Abstract
Candida albicans colonizes human mucosa and prosthetic surfaces associated with artificial joints, catheters, and dentures. In the oral cavity, C. albicans coexists with numerous bacterial species, and evidence suggests that bacteria may modulate fungal growth and biofilm formation. Streptococcus gordonii is found on most oral cavity surfaces and interacts with C. albicans to promote hyphal and biofilm formation. In this study, we investigated the role of the hyphal-wall protein Als3p in interactions of C. albicans with S. gordonii. Utilizing an ALS3 deletion mutant strain, it was shown that cells were not affected in initial adherence to the salivary pellicle or in hyphal formation in the planktonic phase. However, the Als3− mutant was unable to form biofilms on the salivary pellicle or deposited S. gordonii DL1 wild-type cells, and after initial adherence, als3Δ/als3Δ (ΔALS3) cells became detached concomitant with hyphal formation. In coaggregation assays, S. gordonii cells attached to, and accumulated around, hyphae formed by C. albicans wild-type cells. However, streptococci failed to attach to hyphae produced by the ΔALS3 mutant. Saccharomyces cerevisiae S150-2B cells expressing Als3p, but not control cells, supported binding of S. gordonii DL1. However, S. gordonii Δ(sspA sspB) cells deficient in production of the surface protein adhesins SspA and SspB showed >50% reduced levels of binding to S. cerevisiae expressing Als3p. Lactococcus lactis cells expressing SspB bound avidly to S. cerevisiae expressing Als3p, but not to S150-2B wild-type cells. These results show that recognition of C. albicans by S. gordonii involves Als3 protein-SspB protein interaction, defining a novel mechanism in fungal-bacterial communication.
Candida species are the fourth most common causative agents of nosocomial bloodstream infections (2, 47, 54). Crude mortality rates for Candida infections exceed 50% (10, 52), and attributable mortality rates vary between 5 and 48% (3, 10, 13). Candida albicans accounts for 62% of invasive candidiasis infections (46, 47) and is commonly isolated from the oral cavity, gastrointestinal tract, and vagina. The oral carriage rate of C. albicans in healthy subjects ranges from 25 to 60% (28, 42, 48). In the oral cavity, there are estimated to be approximately 700 different species of microorganisms present (45). C. albicans is able to interact physically, by coaggregation, or chemically, through small-molecule signaling, with some of these other microorganisms (1, 18, 20, 29, 33). Interactions of C. albicans with bacteria may be antagonistic, e.g., with Pseudomonas aeruginosa (20), or synergistic, e.g., with Streptococcus gordonii (1), resulting in the formation of diverse polymicrobial communities.
Streptococcus gordonii is a primary colonizer of the oral cavity and may be isolated from mucosal or hard surfaces present there (17, 41). It has previously been shown that S. gordonii, and other viridans streptococci, can coaggregate with C. albicans cells both in vitro and in vivo (21, 29, 57). The interactions between oral streptococci and C. albicans are recognized as contributing to formation of enhanced biofilms (1), which may occur on dentures, leading to denture stomatitis (42). Oral streptococci express a range of cell surface polypeptides, many of which act as adhesins to promote colonization (31, 38). The antigen (Ag) I/II family of polypeptides are cell wall-anchored proteins produced by most indigenous species of oral streptococci (4). These adhesins have been shown to bind a wide range of host cell proteins, including fibronectin (49) and salivary agglutinin gp-340 (5, 12, 27). In addition, the Ag I/II family polypeptide SspB from S. gordonii has been shown to interact directly with other microorganisms, including Actinomyces naeslundii (27), Porphyromonas gingivalis (11), and C. albicans (1, 22). It is thus proposed that oral streptococci may promote colonization by these other microorganisms by providing alternative surfaces to adhere to (30) and possibly metabolic benefits (25).
Candida albicans is a pleomorphic fungus, with the two most commonly identified morphologies being yeast cells and hyphae. Hyphal-filament formation may be induced by many factors, including pH, serum, temperature, nutrient availability, and diffusible cell signaling molecules (53). In a mixed-species biofilm model, S. gordonii enhances hyphal formation, and there is evidence that this may be mediated, at least in part, by soluble factors released by streptococci (1). Within mixed-species biofilms of S. gordonii and C. albicans, streptococci were found associated with yeast cells, pseudohyphae, and hyphae, but preferentially with hyphal filaments (1).
The hyphal cell wall comprises a mixture of chitin, β-1,3 glucans, and β-1,6 glucans, as well as a vast array of proteins (7). One of the major families of C. albicans adhesins is the ALS (agglutinin-like sequence) group of cell wall glycoproteins (24). The family comprises 8 members, several of which have adhesive functions involved in host-pathogen interactions (24). One of these adhesins, Als3p, is a hypha-specific protein (9, 23) and has been shown to be required for mature-biofilm formation, binding extracellular matrix, adhesion to host cells, and internalization of C. albicans by endothelial cells (24, 50, 56). There is also evidence that the Als5 protein is involved in recognition of S. gordonii by C. albicans (32).
In this study, we investigated the role of hypha-specific Als3p in early-stage biofilm formation and in intergeneric interactions of C. albicans with S. gordonii. The results suggest that Als3p interacts directly with SspB on the surface of S. gordonii, a binding event that may then enable additional concerted adhesin-receptor interactions to become established.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study were S. gordonii DL1 (Challis) wild type (WT), S. gordonii UB1360 Δ(sspA sspB) (19), Lactococcus lactis MG1363, and L. lactis UB1586(pUB1000-sspB), a strain constitutively expressing heterologous SspB (27). Streptococci were routinely maintained on BHY agar (37 g/liter brain heart Infusion [Difco], 5 g/liter yeast extract, and 1.5% agar). Liquid cultures were grown statically in BHY broth in capped bottles at 37°C. S. gordonii UB1360 cultures were supplemented with spectinomycin (100 μg/ml). Lactococci were cultivated on M17 medium (Difco) containing 0.5% glucose and 2% agar. Liquid cultures were grown statically in M17-glucose at 30°C in capped tubes. Strain UB1586 containing plasmid pUB1000-sspB was grown in the presence of erythromycin (5 μg/ml). The yeast strains used in this study were C. albicans strain NGY152 (CAI-4/CIp10) (6, 37) or 1843 als3Δ/als3Δ (55) and Saccharomyces cerevisiae S150-2B containing plasmid pADH or pADH-ALS3, constitutively expressing heterologous ALS3 under the alcohol dehydrogenase (ADH) promoter (50). C. albicans NGY152 expresses URA3 in a CAI-4 (Ura-negative) background and was used as a control strain for comparison with the als3Δ/als3Δ (ΔALS3) mutant. C. albicans strains were maintained aerobically on Sabouraud dextrose agar (Difco) at 37°C, and broth cultures were grown in yeast extract-peptone-dextrose (YPD) medium (10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose) at 37°C with orbital shaking at 200 rpm. S. cerevisiae containing pADH or pADH-ALS3 was maintained on synthetic medium lacking uracil (CSM-glu) (0.077% CSM-ura [Formedium], 0.67% yeast nitrogen base [Difco], 2% glucose, and 2.5% agar). Liquid cultures were grown aerobically at 30°C with shaking. C. albicans biofilm formation and hyphal induction were performed at 37°C in YPT-Glc medium (yeast nitrogen base in 10 mM NaH2PO4-Na2HPO4 buffer, pH 7.0, containing 0.05% Bacto tryptone and 0.5% glucose) (1).
Saliva preparation.
Unstimulated whole human saliva was collected from at least 5 healthy volunteers with Institutional Review Board approval. Saliva was pooled and mixed with dithiothreitol (2.5 mM) before clarification by centrifugation (10,000 × g; 10 min). The supernatant was diluted to 25% in distilled water (dH2O) and filter sterilized through a 0.22-μm nitrocellulose membrane. Aliquots of prepared saliva were stored at −20°C.
Biofilm formation.
Sterile glass coverslips (13-mm diameter) were incubated with filter-sterilized 10% saliva in distilled water for 16 h at 4°C and washed twice with dH2O. For monospecies biofilms, late-stationary-phase cells of S. gordonii in BHY medium were harvested by centrifugation (5,000 × g; 5 min) and suspended in YPT-Glc at an optical density at 600 nm (OD600) of 0.1 (∼5 × 107 CFU/ml). Portions (0.5 ml) were added to wells of 24-well polystyrene tissue culture plates containing saliva-coated coverslips, and the plates were incubated at 37°C with gentle shaking at 50 rpm. After 1 h of incubation, nonattached bacteria were aspirated and the coverslips were gently washed twice with YPT-Glc. Fresh YPT-Glc medium was added to the wells, and the biofilms were grown for up to 6 h, with the coverslips recovered at intervals for microscopic or biomass analysis. C. albicans cells were grown for 16 h in YPD medium at 37°C and harvested by centrifugation (5,000 × g; 5 min). The cells were suspended in YPT-Glc at an OD600 of 0.1 (3 × 106 CFU/ml), and portions (0.5 ml) were added to wells containing saliva-coated coverslips. The plates were then incubated for various times at 37°C with gentle movement (50 rpm) to promote biofilm formation.
To produce mixed-species biofilms, C. albicans suspensions prepared as described above were added to coverslips that had previously been incubated with S. gordonii cells for 1 h and washed. After inoculation with C. albicans, the plates were incubated at 37°C for up to 6 h. At intervals, duplicate coverslips were removed, washed with PBS, and air dried. The biofilms were then stained with crystal violet (CV) and visualized by light microscopy. Quantification of biofilm formation was achieved by releasing crystal violet with 10% acetic acid and measuring the absorbance at 595 nm.
Coaggregation assays.
Streptococci or lactococci were cultured for 16 h under their respective growth conditions. Bacteria were harvested by centrifugation (5,000 × g; 5 min), suspended in 1.5 mM fluorescein isothiocyanate (FITC) solution in 0.05 M Na2CO3 containing 0.1 M NaCl (pH 7.5), and incubated at 20°C for 30 min. The fluorescently labeled cells were washed thoroughly by alternate centrifugation and suspension in TNMC buffer (1 mM Tris-HCl, pH 8.0, containing 0.15 M NaCl, 0.1 mM MgCl2, and 0.1 mM CaCl2) (8) to remove excess FITC, and suspended in TNMC at an OD600 of 0.5. C. albicans filamentation was induced by incubation of cells in YPT-Glc for 3 h at 37°C. S. cerevisiae was grown in CSM-ura medium for 16 h at 30°C with vigorous aeration. Yeast or hyphal cells were collected by centrifugation, washed with TNMC buffer, and suspended in TNMC at an OD600 of 1.0. For coaggregation assays, FITC-labeled bacterial suspension (1 ml) was mixed with yeast cell suspension (1 ml) and incubated for 1 h at 37°C (C. albicans) or at 30°C (S. cerevisiae) with shaking. Portions of the suspensions were then deposited onto microscope slides and visualized by light or fluorescence microscopy. Degrees of coaggregation of S. gordonii or L. lactis with C. albicans were assigned to one of three categories: 2+, extensive attachment of bacteria to hyphae or yeast cells with bacterial-cell clumping; 1+, alignment of bacterial cells along hyphae in distinct patches; 0, sparse or no interactions between bacteria and hyphae. The numbers of hyphae with degrees of bacterial binding, expressed as percentages of the total number of hyphae counted, were determined from 2 independent experiments. We found that the C. albicans strains NGY152 (CAI4-4/CIp10) and SC5314 (wild type) behaved identically in all assays. For S. cerevisiae coaggregations, the yeast cells binding bacteria were counted and expressed as the proportion of the total number of yeast cells visualized from two independent experiments. For aggregation assays, between 50 and 100 hyphal cells and between 300 and 500 S. cerevisiae cells were counted for each coaggregation pairing.
Yeast cell wall extracts.
Late-stationary-phase S. cerevisiae cells or C. albicans cells, induced to form hyphal filaments, were harvested by centrifugation and washed in Tris-buffered saline (TBS) (10 mM Tris-HCl containing 0.15 M NaCl, pH 8.0). The cell pellets were suspended in 1 M sorbitol containing 40 mM 2-mercaptoethanol and 125 U/ml lyticase (Sigma) (1 ml), and incubated for 4 h at 37°C with shaking. Crude cell wall fractions were separated from cell debris by centrifugation (3,000 × g; 10 min; 4°C). The supernatant containing cell wall proteins was then centrifuged (5,000 × g; 5 min), and portions of the supernatant were mixed with sample buffer (50 mM Tris-HCl, pH 6.8, containing 1% SDS) and subjected to SDS-PAGE. Proteins were stained with Coomassie blue R250 or electroblotted onto a nitrocellulose membrane (Hybond). The blots were probed with monoclonal antibody to Als3p (3-A5) (9) diluted 1:1,000. Antibody binding was detected with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Dako), and the blots were developed with 4-chloro-1-naphthol.
L. lactis expression of SspB.
Lactococcal cells were harvested from late-exponential-phase cultures and suspended in TNMC buffer at an OD600 of 0.5. Anti-Ag I/II antibody (26) diluted 1:500 or irrelevant rabbit antiserum (1:500) was mixed with 0.5 ml cells and incubated for 1 h at 37°C. Bacteria were harvested, washed three times with TNMC buffer to remove nonspecific antibody, and then incubated with FITC-conjugated anti-rabbit antibody (Dako) (diluted 1:1,000) for 30 min. Cells were collected by centrifugation and washed three times, and portions were applied to glass slides and visualized by light or fluorescence microscopy. Western immunoblot analysis of cell surface protein extracts of L. lactis expressing SspB reactive with Ag I/II antibody has been previously described (26).
Statistics.
Data were processed using PRISM software (Graph Pad). An unpaired Student's t test was used to perform statistical analyses at a confidence level with a P value of <0.05.
RESULTS
Role of Als3p in early-stage biofilm formation.
S. gordonii forms dual-species biofilms with C. albicans in which streptococcal cells clearly associate more avidly with hyphal filaments than with yeast cells (1). Therefore, we hypothesized that a hypha-specific component or components provided receptors for binding by S. gordonii. The Als3p protein is one of several hypha-specific glycoproteins involved in adhesion to host tissues and tissue proteins and has been shown to be required for mature-biofilm formation (39, 56). Accordingly, we first investigated the role of Als3p in early-stage biofilm formation in our system, and then the interaction of Als3p with S. gordonii. Glass coverslips were coated with human salivary proteins as described in Materials and Methods and incubated with suspensions of C. albicans NGY152 (CAI4/Clp10) (here referred to as WT) or the 1843 als3Δ/als3Δ mutant (here designated ΔALS3). Biofilms were developed at 37°C in YPT-Glc medium with gentle agitation and visualized by light microscopy at intervals over 6 h. In the first hour, similar numbers of WT and ΔALS3 cells attached to the surface, and hyphal formation was clearly initiated in similar proportions of cells (Fig. 1 A). At 3 h, similar patterns of colonization were observed for the two strains, with cells forming hyphal filaments up to 20 μm in length and relatively evenly distributed across the salivary-pellicle surface (Fig. 1A). At 6 h, C. albicans WT formed a dense biofilm consisting of networks of hyphal filaments, pseudohyphae, and yeast cells. However, the ΔALS3 mutant had not formed a biofilm, and only a few hyphae and yeast cells remained attached to the pellicle-coated surface (Fig. 1A). These results suggested that although biofilm formation was initiated in the ΔALS3 mutant, most of the cells developing hyphae then failed to remain attached to the surface and became dispersed. Quantification of the biofilms confirmed the visual observations showing little or no difference in biomass between the two strains after 1 h or 3 h of growth. However, the biomass of the ΔALS3 mutant biofilm was significantly reduced at 6 h compared to the WT parent strain (P < 0.05) (Fig. 1C). To confirm that the ΔALS3 mutant was not deficient in growth, biofilms were produced in which C. albicans cells were allowed to adhere to the saliva-coated coverslip for 1 h and then nonattached cells were removed and replaced with fresh medium. The OD600 of planktonic-phase samples taken at intervals up to 6 h increased markedly for the ΔALS3 mutant compared to the WT strain (Fig. 1D). These results suggested that WT cells adhered to the substratum with only a small fraction of cells becoming detached, whereas the ΔALS3 mutant hyphae became detached from the surface and continued to undergo growth and hyphal development in the planktonic phase.
FIG. 1.
Als3p in mono- or dual-species biofilms. (A) C. albicans wild type or C. albicans als3Δ/als3Δ biofilms that formed on saliva-coated glass at 1 h, 3 h, or 6 h stained with CV and visualized by microscopy. The images are representative of experiments performed in triplicate. Scale bars, 40 μm. (B) C. albicans wild type or C. albicans als3Δ/als3Δ biofilms that formed on S. gordonii DL1 cells at 1 h, 3 h, or 6 h stained with crystal violet and visualized by microscopy. The images are representative of experiments performed in triplicate. Scale bars, 25 μm. (C) Biomasses of biofilms of C. albicans WT or C. albicans ΔALS3 assessed by CV release. (D) Cell densities of planktonic-phase samples from developing C. albicans biofilms. The data are means and standard deviations (SD) from experiments performed in triplicate (*, P < 0.05).
Saliva-coated coverslips were then incubated with S. gordonii DL1 cells, which readily form a confluent layer under these conditions (1), and biofilms of C. albicans were allowed to develop on this bacterial base layer. In these dual-species biofilms, C. albicans WT hyphal filaments were more prolific than those formed on the salivary-pellicle base, and dense networks of hyphae were associated with the S. gordonii monolayer (Fig. 1B). As before, the ΔALS3 mutant showed levels of attachment to similar those of the WT after 1 h or 3 h of incubation. However, after 6 h there were virtually no hyphal filaments or yeast cells of the ΔALS3 mutant attached to the streptococcal cell monolayer (Fig. 1B). These experiments showed that the ΔALS3 mutant became detached not only from a pellicle-coated surface, but also from a streptococcal monolayer. The detachment occurred concomitantly with the elongation of preexisting hyphal cells as opposed to the initiation of emerging germ tube filaments.
Als3p is necessary for S. gordonii binding to C. albicans hyphae.
To further characterize the physical interactions occurring between C. albicans Als3p and S. gordonii DL1 cells, a fluorescence-based coaggregation assay was developed. Fluorescently labeled S. gordonii cells were incubated with C. albicans cells that had been induced to form hyphal filaments for 3 h. Because of heterogeneity within the population of C. albicans with respect to the degree of hyphal formation, and in levels of bacterial attachment to hyphal filaments, we categorized the hyphal-bacterial interactions into three groupings: 2+, streptococci forming dense accumulations around hyphae (Fig. 2 A and B); 1+, distinct patches of streptococci aligned alongside regions of hyphae (Fig. 2C and D); 0, few or no streptococcal cells associated with hyphae (Fig. 2E and F). In Fig. 2G, summarizing these results, the bars above the baseline represent percentages of hyphae with streptococci bound, while the bars below the baseline show percentages of hyphae scoring 0. On the basis of this assessment, it was estimated that 60% of wild-type hyphal filaments demonstrated 2+ binding of streptococci, 25% showed 1+ binding, and 15% of hyphae had no streptococci bound to them (Fig. 2G). For the C. albicans ΔALS3 strain, coaggregation was ablated, with 97% of hyphal filaments having no streptococcal cells attached and thus scoring 0 in the assay (Fig. 2G).
FIG. 2.
Coaggregation of C. albicans WT or ΔALS3 with S. gordonii. (A to F) Fluorescently (FITC) labeled S. gordonii DL1 cells were incubated with C. albicans hypha-forming cells for 1 h. Coaggregated microorganisms were visualized by phase-contrast microscopy (A, C, and E) or by fluorescence microscopy (B, D, and F). Scale bars, 25 μm. Images A to F are representative of samples of S. gordonii DL1 and C. albicans WT exhibiting population heterogeneity with respect to the ability of hyphae to bind bacteria. (G) Percentages of total C. albicans hyphae with attached S. gordonii DL1 determined on the basis of the following binding levels (see Materials and Methods): 2+, hyphae completely surrounded by streptococci (B); 1+, hyphae with areas where streptococcal cells are attached (D); 0, little or no bacterial binding (F). The data are means ± SD from duplicate experiments.
S. gordonii SspB interacts with C. albicans hyphae.
Previous studies have suggested that the S. gordonii antigen I/II proteins SspA and SspB were involved in binding to C. albicans cells (1). Coaggregation assays performed between C. albicans wild-type hyphae and S. gordonii UB1360 Δ(sspA sspB) showed only small, and statistically insignificant, differences in mutant-cell binding levels compared with the S. gordonii DL1 wild type (data not shown). This is probably because other S. gordonii surface components are able to interact with C. albicans (21). We then tested the ability of a surrogate host bacterium, L. lactis expressing SspB, to coaggregate with C. albicans hyphae. Cell wall localization of SspB in L. lactis UB1586 had previously been demonstrated (27), although expression of the protein on the cell surface had not. Accordingly, we confirmed surface expression of SspB by immunofluorescence experiments (Fig. 3). Antibodies reactive with SspB bound only to L. lactis UB1568 cells containing pUB1000-sspB and not to control cells (Fig. 3A to D).
FIG. 3.
Expression of heterologous proteins in L. lactis or S. cerevisiae. Lactococcal cells were incubated with antibodies to SspB, and antibody reactivity was detected with FITC-labeled anti-rabbit secondary antibody. (A and C) Light microscopy. (B and D) Fluorescence microscopy. (A and B) L. lactis MG1363 (control) cells nonreactive with antibody. (C and D) L. lactis expressing SspB. (E) Western blots of lyticase-extracted fungal cell wall proteins reacted with monoclonal antibody to Als3p and antibody binding detected with HRP-linked anti-mouse secondary antibody. Lane 1, S. cerevisiae wild type; lane 2, S. cerevisiae expressing Als3p; lane 3, C. albicans NGY152 wild type; lane 4, C. albicans als3Δ/als3Δ.
In coaggregation assays of L. lactis MG1363 wild-type cells with C. albicans WT, low levels of interaction were observed, with 45% of hyphae showing no bound lactococci and 52% with a 1+ level of coaggregation (Fig. 4). However, with L. lactis expressing SspB, ∼50% of wild-type hyphae showed 2+ coaggregation levels, significantly more than with L. lactis WT (P < 0.05), and ∼25% showed 1+ binding (Fig. 4), thus confirming a direct role for SspB in hyphal-filament recognition. On the other hand, cells of L. lactis expressing SspB were deficient in binding to hyphae formed by the C. albicans ΔALS3 mutant. Less than 5% of ΔALS3 mutant hyphae avidly bound L. lactis expressing SspB, and 60% of hyphae showed no binding (P < 0.05) (Fig. 4). There were lower levels of coaggregation between C. albicans ΔALS3 and L. lactis WT than between C. albicans WT and L. lactis WT (Fig. 4). This suggests that Als3p might also weakly recognize an irrelevant lactococcal surface component.
FIG. 4.
Coaggregation of L. lactis expressing SspB with C. albicans hyphae. (B to I) FITC-labeled L. lactis MG1363 (Ll control) or L. lactis MG1363(pUB1000-sspB) (LlSspB) was incubated in suspension with hypha-forming cells of C. albicans NGY152 (CaWT) or the 1843 als3Δ/als3Δ mutant (CaΔALS3). Aggregates were visualized by light or fluorescence microscopy. (A) Percentages of hyphae with attached bacteria were calculated on the basis of bacterial binding levels, as described in the legend to Fig. 2. The data are means ± standard errors (SE) from 4 independent experiments (*, P < 0.05).
SspB interacts with Als3p.
The previous results suggested that S. gordonii SspB might interact directly with C. albicans Als3p. Therefore, we further characterized the interactions by utilizing S. cerevisiae expressing heterologous Als3p. Expression of Als3p in this host was confirmed following immunoblot analysis of proteins released from cell walls following lyticase treatment. Blots of cell wall protein extracts were probed with monoclonal antibody to Als3p (9), and this detected a band with an approximate molecular mass of 105 kDa (Fig. 3E). A band of ∼120 kDa was present in extracts from C. albicans wild-type hyphal cell walls, corresponding well to the predicted mass of ∼124 kDa (Fig. 3E). No band was observed on immunoblots of cell wall extracts obtained from the C. albicans als3Δ/als3Δ mutant (Fig. 3E). Als3p is predicted to be heavily glycosylated (23), and thus, the differences in observed masses between S. cerevisiae recombinant Als3p and C. albicans Als3p might result from differential glycosylation.
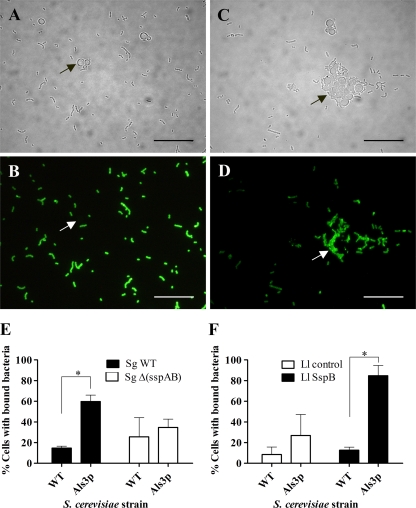
Recombinant S. cerevisiae cells expressing Als3p were found to bind S. gordonii DL1 wild-type cells avidly, with 60% of S. cerevisiae cells binding S. gordonii compared with <10% of S. cerevisiae WT cells (Fig. 5 E). Deletion of the sspA and sspB genes in S. gordonii UB1360 led to significantly reduced numbers of S. gordonii cells attaching to S. cerevisiae ALS3+ (Fig. 5E). We then determined if L. lactis expressing SspB interacted with S. cerevisiae. L. lactis cells expressing SspB bound only weakly to wild-type S. cerevisiae cells (Fig. 5A, B, and F). However, SspB-expressing cells of L. lactis interacted strongly with ∼85% of S. cerevisiae cells expressing Als3p (Fig. 5C, D, and F). Wild-type L. lactis interacted with only ∼25% of S. cerevisiae cells expressing Als3p (Fig. 5F). Taken collectively, these data show that S. gordonii SspB interacts directly with C. albicans Als3p, thus providing a molecular mechanism for mediating binding of S. gordonii cells to C. albicans hyphal filaments.
FIG. 5.
Coaggregation between bacterial strains and S. cerevisiae expressing Als3p. FITC-labeled streptococci or lactococci were incubated with S. cerevisiae S150-2B parent cells or with cells constitutively expressing Als3p. Coaggregates were visualized by light or fluorescence microscopy. (A and B) S. cerevisiae S150-2B control cells mixed with L. lactis expressing SspB. (C and D) S. cerevisiae S150-2B expressing Als3p mixed with L. lactis expressing SspB, showing coaggregation. The arrows indicate yeast cells with or without adhered bacteria. (E) Percentages of S. cerevisiae parent (WT) or Als3p-expressing cells interacting with S. gordonii DL1 (Sg WT) or S. gordonii Δ(sspA sspB) [Sg Δ(sspAB)]. (F) Percentages of S. cerevisiae parent (WT) or Als3p-expressing cells interacting with L. lactis MG1363 control cells (Ll control) or L. lactis MG1363 expressing SspB (Ll SspB). The data are means and SD from 2 independent experiments (*, P < 0.05).
DISCUSSION
Attachment of microorganisms to oral or dental surfaces, and the subsequent formation of biofilms, may lead to persistent oral colonization by C. albicans. These biofilms may subsequently act as reservoirs of C. albicans cells that could become dispersed and lead to systemic disease (51). Within the oral cavity, attachment to a surface is important for microorganisms to avoid being cleared by salivary flow (30, 43). It is essential to understand the complex physical and chemical interactions occurring between C. albicans and the host, and other microorganisms present in biofilm communities. Mature C. albicans biofilms comprise a mixture of yeast cells, pseudohyphae, and hyphal filaments. The last form a network of interlocked branches that is thought to give the biofilm structure and rigidity. The current notion is that C. albicans surface proteins, some of which are hypha specific (7), are necessary for initiation of biofilm formation and for interhyphal communication within biofilms (24). Thus, when genes encoding the hyphal-wall protein Hwp1 and the related adhesins Hwp2 and Rbt1 were deleted, biofilm formation was inhibited (14, 39, 40). The surface protein adhesin Eap1, which is not hypha specific, also contributes to biofilm formation (35). The hypha-specific surface protein Als3p is involved in hyphal aggregation, and als3Δ/als3Δ null mutants formed very weak biofilms after 48 h in a catheter model (56).
In this study, we have shown that als3Δ/als3Δ mutant cells were able to attach to a saliva-coated surface in a manner similar to that of C. albicans WT parent cells and formed very early-stage biofilms up to 3 h. This result might have been anticipated, because initial attachment of yeast cells must involve adhesins other than Als3p, which is a hypha-specific protein. At 3 h, short hyphal filaments of WT and ΔALS3 strains could be seen associated with the saliva-coated surface, but by 6 h, the als3Δ/als3Δ mutant cells forming hyphae had become detached from the surface. This suggests that the initial adherence interactions were not sufficient to sustain biofilm formation or that the mediators of initial attachment were downregulated upon hyphal formation and Als3p expression. Interestingly, hyphal development by the ΔALS3 strain continued to occur in the planktonic phase, confirmed by growth and development measurements (Fig. 1D), but these hypha-producing cells did not subsequently become attached to the substratum. This suggests a critical role for Als3p in hyphal-filament adherence to the substratum. Recent studies (40) have shown that an als1Δ/als1Δ als3Δ/als3Δ double mutant, or an hwp1Δ/hwp1Δ mutant, was unable to form biofilms individually. However, when the two mutants were coincubated, biofilms were formed that were as thick as those formed by the WT strain (40). These observations indicate that there may be complementary adhesin functions between Hwp1p, Als1p, and Als3p.
A majority of studies investigating C. albicans adherence and biofilm formation have been concerned with monospecies cultivation. However, within the natural environment of the oral cavity, hundreds of different species of bacteria are present that potentially influence colonization by C. albicans. For example, S. gordonii enhances biofilm formation when it is in association with C. albicans and promotes hyphal-filament production (1). It is suggested that close proximity of the two microorganisms may aid exchange and sensing of diffusible signals. Since Als3p is reported to bind a range of molecules (24), we investigated whether a complementary adhesin mechanism, as described above, might operate between the als3Δ/als3Δ mutant cells and S. gordonii. However, in dual-species biofilm formation with S. gordonii, the ΔALS3 strain forming hyphae became detached as before (Fig. 1B), suggesting that Als3p might be a protein receptor for binding of S. gordonii to hyphae.
Previous studies investigating the physical interactions between S. gordonii and C. albicans have focused mainly upon the streptococcal adhesins (21, 22). These studies have suggested that the antigen I/II family polypeptides SspA and SspB and the surface fibrillar polypeptide CshA (36) were involved in adherence of C. albicans to immobilized streptococcal cells. In coaggregation assays, S. gordonii wild-type cells demonstrated a preference for binding hyphal filaments, suggesting that a hypha-specific component may act as a receptor for S. gordonii. Since there was little or no coaggregation between S. gordonii and the ΔALS3 mutant, it seemed likely that a major receptor was Als3p (Fig. 2G). An alternative possibility was that deletion of ALS3 affected the expression or presentation of other hyphal surface molecules. We therefore utilized an S. cerevisiae strain expressing Als3p in coaggregation assays with S. gordonii to avoid these potential issues and corroborate the current findings. This clearly showed that S. gordonii interacted with Als3p, complementing previous studies by Klotz et al. suggesting that Als5p bound S. gordonii (32).
To identify the S. gordonii factor recognizing Als3p, we utilized an L. lactis strain expressing SspB on the cell surface, based on evidence from mutagenesis studies that the SspB and SspA proteins were involved in C. albicans recognition (22). The SspB polypeptide has been shown to interact with P. gingivalis and to promote biofilm formation by this oral anaerobic and pathogenic bacterium (11). The SspB protein also mediates coaggregation of S. gordonii with A. naeslundii to form early dental plaque biofilms (27). The SspB-expressing strain of L. lactis, but not control L. lactis, mediated strong coaggregation with S. cerevisiae expressing Als3p. These results identify the first molecular mechanism for binding of a Gram-positive bacterium to C. albicans. We have not yet investigated the potential role of the CshA fibrillar protein or SspA in interaction with Als3p. Since deletion of the sspA and sspB genes in S. gordonii did not ablate coaggregation with C. albicans wild-type hyphae or S. cerevisiae expressing Als3p, it seems likely that other surface components of S. gordonii may also interact with Als3p. The possibility that alternative streptococcal adhesins are involved in currently under investigation.
A feature of S. gordonii coaggregation with C. albicans was that, in addition to adhering to hyphal filaments, the bacteria adhered to each other. Since the streptococcal strain utilized here does not self-aggregate, these observations suggest that upon attachment to hyphae, the bacteria acquired the ability to recruit additional streptococcal cells. One explanation might be that adherence of streptococci to hyphae led to upregulation or unmasking of cell surface factors that promoted streptococcal cell-cell aggregation. Another possibility is that a diffusible signal from C. albicans led to cell surface expression of streptococcal self-aggregation factors. These effects may be relevant to the development of biofilm architecture, promoting the formation of societies of streptococci that can be seen among the C. albicans hyphae (1). Discrete groupings or societies of microorganisms are frequently observed in dual-species biofilms (34), and these could be initiated and sustained by self-aggregation mechanisms.
In summary, we have investigated the physical interactions occurring between C. albicans and S. gordonii and demonstrated that the hypha-specific protein Als3p is a major receptor for streptococcal attachment. In addition, we provide evidence that the S. gordonii SspB protein interacts directly with Als3p, though this should be confirmed by further investigation utilizing protein-protein interaction technologies. The region of Als3p that is recognized by S. gordonii is not known but is currently under study. It has recently been suggested that a tridecapeptide structure present within the N termini of Als1p, Als3p, and Als5p has amyloid-forming potential in catalyzing the formation of parallel β-sheets in Als protein-protein interactions (44). This may, in part, account for the aggregative properties of Als3p that presumably contribute to the frequently reported clumping of hyphae. Interestingly, the SspB polypeptide sequence also contains three potential β-aggregation sequences, predicted by the TANGO algorithm (15). One of these (742 to 746 amino acids [aa]) is within the central V region of the protein and, on the basis of the three-dimensional (3D) crystal structure (4), is predicted to be at the opening of the receptor-binding pocket (16). It remains to be determined if these self-propagating sequences are involved in interactions of Als3p with SspB.
Acknowledgments
We thank Lois Hoyer for supplying monoclonal antibody and for helpful advice. We also thank Scott Filler for the provision of S. cerevisiae strains, Caroline Bamford and Angela Nobbs for helpful discussions, and Jane Brittan and Lindsay Dutton for excellent technical assistance.
This work was supported by NIH (NIDCR) grant R01-DE016690 awarded to H.F.J. and M.M.V.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 30 August 2010.
REFERENCES
- 1.Bamford, C. V., A. d'Mello, A. H. Nobbs, L. C. Dutton, M. M. Vickerman, and H. F. Jenkinson. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77:3696-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S. N., T. G. Emori, D. H. Culver, R. P. Gaynes, W. R. Jarvis, T. Horan, J. R. Edwards, J. Tolson, T. Henderson, W. J. Martone, and the National Nosocomial Infections Surveillance System. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. Am. J. Med. 91:S86-S89. [DOI] [PubMed] [Google Scholar]
- 3.Blot, S. I., and K. H. Vandewoude. 2003. Estimating attributable mortality of candidemia: clinical judgement vs matched cohort studies. Eur. J. Clin. Microbiol. Infect. Dis. 22:132-133. [DOI] [PubMed] [Google Scholar]
- 4.Brady, L. J., S. E. Maddocks, M. R. Larson, N. Forsgren, K. Persson, C. C. Deivanayagam, and H. F. Jenkinson. 2010. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77:276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady, L. J., D. A. Piacentini, P. J. Crowley, P. C. Oyston, and A. S. Bleiweis. 1992. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 60:1008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, A., D. M. MacCallum, A. J. Brown, N. A. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaffin, W. L. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, D. A., S. H. Oh, X. Zhao, H. Zhao, J. T. Hutchins, J. H. Vernachio, J. M. Patti, and L. L. Hoyer. 2009. Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J. Microbiol. Methods 78:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Concia, E., M. A. Anna, and M. Conti. 2009. Epidemiology, incidence and risk factors for invasive candidiasis in high-risk patients. Drugs 69:5-14. [DOI] [PubMed] [Google Scholar]
- 11.Daep, C. A., D. M. James, R. J. Lamont, and D. R. Demuth. 2006. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect. Immun. 74:5756-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demuth, D. R., E. E. Golub, and D. Malamud. 1990. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J. Biol. Chem. 265:7120-7126. [PubMed] [Google Scholar]
- 13.Eggimann, P., J. Garbino, and D. Pittet. 2003. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3:685-702. [DOI] [PubMed] [Google Scholar]
- 14.Ene, I. V., and R. J. Bennett. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot. Cell 8:1909-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Escamilla, A. M., F. Rousseau, J. Schymkowitz, and L. Serrano. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22:1302-1306. [DOI] [PubMed] [Google Scholar]
- 16.Forsgren, N., R. J. Lamont, and K. Persson. 2009. Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci. 18:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 18.Grimaudo, N., W. E. Nesbitt, and W. B. Clark. 1996. Coaggregation of Candida albicans with Actinomyces species. Oral Microbiol. Immunol. 11:59-61. [DOI] [PubMed] [Google Scholar]
- 19.Heddle, C., A. H. Nobbs, N. S. Jakubovics, M. Gal, J. P. Mansell, D. Dymock, and H. F. Jenkinson. 2003. Host collagen signal induces antigen I/II adhesin and invasin gene expression in oral Streptococcus gordonii. Mol. Microbiol. 50:597-607. [DOI] [PubMed] [Google Scholar]
- 20.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, A. R., R. McNab, and H. F. Jenkinson. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, A. R., C. Gilbert, J. M. Wells, and H. F. Jenkinson. 1998. Binding properties of Streptococcus gordonii SspA and SspB (Antigen I/II Family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer, L. L., T. L. Payne, M. Bell, A. M. Myers, and S. Scherer. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451-459. [DOI] [PubMed] [Google Scholar]
- 24.Hoyer, L. L., C. B. Green, S. H. Oh, and X. Zhao. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family: a sticky pursuit. Med. Mycol. 46:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubovics, N. S., S. R. Gill, M. M. Vickerman, and P. E. Kolenbrander. 2008. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol. Ecol. 66:637-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakubovics, N. S., S. W. Kerrigan, A. H. Nobbs, N. Strömberg, C. J. van Dolleweerd, D. M. Cox, C. G. Kelly, and H. F. Jenkinson. 2005. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 73:6629-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakubovics, N. S., N. Strömberg, C. J. van Dolleweerd, C. G. Kelly, and H. F. Jenkinson. 2005. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55:1591-1605. [DOI] [PubMed] [Google Scholar]
- 28.Jenkinson, H. F., and L. J. Douglas. 2002. Interactions between Candida species and bacteria in mixed infections. p. 357-373. In K. A. Brogden and J. M. Guthmiller (ed.), Polymicrobial disease. ASM Press, Washington DC. [PubMed]
- 29.Jenkinson, H. F., H. C. Lala, and M. G. Shepherd. 1990. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkinson, H. F., and R. J. Lamont. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589-595. [DOI] [PubMed] [Google Scholar]
- 31.Jenkinson, H. F. 1994. Cell surface protein receptors in oral streptococci. FEMS Microbiol. Lett. 121:133-140. [DOI] [PubMed] [Google Scholar]
- 32.Klotz, S. A., N. K. Gaur, R. De Armond, D. Sheppard, N. Khardori, J. E. Edwards, P. N. Lipke, and M. El-Azizi. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med. Mycol. 45:363-370. [DOI] [PubMed] [Google Scholar]
- 33.Kolenbrander, P. E., N. Ganeshkumar, F. J. Cassels, and C. V. Hughes. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 7:406-413. [DOI] [PubMed] [Google Scholar]
- 34.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, F., M. J. Svarovsky, A. J. Karlsson, J. P. Wagner, K. Marchillo, P. Oshel, D. Andes, and S. P. Palecek. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNab, R., H. Forbes, P. S. Handley, D. M. Loach, G. W. Tannock, and H. F. Jenkinson. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 38.Nobbs, A. H., R. J. Lamont, and H. F. Jenkinson. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobile, C. J., J. E. Nett, D. R. Andes, and A. P. Mitchell. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobile, C. J., H. A. Schneider, J. E. Nett, D. C. Sheppard, S. G. Filler, D. R. Andes, and A. P. Mitchell. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 42.Odds, F. C. 1988. Candida and candidosis. Bailliere Tindall, London, United Kingdom.
- 43.O'Sullivan, J. M., H. F. Jenkinson, and R. D. Cannon. 2000. Adhesion of Candida albicans to oral streptococci is promoted by selective adsorption of salivary proteins to the streptococcal cell surface. Microbiology 146:41-48. [DOI] [PubMed] [Google Scholar]
- 44.Otoo, H. N., K. G. Lee, W. Qiu, and P. N. Lipke. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 46.Pfaller, M. A., L. Boyken, S. A. Messer, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2005. Comparison of results of voriconazole disk diffusion testing for Candida species with results from a central reference laboratory in the Artemis global antifungal surveillance program. J. Clin. Microbiol. 43:5208-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sánchez-Vargas, L. O., N. G. Ortiz-López, M. Villar, M. D. Moragues, J. M. Aguirre, M. Cashat-Cruz, J. L. Lopez-Ribot, L. A. Gaitán-Cepeda, and G. Quindós. 2005. Oral Candida isolates colonizing or infecting human immunodeficiency virus-infected and healthy persons in Mexico. J. Clin. Microbiol. 43:4159-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sciotti, M. A., I. Yamodo, J. P. Klein, and J. A. Ogier. 1997. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol. Lett. 153:439-445. [DOI] [PubMed] [Google Scholar]
- 50.Sheppard, D. C., M. R. Yeaman, W. H. Welch, Q. T. Phan, Y. Fu, A. S. Ibrahim, S. G. Filler, M. Zhang, A. J. Waring, and J. E. Edwards, Jr. 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480-30489. [DOI] [PubMed] [Google Scholar]
- 51.Uppuluri, P., A. K. Chaturvedi, A. Srinivasan, M. Banerjee, A. K. Ramasubramaniam, J. R. Köhler, D. Kadosh, and J. L. Lopez-Ribot. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viudes, A., J. Pemán, E. Cantón, P. Úbeda, J. Lopez-Ribot, and M. Gobernado. 2002. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J. Clin. Microbiol. Infect. Dis. 21:767-774. [DOI] [PubMed] [Google Scholar]
- 53.Whiteway, M., and C. Bachewich. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wisplinghoff, H., T. Bischoff, S. Tallent, H. Seifert, R. Wenzel, and M. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, X., S. H. Oh, G. Cheng, C. B. Green, J. A. Nuessen, K. Yeater, R. P. Leng, A. J. Brown, and L. L. Hoyer. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415-2428. [DOI] [PubMed] [Google Scholar]
- 56.Zhao, X., K. J. Daniels, S. H. Oh, C. B. Green, K. M. Yeater, D. R. Soll, and L. L. Hoyer. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zijnge, V., M. B. van Leeuwen, J. E. Degener, F. Abbas, T. Thurnheer, R. Gmür, and H. J. Harmsen. 2010. Oral biofilm architecture on natural teeth. PLoS One. 5:e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]