Abstract
Hantaviruses, members of the Bunyaviridae family, are emerging category A pathogens that initiate the translation of their capped mRNAs by a novel mechanism mediated by viral nucleocapsid protein (N). N specifically binds to the mRNA 5′ m7G cap and 40S ribosomal subunit, a complex of 18S rRNA and multiple ribosomal proteins. Here, we show that N specifically interacts with the ribosomal protein S19 (RPS19), located at the head region of the 40S subunit. We suggest that this N-RPS19 interaction facilitates ribosome loading on capped mRNAs during N-mediated translation initiation.
Hantaviruses, members of the Bunyaviridae family, are emerging category A pathogens that cause hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS) in humans, with mortality rates of 12% and 60%, respectively (1, 6, 12, 26-28). The hantaviral genome is composed of three negative-sense genomic RNA segments, S, M, and L, that encode nucleocapsid protein (N), glycoprotein precursor (GPC), and RNA-dependent RNA polymerase (RdRp), respectively (7, 8, 29). Glycoprotein precursor is cleaved into two glycoproteins, Gn and Gc. Multiple studies have suggested that N is involved in diverse viral functions, including the encapsidation and packaging of viral RNA (10, 13, 18, 20, 23, 30-33), apoptosis (22), and replication of the viral genome (2, 3, 5, 9, 11, 23, 25). We have recently shown that N binds to the 5′ caps of cellular mRNAs and protects their degradation in cellular P bodies (14). The rescued 5′-capped oligoribonucleotides of host cell mRNAs are stored by N in P bodies that are later used as primers by viral RdRp during transcription initiation. This suggests that N has a role in cap snatching (14). We have also reported that N augments the translation of capped mRNAs (15) and preferentially favors the translation of viral transcripts due to high-affinity binding of N with the viral mRNA 5′ untranslated region (UTR) (19). N-mediated translation is initiated by the specific interaction between N and the 40S ribosomal subunit and the subsequent loading of 40S subunits onto the 5′ mRNA terminus. Since the 40S ribosomal subunit is a massive ribonucleoprotein complex composed of 18S rRNA and 32 ribosomal proteins (34), it was evident that we needed to identify the exact component of the 40S subunit that specifically binds to N.
We purified the 40S subunit from HeLa cells as previously reported (15). Sin Nombre hantavirus (SNV) nucleocapsid protein was expressed in Escherichia coli and purified as a C-terminal His-tagged fusion protein, as previously described (14-18, 20, 21). To determine whether N interacts with 18S rRNA or protein components of the 40S subunit or both, immunoprecipitation experiments were undertaken. Since the 40S subunit is a very stable protein-RNA complex, pulling down N, either by an immunoprecipitation assay using an anti-N antibody or by a His tag using Ni-nitrilotriacetic acid (NTA) beads, copurified the whole 40S subunit, thereby creating hurdles in the identification of the exact component of the 40S subunit that specifically interacts with N. Conversely, immunoprecipitation of the 40S subunit with an antibody against any 40S ribosomal protein resulted in copurification of N. Therefore, neither approach permitted the identification of the exact component of the 40S subunit that specifically interacts with N.
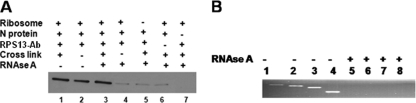
As an alternative approach, 15 μg of purified 40S ribosomal subunits were mixed with 0.5 μg of purified N in HEPES buffer, pH 7.4, and incubated with 0.5% formaldehyde at room temperature for 5 min to allow the protein-protein and protein-RNA cross-linking to occur. The cross-linking reaction was terminated by the addition of glycine to a final concentration of 250 mM, followed by incubation at room temperature for 5 min. Half of the cross-linked mixture was immunoprecipitated with an antibody against ribosomal protein S13 (RPS13). Immunoprecipitated material was examined by Western blotting, using anti-N antibody. As expected, the immunoprecipitated material contained the copurified N (Fig. 1A, lane 1). Similar results were obtained without prior cross-linking of N-40S subunit complexes (Fig. 1A, lane 2). In a control experiment, the remaining half of cross-linked N-40S subunit mixture was digested with RNase A (100 μg/ml) at room temperature for 4 h to degrade the 18S rRNA prior to immunoprecipitation with the RPS13 antibody. Real-time PCR analysis using four sets of primers targeted to different regions of 18S rRNA confirmed the complete degradation of 18S rRNA (Fig. 1B). It was expected that immunoprecipitation with the RPS13 antibody would not copurify N if it selectively bound to 18S rRNA and not to ribosomal proteins. However, it is clear from Fig. 1A (lane 3) that immunoprecipitation with RPS13 antibody copurified N, suggesting that N interacts with ribosomal proteins. When the same experiment was performed without cross-linking (Fig. 1A, lane 4), immunoprecipitation with RPS13 antibody did not copurify N due to the loss of structural integrity of the 40S subunit after RNase A treatment. Taken together, these analyses clearly demonstrate that N interacts with ribosomal proteins, although the possibility that N bound to both ribosomal proteins and 18S rRNA cannot be excluded.
FIG. 1.
(A) Purified 40S subunits were incubated with bacterially expressed and purified N. The ribosome-N mixture was cross-linked with formaldehyde and immunoprecipitated with anti-RPS13 antibody (Ab). Immunoprecipitated material was examined by Western blotting using anti-N antibody (lane 1). The same experiment was performed without cross-linking (lane 2). The experiment performed for lane 3 was same as that for lane 1, except the reaction mixture was treated with RNase A prior to immunoprecipitation. The experiment performed for lane 4 was same as that for lane 3, except that N-40S subunit complexes were not cross-linked. Control immunoprecipitation experiments lacking the ribosome (lane 5) and the anti-RPS13 antibody (lane 6) show the nonspecific binding of N to the beads. However, lane 7 does not show such nonspecific background, due to the lack of N protein in the reaction mixture. (B) To confirm that 18S rRNA was completely digested with RNase A treatment in the samples used in panel A, total RNA was purified from both RNase A-treated and untreated samples and reverse transcribed using random primers. The cDNA was used as the template, and a PCR analysis was performed using four sets of primers targeted to different regions of 18S rRNA. The PCR products generated using primer pairs 5′-ATGCTTGTCTCAAAGATTAAG-3′ and 5′-CGAAAGAGTCCTGTATTGTTA-3′ (lanes 1 and 5), 5′-AGGCCCTGTAATTGGAATGAG-3′ and 5′-TGCGCCGGTCCAAGAATTTCA-3′ (lanes 2 and 6), 5′-AGACGGACCAGAGCGAAAGCA-3′ and 5′-AGCATGCCAGAGTCTCGTTCG-3′ (lanes 3 and 7), and 5′-AACTAGTTACGCGACCCCCGA-3′ and 5′-TAATGATCCTTCCGCAGGTTC-3′ (lanes 4 and 8) are shown.
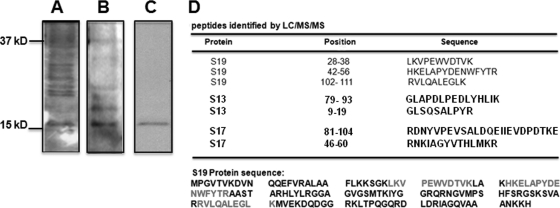
Since the 40S ribosomal subunit is a complex of 32 ribosomal proteins, we used far-Western blot analysis to identify the exact protein that selectively interacts with N. Purified 40S ribosomal subunits were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane, followed by incubation with phosphate-buffered saline (PBS) containing 330 nM purified N for 1 h. Nonspecific background was decreased by a gradual increase in NaCl concentration. Membrane was incubated with anti-N antibody, and bands were detected with alkaline phosphatase-conjugated secondary antibody. Under low-stringency conditions (140 mM NaCl), N interacted nonspecifically with multiple ribosomal proteins (Fig. 2A). At 200 mM NaCl, there was a slight decrease in nonspecific binding of N with ribosomal proteins (Fig. 2B). However, when the NaCl concentration was raised to 500 mM, nonspecific background was significantly reduced, and a single band was detected on the PVDF membrane (Fig. 2C). This band was precisely excised from the PVDF membrane and digested with trypsin (4). The resulting peptides were extracted and separated on a microcapillary C18 column coupled to the nanospray ionization source of a Deca XP Plus ion trap mass spectrometer (MS; Thermo Finnigan). Full MS as well as MS/MS spectra were obtained, and the BioWorks 3.2 program based on the SEQUEST algorithm was used for data analysis. This liquid chromatography (LC)/MS/MS analysis identified peptides that matched three ribosomal proteins, RPS19, RPS17, and RPS13 (Fig. 2D), all structural constituents of the 40S subunit.
FIG. 2.
Far-Western blot analysis showing the interaction of N with ribosomal proteins from the 40S subunit. To reduce the nonspecific background, the experiment was carried out at increasing salt concentrations. Representative Western blots at three NaCl concentrations, 140 mM (A), 200 mM (B), and 500 mM (C), are shown. The single band from the PVDF membrane, shown in panel C, was excised and analyzed by mass spectrometry. The LC/MS/MS study detected peptides matching three ribosomal proteins, RPS13, RPS17, and RPS19 (D). The exact locations of the peptides in RPS19 are shown at the bottom.
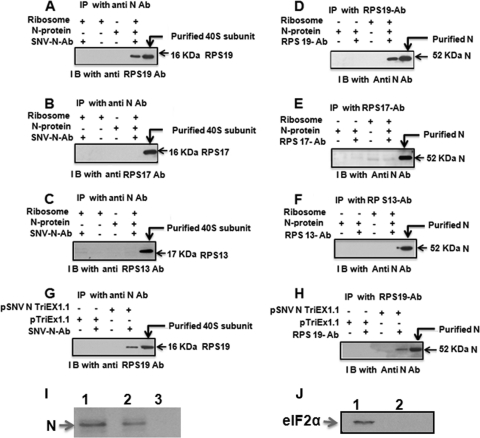
We used coimmunoprecipitation analysis to determine which of the three target proteins specifically interacts with N. Since 18S rRNA bridges the interaction between multiple ribosomal proteins in the 40S subunit, we incubated the purified 40S subunit with RNase A to completely degrade the 18S rRNA and dissociate the structural integrity of the 40S subunit. The mixture of ribosomal proteins obtained after the degradation of 18S rRNA was incubated with purified N and immunoprecipitated with an anti-N antibody. Immunoprecipitated material was examined by Western blot analysis, using an antibody against RPS13, RPS17, or RPS19. This analysis showed that N interacts with RPS19 and not with RPS13 or RPS17 (Fig. 3A to C). Conversely, we carried out immunoprecipitation with an antibody against RPS13, RPS17, or RPS19 and asked whether N copurifies with immunoprecipitated material. This analysis again demonstrated that N interacts with RPS19 and not with RPS13 or RPS17.
FIG. 3.
Purified 40S ribosomal subunits were treated with RNase A and incubated with bacterially expressed and purified N. The reaction mixture was immunoprecipitated (IP) with anti-N antibody and examined by anti-RPS19 antibody (A), anti-RPS17 antibody (B), or anti-RPS13 antibody (C). Conversely, immunoprecipitation was carried out with anti-RPS19 antibody (D), anti-RPS17 antibody, (E) or anti-RPS13 antibody (F) followed by Western immunoblot (I B) analysis with anti-N antibody. Cell lysate from HeLa cells transfected with N expression plasmid (pSNV N TriEx1.1) or empty vector (pTriEx1.1) was treated with RNase A to degrade the total RNA, including 18S rRNA. After RNase A treatment, cell lysate was immunoprecipitated with anti-N antibody, and the immunoprecipitated material was examined by Western blot analysis using anti-RSP19 antibody (G). Conversely, immunoprecipitation was carried out with anti-RPS19 antibody, followed by Western blot analysis using anti-N antibody (H). In panel I, the 40S ribosomal subunit was purified from HeLa cells expressing N. The purified 40S subunit was run on SDS-PAGE and transferred to a PVDF membrane. Western blot analysis was carried out using anti-N antibody. Bacterially expressed and purified N was used as the positive control (lane 1). Purified 40S subunits from HeLa cells transfected with N expression plasmid (lane 2) or transfected with the empty vector (lane 3) clearly show that N remains associated with the 40S subunit. Purified 40S subunits from panel I were examined for the presence of eIF2α by Western blotting using anti-eIF2α antibody (J). Cell lysate from HeLa cells expressing N was used as the positive control (lane 1). Purified 40S subunits prepared from HeLa cells expressing N (lane 2) clearly show that eIF2α is not copurified along with 40S subunits.
These in vitro studies were followed by in vivo studies of cells to confirm the N-RPS19 interaction. HeLa cells were transfected with an N expression plasmid and lysed 48 h posttransfection. The cell lysate was incubated with RNase A to degrade the total RNA, including 18S rRNA, and disrupt the 40S subunit. Degradation of 18S rRNA was confirmed by reverse transcription-PCR (RT-PCR) analysis (not shown). After RNase A treatment, the cell lysate was immunoprecipitated with anti-N antibody, and the immunoprecipitated material was examined by Western blotting, using an anti-RSP19 antibody. As shown in Fig. 3G, RPS19 copurified with immunoprecipitated N protein. Conversely, immunoprecipitation of the RNase-treated HeLa cell lysate with an anti-RPS19 antibody copurified N (Fig. 3H). This in vivo study further confirms the specific interaction between N and RPS19. A similar in vivo analysis performed with RPS13 and RPS17 showed that N did not interact with these two proteins in vivo (not shown), consistent with the in vitro data.
To determine the relative affinity of N with 40S subunits, we purified 40S subunits from HeLa cells expressing N. Purified 40S subunits were boiled in SDS loading buffer, and ribosomal proteins were separated by SDS-PAGE and transferred to a PVDF membrane. Western blot analysis with an anti-N antibody showed the presence of N in the purified 40S subunit sample, suggesting that N remains tightly associated with the 40S subunit and copurifies with it (Fig. 3I). As expected, a similar Western blot analysis showed that purified 40S subunits lack initiation factor eIF2 (Fig. 3J), consistent with the removal of all initiation factors by the protocol used for the purification of the 40S subunit (24). This observation suggests that the interaction of N with RPS19 is stronger than the interaction of many eukaryotic initiation factors with the 40S subunit. Since N is highly expressed in comparison to viral RdRp and glycoproteins Gn and Gc in virus-infected cells, it is likely that high-affinity N-RPS19 interaction reserves a population of 40S subunits in the cell cytoplasm that remains dedicated for the translation of hantaviral mRNAs. It would be interesting to examine whether virus-directed ribosome selection that favors viral multiplication exists in virus-infected cells.
Since N is an RNA binding protein, its interaction with 18S rRNA cannot be excluded. It is possible that simultaneous binding of N with RPS19 and 18S rRNA stabilizes the interaction of N with the 40S subunit. It is likely that the N-RPS19 interaction mediates the loading of the 40S ribosomal subunit on capped mRNA during N-mediated translation initiation. On the basis of the crystal structure of RPS19 from Pyrococcus abyssi and cryo-electron microscopy (cryo-EM) mapping, the location of RPS19 has been mapped to the head region of the 40S subunit (34), suggesting that N is recruited to the head region of the 40S subunit. However, the formation of the translation-competent 80S ribosome requires the loading of the 60S subunit at the head region of the 40S subunit. Further studies are required to determine if N dissociates from the 40S subunit during translation initiation or elongation. It is probable that hantaviruses have evolved with this unique translation initiation strategy to lure the host cell translation machinery to preferentially translate their mRNAs.
Acknowledgments
This work was supported by research grant 5R21AI083672-02 from the NIH and grant P20RR016443 from the NCRR COBRE program.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakqori, G., G. Kochs, O. Haller, and F. Weber. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J. Gen. Virol. 84:1207-1214. [DOI] [PubMed] [Google Scholar]
- 3.Bridgen, A., and R. M. Elliott. 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc. Natl. Acad. Sci. U. S. A. 93:15400-15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez, J., and S. M. Mische. 2001. Enzymatic digestion of proteins on PVDF membranes. Curr. Protoc. Protein Sci. Chapter 11:Unit 11.2. [DOI] [PubMed] [Google Scholar]
- 5.Flick, K., J. W. Hooper, C. S. Schmaljohn, R. F. Pettersson, H. Feldmann, and R. Flick. 2003. Rescue of Hantaan virus minigenomes. Virology 306:219-224. [DOI] [PubMed] [Google Scholar]
- 6.Glass, G. E., T. L. Yates, J. B. Fine, T. M. Shields, J. B. Kendall, A. G. Hope, C. A. Parmenter, C. J. Peters, T. G. Ksiazek, C. S. Li, J. A. Patz, and J. N. Mills. 2002. Satellite imagery characterizes local animal reservoir populations of Sin Nombre virus in the southwestern United States. Proc. Natl. Acad. Sci. U. S. A. 99:16817-16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hepojoki, J., T. Strandin, A. Vaheri, and H. Lankinen. 2010. Interactions and oligomerization of hantavirus glycoproteins. J. Virol. 84:227-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huiskonen, J. T., J. Hepojoki, P. Laurinmaki, A. Vaheri, H. Lankinen, S. J. Butcher, and K. Grunewald. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84:4889-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikegami, T., C. J. Peters, and S. Makino. 2005. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson, C. B., J. Gallegos, P. Ferro, W. Severson, X. Xu, C. S. Schmaljohn, and P. Fero. 2001. Purification and characterization of the Sin Nombre virus nucleocapsid protein expressed in Escherichia coli. Protein Expr. Purif. 23:134-141. [DOI] [PubMed] [Google Scholar]
- 11.Kohl, A., T. J. Hart, C. Noonan, E. Royall, L. O. Roberts, and R. M. Elliott. 2004. A bunyamwera virus minireplicon system in mosquito cells. J. Virol. 78:5679-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeDuc, J. W., J. E. Childs, and G. E. Glass. 1992. The hantaviruses, etiologic agents of hemorrhagic fever with renal syndrome: a possible cause of hypertension and chronic renal disease in the United States. Annu. Rev. Public Health 13:79-98. [DOI] [PubMed] [Google Scholar]
- 13.Mir, M. A., B. Brown, B. Hjelle, W. A. Duran, and A. T. Panganiban. 2006. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J. Virol. 80:11283-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mir, M. A., W. A. Duran, B. L. Hjelle, C. Ye, and A. T. Panganiban. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U. S. A. 105:19294-19299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mir, M. A., and A. T. Panganiban. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J. 27:3129-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mir, M. A., and A. T. Panganiban. 2006. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 80:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mir, M. A., and A. T. Panganiban. 2006. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA 12:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir, M. A., and A. T. Panganiban. 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 79:1824-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mir, M. A., and A. T. Panganiban. 2010. The triplet repeats of the Sin Nombre hantavirus 5′ untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation. J. Virol. 84:8937-8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir, M. A., and A. T. Panganiban. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78:8281-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mir, M. A., S. Sheema, A. Haseeb, and A. Haque. 2010. Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J. Biol. Chem. 285:11357-11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ontiveros, S. J., Q. Li, and C. B. Jonsson. 2010. Modulation of apoptosis and immune signaling pathways by the Hantaan virus nucleocapsid protein. Virology 401:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborne, J. C., and R. M. Elliott. 2000. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946-9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinschewer, D. D., M. Perez, and J. C. de la Torre. 2003. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 77:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmaljohn, C. M. 1996. Molecular biology of hantaviruses. Plenum Press, New York, NY.
- 28.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Schmaljohn, C. S., and C. B. Jonsson. 2001. Replication of hantaviruses, p. 15-32. In S. A. Nichol (ed.), Hantaviruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 30.Schmaljohn, C. S., M. D. Parker, W. H. Ennis, J. M. Dalrymple, M. S. Collett, J. A. Suzich, and A. L. Schmaljohn. 1989. Baculovirus expression of the M genome segment of Rift Valley fever virus and examination of antigenic and immunogenic properties of the expressed proteins. Virology 170:184-192. [DOI] [PubMed] [Google Scholar]
- 31.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 32.Severson, W., X. Xu, M. Kuhn, N. Senutovitch, M. Thokala, F. Ferron, S. Longhi, B. Canard, and C. B. Jonsson. 2005. Essential amino acids of the hantaan virus N protein in its interaction with RNA. J. Virol. 79:10032-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, D. J., B. Devkota, A. D. Huang, M. Topf, E. Narayanan, A. Sali, S. C. Harvey, and J. Frank. 2009. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17:1591-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]