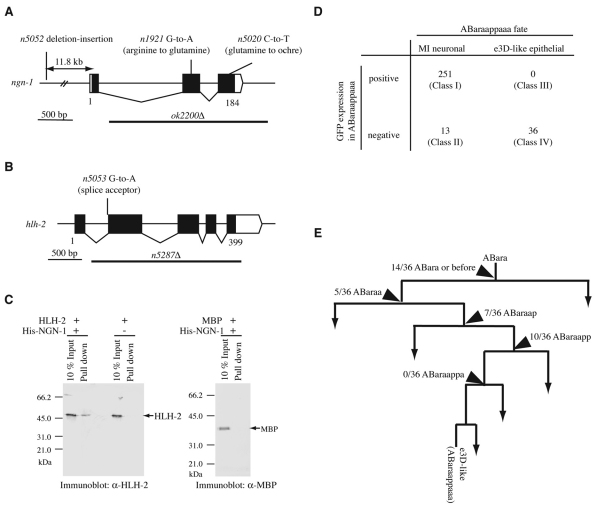
Fig. 3.
NGN-1 binds to HLH-2 and acts cell-autonomously. (A,B) Gene structures of (A) ngn-1 and (B) hlh-2, and mutations associated with each mutant are shown. The black boxes indicate exons; white boxes indicate untranslated regions. (C) In vitro pull-down experiments between hexa-histidine-tagged NGN-1 (His-NGN-1) and HLH-2. Proteins included in each reaction are indicated. The amounts of HLH-2 protein (left panel) and MBP protein (right panel) in the initial reaction mixture (10% input) and in the bound fraction (pull down) were determined by western blot analyses. The numbers indicate the positions of molecular weight markers. (D) Mosaic analysis of ngn-1. Mosaic animals were grouped into four classes based on cell fates and the presence of the extrachromosomal array in ABaraappaaa (see text). The number of mosaic animals in each class is indicated. (E) Determination of the site of the extrachromosomal array loss in each of the 36 Class IV animals. A region of the cell lineage showing the origin of ABaraappaaa is shown. The numbers represent the fraction of Class IV animals in which the extrachromosomal array was lost at the corresponding cell division.
These findings demonstrate that ngn-1 is required to establish bilateral asymmetry in this cell lineage.
n5053 is an allele of hlh-2, the C. elegans ortholog of E2A/Daughterless
We mapped n5053 to a 2 map-unit interval of LG I between dpy-5 and unc-13. This genomic region contains the gene hlh-2, the C. elegans ortholog of E2A/Daughterless (Krause et al., 1997). Because E2A (Tcf3 – Mouse Genome Informatics) in mammals and Daughterless in Drosophila encode bHLH proteins that form heterodimers with other bHLH proteins, including mammalian Neurod1 (Longo et al., 2008; Poulin et al., 1997) and Drosophila Atonal (Jarman et al., 1993), respectively, both of which are closely related to the neurogenin subfamily, we examined whether n5053 is an allele of hlh-2. n5053 mutants carry a G-to-A transition mutation that alters the splice acceptor sequence preceding the second exon of hlh-2 (Fig. 3B). We generated an hlh-2 genomic clone that contains 12.2 kb 5′ upstream and 5.2 kb 3′ downstream region of the hlh-2 coding sequence and found that n5053 mutants transformed with this hlh-2 clone were rescued for the MI transformation as well as the embryonic lethality (see Table S1B,C in the supplementary material). In addition, RNAi treatment of hlh-2 caused MI transformation (see Table S2 in the supplementary material). Furthermore, we isolated an hlh-2 deletion allele, hlh-2(n5287Δ), that removes the entire coding sequence except for the first exon, thus presumably defining a null allele of hlh-2 (Fig. 3B; see Fig. S4 in the supplementary material for details concerning this mutation). In animals heterozygous for hlh-2(n5287Δ), MI was transformed into an e3D-like cell (Table 1A,B), and animals homozygous for hlh-2(n5287Δ) displayed embryonic lethality. These results indicate that hlh-2 is a haplo-insufficient locus required for establishing bilateral asymmetry in this cell lineage.
A physical interaction between NGN-1 and HLH-2 has been shown previously using yeast two-hybrid studies (Grove et al., 2009). We examined whether NGN-1 and HLH-2 directly interact using an in vitro pull-down assay with purified proteins and found that His-tagged NGN-1 proteins associated with HLH-2 proteins but not with a control protein, MBP (Fig. 3C). These results suggest that NGN-1 and HLH-2 form a transcriptional heterodimer required to establish bilateral asymmetry in this cell lineage.
Previous studies indicated that HLH-2 is involved in LIN-12/Notch-mediated lateral signaling in C. elegans (Karp and Greenwald, 2003). We therefore asked whether mutations in genes encoding components of LIN-12/Notch signaling pathways, lin-12 and glp-1 (Austin and Kimble, 1989; Yochem et al., 1988), affect the MI-e3D bilateral asymmetry. We observed that in lin-12 and glp-1 mutants, both MI and e3D were normally present (see Table S3 in the supplementary material).
ngn-1 acts cell autonomously to establish a left-right asymmetry
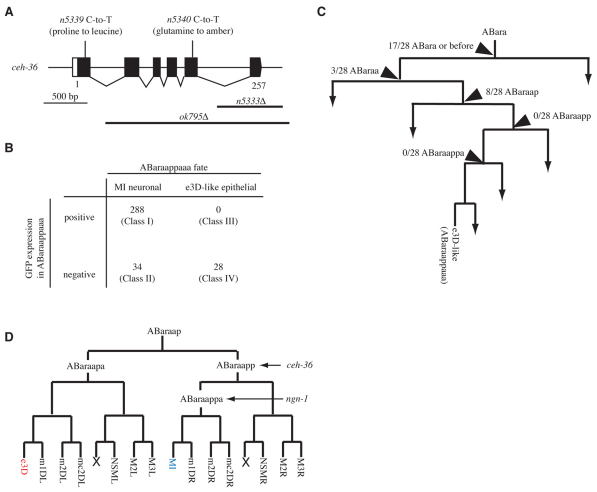
To identify when and where ngn-1 functions to generate MI, we performed mosaic analysis of ngn-1. Specifically, we generated ngn-1(n1921) mutant animals carrying an extrachromosomal array containing the ngn-1 rescuing construct marked with the cell-autonomous gfp markers sur-5::gfp (Yochem et al., 1998) and unc-119::gfp (Maduro and Pilgrim, 1995). Extrachromosomal arrays in C. elegans are mitotically unstable, resulting in the generation of clones of cells that lose the arrays. We examined ngn-1(n1921) animals carrying the array using Nomarski optics equipped with epifluorescence and examined ABaraappaaa, which normally becomes MI in wild-type animals to determine: (1) the fate of ABaraappaaa, i.e. an MI neuronal fate or an e3D-like epithelial fate, judged by the size and the morphology of its nucleus; and (2) the presence or the absence of the array in the ABaraappaaa cell, judged by GFP fluorescence. Based on these two criteria, each animal was classified into one of four categories: Class I animals, ABaraappaaa differentiated into an MI neuron that retained the array; Class II animals, ABaraappaaa differentiated into an MI neuron that lacked the array; Class III animals, ABaraappaaa was transformed into an e3D-like cell that retained the array; and Class IV animals, ABaraappaaa was transformed into an e3D-like cell that lacked the array (Fig. 3D). If ngn-1 acts cell autonomously, then ABaraappaaa transformed into an e3D-like cell should not retain the array, resulting in the absence of the Class III animals. Of 300 animals examined, 36 animals contained ABaraappaaa transformed into an e3D-like cell. All of these 36 animals were classified as Class IV, in which ABaraappaaa did not retain the array; no animals were categorized as Class III (Fig. 3D). The remaining 264 animals contained an ABaraappaaa that differentiated into an MI neuron. Of these animals, 251 animals were classified as Class I, in which ABaraappaaa retained the array, which is consistent with the cell-autonomous action of ngn-1. The remaining 13 animals were categorized as Class II, in which ABaraappaaa differentiated as normal into an MI neuron despite the absence of the array. These 13 Class II animals presumably at least in part reflect the incomplete penetrance of the ngn-1(n1921) mutation (Table 1A,B). In short, our data support a strong association between the specification of the MI neuron fate and the presence of the array in ABaraappaaa (P<0.001) and indicate that ngn-1 acts cell-autonomously to generate bilateral asymmetry in the cell lineage.
We further examined the 36 animals of Class IV, in which ABaraappaaa was transformed into an e3D-like cell that had lost the array. We determined the cell division at which the array had been mitotically lost by scoring the presence or absence of the array in cells that shared a precursor cell with ABaraappaaa at some point of their lineage history. We identified 10 animals that had an extra e3D-like cell and had lost the array at the cell division of ABaraapp (Fig. 3E). This observation indicates that the presence of a functional ngn-1 gene in ABaraapp is insufficient to generate an MI neuron and thus that to generate MI a functional ngn-1 gene is necessary at or later than the stage of ABaraappa, the MI grandmother cell. By contrast, we identified no animals that had lost the array at the next cell division, that of the MI grandmother cell, ABaraappa (Fig. 3E). This result suggests that in all animals in which the array was present in the MI grandmother cell, ABaraappaaa differentiated as normal into an MI neuron whether the array was (Class I animals) or was not (Class II animals) transmitted to the presumptive MI neuron. Thus, the presence of the array in the MI grandmother cell was sufficient to rescue the MI transformation of ngn-1(n1921) mutants. Together, these findings indicate that the presence of the ngn-1 wild-type gene in the MI grandmother cell is both necessary and sufficient to generate an MI neuron and establish left-right asymmetry in the cell lineage.
Expression of ngn-1 and hlh-2 is left-right asymmetric
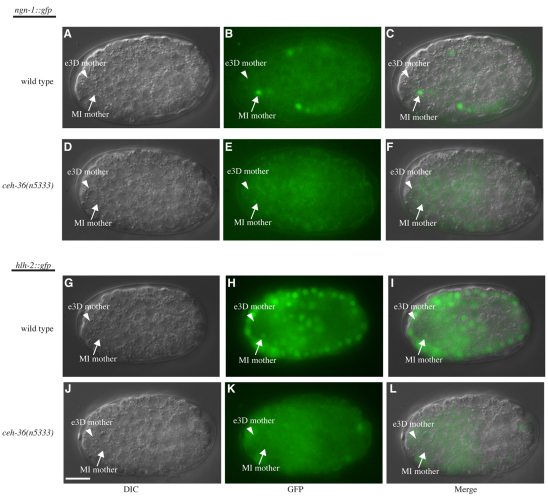
To identify when and where ngn-1 is expressed during embryogenesis, we generated a translational ngn-1::gfp fusion gene by an in-frame insertion of a gfp-coding sequence into the 3′ end of the ngn-1-coding sequence flanked by the 12.4 kb 5′ upstream and 1.5 kb 3′ downstream genomic sequence. We integrated this ngn-1::gfp transgene into the C. elegans genome and found that the ngn-1::gfp transgene rescued the MI transformation of ngn-1(n1921) mutants (see Table S1A in the supplementary material), indicating that this ngn-1::gfp is functional. Because mosaic analysis of ngn-1 suggested the cell-autonomous action of ngn-1, we asked whether ngn-1 is expressed in the cell lineage that gives rise to MI. We observed developing embryos carrying ngn-1::gfp and found that NGN-1::GFP protein was present in the MI mother cell (Fig. 4A-C). By contrast, NGN-1::GFP was not detectable in the e3D mother cell (Fig. 4A-C), indicating that expression of ngn-1 is left-right asymmetric within this asymmetric cell lineage.
Fig. 4.
Expression of ngn-1 and hlh-2 is left-right asymmetric. (A) Nomarski image of a wild-type embryo carrying ngn-1::gfp. (B) Fluorescence image of the same embryo. (C) Merged image of A and B. (D) Nomarski image of a ceh-36(n5333) mutant embryo carrying ngn-1::gfp. (E) Fluorescence image of the same embryo. (F) Merged image of D and E. (G) Nomarski image of a wild-type embryo carrying hlh-2::gfp. (H) Fluorescence image of the same embryo. (I) Merged image of G and H. (J) Nomarski image of a ceh-36(n5333) embryo carrying hlh-2::gfp. (K) Fluorescence image of the same embryo. (L) Merged image of J and K. The arrows indicate the MI mother cell, ABaraappaa; the arrowheads indicate the e3D mother cell, ABaraapaaa. Anterior is towards the left; ventral is towards the top. Scale bar: 5 μm.
Immunostaining experiments using an HLH-2 antibody had demonstrated previously that HLH-2 is present in all cells up to the 100- to 200-cell stage of embryonic development (Krause et al., 1997). To test specifically whether hlh-2 is expressed in the MI mother cell, we generated a translational hlh-2::gfp fusion gene by an in-frame insertion of a gfp-coding sequence to the 3′ end of the hlh-2-coding sequence flanked by the 18.7 kb 5′ upstream and 10.6 kb 3′ downstream genomic sequence. We integrated this hlh-2::gfp transgene into the C. elegans genome and found that hlh-2::gfp rescued the MI transformation as well as the embryonic lethality of hlh-2(n5053) mutants (see Table S1B,C in the supplementary material), indicating that hlh-2::gfp is functional. We observed developing embryos carrying hlh-2::gfp and found that HLH-2::GFP was localized asymmetrically: HLH-2::GFP was detectable in the MI mother cell but not in the e3D mother cell (Fig. 4G-I). These results indicate that both ngn-1 and hlh-2 are expressed asymmetrically, and suggest that this left-right asymmetric expression of ngn-1 and hlh-2 establishes bilateral asymmetry in the cell lineage.
ceh-36, an Otx/Otd homeodomain gene, is required for establishing the MI-e3D bilateral asymmetry
To identify factors that regulate the asymmetric expression of ngn-1 and hlh-2, we performed additional genetic screens for mutations that cause left-right symmetry in this normally asymmetric cell lineage. We mutagenized wild-type animals carrying the e3D cell-fate reporter, D2096.6::pes-10::gfp, and screened for mutants in which an extra e3D-like cell was present or e3D was absent (R.E.E. and H.R.H., unpublished). Among the isolates we recovered were three allelic mutations (n5333, n5339 and n5340) that caused the presence of an extra e3D-like cell (Table 1B; Fig. 2L). We introduced the MI cell-fate reporter sams-5::gfp into these mutants and observed that these mutants failed to generate MI (Table 1A; Fig. 2H), indicating that these mutations transform MI into an e3D-like cell (Fig. 1B).
We mapped n5333 to a 100 kb region of LG X that includes the gene ceh-36. We generated a 5.7 kb ceh-36 genomic clone that contains 2.5 kb 5′ upstream and 1.1 kb 3′ downstream sequence of the ceh-36-coding region and found that this ceh-36 clone rescued the MI transformation of n5333 (see Table S4 in the supplementary material). ceh-36 encodes a 257 amino acid protein similar to members of the Otx/Otd homeodomain subfamily (Chang et al., 2003; Koga and Ohshima, 2004; Lanjuin et al., 2003). We determined the DNA sequence of ceh-36 in our mutants and identified a mutation in each case (Fig. 5A). We also characterized a ceh-36 deletion allele, ceh-36(ok795Δ), which lacks the entire coding sequence of ceh-36 except for the first exon, thus likely defining a null allele of ceh-36 (Fig. 5A). We observed that ceh-36(ok795Δ) mutants displayed larval lethality and exhibited MI transformation (Table 1A,B). These findings establish that ceh-36 is required for establishing asymmetry in this cell lineage.
Fig. 5.
ceh-36 acts cell-autonomously to establish a bilateral asymmetry. (A) Gene structure of ceh-36 and mutations associated with each mutant are shown. The black boxes indicate exons; white boxes indicate untranslated regions. (B) Mosaic analysis of ceh-36. Mosaic animals were grouped into four classes based on cell fates and the presence of the extrachromosomal array in ABaraappaaa (see text). The number of mosaic animals in each class is indicated. (C) Determination of the site of the extrachromosomal array loss in each of the 28 Class IV animals. A region of the cell lineage showing the origin of ABaraappaaa is shown. The numbers represent the fraction of the Class IV animals in which the extrachromosomal array was lost at the corresponding cell division. (D) Cell lineage diagram indicates the sites of actions of ceh-36 and ngn-1. The wild-type ceh-36 gene in the MI great grandmother cell (ABaraapp) is necessary and sufficient to rescue the MI transformation of ceh-36(n5333) mutants. The wild-type ngn-1 gene in the MI grandmother cell (ABaraappa) is necessary and sufficient to rescue the MI transformation of ngn-1(n1921) mutants.
Establishment of the MI-e3D asymmetry does not require genes that specify non-anatomical neuronal bilateral asymmetries
Some aspects of left-right asymmetry in the C. elegans nervous system are not apparent anatomically: left-right pairs of the ASE gustatory neurons and the AWC olfactory neurons are each morphologically similar but distinct both functionally (Pierce-Shimomura et al., 2001; Wes and Bargmann, 2001) and in their patterns of gene expression (Ortiz et al., 2006; Troemel et al., 1999; Yu et al., 1997).
We tested whether genes that specify the asymmetry of the ASE neurons and the AWC neurons are required to establish the MI-e3D bilateral asymmetry. We observed that in cog-1, lim-6, lin-49 and lsy-6 mutants, in which the asymmetry of the ASE neurons is lost (Chang et al., 2003; Johnston and Hobert, 2003), and in inx-19, nsy-4 and unc-43 mutants, in which the asymmetry of the AWC neurons is lost (Chuang et al., 2007; Troemel et al., 1999; Vanhoven et al., 2006), both MI and e3D were correctly specified (see Table S5A,B in the supplementary material). These results indicate that ceh-36, ngn-1 and hlh-2 represent components of a novel pathway that establishes a bilateral asymmetry in the C. elegans nervous system.
ceh-36 acts cell autonomously to establish a left-right asymmetry
To identify when and where ceh-36 functions to generate MI, we performed a mosaic analysis of ceh-36 similar to our ngn-1 mosaic analysis. We generated ceh-36(n5333) mutant animals carrying an extrachromosomal array containing the ceh-36 rescuing construct marked with the cell-autonomous gfp markers sur-5::gfp (Yochem et al., 1998) and unc-119::gfp (Maduro and Pilgrim, 1995), and determined in each animal: (1) the fate of ABaraappaaa, i.e. an MI neuronal fate or an e3D-like fate; and (2) the presence or the absence of the array in the ABaraappaaa cell. Based on these criteria, we classified each animal into one of four categories, as we did in our ngn-1 mosaic analysis (Fig. 5B). Of 350 animals examined, 28 animals contained ABaraappaaa transformed into an e3D-like cell. All of these 28 animals were classified as Class IV; no animals were categorized as Class III. The remaining 322 animals contained an ABaraappaaa that differentiated into an MI neuron. Of these animals, 288 animals were classified as Class I, and the remaining 34 animals were categorized as Class II (Fig. 5B). In short, our data support a strong association between the specification of the MI neuron fate and the presence of the array in ABaraappaaa (P<0.001) and indicate that ceh-36 acts cell-autonomously to generate bilateral asymmetry in the cell lineage.
We further examined the 28 animals of Class IV, in which ABaraappaaa was transformed into an e3D-like cell that had lost the array and determined the cell division at which the array had been mitotically lost. We identified eight animals that had an extra e3D-like cell and had lost the array at the cell division of ABaraap (Fig. 5C). By contrast, we identified no animals that had lost the array at the next cell division, that of the MI great grandmother cell, ABaraapp (Fig. 5C). Together, these findings indicate that the presence of the ceh-36 wild-type gene in the MI great grandmother cell is both necessary and sufficient to rescue the MI transformation defect of ceh-36(n5333).
Left-right asymmetric expression of ngn-1 and hlh-2 is abolished in ceh-36 mutants
Our mosaic analyses suggest that the site of ceh-36 action is one cell division earlier than that of ngn-1 action: our ceh-36 mosaic analysis indicates that the presence of the ceh-36 wild-type gene in the MI great grandmother cell (ABaraapp) is both necessary and sufficient to generate MI, while our ngn-1 mosaic analysis indicates that the presence of the ngn-1 wild-type gene in the MI grandmother cell (ABaraappa) is both necessary and sufficient to generate MI (Fig. 5D). Because these observations suggest that ceh-36 acts upstream of or in parallel to ngn-1, we examined expression of ngn-1 in ceh-36(n5333) mutant embryos. We observed developing ceh-36(n5333) mutant embryos carrying ngn-1::gfp and found that NGN-1::GFP was not detectable in the MI mother cell (Fig. 4D-F). We also asked whether expression of hlh-2 in the MI mother cell requires ceh-36. We observed developing ceh-36(n5333) mutant embryos carrying hlh-2::gfp and found that HLH-2::GFP was not detectable in the MI mother cell (Fig. 4J-L). These observations indicate that establishment of left-right asymmetric expression of ngn-1 and hlh-2 requires ceh-36 and that ceh-36 acts upstream of ngn-1 and hlh-2 to establish left-right asymmetry in the cell lineage.
Expression of ceh-36 is left-right asymmetric
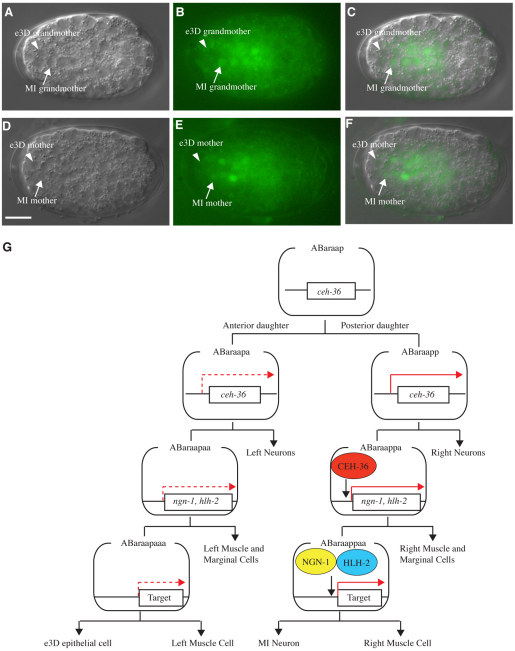
To identify when and where ceh-36 is expressed during embryogenesis, we generated a translational ceh-36::gfp fusion gene by an in-frame insertion of a gfp-coding sequence into the 3′ end of the ceh-36-coding sequence flanked by the 2.5 kb 5′ upstream and 1.1 kb 3′ downstream genomic sequence. We integrated this ceh-36::gfp transgene into the C. elegans genome and found that the ceh-36::gfp transgene rescued the MI transformation of ceh-36(n5333) mutants (see Table S4 in the supplementary material), indicating that this ceh-36::gfp is functional. We observed developing wild-type embryos carrying the ceh-36::gfp transgene and found that CEH-36::GFP was present in the MI grandmother cell but not in the e3D grandmother cell (Fig. 6A-C). CEH-36::GFP in the MI grandmother cell became detectable 25 minutes after this cell was generated. We also found that CEH-36::GFP was present in the MI mother cell but not in the e3D mother cell (Fig. 6D-F). These results indicate that the ceh-36/ngn-1/hlh-2 transcriptional cascade acts specifically on the right of the cell lineage but not on the left to generate MI and establish left-right asymmetry in this cell lineage.
Fig. 6.
Expression of ceh-36 is left-right asymmetric. (A-C) Expression of ceh-36 at the stage of the MI grandmother cell. (A) Nomarski image of an embryo carrying ceh-36::gfp. (B) Fluorescence image of the same embryo. (C) Merged image of A and B. The arrows indicate the MI grandmother cell, ABaraappa; the arrowheads indicate the e3D grandmother cell, ABaraapaa. (D-F) Expression of ceh-36 at the stage of the MI mother cell. (D) Nomarski image of an embryo carrying ceh-36::gfp. (E) Fluorescence image of the same embryo. (F) Merged image of D and E. The arrows indicate the MI mother cell, ABaraappaa; the arrowheads indicate the e3D mother cell, ABaraapaaa. Anterior is towards the left; ventral is towards the top. Scale bar: 5 μm. (G) A model for the establishment of the left-right asymmetric cell lineage. The solid and dotted arrows in red indicate the presence and absence of transcriptional induction, respectively. `Target' represents a locus induced by a heterodimer of NGN-1 and HLH-2.
DISCUSSION
Bilateral asymmetry is a conserved and fundamental feature of nervous systems. Despite its importance, the molecular and cellular mechanisms that establish neuroanatomical asymmetry have been largely elusive. Although Otx homeodomain proteins and proneural bHLH transcription factors have been shown to promote neurogenesis, they have not previously been shown to establish bilateral asymmetry. In this study, we show that two proneural bHLH genes, ngn-1 and hlh-2, and the Otx homeodomain gene ceh-36 are required to generate the single left-right unpaired MI motoneuron. Our results indicate that CEH-36 proteins are present in the MI grandmother cell but not in the e3D grandmother cell. ceh-36 is required for the asymmetric expression of ngn-1 and hlh-2. We propose that the asymmetric localization of CEH-36 proteins promotes asymmetric expression of ngn-1 and hlh-2 in the MI grandmother cell but not in the e3D grandmother cell. The ngn-1 and hlh-2 products generated in the MI grandmother cell are then transmitted to the MI mother cell, leading to the formation of a heterodimer between NGN-1 and HLH-2 in the MI mother cell but not in the e3D mother cell. This asymmetric localization of NGN-1 and HLH-2 then induces an asymmetric neurogenic program, giving rise to the MI neuron on the right side of the cell lineage and the e3D epithelial cell on the left (Fig. 6G).
Our ceh-36 mosaic analysis indicates that the presence of the ceh-36 wild-type gene in the MI great grandmother cell (ABaraapp) is necessary and sufficient to generate an MI neuron. This observation suggests that transcription of ceh-36 initiates in this cell (Fig. 6G). Our expression analysis of ceh-36 supports this notion: we observed a CEH-36::GFP functional protein localized to the nucleus of the MI grandmother cell 25 minutes after its generation. Because fluorophore formation of the variant of GFP we used requires at least 30 minutes (Heim et al., 1995), and because the MI great grandmother cell divides about 30 minutes after its generation to give rise to the MI grandmother cell, it is highly likely that transcription of ceh-36 indeed initiates in the MI great grandmother cell. By contrast, our expression analysis of ceh-36 indicates that CEH-36::GFP was not detectable in the e3D grandmother cell. This observation suggests that ceh-36 is not transcribed in the e3D great grandmother cell, ABaraapa. Although post-transcriptional regulation remains a formal possibility, we suggest that the developmental program establishing the MI-e3D bilateral asymmetry is specified by the asymmetric transcription of ceh-36 in the MI great grandmother cell, ABaraapp, but not in the e3D great grandmother cell, ABaraapa (Fig. 6G). Thus, the determination of the left-right asymmetry of this cell lineage occurs at least three cell generations prior to the generation of MI and e3D within the two sister cells, ABaraapa and ABaraapp, that first separate the left and right branches of cell lineage. We suggest that this initial apparently cryptic asymmetry between ABaraapa and ABaraapp is transduced to the post-mitotic MI neuron by a CEH-36/NGN-1/HLH-2 transcriptional cascade that acts through multiple rounds of cell division on the right side.
Our results indicate that mutations in genes required to establish the non-anatomical bilateral asymmetries of the AWC and ASE neurons do not affect the MI-e3D anatomical asymmetry. Thus, the ceh-36/ngn-1/hlh-2 transcriptional cascade is a novel pathway involved in establishing bilateral asymmetry in the C. elegans nervous system. The AWC bilateral asymmetry is established after the generation of the two post-mitotic AWC neurons through an interaction between these cells (e.g. Chuang et al., 2007). Likewise, the ASE bilateral asymmetry is established by a regulatory pathway that acts within the two post-mitotic ASE neurons (Johnston et al., 2005). By contrast, our results indicate that ceh-36 and ngn-1 act within dividing cells that are progenitors to MI and e3D in order to establish the MI-e3D bilateral asymmetry. Thus, our studies reveal a novel mechanism in which a CEH-36/NGN-1/HLH-2 pathway acts through three rounds of cell division to establish an anatomical bilateral asymmetry manifested by post-mitotic differentiated cells.
Our finding that ngn-1 and ceh-36 are required to generate the MI neuron demonstrates that the role of neurogenin and Otx genes in promoting neurogenesis is evolutionarily conserved from C. elegans to mammals (Fode et al., 1998; Ma et al., 1998; Omodei et al., 2008). Do these genes in vertebrates also act to establish bilateral asymmetry in the nervous system? The epithalamus of the diencephalon displays anatomical asymmetries in many vertebrate species (Concha and Wilson, 2001). For example, in zebrafish, the habenular nuclei in the dorsal diencephalon display anatomical left-right asymmetry (Concha et al., 2000). It has been shown that the establishment of this left-right difference in the habenular structure requires asymmetry in the timing of neurogenesis (Aizawa et al., 2007) and that the habenula express the zebrafish ngn1 gene (Mueller and Wullimann, 2003). In addition, the zebrafish ngn3 gene is asymmetrically expressed in the anterior-ventral diencephalon, with stronger expression on the left side than on the right (Wang et al., 2001). Furthermore, in mammals, Otx2 is required for the generation of the mesencephalic dopaminergic neurons, in which it promotes expression of ngn2 in the mesencephalic dopaminergic progenitors (Omodei et al., 2008). It has been shown that ngn2 is also required for the generation of the mesencephalic dopaminergic neurons, including those located in the retro-rubral area (Andersson et al., 2006), and that the retro-rubral area displays bilateral asymmetry in the number of the dopaminergic neurons (Zaborszky and Vadasz, 2001). Given our results, these observations raise the intriguing possibility that an evolutionarily conserved transcriptional cascade composed of an Otx homeodomain gene and a neurogenin bHLH gene is involved in establishment of nervous system bilateral asymmetry in many animals, including mammals.
Supplementary Material
Acknowledgments
We thank Yuji Kohara for ngn-1 and hlh-2 cDNA; Michael Krause for the HLH-2 antibody; Andy Fire for expression vectors; the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR), for strains; Na An, Beth Castor, Elissa Murphy, Rita Droste, Tove Ljungars and Nikhil Bhatla for technical assistance; and Brendan Galvin and Daniel Denning for critical reading of the manuscript. S.N. was supported in part by an MIT Praecis Presidential Fellowship and a McGovern Institute Schoemaker Graduate Fellowship. H.R.H. is an Investigator of the Howard Hughes Medical Institute and the David H. Koch Professor of Biology at MIT. This work was supported by the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.058834/-/DC1
References
- Aizawa H., Goto M., Sato T., Okamoto H. (2007). Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev. Cell 12, 87-98 [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N. (1976). The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275, 299-325 [DOI] [PubMed] [Google Scholar]
- Andersson E., Jensen J. B., Parmar M., Guillemot F., Bjorklund A. (2006). Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development 133, 507-516 [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J. (1989). Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell 58, 565-571 [DOI] [PubMed] [Google Scholar]
- Avery L., Horvitz H. R. (1987). A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51, 1071-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth K. A., Miklosi A., Watkins J., Bianco I. H., Wilson S. W., Andrew R. J. (2005). fsi zebrafish show concordant reversal of laterality of viscera, neuroanatomy, and a subset of behavioral responses. Curr. Biol. 15, 844-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R. J., Jr, Hobert O. (2003). A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17, 2123-2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. F., Vanhoven M. K., Fetter R. D., Verselis V. K., Bargmann C. I. (2007). An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell 129, 787-799 [DOI] [PubMed] [Google Scholar]
- Concha M. L., Wilson S. W. (2001). Asymmetry in the epithalamus of vertebrates. J. Anat. 199, 63-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha M. L., Burdine R. D., Russell C., Schier A. F., Wilson S. W. (2000). A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28, 399-409 [DOI] [PubMed] [Google Scholar]
- Fode C., Gradwohl G., Morin X., Dierich A., LeMeur M., Goridis C., Guillemot F. (1998). The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20, 483-494 [DOI] [PubMed] [Google Scholar]
- Gaudet J., Mango S. E. (2002). Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821-825 [DOI] [PubMed] [Google Scholar]
- Grove C. A., De Masi F., Barrasa M. I., Newburger D. E., Alkema M. J., Bulyk M. L., Walhout A. J. (2009). A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138, 314-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y. (1995). Improved green fluorescence. Nature 373, 663-664 [DOI] [PubMed] [Google Scholar]
- Hobert O., Johnston R. J., Jr, Chang S. (2002). Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat. Rev. Neurosci. 3, 629-640 [DOI] [PubMed] [Google Scholar]
- Jansen G., Hazendonk E., Thijssen K. L., Plasterk R. H. (1997). Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17, 119-121 [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Grau Y., Jan L. Y., Jan Y. N. (1993). atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73, 1307-1321 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O. (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845-849 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Jr, Chang S., Etchberger J. F., Ortiz C. O., Hobert O. (2005). MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 102, 12449-12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Greenwald I. (2003). Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17, 3100-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D. (1979). The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70, 396-417 [DOI] [PubMed] [Google Scholar]
- Koga M., Ohshima Y. (2004). The C. elegans ceh-36 gene encodes a putative homemodomain transcription factor involved in chemosensory functions of ASE and AWC neurons. J. Mol. Biol. 336, 579-587 [DOI] [PubMed] [Google Scholar]
- Krause M., Park M., Zhang J. M., Yuan J., Harfe B., Xu S. Q., Greenwald I., Cole M., Paterson B., Fire A. (1997). A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124, 2179-2189 [DOI] [PubMed] [Google Scholar]
- Lanjuin A., VanHoven M. K., Bargmann C. I., Thompson J. K., Sengupta P. (2003). Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5, 621-633 [DOI] [PubMed] [Google Scholar]
- Levin M. (2005). Left-right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 122, 3-25 [DOI] [PubMed] [Google Scholar]
- Liu L. X., Spoerke J. M., Mulligan E. L., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., et al. (1999). High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9, 859-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo A., Guanga G. P., Rose R. B. (2008). Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 47, 218-229 [DOI] [PubMed] [Google Scholar]
- Ma Q., Chen Z., del Barco, Barrantes I., de la Pompa J. L., Anderson D. J. (1998). neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20, 469-482 [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D. (1995). Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141, 977-988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller T., Wullimann M. F. (2003). Anatomy of neurogenesis in the early zebrafish brain. Dev. Brain Res. 140, 137-155 [DOI] [PubMed] [Google Scholar]
- Omodei D., Acampora D., Mancuso P., Prakash N., Di Giovannantonio L. G., Wurst W., Simeone A. (2008). Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development 135, 3459-3470 [DOI] [PubMed] [Google Scholar]
- Ortiz C. O., Etchberger J. F., Posy S. L., Frokjaer-Jensen C., Lockery S., Honig B., Hobert O. (2006). Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173, 131-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A., Huang K. L., Neveu J., Preat T. (2004). Neuroanatomy: brain asymmetry and long-term memory. Nature 427, 605-606 [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J. T., Faumont S., Gaston M. R., Pearson B. J., Lockery S. R. (2001). The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410, 694-698 [DOI] [PubMed] [Google Scholar]
- Poulin G., Turgeon B., Drouin J. (1997). NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17, 6673-6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H., Hamada H. (2006). The left-right axis in the mouse: from origin to morphology. Development 133, 2095-2104 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119 [DOI] [PubMed] [Google Scholar]
- Toga A. W., Thompson P. M. (2003). Mapping brain asymmetry. Nat. Rev. Neurosci. 4, 37-48 [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Sagasti A., Bargmann C. I. (1999). Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387-398 [DOI] [PubMed] [Google Scholar]
- Vanhoven M. K., Bauer Huang S. L., Albin S. D., Bargmann C. I. (2006). The claudin superfamily protein NSY-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron 51, 291-302 [DOI] [PubMed] [Google Scholar]
- Wang X., Chu L. T., He J., Emelyanov A., Korzh V., Gong Z. (2001). A novel zebrafish bHLH gene, neurogenin3, is expressed in the hypothalamus. Gene 275, 47-55 [DOI] [PubMed] [Google Scholar]
- Wes P. D., Bargmann C. I. (2001). C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698-701 [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. (1988). The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 335, 547-550 [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. (1998). A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149, 1323-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D. L. (1997). Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94, 3384-3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborszky L., Vadasz C. (2001). The midbrain dopaminergic system: anatomy and genetic variation in dopamine neuron number of inbred mouse strains. Behav. Genet. 31, 47-59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.