Abstract
Pseudohyphal growth and meiosis are two differentiation responses to nitrogen starvation of diploid Saccharomyces cerevisiae. Nitrogen starvation in the presence of fermentable carbon sources is thought to induce pseudohyphal growth, whereas nitrogen and sugar starvation induces meiosis. In contrast to the genetic background routinely used to study pseudohyphal growth (Σ1278b), nonfermentable carbon sources stimulate pseudohyphal growth in the efficiently sporulating strain SK1. Pseudohyphal SK1 cells can exit pseudohyphal growth to complete meiosis. Two stimulators of meiosis, Ime1 and Ime2, are required for pseudohyphal growth of SK1 cells in the presence of nonfermentable carbon sources. Epistasis analysis suggests that Ime1 and Ime2 act in the same order in pseudohyphal growth as in meiosis. The different behaviors of strains SK1 and Σ1278b are in part attributable to differences in cyclic AMP (cAMP) signaling. In contrast to Σ1278b cells, hyperactivation of cAMP signaling using constitutively active Ras2G19V inhibited pseudohyphal growth in SK1 cells. Our data identify the SK1 genetic background as an alternative genetic background for the study of pseudohyphal growth and suggest an overlap between signaling pathways controlling pseudohyphal growth and meiosis. Based on these findings, we propose to include exit from pseudohyphal growth and entry into meiosis in the life cycle of S. cerevisiae.
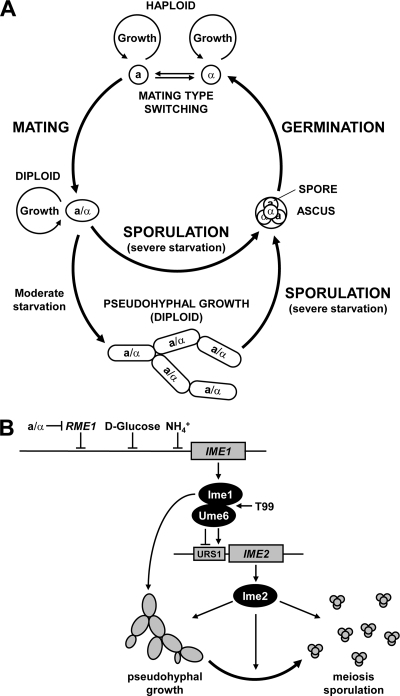
Diploid cells of the budding yeast Saccharomyces cerevisiae choose between two developmental responses to nitrogen starvation, namely, pseudohyphal growth and meiosis (30). Pseudohyphal growth allows sessile S. cerevisiae cells to forage for nutrients and is a growth form distinct from the vegetative, yeast-like growth form. Pseudohyphal growth is induced by nitrogen starvation in the presence of fermentable carbon sources (30). Pseudohyphal growth is characterized by an elongated cell shape, adhesion of cells to each other after cell division has been completed, a switch from a bipolar to a unipolar budding pattern, and prolongation of the G2 phase of the cell cycle to allow daughter cells to grow to the size of their mothers (45). This increased cell size allows newly born pseudohyphal daughters to immediately enter the cell cycle and to bud in synchrony with their mothers. Starvation for nitrogen and fermentable carbon sources is a prerequisite for induction of meiosis. In meiosis (also called sporulation), single diploid cells form an ascus containing four haploid, stress- and starvation-resistant spores. Meiosis is temporally divided into at least early, middle, and late phases of gene expression (16, 74). After premeiotic DNA replication, cells go through two meiotic divisions (meiosis I and II), initiation of prospore wall growth at sites near the spindle pole bodies (SPBs), nuclear division, and maturation of the spore walls to form mature asci (47).
The protein kinase A (PKA) pathway and the mating and filamentation mitogen-activated protein kinase (MAPK) pathway control pseudohyphal growth. Both pathways stimulate pseudohyphal growth by stimulating expression of the cell surface flocculin Flo11 (51). The PKA pathway is activated by the glucose sensor Gpr1 (55, 102) and by the high-affinity ammonium permease Mep2 (7, 53, 83). Gpa2 activates adenylate cyclase, which in turn activates PKA (46, 54). In S. cerevisiae, three catalytic subunits of PKA, Tpk1, Tpk2, and Tpk3, regulate pseudohyphal growth. Tpk2 directly interacts with and inhibits the transcriptional repressor of FLO11, Sfl1 (77). Phosphorylation of the transcription factor Flo8 by Tpk2 stimulates binding of Flo8 to the FLO11 promoter and activation of FLO11 (71, 72). In contrast to TPK2, deletion of TPK1 or TPK3 enhances pseudohyphal growth (71, 77), suggesting that Tpk1 and Tpk3 are inhibitors of pseudohyphal growth. Substrates for Tpk1 or Tpk3 involved in repression of pseudohyphal growth have not been identified. The mating and filamentation MAPK cascade (56), consisting of the MAPK kinase kinase Ste11, the MAPK kinase Ste7, the MAPK Kss1, and the scaffold Ste5, is regulated by the cell surface mucin Msb2 (20). Msb2 recruits general signaling proteins, such as Sho1, the isoprenylated, plasma membrane-tethered protein Cdc42, and its p21-activated kinase, Ste20, to the filamentation MAPK cascade (20). The MAPK pathway controls the activity of the heterodimeric transcription factor Ste12-Tec1 (29, 82), which activates expression of FLO11 and regulates cell elongation.
Entry into meiosis is governed by a transcriptional cascade controlling expression of early meiotic genes (EMGs) (47). Starvation induces expression of IME1 (85). Ime1 carries a transcriptional activation domain (98), which activates transcription of EMGs (99), including IME2, after binding of Ime1 to the DNA-binding protein Ume6 (11, 79). A two-hybrid interaction between Ume6 and Ime1 is stimulated by several protein kinases, including the glycogen synthase kinase 3β homologs Rim11, Mck1, and Mrk1 and the protein kinase Rim15 (57, 58, 106, 110). Binding of Ime1 to Ume6 induces degradation of Ume6 (59). The protein kinase Ime2 is required for full expression of EMGs (99), stimulates its own expression through an upstream activating site (96), and promotes meiotic DNA replication by directly phosphorylating Rfa2 (17, 18). Sic1 phosphorylation by Ime2 triggers its proteasomal destruction and entry into meiotic S phase (21, 89). IME2 is also required for expression of middle meiotic genes (4, 64, 69) and for reestablishment of repression of EMGs in the middle meiotic phase (47). Nutrient-rich conditions repress transcription of IME1. The PKA pathway represses expression of IME1 in the presence of glucose (60, 62) and inhibits phosphorylation of Ime1 by Rim11 (80). Cells expressing constitutively active Ras2G19V, Gpa2R273A, or Gpa2G132V (22, 105, 111) or deleted for the regulatory subunit of PKA, BCY1, do not sporulate (12).
Using an efficiently sporulating strain, SK1 (42), we report that elements of the early meiotic cascade, such as Ime1, binding of Ime1 to Ume6, and Ime2, are required for pseudohyphal growth of SK1 cells. In contrast to the genetic background routinely used to study pseudohyphal growth, i.e., strain Σ1278b (30), nonfermentable carbon sources stimulate pseudohyphal growth of SK1 cells independent of their utilization in respiration. Pseudohyphal SK1 cells can complete meiosis. Differences in cyclic AMP (cAMP) signaling may explain, in part, the different behaviors of these two strains. Whereas constitutively active Ras2G19V stimulates pseudohyphal growth of Σ1278b cells, it inhibits pseudohyphal growth of SK1 cells. Our work establishes the SK1 genetic background as a tool for the study of mechanisms controlling the life choice decision between pseudohyphal growth and sporulation of dimorphic yeasts and filamentous fungi.
MATERIALS AND METHODS
Plasmid constructions.
To obtain plasmid pRS316-T99N-UME6-lexA, the ∼4.4-kbp SpeI/HindIII fragment of pKB193 (11) was cloned into SpeI- and HindIII-digested pRS316 (97). To cure the T99N mutation in UME6, the 263-bp BamHI/NheI fragment of pRS316-T99N-UME6-lexA was replaced with a similar fragment from pCITE-4a(+)-HA-UME6 (M. Schröder and R. J. Kaufman, unpublished data) encoding wild-type (WT) Ume6. pMW2 (108) (CEN URA3 RAS2G19V) was used to express constitutively active Ras2. The plasmids used for this study are listed in Table 1.
TABLE 1.
Plasmids used for this study
| Plasmid | Features | Reference |
|---|---|---|
| pCITE-4a(+)-HA-UME6 | UME6-His6bla | Schröder and Kaufman, unpublished data |
| pHS103 | 2μm URA3 IME1 bla | 99 |
| pHS105 | 2μm URA3 IME2 bla | 99 |
| pIL30 | FG(TyA)lacZ::URA3 | 66 |
| pKB193 | 2μm URA3 T99N-UME6-lexA bla | 11 |
| pMW2 | CEN URA3 RAS2G19V bla | 108 |
| pRS316 | CEN6 ARSH4 URA3 bla | 97 |
| pRS316-UME6-lexA | CEN6 ARSH4 URA3 UME6-lexA bla | This study |
| pRS316-T99N-UME6-lexA | CEN6 ARSH4 URA3 T99N-UME6-lexA bla | This study |
Yeast methods.
Yeast strains (Table 2) were transformed by the LiOAc method (15). IME1 and IME2 were deleted by PCR-based gene deletion (32, 107), using the oligodeoxynucleotides listed in Table 3. Mating type was determined by PCR (38). Respiration-deficient ρ0 cells were generated by treatment with 20 μg/ml ethidium bromide as described previously (24). Growth was monitored as described before (91). Pseudohyphal growth was assayed on synthetic low-ammonium (SLA) medium (91) plates containing a 2% (wt/vol) concentration of the indicated carbon source and, as required, amino acids or uracil to complement auxotrophies, at 5 to 10 mg/liter for SK1 cells. Ethanol was used in sealed containers containing a 2% (wt/vol) ethanol reservoir. WT and mutant strains were matched for auxotrophic mutations. In experiments in which mutant strains carried WT metabolic genes, the corresponding amino acids or uracil was added at the same concentration to plates for both the WT and mutant strains. Uracil was provided at 5 mg/liter in SLA plates for Σ1278b cells (30, 54, 55). Cells were streaked onto SLA plates to obtain single colonies. Pseudohyphal growth and agar invasion were scored after growth at 30°C for the times indicated in the figure legends. Pilot experiments revealed no difference in pseudohyphal growth on plates supplemented with 2.5 to 10 mg/liter of the amino acids or uracil required to complement auxotrophies. l-Lysine was included in all plates because the ho::LYS2 allele produces a weak Lys+ phenotype. cAMP (Calbiochem, Merck, Darmstadt, Germany) was used at 5 mM. To document the filamentation phenotype, pictures from at least four representative colonies were taken under bright-field illumination at a magnification of ×40 to ×100 with an inverted microscope (Inverso; Fisher Scientific), an eyepiece camera (Globecam D; Fisher Scientific), and imaging software (Image Driving software; Fisher Scientific). Pictures from asci were taken at a magnification of ×400. Time-lapse video microscopy was performed on the same microscope, using AMCap software. Plates were incubated at room temperature for time-lapse video microscopy. Wet tissues were placed into plates, and plates were sealed with Parafilm to minimize evaporation. Agar invasion was determined after washing away cells above the agar by gently scraping plates with a spreader under running distilled water for ∼1 min. The remaining cells were photographed at a magnification of ×40 to ×100. Haploid invasive growth was assayed after growth for 3 days at 30°C on yeast extract-peptone-dextrose (YPD) plates. Plates were photographed before and after washing the cells from the agar surface to document total and invasive growth, respectively. Activity of the FG(TyA)::lacZ reporter was measured and standardized to total cellular protein activity as described before (66, 91). Cell length and width were measured using the straight tool in ImageJ. Bud and birth scars were stained as described before (25, 75). Briefly, cells were grown for 18 h on SLA acetate plates and washed off plates. A total of 1 × 104 cells were resuspended in 25 μl of 1-mg/ml calcofluor white M2R (dissolved in water) and 50 μl of 1-mg/ml fluorescein isothiocyanate-wheat germ agglutinin (FITC-WGA) dissolved in phosphate-buffered saline (PBS; 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 2.7 mM KCl, and 137 mM NaCl [pH 7.2]), incubated for 15 min at room temperature, and washed three times with PBS. Cells were visualized using a Nikon Eclipse TE 300 microscope and a 60× A/1.4 oil objective. Calcofluor white-stained bud scars were observed using a DAPI (4′,6-diamidino-2-phenylindole) filter, and FITC-WGA-stained birth and bud scars were observed using an FITC filter.
TABLE 2.
Yeast strains used for this studya
| Strain and genetic background | Genotype | Reference |
|---|---|---|
| SK1 genetic background | ||
| AMP 109 | a/α | 10 |
| AMP 115 | a/α ime1-12::TRP1/ime1-12::TRP1 | 10 |
| AMP 1618 | α IME2-20-lacZ::LEU2 rme1Δ5::LEU2 met4 | 106 |
| AMP 1619 | aIME2-20-lacZ::LEU2 rme1Δ5::LEU2 arg6 | 91 |
| KSY 162 | a/α ime2-K97R-myc::TRP1/ime2-K97R-myc::TRP1 trp1ΔFA::hisG/trp1ΔFA::hisG his3-11,15/his3-11,15 ho::hisG/ho::hisG | 65 |
| KSY 187 | a/α IME2-myc::TRP1/IME2-myc::TRP1 trp1ΔFA::hisG/trp1ΔFA::hisG his3-11,15/his3-11,15 ho::hisG/ho::hisG | 65 |
| MSY 133-29 | ahis3ΔSK rme1Δ5::LEU2 | This study |
| MSY 133-34 | ahis3ΔSK rme1Δ5::LEU2 | This study |
| MSY 135-12 | amet4 rme1Δ5::LEU2 | This study |
| MSY 135-43 | amet4 rme1Δ5::LEU2 | This study |
| MSY 136-40 | α arg6 rme1Δ5::LEU2 | 92 |
| MSY 138-17 | α his3ΔSK rme1Δ5::LEU2 | 92 |
| MSY 184-55 | aarg6 rme1Δ5::LEU2 ume6-5::LEU2 | This study |
| MSY 185-65 | α his3ΔSK rme1Δ5::LEU2 ume6-5::LEU2 | This study |
| MSY 186-68 | α arg6 rme1Δ5::LEU2 ume6-5::LEU2 | 92 |
| MSY 188-119 | ahis3ΔSK rme1Δ5::LEU2 ume6-5::LEU2 | This study |
| MSY 202-14 | α arg6 rme1Δ5::LEU2 ime2-2::LEU2 | This study |
| MSY 203-22 | amet4 rme1Δ5::LEU2 ime2-2::LEU2 | This study |
| MSY 203-27 | amet4 rme1Δ5::LEU2 ime2-2::LEU2 | This study |
| MSY 203-33 | amet4 rme1Δ5::LEU2 ime2-2::LEU2 | This study |
| MSY 206-36 | α his3ΔSK rme1Δ5::LEU2 ime2-2::LEU2 | This study |
| MSY 552-17 | α his3ΔSK rme1Δ5::LEU2 ime1Δ::hphMX4 | This study |
| MSY 558-38 | α his3ΔSK rme1Δ5::LEU2 | This study |
| S497 | a/α his3-11,15/his3-11,15 trp1ΔFA/trp1ΔFA ho::hisG/ho::hisG | 88 |
| S635 | a/α IME2ΔC241-HA6-kanMX/IME2ΔC241-HA6-kanMX his3-11,15/his3-11,15 trp1ΔFA/trp1ΔFA ho::hisG/ho::hisG | 88 |
| Σ1278b genetic background | ||
| MLY 61 a/α | a/α ura3-52/ura3-52 | 54 |
| MLY 187 a/α | a/α ura3-52/ura3-52 ras2::G418/ras2::G418 | 55 |
| MLY 232 a/α | a/α ura3-52/ura3-52 gpr1::G418/gpr1::G418 | 55 |
| MSY 699-01 a/α | a/α ura3-52/ura3-52 ime1Δ::kanMX2/ime1Δ::hphMX4 | This study |
| MSY 694-51 a/α | a/α ura3-52/ura3-52 ime2Δ::kanMX2/ime2Δ::hphMX4 | This study |
All haploid SK1 strains have the additional alleles ura3, leu2::hisG, trp1::hisG, and lys-2 ho::LYS2, and all diploid SK1 strains are homozygous for these alleles, if not noted otherwise. The alleles arg6 (68), his3-11,15 (Saccharomyces Genome Database), his3ΔSK (68), ho::hisG (2), ho::LYS2 (2), ime1-12::TRP1 (99), ime2-2::LEU2 (64), ime2::kanMX (88), IME2-20-lacZ::LEU2 (106), IME2-myc::TRP1 (4), ime2-K97R-myc::TRP1 (4), IME2ΔC241-HA6-kanMX (88), leu2::hisG (2), lys2 (2), met4 (68), rme1Δ5::LEU2 (19), trp1::hisG (2), trp1ΔFA::hisG (37), ume6-5::LEU2 (101), ura3 (2), and ura3-52 (78) have been described before.
TABLE 3.
Oligodeoxynucleotides used for this study
| Oligonucleotide | Sequence |
|---|---|
| ime1Δ, 5′ | GCTTTTCTATTCCTCTCCCCACAAACAAAATGCAAGCGGATATGCATGGACAGCTGAAGCTTCGTACGC |
| ime1Δ, 3′ | TGAATGGATATATTTTGAGGGAAGGGGGAAGATTGTAGTACTTTTCGAGAAGGCCACTAGTGGATCTG |
| ime2Δ, 5′ | CGGTTAAGGTGGCTGTCTAGAGAATATAAACCTGTATTTATTTACCAGGCAGGCCACTAGTGGATCTG |
| ime2Δ, 3′ | CTGAGCCGGGTAACCGAACACAAAGATCTCGTTCTACTTTTTTTGACCTCAAGCTTCGTACGCTGCAGG |
Measurement of sporulation.
To determine sporulation, a fresh stationary-phase culture grown in rich medium (YPD; 2% [wt/vol] glucose, 2% [wt/vol] peptone, 1% [wt/vol] yeast extract) was used to inoculate rich acetate medium (YPAc; 2% [wt/vol] KOAc, 1% [wt/vol] yeast extract, 2% [wt/vol] peptone). These cells were grown to mid-log phase in baffled flasks, collected by centrifugation at 3,000 × g for 2 min, washed once with water, resuspended in complete sporulation medium (C-SPO) (106), and grown at 30°C with shaking for the required amount of time. Cells were visualized under a phase-contrast microscope, and the percentage of asci was determined for 3 replicates.
Tetrad dissection.
Asci were washed off plates from areas containing small colonies consisting predominantly of asci formed by pseudohyphal cells by use of sterile water collected by centrifugation, and the cell wall was digested with Glusulase for 15 min at 30°C. Spores were dissected using a tetrad dissection microscope (Singer Instruments, Watchet, United Kingdom), placed onto a YPD plate, and allowed to germinate and grow for 2 days at 30°C. Pictures were taken with a GelDoc 2000 system (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom).
Heat shock treatment.
Cells were grown to mid-log phase in liquid YPD medium at 25°C (for glycogen and trehalose determinations) or 30°C (for Northern analysis) with shaking before being shifted to 37°C (for glycogen and trehalose determinations) or 39°C (for Northern analysis) for the indicated times. Samples were taken immediately and processed as described below.
Metabolite determinations.
Glycogen and trehalose concentrations were determined as described by Parrou et al. (73). Briefly, cells were collected by centrifugation and washed once with ice-cold water, and the pellet was frozen immediately at −20°C. Cell pellets were resuspended in 250 μl 0.25 M Na2CO3 and heated at 95°C for 2 h with occasional mixing. The pH was adjusted to 5.2 by addition of 150 μl 1 M acetic acid and 600 μl 0.2 M sodium acetate buffer, pH 5.2. The suspension was split into two equal parts. The first of these was incubated overnight at 57°C with continuous shaking in the presence of 100 μg α-amyloglucosidase from Aspergillus niger (Sigma, St. Louis, MO), freshly prepared as a 10-mg/ml stock dissolved in 0.2 M sodium acetate buffer, pH 5.2. The second half of the suspension was incubated overnight at 37°C with 3 mU trehalase (0.25 U/ml; Sigma). Liberated glucose was measured using a GO glucose assay kit (Sigma) as directed. To measure the glucose concentration in plates, small sections of the most densely grown areas were cut out with a scalpel and placed into a syringe attached to a 0.22-μm filter. Liquid was expelled by applying pressure to the plunger. The glucose concentration was measured using a glucose meter (Roche Diagnostics).
cAMP assay.
cAMP concentrations were measured essentially as described before (23, 67, 84, 90). In brief, cells grown to mid-log phase on acetate were collected by centrifugation and washed with ice-cold water. The cell pellet was resuspended in 6% ice-cold trichloroacetic acid. Acid-washed glass beads (diameter = 0.4 to 0.6 mm) were added before the suspension was snap-frozen in liquid nitrogen and thawed on ice. Cells were then lysed using a Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at 6,000 rpm twice for 30 s each, with a 1-min break between, at 4°C. Following removal of cell debris by centrifugation (2,000 × g for 15 min, 4°C), HCl was added to the supernatant to a final concentration of 10 mM. The sample was extracted four times with diethyl ether and subsequently dried in a speed vacuum. The lyophilized cAMP was resuspended in assay buffer from the cAMP Biotrak enzyme immunoassay (EIA) system (GE Healthcare, Little Chalfont, United Kingdom), and the cAMP concentration was measured by use of this kit as directed. Samples were standardized against the cell number.
Northern analysis.
RNA analysis by Northern blotting was performed as described previously (92). Probes for HSP12, HSP26, HSP82, and FLO11 (MUC1) were generated by PCR, using genomic DNA as the template. The probes for HOP1, IME1, IME2, INO1, SPO13, and the loading control pC4/2 have been described elsewhere (48, 92). All mRNAs were quantitated by phosphorimaging on a Typhoon 9400 system (GE Healthcare).
RESULTS
Nonfermentable carbon sources stimulate pseudohyphal growth.
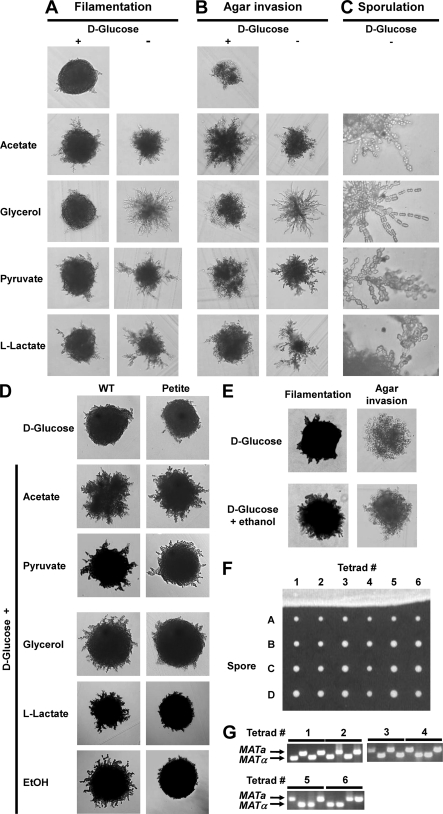
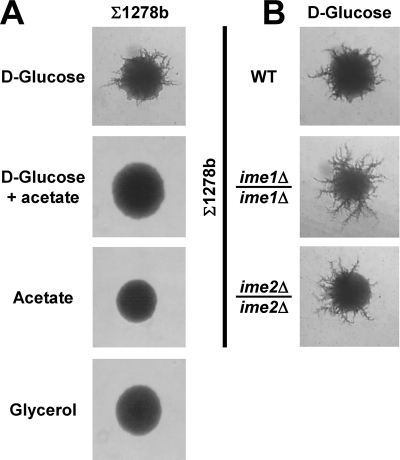
We previously reported that a/α diploid cells defective in a nutrient-regulated signaling pathway respond inappropriately to a meiotic stimulus by initiating pseudohyphal growth (91). These observations prompted us to investigate whether diploid a/α WT cells with the SK1 genetic background induce pseudohyphal growth when exposed to starvation conditions known to induce meiosis. To test this hypothesis, we grew diploid a/α SK1 cells on plates containing nonfermentable carbon sources and a limiting ammonium sulfate concentration (50 μM). In these plate assays, pseudohyphal growth is characterized by multiple projections of cells radiating away from the colony center (30). Diploid WT SK1 cells formed few pseudohyphae and were modestly capable of invading the agar when grown on glucose (Fig. 1A and B). Nonfermentable carbon sources, such as acetate, glycerol, pyruvate, and l-lactate, stimulated formation of branched pseudohyphae and agar invasion (Fig. 1A and B). Pseudohypha formation and agar invasion were also enhanced on plates containing both glucose and a nonfermentable carbon source (Fig. 1A and B). This stimulation could be observed as early as 1 day after inoculation of the plates (not shown). Furthermore, ∼1.6% (wt/vol) glucose remained even in the most densely grown areas of plates after 10 days of growth, suggesting that nonfermentable carbon sources stimulate pseudohyphal growth in the presence of glucose. Based on their appearance, pseudohyphae formed on nonfermentable carbon sources appear to share many morphological features with previously described pseudohyphae (30), such as cell adhesion after completion of cytokinesis, agar invasion, and directional growth (Fig. 1).
FIG. 1.
Nonfermentable carbon sources stimulate pseudohyphal growth in WT a/α diploid SK1 strains (AMP 1618 × AMP 1619 transformed with pRS316). Identical results were obtained with another diploid WT strain (AMP 109) (cf. Fig. 1 and 2). Colony morphology (A), agar invasion (B), and ascus formation (C) are shown after growth for 7 days. (D) Stimulation of pseudohyphal growth in respiration-deficient petite cells. The colony morphology after 7 days of growth is shown. (E) Stimulation of pseudohyphal growth by ethanol. (F) Asci formed by pseudohyphal cells contain four viable spores. Asci formed by a WT strain (AMP109) on SLA medium supplemented with 2% KOAc were dissected with a tetrad dissection microscope, and spores were placed onto a YPD plate and allowed to germinate and grow for 2 days. (G) PCR genotyping of the mating type locus reveals a 2:2 segregation pattern for MATa and MATα.
Glucose strongly inhibits utilization of alternative carbon sources by inhibiting expression of several enzymes of the citric acid cycle and the respiratory chain (93). However, limiting nitrogen concentrations under pseudohyphal growth conditions may derepress these genes. For example, rapamycin treatment, which mimics nitrogen starvation, induces glucose-repressible genes in glucose-grown cultures (35). To establish whether respiratory metabolism of nonfermentable carbon sources is required for stimulation of pseudohyphal growth by these carbon sources, we produced respiration-deficient petite cells. We confirmed the loss of respiratory function by the inability of petite cells to grow on plates containing only acetate as a carbon source. Acetate and pyruvate stimulated pseudohyphal growth in petite cells in the presence of glucose (Fig. 1D). In contrast, glycerol, ethanol, and l-lactate did not stimulate pseudohyphal growth of petite cells. These three carbon sources require NAD+-dependent oxidation reactions in order to be metabolized (93), indicating that less efficient regeneration of NAD+ from NADH may interfere with stimulation of pseudohyphal growth by these carbon sources in petite cells. Taken together, these data indicate that nonfermentable carbon sources stimulate pseudohyphal growth independent of their use as respiratory energy sources. These results also suggest that the less pronounced pseudohyphal growth seen on glucose as a sole carbon source (Fig. 1A) may be caused by the accumulation of glycolytic waste products, especially ethanol, during fermentative growth. In support of this hypothesis, we found that ethanol stimulates pseudohyphal growth and agar invasion (Fig. 1E) (52).
Microscopic examination of pseudohyphae on plates containing only nonfermentable carbon sources at a higher magnification revealed that pseudohyphal cells exited pseudohyphal growth and formed asci (Fig. 1C). Cells both above and below the agar surface sporulated as early as 3 days after inoculation of plates (not shown). All pseudohyphae of all colonies sporulated. All spores isolated from asci formed by pseudohyphal cells on acetate were viable (Fig. 1F). The mating type locus displayed a 2:2 segregation pattern (Fig. 1G), indicating normal execution of both meiotic divisions. No asci were observed in the presence of glucose. Asci sometimes displayed a linear arrangement of two to four spores, especially in cells grown on glycerol (Fig. 1C). All spores from linear asci were viable and displayed a 2:2 segregation pattern for the mating type locus (not shown). Taken together, these data show that nonfermentable carbon sources stimulate pseudohyphal growth and that pseudohyphal cells can enter and successfully complete meiosis.
Ime1 and the protein kinase activity of Ime2 are required for pseudohyphal growth on nonfermentable carbon sources.
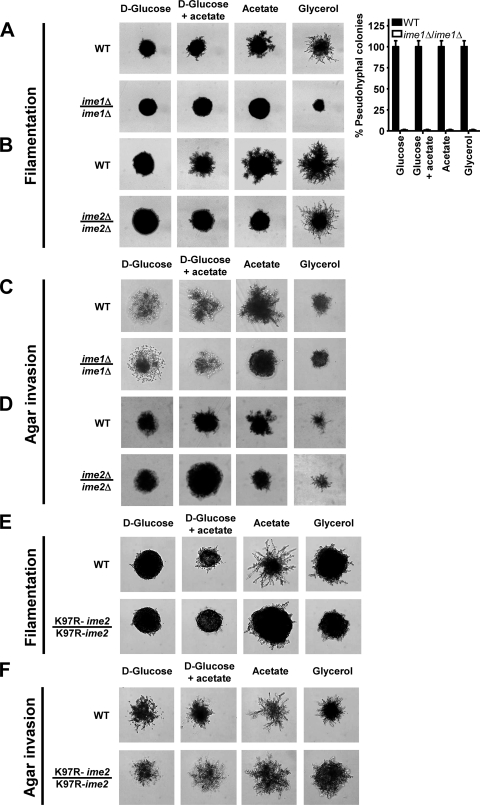
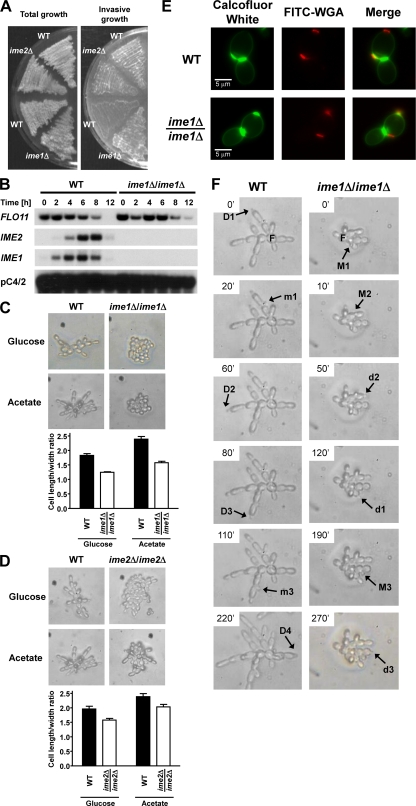
Sporulation of pseudohyphae precluded investigation of cellular characteristics of pseudohyphal cells, such as cell elongation. Both Ime1 and Ime2 control entry into meiosis. Sporulation of ime1Δ/ime1Δ and ime2Δ/ime2Δ cells is decreased several hundredfold compared to that of WT strains (43, 99). Functions for IME1 or IME2 outside meiosis have not been reported. Hence, we characterized pseudohypha formation and agar invasion of ime1Δ/ime1Δ and ime2Δ/ime2Δ strains. Surprisingly, pseudohypha formation was nearly completely absent in ime1Δ/ime1Δ cells (Fig. 2A). Deletion of IME1 severely decreased the pseudohyphal morphology of nearly all colonies on a plate, independent of the position of the colony on the plate (Fig. 2A). However, ime1Δ/ime1Δ cells were able to invade the agar (Fig. 2C), showing that filamentation and agar invasion are genetically separable phenotypes. Similarly, filamentation, but not agar invasion, was defective in ime2Δ/ime2Δ cells grown on glucose, a mixture of glucose and acetate, or acetate, but not glycerol (Fig. 2B and D). Deletion of IME2 decreased the pseudohyphal morphology of nearly all colonies on plates containing glucose, glucose plus acetate, or acetate as the carbon source (data not shown). However, the pseudohyphal growth defect of ime2Δ/ime2Δ cells was not as severe as the pseudohyphal growth defect of ime1Δ/ime1Δ cells (Fig. 2A and B). The roles of IME1 and IME2 in pseudohyphal growth were independent of the ability of yeast to respire (not shown). Overexpression of Ime1 and Ime2 from multicopy (2μm) plasmids enhanced pseudohyphal growth (Fig. 3A) and had no effect on agar invasion (Fig. 3D). This finding is consistent with decreased pseudohypha formation of ime1Δ/ime1Δ and ime2Δ/ime2Δ strains (Fig. 2). Cells carrying the protein kinase-defective K97R-ime2 allele displayed a defect in pseudohypha formation similar to that with the IME2 deletion (Fig. 2E and F), showing that the protein kinase activity of Ime2 is required for pseudohyphal growth and meiosis. Deletion of the C terminus of Ime2 (Ime2ΔC241), which controls the mitotic stability of Ime2 (88), did not affect pseudohyphal growth (not shown). Taken together, these data show that Ime1 and Ime2 are required for pseudohypha formation by SK1 cells.
FIG. 2.
Ime1 and the protein kinase activity of Ime2 are required for pseudohyphal growth. The colony morphology of WT (AMP 109) and ime1Δ/ime1Δ (AMP 115) strains (A) and of WT (MSY 135-43 × MSY 136-40) and ime2Δ/ime2Δ (MSY 202-14 × MSY 203-27) strains (B) is shown. Bar graphs show percentages of pseudohyphal colonies. For each strain and carbon source, >200 colonies were classified as pseudohyphal or nonpseudohyphal. Error bars represent standard errors. (C and D) Agar invasion by WT (AMP 109) and ime1Δ/ime1Δ (AMP 115) strains (C) and by WT (MSY 135-12 × MSY 138-17) and ime2Δ/ime2Δ (MSY 203-22 × MSY 206-36) strains (D). Filamentation (E) and agar invasion (F) of WT (KSY 187) and K97R-ime2/K97R-ime2 (KSY 162) strains were scored after 7 days of growth.
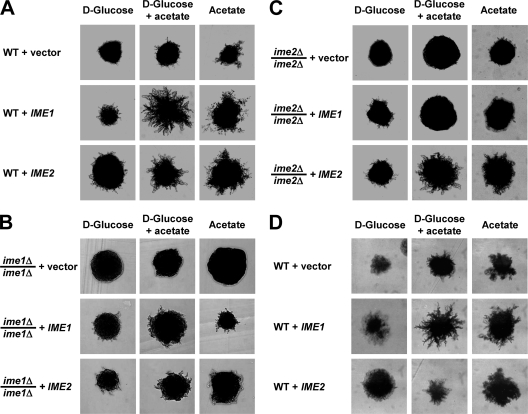
FIG. 3.
IME1 acts through IME2 to stimulate pseudohyphal growth. Colony morphology is shown for WT (MSY 135-12 × MSY 138-17) (A), ime1Δ/ime1Δ (AMP 115) (B), and ime2Δ/ime2Δ (MSY 203-22 × MSY 206-36) (C) strains transformed with empty vector (pRS426) or 2μm plasmids expressing IME1 (pHS103) or IME2 (pHS105) from their endogenous promoters. Similar results were obtained with an rme1Δ/rme1Δ strain (MSY 135-12 × MSY 138-17) and an RME1/RME1 strain (AMP 109). For simplicity, only MSY 135-12 × MSY 138-17 is shown in panel A. (D) Agar invasion of the strains in panel A. Filamentation and agar invasion were scored after 7 days of growth.
Ime1 acts through Ime2 to stimulate pseudohyphal growth.
In meiosis, Ime1 activates transcription of EMGs, including IME2. Ime2 stimulates expression of EMGs, including its own expression, independent of IME1 (64, 96). Ime2 is also a negative regulator of IME1 expression (81, 95, 96, 99) and Ime1 stability (34). To establish whether IME1 and IME2 act in the same order in pseudohyphal growth and meiosis, we investigated whether overexpression of Ime1 or Ime2 from 2μm plasmids would rescue the pseudohyphal growth defects of ime1Δ/ime1Δ and ime2Δ/ime2Δ strains. Expression of Ime1 in ime1Δ/ime1Δ cells and of Ime2 in ime2Δ/ime2Δ cells restored pseudohyphal growth on nonfermentable carbon sources (Fig. 3B and C). Expression of Ime2 in ime1Δ/ime1Δ strains partially restored pseudohypha formation (Fig. 3B), whereas expression of Ime1 in ime2Δ/ime2Δ strains had no effect on acetate (Fig. 3C), suggesting that activation of IME2 by Ime1 is required for pseudohyphal growth on acetate. The partial restoration of pseudohyphal growth by overexpression of Ime2 in ime1Δ/ime1Δ cells is consistent with an Ime2-independent function of Ime1 in pseudohyphal growth. The more severe pseudohyphal growth defects of ime1Δ/ime1Δ cells than those of ime2Δ/ime2Δ cells (Fig. 2) also suggest an Ime2-independent role for Ime1 in pseudohyphal growth. Agar invasion was not altered by overexpression of Ime1 or Ime2 in the WT, ime1Δ/ime1Δ, or ime2Δ/ime2Δ strain (Fig. 3D and not shown), providing additional evidence that Ime1 and Ime2 are not involved in regulation of agar invasion.
Evidence that binding of Ime1 to Ume6 is involved in pseudohyphal growth on nonfermentable carbon sources.
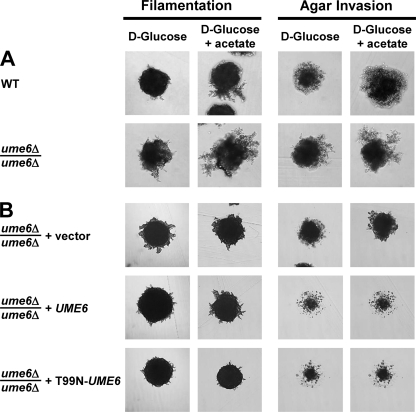
Ime1 activates transcription of EMGs, including IME2, through the DNA-binding protein Ume6 (11, 59, 79). Ume6 also recruits two transcriptional repression complexes to EMG promoters, namely, the ISW2 chromatin remodeling complex (31) and the Rpd3-Sin3 histone deacetylase (HDAC) (41). Deletion of UME6 derepresses EMG transcription, including that of IME2, under nutrient-rich conditions (92, 101, 109) but also abrogates activation of these genes during starvation. A T99N mutation in Ume6 interferes with activation of EMGs by Ime1. This mutation decreased the interaction between Ime1 and Ume6 ∼35-fold in a two-hybrid assay (110). The T99N mutation interferes with association of Ime1 with EMG promoters (11, 39, 79, 92) and with degradation and removal of Ume6 from EMG promoters by Ime1 (59). These data suggested that Ume6 also has a role in pseudohyphal growth. Deletion of UME6 derepressed filamentation (Fig. 4A). Agar invasion by ume6Δ/ume6Δ cells was enhanced compared to WT cells (Fig. 4A). This function of Ume6 in agar invasion is likely to be independent of IME1 and IME2, because deletion or overexpression of these two genes had no effect on agar invasion (Fig. 2 and 3). Expression of WT Ume6 and T99N-Ume6 complemented the enhanced agar invasion of ume6Δ/ume6Δ strains (Fig. 4B). T99N-Ume6 inhibited filamentation more severely than WT Ume6 did (Fig. 4B). This behavior of the T99N-Ume6 mutant is consistent with repression of IME2 by the two transcriptional repression complexes recruited to the IME2 promoter by Ume6 and, at the same time, with abrogation of the Ime1-Ume6 interaction by this mutation. These data indicate that the early meiotic cascade consisting of Ime1, recruitment of Ime1 to URS1 by Ume6, and Ime2 is required for both pseudohyphal growth and meiosis but not for agar invasion.
FIG. 4.
Recruitment of Ime1 to early meiotic gene promoters, including the IME2 promoter, by Ume6 is required for pseudohyphal growth. (A) Pseudohyphal growth is derepressed in a ume6Δ/ume6Δ (MSY 184-55 × MSY 185-65) strain compared to that in a WT strain (MSY 133-34 × MSY 136-40). (B) Abrogation of the interaction between Ume6 and Ime1 in cells expressing the T99N-UME6 allele as the sole source of Ume6 results in a pseudohyphal growth defect. Filamentation and agar invasion phenotypes of ume6Δ/ume6Δ (MSY 186-68 × MSY 188-119) strains carrying empty vector (pRS316) or plasmids expressing WT Ume6 (pRS316-UME6-lexA) or T99N-Ume6 (pRS316-T99N-UME6-lexA) are shown. Filamentation and agar invasion were scored after 7 days of growth.
Ime1 is required for cell elongation, bud site selection, and budding of daughter cells before their mother cells.
Next, we characterized cellular features that distinguish yeast-form and pseudohyphal cells to obtain more detailed insight into the roles of Ime1 and Ime2 in pseudohyphal growth. In contrast to yeast-form cells, pseudohyphal cells invade agar because they overexpress Flo11 (51). Compared to yeast-form cells, pseudohyphal cells are elongated, adhere to each other after cell division has been completed, and switch from a bipolar to a unipolar budding pattern. Yeast-form daughters are born smaller than their mothers. Pseudohyphal daughters are born with a size comparable to the size of their mothers, allowing them to bud at the same time as, or even slightly before, their mothers (45).
Deletion of IME1 or IME2 did not affect agar invasion of diploid cells (Fig. 2), indicating that Ime1 and Ime2 act independent of Flo11. Flo11 is also required for haploid invasive growth (33). Deletion of IME1 and IME2 did not decrease haploid invasive growth (Fig. 5A). Northern blotting confirmed that FLO11 expression was largely unperturbed in ime1Δ/ime1Δ cells (Fig. 5B). Consistent with these observations, we found that deletion of IME1 or IME2 did not significantly decrease expression of an FG(TyA)::lacZ reporter (not shown) whose activation correlates well with the activity of the filamentation MAPK cascade in pseudohyphal cells (66).
FIG. 5.
Ime1 and Ime2 act independent of Flo11 expression and the filamentation MAPK cascade. (A) Haploid invasive growth is not defective in ime1Δ and ime2Δ strains. Invasive growth was scored after 3 days of growth on YPD plates. The strains used were upper WT (MSY 135-12), ime2Δ (MSY 203-27), lower WT (MSY 558-38), and ime1Δ (MSY 552-17). (B) Deletion of IME1 does not affect transcription of FLO11. RNA samples isolated from a WT (AMP 109) or ime1Δ/ime1Δ (AMP 115) strain grown to mid-log phase on YPAc and shifted for the indicated times to C-SPO medium were analyzed by Northern blotting. (C and D) The microscopic appearance of microcolonies is shown for WT (AMP 109) andime1Δ/ime1Δ (AMP 115) strains (C) and for WT (MSY 136-40 × MSY 135-12) and ime2Δ/ime2Δ (MSY 202-14 × MSY 203-27) strains (D) grown for 12 to 24 h on SLA plates containing glucose or acetate as a carbon source. The P values for all pairwise strain and medium comparisons are <0.01. (E) Examples of bud site selection in WT (AMP 109) and ime1Δ/ime1Δ (AMP 115) cells grown on SLA acetate plates at 30°C for 18 h. Bud scars were stained with calcofluor white and are false-colored in green. Bud and birth scars stained with FITC-WGA are false-colored in red to reveal the polarity of the cells. Note that calcofluor white does not stain the birth scar and that FITC-WGA does not stain the chitin ring between the mother and its growing bud. (F) Order of budding of mother and daughter cells in WT (AMP 109) and ime1Δ/ime1Δ (AMP 115) strains grown on SLA acetate plates. Abbreviations: D, daughter; M, mother. Uppercase letters represent the cell budding first, and lowercase letters represent the cell budding last. The numbers identify mother-daughter pairs. The cells from which the colonies originated are labeled with an “F.” These cells are spherical and display a random, nonpolar budding pattern.
Microscopic examination of microcolonies revealed that ime1Δ/ime1Δ cells were not as elongated as WT cells when grown on SLA plates with glucose or acetate as the carbon source (Fig. 5C). Acetate enhanced cell elongation in both strains. ime2Δ/ime2Δ and K97R-ime2/K97R-ime2 cells (Fig. 5D and data not shown) were also not as elongated as their corresponding WT cells. The elongation defects of the ime2Δ/ime2Δ and K97R-ime2/K97R-ime2 strains were less severe than the elongation defect of the ime1Δ/ime1Δ strain. These elongation phenotypes are consistent with the less severe pseudohyphal growth phenotype of ime2Δ/ime2Δ colonies after growth for several days (Fig. 2A) and with the stimulation of pseudohyphal growth by acetate (Fig. 1A). These data show that IME1 and IME2 are required for elongation of pseudohyphal cells.
ime1Δ/ime1Δ and ime2Δ/ime2Δ microcolonies are more globular than microcolonies of WT cells. This suggested that another characteristic of pseudohyphal cells that contributes to directional growth is affected by deletion of IME1 and IME2. To characterize whether the budding patterns of WT and ime1Δ/ime1Δ strains are bi- or unipolar, we stained bud scars with calcofluor white after growth of both strains for ∼18 h on SLA acetate plates. For both strains, we observed cells with bud scars at both poles (not shown). Calcofluor white staining did not reveal a difference in bud site selection between the two strains (not shown). Bipolar budding cells display a strong bias toward the pole opposite their birth site in their first few divisions (14). For this reason, it may be difficult to distinguish between bi- and unipolar budding patterns by staining with calcofluor white only. To reveal more subtle differences in bud site selection, we costained cells with FITC-WGA. In addition to bud scars, FITC-WGA also stains birth scars (25). Identification of the birth scar allowed classification of budding events into events at the poles distal and proximal to the birth scar (Fig. 5E). This classification revealed that WT and ime1Δ/ime1Δ cells grown for 18 h on SLA acetate plates budded exclusively at the distal pole in their first division (Table 4). This bias for the distal pole persisted for the first few divisions in both strains. However, this bias appeared to decrease faster in the ime1Δ/ime1Δ strain. The second bud was formed at the birth pole in ∼17% of ime1Δ/ime1Δ cells, whereas only ∼4.7% of WT cells chose the birth pole for their second bud (P < 0.05). This small increase in selection of the proximal pole in the second cell cycle may suffice to explain the more globular growth of ime1Δ/ime1Δ cells.
TABLE 4.
Distribution of bud scars relative to the birth site in WT (AMP 109) and ime1Δ/ime1Δ (AMP 115) cellsa
| No. of bud scars on cells | No. of cells with only distal bud scars | No. of cells with ≥1 proximal bud scar | % of cells with only distal bud scars | % of cells with ≥1 proximal bud scar |
|---|---|---|---|---|
| WT cells | ||||
| 1 | 157 | 0 | 100 | 0 |
| 2 | 61 | 3 | 95 | 4.7 |
| 3 | 15 | 12 | 56 | 44 |
| 4 | 5 | 6 | 45 | 55 |
| 5 | 0 | 3 | 0 | 100 |
| 6 | 0 | 3 | 0 | 100 |
| ime1Δ/ime1Δ cells | ||||
| 1 | 106 | 0 | 100 | 0 |
| 2 | 53 | 11 | 82 | 17 |
| 3 | 14 | 14 | 50 | 50 |
| 4 | 3 | 10 | 23 | 77 |
| 5 | 1 | 6 | 14 | 86 |
| 6 | 0 | 5 | 0 | 100 |
| 7 | 0 | 0 | ||
| 8 | 0 | 1 | 0 | 100 |
Cells were grown for 18 h on SLA acetate plates at 30°C. Cells were then stained with calcofluor white and FITC-WGA to reveal bud and birth scars, as described in Materials and Methods. Only the difference in selection of the second bud site was significantly different (P < 0.05, assuming that the number of bud scars formed at the proximal pole follows a Poisson distribution).
Time-lapse video microscopy revealed that pseudohyphal WT daughter cells budded before or around the same time as their mothers (12 of 14 mother-daughter pairs). Mothers could bud much later (>60 min) than their daughters (4 of 12 mother-daughter pairs in which the daughter budded first). In contrast, ime1Δ/ime1Δ daughter cells budded after their mothers (10 of 10 mother-daughter pairs) (Fig. 5F). This reversal in the budding order is a second factor that contributes to the globular growth of ime1Δ/ime1Δ cells.
Comparison of the roles of nonfermentable carbon sources in pseudohyphal growth in the SK1 and Σ1278b genetic backgrounds.
Our data show that stimulation of pseudohyphal growth by nonfermentable carbon sources in the SK1 genetic background requires IME1 and IME2. Next, we wished to extend these findings to the Σ1278b genetic background, which is used more routinely to study pseudohyphal growth (30, 54). In contrast to the case for the SK1 genetic background, nonfermentable carbon sources inhibited pseudohyphal growth in diploid a/α Σ1278b WT cells (Fig. 6A). Likewise, IME1 and IME2 were not required for pseudohypha formation by diploid a/α Σ1278b WT cells on glucose (Fig. 6B).
FIG. 6.
Characterization of the roles of IME1 and IME2 in pseudohyphal growth in the Σ1278b background. (A) Inhibition of pseudohyphal growth by nonfermentable carbon sources in the Σ1278b genetic background. The strain used was MLY 61 a/α. (B) IME1 (MSY 699-01 a/α) and IME2 (MSY 694-51 a/α) are not required for pseudohyphal growth on glucose in Σ1278b cells. In both panels, the colony morphologies after 7 days of growth on the indicated carbon sources are shown.
Elevated cAMP signaling inhibits pseudohyphal growth on nonfermentable carbon sources.
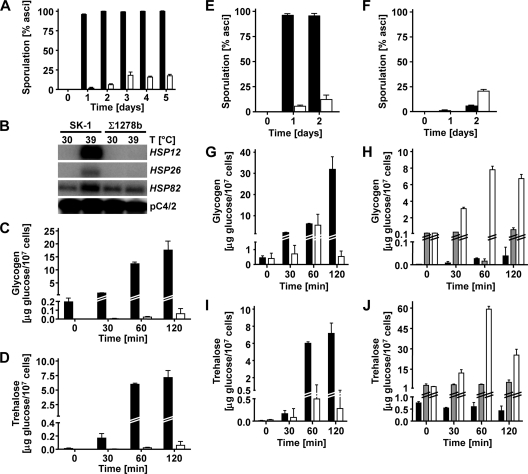
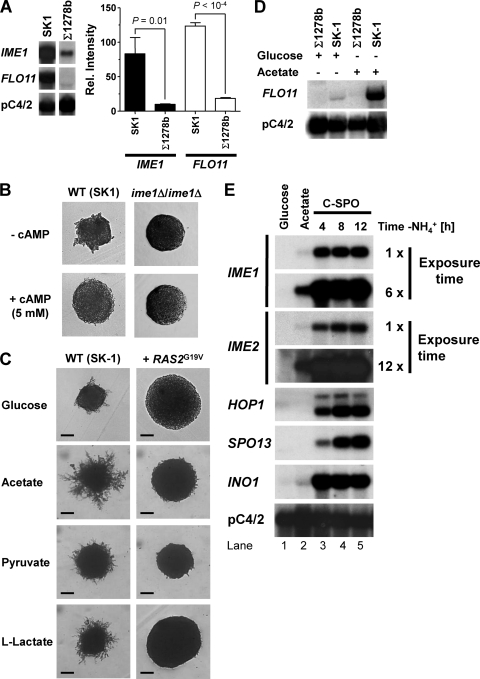
Stimulation of pseudohyphal growth in the presence of nonfermentable carbon sources in SK1 cells is different from the behavior of Σ1278b cells (Fig. 1 and 6A). To explore explanations for the different behaviors of these two genetic backgrounds, we decided to focus on the PKA pathway. The PKA pathway is a signaling pathway responding to the quality of the carbon source, is activated by glucose (87), and is involved in the regulation of pseudohyphal growth (26). The PKA pathway is hyperactive in Σ1278b cells compared to another genetic background, SP1 (100). Strains with high constitutive PKA activity fail to express IME1 (60, 62), and PKA signaling inhibits Ime1 (80) and Ime2 (22, 76). These data suggest that elevated cAMP signaling in Σ1278b cells interferes with activation of Ime1 and Ime2, resulting in a pseudohyphal growth defect on nonfermentable carbon sources. To test this hypothesis, we first confirmed that cAMP signaling is constitutively elevated in Σ1278b cells compared to SK1 cells. Consistent with elevated cAMP signaling in Σ1278b cells, we found that growth on nonfermentable carbon sources, sporulation, induction of heat shock genes, and accumulation of storage carbohydrates upon heat shock were decreased in Σ1278b cells (Fig. 7A to D). All of these phenotypes are known to be inhibited by elevated cAMP signaling (13, 100, 104). Deletion of cAMP signaling components such as GPR1 or RAS2 elevated sporulation and storage carbohydrate accumulation in heat-shocked Σ1278b cells (Fig. 7F, H, and J). Expression of constitutively active Ras2G19V in SK1 cells inhibited sporulation and storage carbohydrate accumulation during heat shock (Fig. 7E, G, and I). These data confirm that cAMP signaling contributes to the magnitude of these phenotypes in Σ1278b and SK1 cells. cAMP levels were elevated in Σ1278b cells compared to those in SK1 cells (4.5 ± 0.3 pmol/107 cells versus 3.5 ± 0.2 pmol/107 cells). These data confirm that cAMP signaling is elevated in Σ1278b cells compared to SK1 cells. Steady-state IME1 mRNA levels were significantly decreased in Σ1278b cells (Fig. 8A), consistent with repression of IME1 by elevated cAMP signaling in Σ1278b cells. Addition of cAMP or expression of constitutively active Ras2G19V inhibited pseudohyphal growth in SK1 cells (Fig. 8B and C). FLO11, whose expression is stimulated by an activated cAMP signaling pathway in Σ1278b cells, displayed strongly elevated mRNA levels in SK1 cells (Fig. 8A and D). Furthermore, acetate induced the expression of FLO11 in SK1 cells (Fig. 8D) but not in Σ1278b cells (70), which may contribute to increased pseudohyphal growth and invasiveness of SK1 strains grown on nonfermentable carbon sources (Fig. 1A and B). Deletion of RAS2, GPR1, or GPA2 did not allow Σ1278b cells to form pseudohyphae on nonfermentable carbon sources (not shown), possibly because FLO11 expression requires cAMP signaling in Σ1278b cells (71, 72, 77). Taken together, these data suggest that cAMP signaling inhibits pseudohyphal growth on nonfermentable carbon sources by inhibiting IME1 and IME2.
FIG. 7.
cAMP signaling is hyperactive in Σ1278b cells compared to SK1 cells. (A) Sporulation of a/α diploid SK1 (AMP 109; filled bars) and Σ1278b (MLY 61 a/α; open bars) cells. (B) Induction of heat shock genes in WT cells (AMP 109 and MLY 61 a/α) shifted from 30°C to 39°C for 30 min. (C and D) Accumulation of glycogen (C) and trehalose (D) in WT cells (AMP 109 [filled bars] and MLY 61 a/α [open bars]) shifted from 25°C to 37°C for the indicated times. (E) Expression of constitutively active Ras2G19V from plasmid pMW2 in a WT a/α diploid SK1 strain (AMP 109) inhibits sporulation. Filled bars, AMP 109 plus pRS316; open bars, AMP 109 plus pMW2. (F) Deletion of GPR1 in a WT a/α diploid Σ1278b strain (MLY 61 a/α) increases sporulation. Filled bars, WT (MLY 61 a/α); open bars, gpr1Δ/gpr1Δ strain (MLY 232 a/α). (G and H) Glycogen accumulation is shown for cells shifted from 25°C to 37°C for the indicated times. (G) AMP 109 transformed with pRS316 (filled bars) or pMW2 expressing Ras2G19V (open bars). (H) MLY 61 a/α (black bars), MLY 232 a/α (gpr1Δ/gpr1Δ; gray bars), and MLY 187 a/α (ras2Δ/ras2Δ; open bars). (I and J) Trehalose accumulation in the cells shown in graphs G and H. For each measurement, the average and standard error for two replicates are shown.
FIG. 8.
cAMP signaling represses pseudohyphal growth in SK1 cells. (A) Expression of IME1 and FLO11 in WT SK1 (AMP 109) and Σ1278b (MLY 61 a/α) strains grown on rich acetate medium (2% KOAc, 1% yeast extract, 2% peptone) to exponential growth phase. All bands are from the same blot. P values were derived from an unpaired, two-tailed t test (n = 6). (B and C) Addition of 5 mM cAMP to SLA glucose plates (B) or expression of constitutively active Ras2G19V from plasmid pMW2 (C) inhibits pseudohyphal growth in SK1 cells (AMP 109). Filamentation and agar invasion were scored after 7 days of growth. Bars, 40 μm for glucose and 100 μm for the other carbon sources. (D) Acetate induces expression of FLO11 in a WT SK1 strain (AMP 109). Cells were grown to mid-log phase on YPD or YPAc before isolation of RNA for Northern analysis. The Σ1278b strain was MLY 61 a/α. (E) Comparison of steady-state mRNA levels for IME1, IME2, the EMGs HOP1 and SPO13, and the inositol biosynthetic gene INO1 in an SK1 strain (AMP 1619) grown to exponential growth phase on glucose (lane 1) or acetate (lane 2) or 4, 8, or 12 h after being shifted to sporulation medium (C-SPO medium) (see Materials and Methods).
DISCUSSION
Our work has shown that nonfermentable carbon sources stimulate pseudohyphal growth independent of respiratory function and that pseudohyphal cells formed in the absence of glucose exit pseudohyphal growth to successfully complete meiosis (Fig. 9). Two regulators of entry into meiosis, IME1 and IME2, are required for stimulation of pseudohyphal growth by nonfermentable carbon sources. The use of a different genetic background, SK1, from that used routinely for investigation of pseudohyphal growth, Σ1278b, was critical for making these observations. Pseudohyphae formed by SK1 cells on nonfermentable carbon sources share several features with pseudohyphae formed by Σ1278b cells, for example, cell elongation, daughter cells budding before or at the same time as their mothers, and invasion of the agar. However, pseudohyphal SK1 cells appear to employ a bipolar budding pattern. Bipolar budding diploid cells display a strong bias toward budding at the pole opposite their birth site in the first few divisions (14). This bias (Table 4), together with daughters budding before their mothers (Fig. 5F), may allow SK1 cells to form pseudohyphae. Both of these features are affected by deletion of IME1. The budding order of daughter and mother cells is reversed in ime1Δ/ime1Δ cells (Fig. 5F). The bias for budding distal to the birth site is less pronounced in ime1Δ/ime1Δ cells, especially for the second budding event (Table 4). These buds and their first daughters are directed toward the origins of the colonies, thus explaining why ime1Δ/ime1Δ cells are defective in pseudohyphal growth.
FIG. 9.
Control of cell differentiation by the early meiotic cascade. (A) Revised life cycle of S. cerevisiae incorporating sporulation of pseudohyphal cells. Haploid a and α cells grow and divide in a nutrient-rich environment. Mating type switching allows a cells to switch to an α mating type and vice versa. a and α cells mate to form an a/α diploid cell when exposed to mating pheromones secreted by their opposite mating types. a/α diploid cells grow and divide in a nutrient-rich environment. Severe starvation triggers sporulation of a/α diploids and formation of an ascus harboring four haploid spores. Moderate starvation triggers pseudohyphal growth, which may allow yeasts to forage for nutrients. Severe starvation of pseudohyphae also induces sporulation. After exposure to nutrients and breakdown of the ascus wall, the spores germinate to form haploid a and α cells. (B) Model summarizing how the early meiotic cascade consisting of Ime1, Ume6, and Ime2 regulates cell differentiation in diploid S. cerevisiae cells. Starvation of a/α diploid cells induces expression of Ime1 and conversion of the transcriptional repressor Ume6 to an activator, leading to induction of early meiotic genes, including IME2. Activation of IME1 and IME2 is a general differentiation signal that promotes both pseudohyphal growth and meiosis. Induction of pseudohyphal growth or meiosis by Ime1 is abolished by a T99N mutation in Ume6. Modulation of the differentiation signal generated by IME1 and IME2 by other, uncharacterized events is expected to govern the choice between pseudohyphal growth and meiosis.
Role of Ime1 and Ime2 in pseudohyphal growth.
Studies on Ime1 and Ime2 in meiosis provide clues regarding the extent to which these two proteins may act through the same or similar downstream targets to stimulate pseudohyphal growth on nonfermentable carbon sources. Ime1 acts through Ume6 to activate EMGs, including IME2 (11, 59, 79, 98). Ume6 serves dual, opposing roles on EMG promoters. Under nutrient-rich conditions, it represses EMGs via recruitment of the Rpd3-Sin3 HDAC (41) and the ISW2 chromatin remodeling complex (31), whereas in starvation its interaction with Ime1 is required for activation of EMGs, either because Ime1 bound to EMG promoters via Ume6 provides a transcriptional activation domain (11, 79, 98) or because the interaction between these two proteins is required to remove Ume6 from EMGs (59). To investigate whether this transcriptional switch also operates in pseudohyphal growth, we turned to a T99N mutation in Ume6. This mutation decreases the interaction of Ume6 with Ime1 in a two-hybrid assay (∼35-fold) but does not derepress expression of EMGs prior to induction of meiosis (11), suggesting that the T99N mutation does not affect interaction of Ume6 with the Rpd3-Sin3 HDAC or the ISW2 chromatin remodeling complex. Cells expressing T99N-Ume6 formed less pseudohyphal colonies than cells deleted for UME6 (Fig. 4B). This finding is consistent with the interpretation that derepression of IME2 enhances pseudohyphal growth in ume6Δ/ume6Δ cells. T99N-Ume6 also interfered with pseudohypha formation compared to WT Ume6 (Fig. 4B), which can be explained by the inability of Ime1 to activate IME2 in cells expressing T99N-Ume6. This effect of the T99N-Ume6 mutant was more pronounced on plates containing both glucose and acetate than on plates containing only glucose (Fig. 4B), which is consistent with elevated expression of IME1 in cells grown on nonfermentable carbon sources (Fig. 8E) and after exponential growth on glucose (44). The pseudohyphal growth defect of the T99N-Ume6-expressing cells was also not as severe as the pseudohyphal growth defect displayed by ime1Δ/ime1Δ cells, either because of a residual interaction between T99N-Ume6 and Ime1 or because of the existence of additional, Ume6-independent Ime1 targets. Epistasis analysis (Fig. 3) supports the idea that Ime1 acts largely through inducing expression of Ime2 to stimulate pseudohyphal growth. The behavior of the T99N-Ume6 mutant (Fig. 4B) provides further support for this view. At the same time, the pseudohyphal growth defect of ime1Δ/ime1Δ cells is more severe than the pseudohyphal growth defect of ime2Δ/ime2Δ cells (Fig. 2 and 5), showing that Ime1 also stimulates pseudohyphal growth independent of Ime2. This situation is similar to regulation of entry into meiosis by Ime1 and Ime2, where Ime1 also acts through Ime2 and independent of Ime2 to stimulate entry into meiosis (64). We conclude that all key elements of the early meiotic cascade function in pseudohyphal growth. The transcriptional targets of Ime1 in pseudohyphal growth remain to be identified, but EMGs and genes carrying the Ume6-binding site URS1 in their promoters are likely targets.
Several substrates for Ime2 are known, including Sic1 (21, 94), Cdh1 (8), the middle meiotic gene-specific transcription factor Ndt80 (4, 69), the repressor of middle meiotic genes Sum1 (65), and Rfa2 (17, 18). Stabilization of the G1-S transition-promoting G1 cyclins Cln1 to -3 in grr1Δ/grr1Δ cells stimulates pseudohyphal growth (6). Consequently, pseudohyphal cells are characterized by a shortened G1 and an extended G2 phase (45). In meiosis, Ime2 substitutes for Cdc28 to trigger entry into meiotic S phase by phosphorylating Sic1 and triggering degradation of Sic1 (21), an inhibitor of Cdc28 and of the G1-S transition, suggesting that Ime2 may stimulate pseudohyphal growth by stimulating Sic1 degradation. Nevertheless, future studies are required to identify the Ime2 substrates in pseudohyphal growth.
Sporulation of pseudohyphal cells.
In the absence of glucose, pseudohyphae of SK1 cells exit pseudohyphal growth to successfully complete meiosis (Fig. 1). Several models for conversion of pseudohyphal cells into asci are imaginable. Pseudohyphal cells may exit pseudohyphal growth to form normal yeast- or vegetative-growth-form cells which then enter and execute meiosis. Alternatively, pseudohyphal cells may directly exit pseudohyphal growth and enter meiosis. Near-quantitative conversion of filaments into asci within 3 days and the requirement of IME1 and IME2 for pseudohyphal growth support the second model. Independent of the precise mechanism of switch from pseudohyphal growth to meiosis, the question arises of which stimulus triggers entry into meiosis by pseudohyphae. A further decrease in nutrient content of the medium caused by metabolic activity of the growing pseudohyphae may induce meiosis. Ascus formation by the tip cell of pseudohyphae (Fig. 1C) provides evidence against such a simple nutrient depletion model. Alternatively, entry into meiosis may be under temporal control. Detailed time-lapse studies will be necessary to establish whether pseudohyphal cells enter meiosis directly or via prior formation of yeast-form cells. Asci sometimes displayed a linear arrangement of two to four spores. This ascus morphology has been observed previously in Saccharomyces ludwigii (36) and after sporulation of newly formed zygotes of S. cerevisiae (103).
Role of cAMP signaling in pseudohyphal growth.
Stimulation of pseudohyphal growth in the presence of nonfermentable carbon sources in SK1 cells contrasts with the behavior of Σ1278b cells, in which nonfermentable carbon sources inhibited pseudohyphal growth (Fig. 6A). cAMP signaling activity was elevated in Σ1278b cells compared to SK1 cells (Fig. 7), confirming an earlier report (100). Strains with high constitutive PKA activity fail to express IME1 (60, 62). PKA signaling inhibits Ime1 (80) and Ime2 (22, 76). Therefore, Σ1278b cells may fail to form pseudohyphae on nonfermentable carbon sources because the activation of IME1 and IME2 is defective. Both IME1 and IME2 provide virtually essential functions for pseudohyphal growth on several nonfermentable carbon sources (Fig. 2). Attenuation of cAMP signaling by deletion of RAS2, GPR1, or GPA2 did not rescue pseudohyphal growth of Σ1278b cells on nonfermentable carbon sources (not shown), possibly because FLO11 expression requires an active cAMP signaling pathway in Σ1278b cells (71, 72, 77). In SK1 cells, FLO11 expression was induced strongly by acetate (Fig. 8D). In contrast, acetate does not induce FLO11 in Σ1278b cells (70). This potential uncoupling of FLO11 expression from the PKA pathway in SK1 cells may allow SK1 cells to attenuate PKA signaling on nonfermentable carbon sources (5, 63) in order to activate IME1 and IME2. Two isoforms of the catalytic subunit of PKA, Tpk1 and Tpk3, have negative roles in pseudohyphal growth (71, 77), indicating that Tpk1 or Tpk3 may target IME1 or IME2 to inhibit pseudohyphal growth on nonfermentable carbon sources.
Several lines of evidence suggest that Ime1 and Ime2 act independently of Flo11 in pseudohyphal growth. Haploid invasive growth or agar invasion by diploid cells was not decreased by deletion of IME1 or IME2 (Fig. 5A). Deletion of IME1 did not significantly affect FLO11 mRNA levels (Fig. 5B). ime1Δ/ime1Δ cells and, to a lesser degree, ime2Δ/ime2Δ cells displayed cell elongation defects (Fig. 5C). Therefore, Ime1 and Ime2 may act independent of and in parallel to FLO11 in pseudohyphal growth.
Implications for other yeast species.
Ime2 belongs to a family of conserved MAPKs found in all eukaryotes (27, 28), including mammalian MAK, the Schizosaccharomyces pombe proteins Mde3 and Pit1, and the Ustilago maydis protein kinase Crk1. All family members may function in sexual development. MAK is expressed during spermatogenesis (40, 61), Mde3 and Pit1 are important for the timing of the meiotic divisions and spore morphogenesis (1), Crk1 is required for mating (28), and Ime2 is required for entry into meiotic S phase (21). Our finding that Ime2, Ime1, and Ume6 are required for pseudohyphal growth of S. cerevisiae suggests that Ime2 orthologs function in processes other than meiosis, for example, in hyphal development and filamentous growth of pathogenic yeasts, such as Ustilago maydis, Magnaporthe grisea, and Candida species, that undergo dimorphic transitions important for their virulence (9, 49, 50, 86). Indeed, Crk1 is required for hyperpolarized growth of U. maydis cells with defects in cAMP signaling (27) and for infection of maize plants (28), and Ume6 is required for hyphal extension in Candida albicans (3). Our work provides a motivation to investigate the roles of Ime2 orthologs, and possibly orthologs of other meiotic genes, in pseudohyphal and hyphal growth forms and in the virulence of pathogenic yeast species.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/C513418/1) and the European Commission (MIRG-CT-2005-016411).
We thank Joseph Heitman (Duke University, Durham, NC), Aaron Mitchell (Columbia University, New York, NY), Julian Rutherford (University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom), Edward Winter (Thomas Jefferson University, Philadelphia, PA), and Stefan Irniger (Georg August University, Göttingen, Germany) for providing strains and plasmids. We thank the Newcastle-upon-Tyne yeast group for use of a Singer tetrad dissection microscope, Patrick Hussey for use of a Nikon Eclipse TE300 microscope, and David Cox and Andrei Smertenko for help with fluorescence microscopy.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Abe, H., and C. Shimoda. 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, M., D. S. Thompson, A. Lazzell, P. L. Carlisle, C. Pierce, C. Monteagudo, J. L. Lopez-Ribot, and D. Kadosh. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beullens, M., K. Mbonyi, L. Geerts, D. Gladines, K. Detremerie, A. W. Jans, and J. M. Thevelein. 1988. Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 172:227-231. [DOI] [PubMed] [Google Scholar]
- 6.Blacketer, M. J., P. Madaule, and A. M. Myers. 1995. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics 140:1259-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckstaens, M., B. Andre, and A. M. Marini. 2007. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64:534-546. [DOI] [PubMed] [Google Scholar]
- 8.Bolte, M., P. Steigemann, G. H. Braus, and S. Irniger. 2002. Inhibition of APC-mediated proteolysis by the meiosis-specific protein kinase Ime2. Proc. Natl. Acad. Sci. U. S. A. 99:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borges-Walmsley, M. I., and A. R. Walmsley. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8:133-141. [DOI] [PubMed] [Google Scholar]
- 10.Bowdish, K. S., and A. P. Mitchell. 1993. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2172-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 15:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 13.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chant, J., and J. R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, D.-C., B.-C. Yang, and T.-T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 16.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 17.Clifford, D. M., S. M. Marinco, and G. S. Brush. 2004. The meiosis-specific protein kinase Ime2 directs phosphorylation of replication protein A. J. Biol. Chem. 279:6163-6170. [DOI] [PubMed] [Google Scholar]
- 18.Clifford, D. M., K. E. Stark, K. E. Gardner, S. Hoffmann-Benning, and G. S. Brush. 2005. Mechanistic insight into the Cdc28-related protein kinase Ime2 through analysis of replication protein A phosphorylation. Cell Cycle 4:1826-1833. [DOI] [PubMed] [Google Scholar]
- 19.Covitz, P. A., I. Herskowitz, and A. P. Mitchell. 1991. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-α2. Genes Dev. 5:1982-1989. [DOI] [PubMed] [Google Scholar]
- 20.Cullen, P. J., W. Sabbagh, Jr., E. Graham, M. M. Irick, E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. Sprague, Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 22.Donzeau, M., and W. Bandlow. 1999. The yeast trimeric guanine nucleotide-binding protein α subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol. Cell. Biol. 19:6110-6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eraso, P., and J. M. Gancedo. 1984. Catabolite repression in yeasts is not associated with low levels of cAMP. Eur. J. Biochem. 141:195-198. [DOI] [PubMed] [Google Scholar]
- 24.Fox, T. D., L. S. Folley, J. J. Mulero, T. W. McMullin, P. E. Thorsness, L. O. Hedin, and M. C. Costanzo. 1991. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 194:149-165. [DOI] [PubMed] [Google Scholar]
- 25.Frydlova, I., I. Malcova, P. Vasicova, and J. Hasek. 2009. Deregulation of DSE1 gene expression results in aberrant budding within the birth scar and cell wall integrity pathway activation in Saccharomyces cerevisiae. Eukaryot. Cell 8:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagiano, M., F. F. Bauer, and I. S. Pretorius. 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2:433-470. [DOI] [PubMed] [Google Scholar]
- 27.Garrido, E., and J. Perez-Martin. 2003. The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47:729-743. [DOI] [PubMed] [Google Scholar]
- 28.Garrido, E., U. Voss, P. Müller, S. Castillo-Lluva, R. Kahmann, and J. Pérez-Martín. 2004. The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 18:3117-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavrias, V., A. Andrianopoulos, C. J. Gimeno, and W. E. Timberlake. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255-1263. [DOI] [PubMed] [Google Scholar]
- 30.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 31.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 33.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttmann-Raviv, N., S. Martin, and Y. Kassir. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. U. S. A. 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawthorne, D. C. 1955. The use of linear asci for chromosome mapping in Saccharomyces. Genetics 40:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horecka, J., and Y. Jigami. 1999. The trp1-ΔFA designer deletion for PCR-based gene functional analysis in Saccharomyces cerevisiae. Yeast 15:1769-1774. [DOI] [PubMed] [Google Scholar]
- 38.Huxley, C., E. D. Green, and I. Dunham. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6:236. [DOI] [PubMed] [Google Scholar]
- 39.Inai, T., M. Yukawa, and E. Tsuchiya. 2007. Interplay between chromatin and trans-acting factors on the IME2 promoter upon induction of the gene at the onset of meiosis. Mol. Cell. Biol. 27:1254-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinno, A., K. Tanaka, H. Matsushime, T. Haneji, and M. Shibuya. 1993. Testis-specific Mak protein kinase is expressed specifically in the meiotic phase in spermatogenesis and is associated with a 210-kilodalton cellular phosphoprotein. Mol. Cell. Biol. 13:4146-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 42.Kane, S. M., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853-862. [DOI] [PubMed] [Google Scholar]
- 44.Kawaguchi, H., M. Yoshida, and I. Yamashita. 1992. Nutritional regulation of meiosis-specific gene expression in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 56:289-297. [Google Scholar]
- 45.Kron, S. J., C. A. Styles, and G. R. Fink. 1994. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5:1003-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kübler, E., H. U. Mösch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 47.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 48.Law, D. T., and J. Segall. 1988. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 8:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee, N., C. A. D'Souza, and J. W. Kronstad. 2003. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 41:399-427. [DOI] [PubMed] [Google Scholar]
- 50.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorenz, M. C., N. S. Cutler, and J. Heitman. 2000. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell 11:183-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 57.Malathi, K., Y. Xiao, and A. P. Mitchell. 1999. Catalytic roles of yeast GSK3β/shaggy homolog Rim11p in meiotic activation. Genetics 153:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malathi, K., Y. Xiao, and A. P. Mitchell. 1997. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol. Cell. Biol. 17:7230-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallory, M. J., K. F. Cooper, and R. Strich. 2007. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27:951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsumoto, K., I. Uno, and T. Ishikawa. 1983. Initiation of meiosis in yeast mutants defective in adenylate cyclase and cyclic AMP-dependent protein kinase. Cell 32:417-423. [DOI] [PubMed] [Google Scholar]
- 61.Matsushime, H., A. Jinno, N. Takagi, and M. Shibuya. 1990. A novel mammalian protein kinase gene (mak) is highly expressed in testicular germ cells at and after meiosis. Mol. Cell. Biol. 10:2261-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuura, A., M. Treinin, H. Mitsuzawa, Y. Kassir, I. Uno, and G. Simchen. 1990. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. EMBO J. 9:3225-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mbonyi, K., and J. M. Thevelein. 1988. The high-affinity glucose uptake system is not required for induction of the RAS-mediated cAMP signal by glucose in cells of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 971:223-226. [DOI] [PubMed] [Google Scholar]
- 64.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore, M., M. E. Shin, A. Bruning, K. Schindler, A. Vershon, and E. Winter. 2007. Arg-Pro-X-Ser/Thr is a consensus phosphoacceptor sequence for the meiosis-specific Ime2 protein kinase in Saccharomyces cerevisiae. Biochemistry 46:271-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mösch, H.-U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nazar, R. N., H. G. Lawford, and J.-T. Wong. 1970. An improved procedure for extraction and analysis of cellular nucleotides. Anal. Biochem. 35:305-313. [DOI] [PubMed] [Google Scholar]
- 68.Neigeborn, L., and A. P. Mitchell. 1991. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 5:533-548. [DOI] [PubMed] [Google Scholar]
- 69.Pak, J., and J. Segall. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palecek, S. P., A. S. Parikh, J. H. Huh, and S. J. Kron. 2002. Depression of Saccharomyces cerevisiae invasive growth on non-glucose carbon sources requires the Snf1 kinase. Mol. Microbiol. 45:453-469. [DOI] [PubMed] [Google Scholar]
- 71.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan, X., and J. Heitman. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parrou, J. L., M. A. Teste, and J. François. 1997. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143:1891-1900. [DOI] [PubMed] [Google Scholar]
- 74.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 75.Pringle, J. R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732-735. [DOI] [PubMed] [Google Scholar]
- 76.Purnapatre, K., M. Gray, S. Piccirillo, and S. M. Honigberg. 2005. Glucose inhibits meiotic DNA replication through SCFGrr1p-dependent destruction of Ime2p kinase. Mol. Cell. Biol. 25:440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. U. S. A. 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rose, M., and F. Winston. 1984. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol. Gen. Genet. 193:557-560. [DOI] [PubMed] [Google Scholar]
- 79.Rubin-Bejerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16:2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rubin-Bejerano, I., S. Sagee, O. Friedman, L. Pnueli, and Y. Kassir. 2004. The in vivo activity of Ime1, the key transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae, is inhibited by the cyclic AMP/protein kinase A signal pathway through the glycogen synthase kinase 3-β homolog Rim11. Mol. Cell. Biol. 24:6967-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubinstein, A., V. Gurevich, Z. Kasulin-Boneh, L. Pnueli, Y. Kassir, and R. Y. Pinter. 2007. Faithful modeling of transient expression and its application to elucidating negative feedback regulation. Proc. Natl. Acad. Sci. U. S. A. 104:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rutherford, J. C., G. Chua, T. Hughes, M. E. Cardenas, and J. Heitman. 2008. An Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol. Biol. Cell 19:3028-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sáez, M. J., and R. Lagunas. 1976. Determination of intermediary metabolites in yeast. Critical examination of the effect of sampling conditions and recommendations for obtaining true levels. Mol. Cell. Biochem. 13:73-78. [DOI] [PubMed] [Google Scholar]
- 85.Sagee, S., A. Sherman, G. Shenhar, K. Robzyk, N. Ben-Doy, G. Simchen, and Y. Kassir. 1998. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1985-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sánchez-Martínez, C., and J. Pérez-Martín. 2001. Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis—similar inputs, different outputs. Curr. Opin. Microbiol. 4:214-221. [DOI] [PubMed] [Google Scholar]
- 87.Santangelo, G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:253-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sari, F., M. Heinrich, W. Meyer, G. H. Braus, and S. Irniger. 2008. The C-terminal region of the meiosis-specific protein kinase Ime2 mediates protein instability and is required for normal spore formation in budding yeast. J. Mol. Biol. 378:31-43. [DOI] [PubMed] [Google Scholar]
- 89.Schindler, K., K. R. Benjamin, A. Martin, A. Boglioli, I. Herskowitz, and E. Winter. 2003. The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell. Biol. 23:8718-8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schlanderer, G., and H. Dellweg. 1974. Cyclic AMP and catabolite repression in yeasts. In Schizosaccharomyces pombe glucose lowers both intracellular adenosine 3′:5′-monophosphate levels and the activity of catabolite-sensitive enzymes. Eur. J. Biochem. 49:305-316. [DOI] [PubMed] [Google Scholar]
- 91.Schröder, M., J. S. Chang, and R. J. Kaufman. 2000. The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Genes Dev. 14:2962-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schröder, M., R. Clark, C. Y. Liu, and R. J. Kaufman. 2004. The unfolded protein response represses differentiation through the RPD3-SIN3 histone deacetylase. EMBO J. 23:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schüller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 94.Sedgwick, C., M. Rawluk, J. Decesare, S. Raithatha, J. Wohlschlegel, P. Semchuk, M. Ellison, J. Yates III, and D. Stuart. 2006. Saccharomyces cerevisiae Ime2 phosphorylates Sic1 at multiple PXS/T sites but is insufficient to trigger Sic1 degradation. Biochem. J. 399:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shefer-Vaida, M., A. Sherman, T. Ashkenazi, K. Robzyk, and Y. Kassir. 1995. Positive and negative feedback loops affect the transcription of Ime1, a positive regulator of meiosis in Saccharomyces cerevisiae. Dev. Genet. 16:219-228. [DOI] [PubMed] [Google Scholar]
- 96.Sia, R. A. L., and A. P. Mitchell. 1995. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol. Cell. Biol. 15:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith, H. E., S. E. Driscoll, R. A. L. Sia, H. E. Yuan, and A. P. Mitchell. 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanhill, A., N. Schick, and D. Engelberg. 1999. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19:7529-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strich, R., R. T. Surosky, C. Steber, E. Dubois, F. Messenguy, and R. E. Esposito. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796-810. [DOI] [PubMed] [Google Scholar]
- 102.Tamaki, H., T. Miwa, M. Shinozaki, M. Saito, C. W. Yun, K. Yamamoto, and H. Kumagai. 2000. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267:164-168. [DOI] [PubMed] [Google Scholar]
- 103.Thomas, J. H., and D. Botstein. 1987. Ordered linear tetrads are produced by the sporulation of newly formed zygotes of Saccharomyces cerevisiae. Genetics 115:229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 106.Vidan, S., and A. P. Mitchell. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wach, A., A. Brachat, R. Pöhlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 108.Ward, M. P., C. J. Gimeno, G. R. Fink, and S. Garrett. 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15:6854-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Washburn, B. K., and R. E. Esposito. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21:2057-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao, Y., and A. P. Mitchell. 2000. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell. Biol. 20:5447-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]