Abstract
Although Plasmodium falciparum apical membrane antigen 1 (AMA1) is a leading malaria vaccine candidate, extensive allelic diversity may compromise its vaccine potential. We have previously shown that naturally acquired antibodies to AMA1 were associated with protection from clinical malaria in this Kenyan population. To assess the impact of allelic diversity on naturally acquired immunity, we first sequenced the ectodomain-encoding region of P. falciparum ama1 from subjects with asymptomatic, mild, and severe malaria and measured allele frequency distributions. We then measured antibodies to three allelic AMA1 proteins (AMA1_3D7, AMA1_FVO, and AMA1_HB3) and used competition enzyme-linked immunosorbent assays (ELISAs) to analyze allele-specific antibodies. Seventy-eight unique haplotypes were identified from 129 alleles sampled. No clustering of allelic haplotypes with disease severity or year of sampling was observed. Differences in nucleotide frequencies in clinical (severe plus mild malaria) versus asymptomatic infections were observed at 16 polymorphic positions. Allele frequency distributions were indicative of balancing selection, with the strongest signature being identified in domain III (Tajima's D = 2.51; P < 0.05). Antibody reactivities to each of the three allelic AMA1 proteins were highly correlated (P < 0.001 for all pairwise comparisons). Although antibodies to conserved epitopes were abundant, 48% of selected children with anti-AMA1 IgG (n = 106) had detectable reactivity to allele-specific epitopes as determined by a competition ELISA. Antibodies to both conserved and allele-specific epitopes in AMA1 may contribute to clinical protection.
Many candidate antigens for subunit malaria vaccines are polymorphic in natural populations, posing challenges for vaccine development. It is important to know how many alleles of a particular candidate will need to be included in a vaccine to induce antibodies with specificity broad enough to recognize the existing antigenic diversity. Populations of Plasmodium falciparum in areas where the disease is highly endemic have high recombination rates (13, 37, 41) and can generate additional haplotypic diversity with every meiotic recombination (54). This is exemplified by apical membrane antigen 1 (AMA1), for which numerous distinct haplotypes are observed, particularly in areas with relatively high malaria transmission intensities (15, 20, 44, 45, 51). These haplotypes are comprised of single-nucleotide polymorphisms, which are distributed throughout the single-locus ama1 gene, but are especially numerous in the portion encoding its surface-accessible ectodomain. Independent studies provide strong evidence that balancing selection is acting to maintain these polymorphisms in the population (15, 20, 44, 45), reflecting the importance of AMA1 as a target of protective immunity. These polymorphisms may need to be incorporated into a vaccine based on AMA1. In animal models, immunization confers better protection against challenge with parasites bearing homologous rather than heterologous alleles of AMA1 (16, 29). Likewise, invasion inhibition is more efficient against parasites bearing homologous alleles (21, 27). Recent studies suggested that the allelic diversity in ama1 could be covered by vaccination with a combination of allelic types (27, 30). However, only a few allelic variants can realistically be included in a vaccine formulation, and it remains to be determined how effective this would be in populations where malaria is endemic, where individuals are repeatedly challenged with parasites bearing diverse ama1 alleles. For example, over 200 unique haplotypes of AMA1 were recently reported for a single geographical location in Mali (51).
We have previously shown that naturally acquired antibodies to AMA1 were associated with protection from clinical malaria in a population in coastal Kenya (42). Here we explore the impact of the allelic diversity of ama1 on naturally acquired antibodies in this population. We compare the allelic diversities observed among parasite isolates obtained from children with asymptomatic infections and mild and severe clinical malaria. We test for signatures of balancing selection acting on the ama1 gene in this population, as reported previously for other populations, and describe antibody responses to proteins representing three allelic versions of AMA1 before, during, and after clinical infections.
MATERIALS AND METHODS
Chonyi community cohort.
The Chonyi community cohort, from a rural village in the Kilifi district on the Kenyan coast, was described in detail previously (39). The study community typically experiences two seasonal peaks in malaria transmission (June to August and November to December) and had an average annual entomological inoculation rate (EIR) of approximately 20 to 100 infective bites/person/year around the time of the community sampling for this study (34). The cohort was recruited at the start of a malaria transmission season in October 2000, and details on recruitment, sampling, follow-up, clinical disease definition, and treatment were reported previously (42, 43, 46, 47). The current study focused on children aged 1 to 10 years (n = 289), with approximately 20% of all children falling within each of the following 2-year age group categories: 1 to 2 years, 3 to 4 years, 5 to 6 years, 7 to 8 years, and 9 to 10 years.
Case-control study.
Some details of the case-control study were reported previously (42). Briefly, a cross-sectional survey was conducted at the start of a malaria transmission season in May 1995, in an area of Kilifi with an EIR of approximately 1.5 to 8 bites/person/year (35). Capillary blood samples were collected from 4,783 children under the age of 5 years. Over the following 8 months, children from this survey who presented to Kilifi District Hospital were identified (passive case detection [n = 165]). Eighty-nine had malaria that was severe enough to require admission to the hospital (severe malaria), while the remainder were attended to in the outpatient department (mild malaria [n = 76]). Controls were selected from children who took part in the cross-sectional survey but did not present to the hospital with malaria. For statistical efficiency, each case of either severe or mild malaria was randomly matched to an average of three controls, using a frequency-based matching method that took into account bed net status and location (i.e., the same controls were shared for severe and mild malaria). To ensure that we ended up with adequate numbers of controls when the samples were retrieved, we assumed that 100 children had severe malaria and 100 had mild malaria and selected 300 controls. Serum samples were insufficient for 2 of the 300 controls (thus, 298 controls in total). Antibody assays were performed on pretransmission season sera for all children in the case-control study (n = 463). Ninety-four children (20%) were aged between 1 and 2 years, 124 (27%) were between 2 and 3 years of age, 118 (25%) were between 3 and 4 years of age, and 127 (27%) were 4 to 5 years old. Additional antibody assays were performed with samples collected at the time of presentation to the hospital with an acute clinical episode of malaria (acute sample [n = 165]) and 3 weeks after this episode (convalescence sample [n = 145]). Parasite isolates during the acute clinical episode were frozen at the ring stages as previously described (7) and used to sequence the ectodomain of AMA1 as described below. Ethical approval was granted by the Kenya National Research Ethics Committee.
Parasite DNA extraction, PCR amplification, and DNA sequencing of ama1.
For the Chonyi cohort, parasite DNA was extracted directly from venous blood samples using QIAamp DNA minikits (Qiagen, United Kingdom). For the case-control study, frozen parasite isolates were thawed and cultured for up to 48 h using standard techniques to allow development from the ring to the late-trophozoite or early-schizont stages to bulk up parasite DNA for extraction as described above. The surface-accessible ectodomain of P. falciparum AMA1 comprising a 1,308-bp segment of the gene was amplified by using a nested PCR approach, and sequencing was performed by using three overlapping pairs of primers as previously described (44, 45). Sequencing employed BigDye version 3.1 dye terminator technology with an ABI Prism 3730 capillary DNA sequencer (Applied Biosystems, Warrington, United Kingdom). Forward and reverse sequence traces from each fragment were aligned and assembled into contiguous sequences by using SeqMan II (DNASTAR, Madison, WI). Individual alleles were then aligned by using the CLUSTAL V method within the MEGALIGN program (DNASTAR, Madison, WI). Only samples that contained a single clear allele of the ama1 gene were included in the analysis.
Population genetic analyses.
To detect whether polymorphisms within P. falciparum ama1 were under natural selection, molecular population genetic analyses were performed on 129 full-ectodomain sequences from the Kilifi district by using the DnaSP v4.50 program (48). Analyses were performed on the total sample of alleles and also for the subsets sampled from children with severe, mild, and asymptomatic malaria. Separate analyses were also performed for ama1 alleles sampled in 1995 (case-control study) and those sampled in 2000 (Chonyi cohort). These analyses included (i) a description of the sequence diversity; (ii) analyses of recombination considering the minimum number of recombination events (RM) (26) and recombination parameter C (25); (iii) analyses of linkage disequilibrium (LD) using the indices D′ and R2 (22, 33); and (iv) tests of neutrality, including Tajima's D (50) and Fu and Li's F (19) using the Plasmodium reichenowi ama1 sequence as an outgroup (28). To account for the recombination observed for the ama1 alleles, critical values at the 5% significance level for the tests of neutrality were determined by using coalescent simulations with 1,000 replicates.
Recombinant antigens and ELISAs.
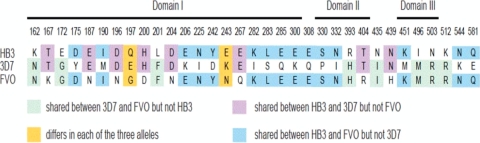
Three AMA1 antigens were expressed in Pichia pastoris and His tagged, as described previously for the FVO allele (29). These antigens were based on the ama1 allelic sequences of the 3D7, FVO, and HB3 cultured lines. The amino acid differences in the surface-exposed ectodomain of AMA1 among the three alleles AMA1_HB3, AMA1_3D7, and AMA1_FVO are shown in Fig. 1. AMA1_HB3 differs from AMA1_3D7 at 23 amino acid residues and from AMA1_FVO at 18 residues, while AMA1_3D7 differs from AMA1_FVO at 23 residues. These alleles were selected since they were representative of the wide genetic diversity observed using the available published P. falciparum ama1 sequences (27).
FIG. 1.
Amino acid differences in the surface-exposed ectodomain of AMA1 between the three alleles AMA1_HB3 (EMBL accession number U33277), AMA1_3D7 (accession number U33274), and AMA1_FVO (accession number AJ277646). AMA1_HB3 differs from AMA1_3D7 at 23 amino acid residues and differs from AMA1_FVO at 18 residues, while AMA1_3D7 differs from AMA1_FVO at 23 residues. Codon positions are numbered according to the numbering reported previously by Hodder et al. (23).
Indirect enzyme-linked immunosorbent assays (ELISAs) were performed by using a previously described protocol (42), and some of the data on the protective efficacy of antibodies to AMA1 (and a larger panel of merozoite antigens) against admission to the hospital with malaria were reported previously (42). Briefly, individual wells of Dynex Immunolon 4HBX ELISA plates (Dynex Technologies Inc.) were coated with 50 ng of antigen per 100 μl of carbonate coating buffer (15 mM Na2CO3, 35 mM NaHCO3 [pH 9.3]). Plates were incubated overnight at 4°C, after which wells were washed four times in PBS-Tween (phosphate-buffered saline-0.05% Tween 20) and blocked for 5 h at room temperature with 1% skimmed milk in PBS-Tween (blocking buffer). Wells were washed again and incubated overnight at 4°C with 100 μl of test sera (1/1,000 dilution in blocking buffer). Plates were then washed four times and incubated for 3 h at room temperature with 100 μl of horseradish peroxidase (HRP)-conjugated rabbit anti-human IgG (Dako Ltd.) at a 1/5,000 dilution in blocking buffer before final washing and detection with H2O2 and O-phenylenediamine (Sigma). The reaction was stopped with 25 μl of 2 M H2SO4 per well, and the absorbance was read at 492 nm. Here, data are presented on samples from all case-control study sera collected (i) at the start of the malaria transmission season (preseason sample [n = 463]), (ii) at the time of presentation to the hospital with either severe or mild malaria (acute sample [n = 165]), and (iii) 3 weeks after the acute clinical episode (convalescence sample [n = 145]).
To determine the specificity of antibodies to allele-specific epitopes within AMA1, competition ELISAs were performed on selected samples (limited by the available amounts of recombinant antigens and volumes of sera) according to an approach previously described (47). All six possible combinations of pairwise competition ELISAs were carried out among the three AMA1 antigens. Briefly, diluted sera were preincubated with an excess (1,000 ng) of competing heterologous antigen for 5 h before their addition to each ELISA plate well coated with 50 ng of homologous antigen and subsequent performance of the indirect ELISA as described above. This ensured that only antibodies to allele-specific epitopes of the homologous antigen were detected, as those to conserved epitopes would have been absorbed in the preincubation step. As a control, an aliquot of the same serum sample was also preincubated with an excess (1,000 ng) of homologous antigen before addition to ELISA plates containing 50 ng of the same antigen. A difference of >0.3 ELISA optical density (OD) units between heterologous and homologous competition assays was counted as significant allele-specific reactivity (43).
Nucleotide sequence accession numbers.
Nucleotide sequences analyzed in this report have been submitted to the EMBL database under accession numbers FN869569 to FN869697.
RESULTS
Sequence diversity.
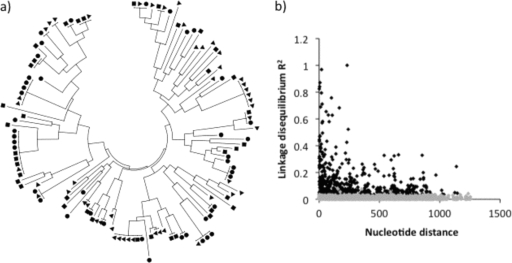
DNA samples from blood containing P. falciparum infections were available from 83 children who were asymptomatically parasitized at the time of sampling in the Chonyi cohort. Of these samples, 34 contained mixed ama1 allele sequences and were excluded from subsequent analyses. In the case-control study, 140 samples were available, of which 60 were excluded (53 due to mixed-allele sequences and 7 with poor-quality sequence traces or DNA that did not amplify). Thus, 129 ama1 alleles were included in the analyses described below. A total of 78 distinct haplotypes (H) were identified, with some of these being shared among children with severe, mild, or asymptomatic malaria. The proportions of distinct haplotypes in samples from each disease category were similar, 32/38 (84%) for severe malaria, 30/42 (71%) for mild malaria, and 36/49 (73%) for asymptomatic parasitemia (haplotype diversities, 0.99, 0.98, and 0.98, respectively). The average pairwise nucleotide diversity per site (π) for the total sample of 129 alleles was 0.016 and was similar across the disease categories. There were 72 polymorphic sites across the entire sequenced region, attributable to a total of 81 mutations (nine of the sites contained 3-nucleotide alleles), and the majority of polymorphic sites were observed in domain I (Table 1). Of the polymorphic sites, 7 were singletons (a minor allele present in only one isolate), whereas 65 had minor alleles that were present in two or more isolates. A dendrogram constructed using the entire sequenced region to visualize the distribution of distinct haplotypes between children with severe, mild, or asymptomatic malaria revealed no clustering among disease categories (Fig. 2 a).
TABLE 1.
Summary of statistics for ama1 and tests for departure from neutralitya
| Region | No. of polymorphic sites | No. of haplotypes | Avg pairwise diversity (n) | Tajima's D (expected upper 95% CI)b | Fu and Li's F (expected upper 95% CI)b |
|---|---|---|---|---|---|
| Total | 72 | 78 | 0.016 | 1.45 (0.72)** | 1.34 (1.00)* |
| Domain I | 42 | 64 | 0.027 | 1.41 (0.95)** | 1.65 (1.35)** |
| Domain II | 11 | 23 | 0.009 | 1.06 (1.34) | 0.80 (1.53) |
| Domain III | 9 | 15 | 0.016 | 2.51 (1.37)** | 1.46 (1.57) |
The total sequenced region included codons 147 to 582, domain I codons 149 to 302, domain II codons 320 to 418, and domain III codons 443 to 509 (23). Ten polymorphic sites were located within the sequenced region but were outside domain I, II, or III.
The expected upper limit of the 95% confidence interval (CI) when Tajima's D and Fu & Li's F were calculated, accounting for recombination (C = 151). Results from coalescent simulations using 1,000 replicates are shown. Asterisks indicate observed values higher than the expected upper limit under neutrality. *, P < 0.05 (95% CI) (as described above); **, P < 0.01 (99% CI) (data not shown).
FIG. 2.
(a) Cluster dendrogram used to visualize 129 AMA1 alleles. There was no evidence of clustering of particular alleles in children presenting with severe malaria (squares), mild malaria (circles), or asymptomatic malaria (triangles). (b) Linkage disequilibrium (LD) calculated by using the R2 index. Black squares indicate significant values, while gray squares indicate nonsignificant values. The LD decreased rapidly with increasing nucleotide distance, indicating a high meiotic recombination rate leading to the generation of many new alleles, as shown in a.
Linkage disequilibrium and recombination.
Linkage disequilibrium was calculated for all nonsingleton polymorphic sites and decreased rapidly with increasing nucleotide distance, indicating a high meiotic recombination rate (Fig. 2b). A minimum of 27 recombination events were predicted to have occurred to give rise to the 78 haplotypes that were observed in the sample of 129 ama1 alleles. The recombination parameter (C) had high values of 0.115 between adjacent nucleotide sites and 151 for the whole sequence.
Tests of neutrality.
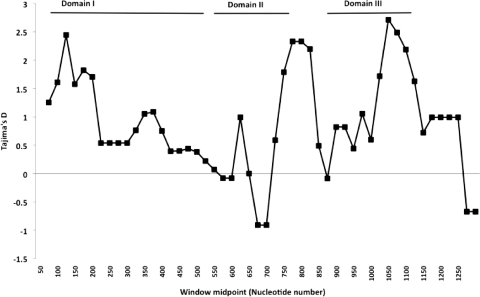
Tajima's D and Fu and Li's F were estimated for the entire sequence and separately for individual domains (Table 1). Positive values for these indices are likely to be due to balancing selection acting to maintain alleles at intermediate frequencies in the population. A sliding-window plot of the observed Tajima's D values across the entire sequenced region is shown for all 129 ama1 alleles in Fig. 3. The most striking result is the strong positive values detected for domain III. A strong signal of selection was also seen in domain I, with positive values of Tajima's D and Fu and Li's F. These tests of neutrality are conservative in the presence of high recombination rates, so coalescent simulations allowing for recombination were used to generate expected estimates of these indices under neutrality, to compare them with observed values. Using the level of recombination estimated above (C = 151), the observed Tajima's D value of 1.452 for the whole sequence was far higher than the upper critical value expected under neutrality (99% confidence limits, −0.94 to 1.04).
FIG. 3.
Sliding-window plot of Tajima's D using a window size of 100 nucleotides and including all 129 ama1 alleles. Positive values generally indicate that balancing selection is acting to maintain alleles at intermediate frequencies.
Polymorphic amino acids.
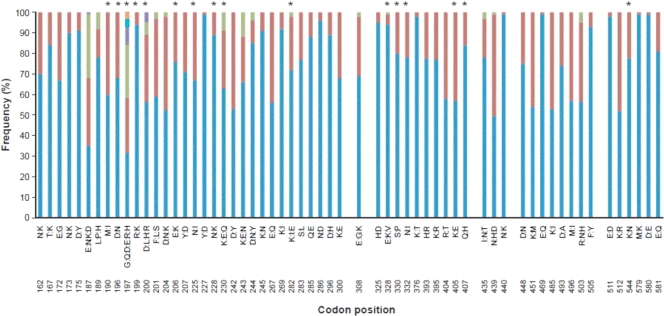
The 72 polymorphic nucleotide sites contained in the sequenced region existed within 62 different codons, 2 of which contained synonymous amino acid replacements. Ten of the remaining 60 codons mapped to sites outside domains I, II, and III. Thus, a total of 50 codon positions (32 in domain I, 10 in domain II, and 8 in domain III) determined the amino acid polymorphisms in domains I, II, and III. The majority encoded two allelic amino acids, while a minority encoded 3 (n = 9), 4 (n = 2), or 6 amino acid residues (codon position 197). The amino acid frequencies at each of these codons are shown in Fig. 4. Particular amino acid residues were observed more commonly for children with asymptomatic infections than for those with clinical malaria (severe and mild malaria), or vice versa, at 16 codon positions (P value of <0.10 by Fisher's exact test) (Table 2). Ten of these codon positions were located in domain I, five were located in domain II, and one was located just outside domain III. After correction for multiple testing by the Bonferroni method (6) (60 comparisons), these differences remained significant only for amino acid residues at codon position 230 (P < 0.001).
FIG. 4.
Amino acid frequencies at each polymorphic site are shown for the entire sequenced region. Blue indicates a common allele, red indicates the second most common allele, and other colors indicate additional rarer alleles at some sites. Asterisks indicate the positions at which the polymorphisms were observed more commonly for children with clinical malaria than for those with asymptomatic infections, or vice versa, at a P value of <0.10 level. After correction for multiple testing by the Bonferroni method (6), this remained significant only for amino acid residues at position 230 (P < 0.001).
TABLE 2.
Amino acid frequencies at particular sites differ in asymptomatic infections compared with clinical episodes
| Codon position | Amino acid | No. of isolates |
P valuea | |
|---|---|---|---|---|
| Asymptomatic (n = 49) | Clinical (n = 80) | |||
| 190 | I | 24 | 27 | 0.098 |
| M | 25 | 53 | ||
| 196 | D | 26 | 62 | 0.006 |
| N | 23 | 18 | ||
| 197 | D | 10 | 23 | 0.096 |
| E | 4 | 7 | ||
| G | 23 | 18 | ||
| H | 1 | 3 | ||
| Q | 9 | 26 | ||
| R | 2 | 3 | ||
| 199 | K | 6 | 2 | 0.053 |
| R | 43 | 78 | ||
| 200 | D | 35 | 38 | 0.045 |
| H | 11 | 31 | ||
| L | 1 | 7 | ||
| R | 2 | 4 | ||
| 206 | E | 42 | 56 | 0.056 |
| K | 7 | 24 | ||
| 225 | I | 10 | 32 | 0.032 |
| N | 39 | 48 | ||
| 228 | K | 10 | 4 | 0.009 |
| N | 39 | 76 | ||
| 230 | E | 24 | 12 | <0.001* |
| K | 24 | 57 | ||
| Q | 1 | 11 | ||
| 282 | E | 1 | 1 | 0.084 |
| I | 8 | 26 | ||
| K | 40 | 53 | ||
| 328 | E | 48 | 74 | 0.050 |
| K | 0 | 6 | ||
| V | 1 | 0 | ||
| 330 | P | 5 | 21 | 0.040 |
| S | 44 | 59 | ||
| 332 | I | 6 | 23 | 0.032 |
| N | 43 | 57 | ||
| 405 | E | 15 | 40 | 0.043 |
| K | 34 | 40 | ||
| 407 | H | 12 | 9 | 0.054 |
| Q | 37 | 71 | ||
| 544 | K | 33 | 67 | 0.049 |
| N | 16 | 13 | ||
P values determined by Fisher's exact test. *, P < 0.001 after correction for multiple testing by the Bonferroni method (60 tests).
Serological data: high correlation between antibodies to three AMA1 alleles.
Serum IgG antibodies to all three recombinant antigens representing allelic forms of AMA1 tested were highly correlated (Table 3). In general, correlations between AMA1_HB3 and AMA1_FVO tended to be marginally higher than those between the other two combinations, consistent with their greater sequence identity (Fig. 1). The correlation coefficients (R2) were notably lower at the acute and convalescence time points than at the preseason time point but were significant at all times (P < 0.001 for each) (Table 3). Antibody levels for each AMA1 antigen increased significantly with age at all time points (P < 0.001 for antibodies to each antigen by a Kruskal-Wallis rank test). In the population of clinical cases as a whole, antibody levels were significantly higher at the time of acute malaria presentation than at preseason time points (P < 0.001 by a Wilcoxon signed-rank test for pairwise comparisons for each antigen). In contrast, antibody levels at convalescence were not significantly different from those sampled at the acute presentation (P > 0.20 by a Wilcoxon signed-rank test for pairwise comparisons for each antigen). The presence of asymptomatic parasitemia was associated with significantly higher levels of antibody to all three allelic versions of AMA1 in the preseason samples (P < 0.001 by a Mann-Whitney test for comparisons for each antigen).
TABLE 3.
Correlation coefficients (R2) between antibody reactivities to AMA1 allelic antigens
| Serum and category |
R2a |
||
|---|---|---|---|
| HB3 vs FVO | HB3 vs 3D7 | 3D7 vs FVO | |
| Pretransmission | |||
| All (n = 463) | 0.96 | 0.95 | 0.92 |
| Severe malaria (n = 89) | 0.93 | 0.86 | 0.83 |
| Mild malaria (n = 76) | 0.95 | 0.94 | 0.91 |
| Acute | |||
| All cases (n = 165) | 0.85 | 0.74 | 0.67 |
| Severe malaria (n = 89) | 0.79 | 0.63 | 0.51 |
| Mild malaria (n = 76) | 0.88 | 0.83 | 0.8 |
| Convalescent | |||
| All cases (n = 165) | 0.87 | 0.76 | 0.73 |
| Severe malaria (n = 89) | 0.85 | 0.62 | 0.56 |
| Mild malaria (n = 76) | 0.88 | 0.87 | 0.85 |
The P value was <0.001 for all comparisons.
Deducing the presence of antibodies to allele-specific epitopes by indirect ELISAs with each antigen separately.
Several definitions were considered for deducing the presence of antibodies to allele-specific epitopes. Counting seropositivity to one allele and seronegativity to the other allele was considered unsuitable because reactivities are commonly close to either side of a cutoff value for positivity. We also considered that small differences in ELISA absorbance readings (<0.3 OD units) cannot be very reliably judged to reflect true differences due to the possibility of assay variability. To be stringent, we therefore chose an arbitrary difference of ≥0.5 ELISA OD units between two allelic versions of AMA1 to define the presence of antibodies to allele-specific epitopes. This definition inevitably excluded samples with low-level ELISA OD readings (<0.5 ELISA OD units) from being counted in this analysis.
Using this definition, among the preseason samples, 47/463 (10%) samples were predicted to contain antibodies to allele-specific epitopes. Of these, 21 had allele-specific antibodies discriminating one pair of AMA1 allelic antigens, 25 discriminated two pairs, and one individual had antibodies discriminating each of the three pairwise combinations of AMA1 allelic types tested (details are provided in Table S1 in the supplemental material). The prevalence of antibodies to allele-specific epitopes did not differ with age (P = 0.53 by Pearson's chi-squared test) or with likelihood of subsequent presentation with clinical malaria (P = 0.69 by Pearson's chi-squared test) but was associated with the presence of asymptomatic parasitemia at the time of sampling (14.2% of 176 children with parasitemia had antibodies to allele-specific epitopes, compared to 7.8% of 287 children who were aparasitemic [P = 0.02 by Pearson's chi-squared test]).
In the acute-phase samples, allele-specific reactivities were detected in 65 (39.3%) of 165 children. Of these 65 children, 25 had antibodies that discriminated one pair of AMA1 alleles, 36 discriminated two pairs, and 4 had antibodies discriminating each of the three pairwise combinations (see Table S1 in the supplemental material). In the convalescence samples, allele-specific reactivities were detected in 47 (32.4%) of 145 children tested. Of these 47 children, 18 had antibodies that discriminated one pair of AMA1 alleles, 28 discriminated two pairs, and 1 had antibodies discriminating each of the three pairwise combinations (Table S1). In the acute and convalescence samples, children with antibodies to allele-specific epitopes tended to be slightly older than those without such antibodies (for acute samples, median of 36.9 versus 32.5 months [P = 0.02]; for convalescence samples, median of 37.2 versus 33.1 months [P = 0.06]).
Antibodies to allele-specific epitopes determined by competition ELISA.
To further investigate antibodies to allele-specific epitopes within AMA1, pairwise competition ELISAs were performed for all six two-way combinations using the three allelic types on a selection of preseason samples. We identified the samples from children who had high antibody levels to all three allelic types of AMA1 tested (n = 141). From these children, we included all children who subsequently presented with clinical episodes of malaria (n = 31; one sample was depleted and thus could not be analyzed) and a randomly selected set of children who did not (n = 76) and compared the prevalences of allele-specific antibodies between these two groups of children.
Overall, 51/106 (48%) of the selected children had detectable antibodies to allele-specific epitopes within AMA1 using the three available allelic antigen types in pairwise competition ELISAs. Twelve children (11%) had antibodies to allele-specific epitopes in one of the six two-way combinations of AMA1 antigens, 24 children (23%) had allele-specific antibodies to two combinations, and 15 children (15%) had allele-specific antibodies to three or more of the six two-way combinations. In this subgroup of children, the prevalences of antibodies to allele-specific epitopes were similar among those who subsequently presented with clinical malaria and those who did not, in all but one pairwise competition ELISA (Table 4).
TABLE 4.
Proportions of children with allele-specific antibodies determined by competition ELISA in preseason samplesa
| Antigen paira | No. (%) of children with antibody |
P value | ||
|---|---|---|---|---|
| Total (n = 106) | Clinical episode during follow-up (n = 30) | No clinical episode during follow-up (n = 76) | ||
| FVO_3D7 | 21 (20) | 14 (18) | 7 (23) | 0.57 |
| FVO_HB3 | 7 (7) | 5 (7) | 2 (7) | 0.99 |
| 3D7_FVO | 23 (22) | 17 (22) | 6 (20) | 0.79 |
| 3D7_HB3 | 19 (18) | 13 (17) | 6 (20) | 0.73 |
| HB3_FVO | 21 (20) | 12 (16) | 9 (30) | 0.10 |
| HB3_3D7 | 29 (27) | 16 (21) | 13 (43) | 0.02* |
The antigen pair consists of coating antigen_competing antigen, which indicates allele-specific antibodies to the coating antigen with respect to the competing antigen. *, P < 0.05 by Pearson's chi-squared test.
DISCUSSION
Although we found a large number of P. falciparum ama1 alleles in this population from coastal Kenya (78 distinct allelic haplotypes out of 129 alleles sampled), an even greater diversity was observed for Nigeria, where 45 haplotypes were identified from 51 alleles sequenced (45). In contrast, a study in Papua New Guinea (PNG) found only 27 haplotypes within domain I in 168 AMA1 alleles sampled (15), considerably fewer than the 64/129 haplotypes observed within domain I in our Kenyan population sample or the 35/51 observed within domain I in the Nigerian population sample (45). In the largest population sample from a single geographical location to date, 214 unique haplotypes of the ectodomain were found in 506 samples from Mali (51). These differences in haplotype diversity are likely to reflect differences in malaria transmission, as has been observed at other polymorphic loci where higher transmission is generally associated with increased diversity (4, 24, 49). This is generally due to the combined effects of a higher effective population size and a higher effective recombination rate (i.e., outcrossing) as well as a possible additional effect of stronger balancing selection in the case of loci encoding antigens such as AMA1.
We found no evidence that any allelic haplotypes were associated with disease severity. However, polymorphisms at 16 codon positions were observed more commonly for children with asymptomatic infections than for those with clinical malaria or vice versa. These differences were significant only at a P value of <0.10, before adjusting for multiple comparisons. Nevertheless, 10 of the 16 codons are located within domain I, and 8 of these 10 codons are located within the binding region of a well-characterized anti-AMA1 monoclonal antibody (MAb) (MAb IF9) that inhibits invasion (9). Mutations at residues 197, 200, 201, 204, 206, 225, and 228 were previously shown to abrogate or reduce the binding of MAb IF9 to AMA1 (10, 11). We found significant differences at a P value of <0.10 between asymptomatic and clinical infections at five of these seven sites. We also found significant differences between asymptomatic and clinical infections at sites that have been shown to make some of the largest molecular interactions with MAb IF9 (principal contact residues 197 and 200), that contribute a smaller surface area to this interaction (residues 190 and 225), or that lie at the periphery of the interface between MAb IF9 and AMA1 (residues 196 and 230) (10). Some of these residues overlap with the antigenic escape residues (AERs) that have been shown to directly mediate antigenic escape from invasion inhibition (18). For example, we found significant differences at a P value of <0.10 at four of five residues that lie within the C1-L loop of domain I (residues 196, 197, 200, and 206) and two of four residues in domain II (residues 330 and 332). Clusters of polymorphisms at both of these sites were shown to mediate antigenic escape (18).
Studies of Toxoplasma gondii revealed that T. gondii AMA1 (TgAMA1) interacts with a rhoptry neck protein (RON4) and two other rhoptry neck proteins (RON2 and RON5) at the tight junction of tachyzoites during invasion (2). Homologues of these rhoptry neck proteins have been identified in P. falciparum (1, 8, 12, 36). Current data suggest that the RON4 complex in P. falciparum binds to the hydrophobic trough within domain I (12). Although the residues in this hydrophobic trough are all conserved, it is surrounded by highly polymorphic residues contained within domains I and II (5, 11). It has been suggested that these surrounding polymorphic residues may serve to “distract” the immune response from a functionally important region of the molecule (5, 12). If the RON4 complex binds to the conserved hydrophobic trough, then the diversity observed in AMA1 would not be of direct consequence but rather would indirectly “protect” this important binding pocket by diverting the immune response to surrounding polymorphic residues. Alternatively, antibodies to nonpolymorphic residues adjacent to the hydrophobic trough could sterically hinder its binding to the rhoptry protein complex, thereby inhibiting invasion. This seems to be the likely mechanism for the potent invasion-inhibitory MAb 4G2, which targets conserved residues within domain II that lie adjacent to the hydrophobic trough (12). In contrast, MAb IF9 has been shown to bind directly to residues within the hydrophobic trough as well as residues from surrounding loops (10). Human antibodies from people exposed to malaria can compete with both MAbs 4G2 and 1F9 for binding to AMA1 (10). Taken together, these data support the view that the hydrophobic trough of AMA1 is a functionally important part of the molecule and that polymorphic residues surrounding this region are selected by protective antibodies.
Our study confirms previous reports from population studies of diversifying selection acting on the ama1 gene (15, 44, 45). In particular, a strong signature of selection was observed for domain III. Previous studies suggested that domain III contains important B-cell epitopes. Mice immunized with a long synthetic peptide from loop I of domain III produced antibodies that were capable of inhibiting parasite growth (38). Human antibodies affinity purified on refolded recombinant domain III inhibited invasion in an allele-specific manner (40). However, these antibodies were isolated from a single donor whose serum was found to have a high titer of antibodies to AMA1 domain III, and this might not be representative of many exposed individuals (40). Previously reported immunoepidemiological studies with different recombinant antigen fragments of AMA1 suggested that antibodies to domain III are relatively rare compared to antibodies to the full ectodomain (14, 47). Experimental studies also indicated that an optimal inhibition of invasion is achieved with antibodies raised against the entire ectodomain, compared to individual subdomains (32). Notably, protective antibodies to AMA1 have been shown to be conformation dependent (3, 16), and many important antibody epitopes may require the contribution of all three domains. Moreover, several T-cell epitopes have been identified in AMA1 (31, 53), raising the possibility that the strong signature of selection observed for domain III could be the result of effective T-cell responses to allele-specific epitopes. We also observed a strong signal of selection in the boundary region between domains II and III. Similar findings were observed for a population sample from Thailand (44), and it is possible that this region is surface exposed in the three-dimensional structure of AMA1.
We found that levels of antibodies to the three allelic recombinant proteins of the AMA1 ectodomain were highly correlated in this cohort of children. This is in keeping with previous studies of naturally acquired (14, 47) and vaccine-induced (17) antibodies to AMA1. Of note, during acute infections and at convalescence, the correlation coefficients between antibodies to the three proteins had declined compared to those at the preseason time point, suggesting that antibodies to allele-specific epitopes had been induced by infection. When we considered differences of >0.5 ELISA OD units to individual AMA1 proteins as indicating fairly high levels of antibodies to allele-specific epitopes, we found the prevalence of such antibodies to be only 10% in the preseason sera, rising to nearly 40% in the acute-phase sera. This compares with data for PNG, where only 7% of sera collected in a cross-sectional survey exhibited different antibody titers against alternative forms of AMA1 (14). While this suggests that antibodies to allele-specific epitopes are not particularly common in the absence of a current infection, it does not exclude the possibility that individuals with similar ELISA OD reactivities to different AMA1 proteins had antibodies to allele-specific as well as shared epitopes. We therefore performed competition ELISAs on a selection of sera that had high antibody titers to all three AMA1 proteins. We found that almost half of these sera contained antibodies to allele-specific epitopes in one or more of the six two-way competition ELISAs.
Overall, our data indicate that naturally acquired antibodies to allele-specific epitopes within AMA1 are frequently detectable but less abundant than those to conserved epitopes. However, our study could not ascertain the relative contributions of antibodies to allele-specific versus conserved epitopes in mediating protection against clinical malaria. In experimental studies, rabbits immunized with a combination of three allelic proteins of AMA1 generated antibodies that were targeted predominantly to conserved or cross-reactive epitopes, and these cross-reactive antibodies were as effective as total anti-AMA1 antibodies (cross-reactive plus allele specific) in growth inhibition assays (30). Conversely, in a recent clinical trial of an AMA1 vaccine in Mali, the only significant protective effect was in reducing the rate of infection by parasites with homologous (rather than heterologous) allelic types, indicating allele-specific immunity and the possible need for a multiallelic formulation (52). These differences notwithstanding, the cumulative body of evidence is in favor of AMA1 as a leading malaria vaccine candidate, and antibodies to both conserved and allele-specific epitopes may contribute to clinical protection.
Supplementary Material
Acknowledgments
This paper was published with the permission of the director of KEMRI.
We thank the parents and children who participated in this study and Brett Lowe (KEMRI-Kilifi).
This work was supported by a research training fellowship awarded to FHAO by the Wellcome Trust, grant 073591.
Editor: J. H. Adams
Footnotes
Published ahead of print on 23 August 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Alexander, D. L., S. Arastu-Kapur, J. F. Dubremetz, and J. C. Boothroyd. 2006. Plasmodium falciparum AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in Toxoplasma gondii. Eukaryot. Cell 5:1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. L., J. Mital, G. E. Ward, P. Bradley, and J. C. Boothroyd. 2005. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 4.Babiker, H. A., J. Lines, W. G. Hill, and D. Walliker. 1997. Population structure of Plasmodium falciparum in villages with different malaria endemicity in east Africa. Am. J. Trop. Med. Hyg. 56:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Bai, T., M. Becker, A. Gupta, P. Strike, V. J. Murphy, R. F. Anders, and A. H. Batchelor. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. U. S. A. 102:12736-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bland, J. M., and D. G. Altman. 1995. Multiple significance tests: the Bonferroni method. BMJ 310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull, P. C., B. S. Lowe, M. Kortok, C. S. Molyneux, C. I. Newbold, and K. Marsh. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat. Med. 4:358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J., O. Kaneko, A. Thongkukiatkul, M. Tachibana, H. Otsuki, Q. Gao, T. Tsuboi, and M. Torii. 2009. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol. Int. 58:29-35. [DOI] [PubMed] [Google Scholar]
- 9.Coley, A. M., N. V. Campanale, J. L. Casey, A. N. Hodder, P. E. Crewther, R. F. Anders, L. M. Tilley, and M. Foley. 2001. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng. 14:691-698. [DOI] [PubMed] [Google Scholar]
- 10.Coley, A. M., A. Gupta, V. J. Murphy, T. Bai, H. Kim, R. F. Anders, M. Foley, and A. H. Batchelor. 2007. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 3:1308-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coley, A. M., K. Parisi, R. Masciantonio, J. Hoeck, J. L. Casey, V. J. Murphy, K. S. Harris, A. H. Batchelor, R. F. Anders, and M. Foley. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 74:2628-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, C. R., C. Withers-Martinez, F. Hackett, and M. J. Blackman. 2009. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog. 5:e1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway, D. J., C. Roper, A. M. Oduola, D. E. Arnot, P. G. Kremsner, M. P. Grobusch, C. F. Curtis, and B. M. Greenwood. 1999. High recombination rate in natural populations of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 96:4506-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortes, A., M. Mellombo, R. Masciantonio, V. J. Murphy, J. C. Reeder, and R. F. Anders. 2005. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 73:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortes, A., M. Mellombo, I. Mueller, A. Benet, J. C. Reeder, and R. F. Anders. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dicko, A., D. J. Diemert, I. Sagara, M. Sogoba, M. B. Niambele, M. H. Assadou, O. Guindo, B. Kamate, M. Baby, M. Sissoko, E. M. Malkin, M. P. Fay, M. A. Thera, K. Miura, A. Dolo, D. A. Diallo, G. E. Mullen, C. A. Long, A. Saul, O. Doumbo, and L. H. Miller. 2007. Impact of a Plasmodium falciparum AMA1 vaccine on antibody responses in adult Malians. PLoS One 2:e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta, S., S. Y. Lee, A. H. Batchelor, and D. E. Lanar. 2007. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. U. S. A. 104:12488-12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, Y. X., and W. H. Li. 1993. Statistical tests of neutrality of mutations. Genetics 133:693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg, S., M. T. Alam, M. K. Das, V. Dev, A. Kumar, A. P. Dash, and Y. D. Sharma. 2007. Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations. Malar. J. 6:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healer, J., V. Murphy, A. N. Hodder, R. Masciantonio, A. W. Gemmill, R. F. Anders, A. F. Cowman, and A. Batchelor. 2004. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol. Microbiol. 52:159-168. [DOI] [PubMed] [Google Scholar]
- 22.Hill, W. G., and A. Robertson. 1968. The effects of inbreeding at loci with heterozygote advantage. Genetics 60:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, E. H., L. A. da Silveira, R. Tonhosolo, F. J. Pereira, W. L. Ribeiro, A. P. Tonon, F. Kawamoto, and M. U. Ferreira. 2001. Geographical patterns of allelic diversity in the Plasmodium falciparum malaria-vaccine candidate, merozoite surface protein-2. Ann. Trop. Med. Parasitol. 95:117-132. [DOI] [PubMed] [Google Scholar]
- 25.Hudson, R. R. 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50:245-250. [DOI] [PubMed] [Google Scholar]
- 26.Hudson, R. R., and N. L. Kaplan. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 29.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusi, K. A., B. W. Faber, A. W. Thomas, and E. J. Remarque. 2009. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One 4:e8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lal, A. A., M. A. Hughes, D. A. Oliveira, C. Nelson, P. B. Bloland, A. J. Oloo, W. E. Hawley, A. W. Hightower, B. L. Nahlen, and V. Udhayakumar. 1996. Identification of T-cell determinants in natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in an adult population exposed to malaria. Infect. Immun. 64:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalitha, P. V., L. A. Ware, A. Barbosa, S. Dutta, J. K. Moch, J. D. Haynes, B. B. Fileta, C. E. White, and D. E. Lanar. 2004. Production of the subdomains of the Plasmodium falciparum apical membrane antigen 1 ectodomain and analysis of the immune response. Infect. Immun. 72:4464-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewontin, R. C. 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49:49-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mbogo, C. M., J. M. Mwangangi, J. Nzovu, W. Gu, G. Yan, J. T. Gunter, C. Swalm, J. Keating, J. L. Regens, J. I. Shililu, J. I. Githure, and J. C. Beier. 2003. Spatial and temporal heterogeneity of Anopheles mosquitoes and Plasmodium falciparum transmission along the Kenyan coast. Am. J. Trop. Med. Hyg. 68:734-742. [PubMed] [Google Scholar]
- 35.Mbogo, C. N., R. W. Snow, E. W. Kabiru, J. H. Ouma, J. I. Githure, K. Marsh, and J. C. Beier. 1993. Low-level Plasmodium falciparum transmission and the incidence of severe malaria infections on the Kenyan coast. Am. J. Trop. Med. Hyg. 49:245-253. [DOI] [PubMed] [Google Scholar]
- 36.Morahan, B. J., G. B. Sallmann, R. Huestis, V. Dubljevic, and K. L. Waller. 2009. Plasmodium falciparum: genetic and immunogenic characterisation of the rhoptry neck protein PfRON4. Exp. Parasitol. 122:280-288. [DOI] [PubMed] [Google Scholar]
- 37.Mu, J., P. Awadalla, J. Duan, K. M. McGee, D. A. Joy, G. A. McVean, and X. Z. Su. 2005. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 3:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller, M. S., A. Renard, F. Boato, D. Vogel, M. Naegeli, R. Zurbriggen, J. A. Robinson, and G. Pluschke. 2003. Induction of parasite growth-inhibitory antibodies by a virosomal formulation of a peptidomimetic of loop I from domain III of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 71:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mwangi, T. W., A. Ross, R. W. Snow, and K. Marsh. 2005. Case definitions of clinical malaria under different transmission conditions in Kilifi District, Kenya. J. Infect. Dis. 191:1932-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nair, M., M. G. Hinds, A. M. Coley, A. N. Hodder, M. Foley, R. F. Anders, and R. S. Norton. 2002. Structure of domain III of the blood-stage malaria vaccine candidate, Plasmodium falciparum apical membrane antigen 1 (AMA1). J. Mol. Biol. 322:741-753. [DOI] [PubMed] [Google Scholar]
- 41.Neafsey, D. E., S. F. Schaffner, S. K. Volkman, D. Park, P. Montgomery, D. A. Milner, Jr., A. Lukens, D. Rosen, R. Daniels, N. Houde, J. F. Cortese, E. Tyndall, C. Gates, N. Stange-Thomann, O. Sarr, D. Ndiaye, O. Ndir, S. Mboup, M. U. Ferreira, S. L. Moraes, A. P. Dash, C. E. Chitnis, R. C. Wiegand, D. L. Hartl, B. W. Birren, E. S. Lander, P. C. Sabeti, and D. F. Wirth. 2008. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 9:R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osier, F. H., G. Fegan, S. D. Polley, L. Murungi, F. Verra, K. K. Tetteh, B. Lowe, T. Mwangi, P. C. Bull, A. W. Thomas, D. R. Cavanagh, J. S. McBride, D. E. Lanar, M. J. Mackinnon, D. J. Conway, and K. Marsh. 2008. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect. Immun. 76:2240-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osier, F. H., S. D. Polley, T. Mwangi, B. Lowe, D. J. Conway, and K. Marsh. 2007. Naturally acquired antibodies to polymorphic and conserved epitopes of Plasmodium falciparum merozoite surface protein 3. Parasite Immunol. 29:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polley, S. D., W. Chokejindachai, and D. J. Conway. 2003. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics 165:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polley, S. D., D. J. Conway, D. R. Cavanagh, J. S. McBride, B. S. Lowe, T. N. Williams, T. W. Mwangi, and K. Marsh. 2006. High levels of serum antibodies to merozoite surface protein 2 of Plasmodium falciparum are associated with reduced risk of clinical malaria in coastal Kenya. Vaccine 24:4233-4246. [DOI] [PubMed] [Google Scholar]
- 47.Polley, S. D., T. Mwangi, C. H. Kocken, A. W. Thomas, S. Dutta, D. E. Lanar, E. Remarque, A. Ross, T. N. Williams, G. Mwambingu, B. Lowe, D. J. Conway, and K. Marsh. 2004. Human antibodies to recombinant protein constructs of Plasmodium falciparum apical membrane antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 23:718-728. [DOI] [PubMed] [Google Scholar]
- 48.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 49.Schoepflin, S., F. Valsangiacomo, E. Lin, B. Kiniboro, I. Mueller, and I. Felger. 2009. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar. J. 8:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takala, S. L., D. Coulibaly, M. A. Thera, A. H. Batchelor, M. P. Cummings, A. A. Escalante, A. Ouattara, K. Traore, A. Niangaly, A. A. Djimde, O. K. Doumbo, and C. V. Plowe. 2009. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci. Transl. Med. 1:2ra5. [DOI] [PMC free article] [PubMed]
- 52.Thera, M. A., O. K. Doumbo, B. Coulibaly, M. B. Laurens, A. Kone, A. Guindo, D. Diallo, T. K. B. Kouriba, I. Diarra, A. Dolo, A. Niangaly, M. Daou, M. Sissoko, D. Traore, K. E. Lyke, S. Takala, O. Godeaux, J. Thonnard, J. Cohen, D. E. Lanar, C. Diggs, L. Soisson, D. G. Heppner, and C. V. Plowe. 2009. Randomized, controlled, phase 2B clinical trial to evaluate the safety, immunogenicity and efficacy of WRAIR's AMA-1 malaria vaccine (FMP2.1) adjuvanted in GSK Biologicals' AS02A vs. rabies vaccine in 1-6 year old children in Bandiagara, Mali. Abstr. Am. Soc. Trop. Med. Hyg. 58th Annu. Meet., Washington, DC.
- 53.Udhayakumar, V., S. Kariuki, M. Kolczack, M. Girma, J. M. Roberts, A. J. Oloo, B. L. Nahlen, and A. A. Lal. 2001. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am. J. Trop. Med. Hyg. 65:100-107. [DOI] [PubMed] [Google Scholar]
- 54.Walliker, D., I. A. Quakyi, T. E. Wellems, T. F. McCutchan, A. Szarfman, W. T. London, L. M. Corcoran, T. R. Burkot, and R. Carter. 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236:1661-1666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.