Abstract
Microbial rhodopsins, a diverse group of photoactive proteins found in Archaea, Bacteria, and Eukarya, function in photosensing and photoenergy harvesting and may have been present in the resource-limited early global environment. Four different physiological functions have been identified and characterized for nearly 5,000 retinal-binding photoreceptors, these being ion transporters that transport proton or chloride and sensory rhodopsins that mediate light-attractant and/or -repellent responses. The greatest number of rhodopsins previously observed in a single archaeon had been four. Here, we report a newly discovered six-rhodopsin system in a single archaeon, Haloarcula marismortui, which shows a more diverse absorbance spectral distribution than any previously known rhodopsin system, and, for the first time, two light-driven proton transporters that respond to the same wavelength. All six rhodopsins, the greatest number ever identified in a single archaeon, were first shown to be expressed in H. marismortui, and these were then overexpressed in Escherichia coli. The proteins were purified for absorption spectra and photocycle determination, followed by measurement of ion transportation and phototaxis. The results clearly indicate the existence of a proton transporter system with two isochromatic rhodopsins and a new type of sensory rhodopsin-like transducer in H. marismortui.
Microbial rhodopsins comprise a large family of seven-transmembrane helical proteins that either mediate light-driven ion transport to harvest solar energy or serve as receptors to mediate phototaxis (13) and possibly photoadaptation (32). In archaea, four rhodopsins responding to different wavelengths of light with distinct functions in Halobacterium salinarum have been identified and characterized (20, 32). These consist of two light-driven ion transporters that pump protons (bacteriorhodopsin [BR]) (21) or chloride (halorhodopsin [HR]) (19, 28) and two sensory rhodopsins that mediate both attractant and repellent phototaxis (sensory rhodopsin I [SRI]) (4, 33) or only repellent phototaxis (sensory rhodopsin II [SRII]) (35).
The two ion-transporting rhodopsins perform light-driven outward proton transport to create a proton-electrochemical potential or inward chloride transport to maintain the osmotic and pH homeostasis of the cell. The photoactivated sensory rhodopsins, on the other hand, undergo light-triggered conformational changes to relay signals to their cognate transducers and consequently activate signaling cascades in a manner similar to that of the two-component system involved in eubacterial chemotaxis (1, 22) to control flagellum rotation and thus swimming direction. Our current understanding of microbial rhodopsins as both ion transporters and photosensory receptors has been based primarily on these four known rhodopsins.
A recently completed genome project for Haloarcula marismortui (3) proposed the existence of six opsin-related genes, the greatest number ever found in a single archaeon. This proposal immediately raised three questions. (i) Are these six rhodopsins biologically expressed and functionally active? (ii) What is the maximum-absorbance-wavelength (λmax) distribution pattern of these six rhodopsins? (iii) Do the two extra rhodopsins, compared to the four in H. salinarum, perform new functions or have new features beyond those of the known system? This study demonstrates several features of this system that are unique compared to the features of currently known systems, and it confirms the six-rhodopsin system and the presence of two additional isochromatic rhodopsins comprising a novel light-driven proton transporter system and a new type of sensory-like rhodopsin signaling complex with a unique transducer.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α was used for cloning, and E. coli C43(DE3) was used for protein expression. The rhodopsin and transducer genes were amplified by PCR from the genomic DNA of H. marismortui. Primers were designed by introducing an NdeI restriction enzyme cutting site into the start codon and a HindIII restriction enzyme cutting site in apposition to the stop codon. The NdeI-HindIII DNA fragments were digested and ligated into NdeI-HindIII-treated pET-21b(+). Consequently, the vector encoded 6 histidines at the C terminus, which resulted in the following N- and C-terminal peptide sequences: 1M—-KLAAALEHHHHHH, where the hyphens represent the protein sequence for the gene of interest and the underlined amino acids represent the introduced HindIII site. The accuracy of all constructs was confirmed using nucleotide sequencing. H. marismortui (ATCC 43049) cells were grown in medium designed for H. marismortui and herein designated halomedium [4.28 M NaCl, 81 mM MgSO4, 1.02 mM Na3C6H5O7, 27 mM KCl, 1% (wt/wt) Oxoid peptone LP0034, 2.30 μM ZnSO4, 1.01 μM MnSO4, 1.40 μM Fe(NH4)SO4, 0.28 μM CuSO4] (23) at 42°C. Different illumination conditions and culture periods were selected for different experiments.
Protein expression and purification.
The protein sample was prepared as described previously and optimized with some modifications (14, 27). Briefly, both rhodopsins and transducers were expressed in E. coli C43(DE3). Freshly transformed E. coli C43(DE3) cells were grown in an LB medium containing 50 mg/liter ampicillin at 37°C. Overexpression of the protein was induced using IPTG (isopropyl-β-d-thiogalactopyranoside plus 10 μM all-trans retinal for rhodopsin expression). Cells were harvested by centrifugation (6,000 × g) and resuspended in buffer A (50 mM Tris, 4 M NaCl, pH 7.8, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 14.7 mM β-mercaptoethanol). Cell disruption was performed with sonication. Membranes were sedimented at 100,000 × g for 1 h at 4°C and solubilized in buffer A containing 1.5% (wt/vol) n-dodecyl-β-d-maltoside (DDM) (Anatrace) for 16 h at 4°C. After centrifugation of the solubilized membranes (50,000 × g, 30 min, 4°C), the supernatant was treated with Ni-nitrilotriacetic acid (NTA) agarose, which had been preequilibrated with buffer B (50 mM Tris, 4 M NaCl, 0.05% DDM, pH 7.8) containing 20 mM imidazole for 6 to 8 h at 4°C with slow rotation. The loaded resin was filled onto a chromatography column and washed extensively with buffer C (buffer B plus 50 mM imidazole). Target proteins were then eluted from the column with buffer D (buffer B plus 250 mM imidazole). Imidazole was removed from these fractions using Amicon against buffer E (50 mM MES [morpholineethanesulfonic acid], 4 M NaCl, pH 5.8, 0.05% DDM) for further assay and long-term storage. The typical yield of purified protein was a 2-mg/liter culture, with minor variations among six rhodopsins.
RT-PCR of rhodopsins and transducers transcript.
For isolation of H. marismortui total RNA, approximately 50 ml of H. marismortui culture (optical density at 600 nm [OD600] = 0.5) were centrifuged at 7,500 × g and 4°C for 15 min. The cell pellet was lysed in 1.2 ml of TRI reagent (Sigma, St. Louis, MO), and the total RNA was isolated by following manufacturer-suggested procedures. Finally, the RNA pellet was resuspended in RNase-free H2O and stored at −80°C until use. For cDNA synthesis, 4 μg of DNase I (Sigma, St. Louis, MO)-treated and ReverTra Ace (Toyobo, Osaka, Japan)-purified total RNA was used in reverse transcription with random hexamers (ReverTra Ace kit; Toyobo). The cDNA products were diluted 20-fold with deionized water and used as a template in reverse transcriptase PCR (RT-PCR). The primers used to differentiate between the six rhodopsins and three transducers are listed in Table SA2 in the supplemental material. The HotStar Taq DNA polymerase (Qiagen, Chatsworth, CA) was used according to suggested procedures. The PCR amplicons were examined using agarose gel electrophoresis.
Flash-laser-induced photocycle measurements.
Nd-YAG laser (532 nm, 6-ns pulse, 40 mJ) flash-absorbance changes were measured in the laboratory of John Spudich by employing a laboratory-constructed laser cross-beam flash spectrometer as previously described (6). The purified proteins were dissolved in buffer E to reach a concentration of 0.3 at a maximum-wavelength (λmax) OD (ODλmax), and transient-absorbance changes were recorded at their corresponding ground-state λmax. The curves represent the loss and recovery of absorbance at the target wavelengths upon green-laser (532-nm) excitation.
Light-driven ion transporter assay.
A light-driven ion transporter assay was conducted and modified as described in previous research (36). Briefly, E. coli cell-expressed rhodopsins were harvested by centrifugation (3,600 × g) at 25°C for 10 min and then washed twice with a nonbuffered solution (10 mM NaCl, 10 mM MgSO4, and 100 μM CaCl2) at room temperature. Finally, the cells were resuspended in a nonbuffer solution adjusted to an OD600 of ∼2 in a dark room, and the samples were activated by illuminating them with a 1-W continuous-light-emitting diode (LED) green laser (532 nm). The real-time ion transporter activity was monitored with an RS-232-linked computerized Eutech Instruments Cyberscan 2100 pH meter.
Chloride binding affinity assay.
The concentration of purified H. marismortui HR (HmHR) in different chloride concentrations was adjusted to determine the chloride binding affinity as described in a previous study, with slight modifications (26). The NaCl concentration was reduced from 4 M to 0.24 mM in 50 mM MES buffer, pH 5.8. At each chloride concentration, the UV/visible-light (Vis) spectrum was recorded and normalized by using the spectrum at A280 as a standard. Differences of absorbance in relative units were plotted against chloride concentration (log10 scale), and then the resultant curve was well fitted to the Hill equation (R square = 0.99) supplied in the Origin 8 program: y = (Vmax × xn)/(kn + xn), where Vmax is maximum velocity, k is the Michaelis constant, and n is the number of cooperative sites. The k value was considered a dissociation constant.
Motile-cell culture.
H. marismortui cells were selected for motility, with swarm plates containing 0.4% (wt/vol) agar in halomedium. Cells at the edge of diffusive cell masses were picked and inoculated into halomedium and shaken at 42°C for at least 5 days to reach saturation. Saturated culture was diluted 1:100 in halomedium and subcultured at 42°C for 48 h and then diluted 1:3 with halomedium immediately preceding microscopic observation.
Phototaxis measurements.
Phototaxis measurements were performed according to a previously described motion analysis algorithm (17).
RESULTS
Primary sequence analysis and biological expression of six opsin genes and transducers in H. marismortui.
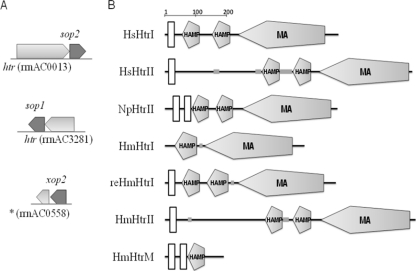
Three photosensory transducer gene candidates (Fig. 1A), hinted at in the original H. marismortui genomic study, were first identified and analyzed from the whole-genome map (see Table SA1 in the supplemental material). Protein domains and structural-feature analysis (Fig. 1B) using BLAST (2, 12, 18) and SMART (16, 29) revealed that the encoded protein of the first htr (GenBank locus tag rrnAC0013), HtrII of H. marismortui (HmHtrII), is a sensory rhodopsin transducer, conjugated upstream of sop2, as with HtrII from H. salinarum (HsHtrII) (34). HmHtrII contained a two-transmembrane region connected by a large extracellular loop, followed by a histidine kinase-binding domain extruding to the cytoplasmic side in the C terminus (15, 37). The protein encoded by the second transducer candidate (htr, rrnAC3281), HmHtrI, conjugated upstream of sop1, was first determined to share 53.8% identity with the cytoplasmic region in the C terminus of HsHtrI (25), but it lacks the two-transmembrane region (Fig. 1B). Later sequence analysis recognized that the two-transmembrane region existed if an upstream alternative start codon, GTG (−291), was adopted, making it a 528-residue protein with 57.3% identity with full-length HsHtrI, designated reHmHtrI (interchangeable with HmHtrI for the rest of this article). For the third transducer gene candidate (rrnAC0558) conjugated downstream of xop2, no known analogue was identified using the same analysis, but the theoretically translated product, HmHtrM, clearly showed a two-transmembrane region with an unusual shortened C terminus containing only a HAMP (stands for histidine kinases, adenylyl cyclases, methyl-binding proteins, and phosphatases) domain and extruded to the cytoplasmic side, a conserved structural pattern for known sensory rhodopsin cognate transducers (Fig. 1B). No matching cognate transducer genes were found for bop, xop1, and hop.
FIG. 1.
Gene candidate identification for cognate transducers. (A) Three htr open reading frames (ORFs) (light shading) conjugated either upstream or downstream of a corresponding opsin gene (dark shading). The gene locus tag is indicated for each htr. *, no annotation was made for this gene from the original genome project. (B) The translated protein sequences from the htr ORFs were analyzed using SMART (http://smart.embl-heidelberg.de). reHmHtrI represents restoration of the missing two-transmembrane region of HmHtrI (see the text for details). The scale bar shows the length of the amino acid sequence. The large open vertical rectangle indicates that this section of the protein sequence was analyzed as a transmembrane region. The small gray rectangle indicates the amino acid sequence predicted to be a low-complexity region. The pentagons illustrate protein-signaling domains. HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins and phosphatases; MA (MCP), methyl-accepting chemotaxis protein.
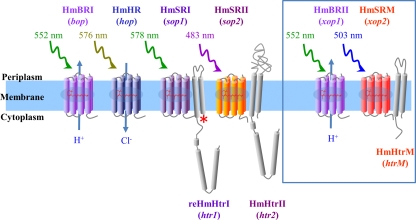
Together with the six annotated opsin genes, a six-rhodopsin three-transducer system, including protein names, was proposed (Fig. 2) (protein names and maximum absorbance values for each rhodopsin were determined and are explained below).
FIG. 2.
Configurations, gene names, and protein names for the six rhodopsins and their cognate transducers from H. marismortui. HmBRI (bop) is a light-driven proton transporter; HmHR (hop) is a light-driven chloride transporter; HmBRII (xop1), originally predicted to be a BR precursor, is, like HmBRI, a light-driven proton transporter; and HmSRM (xop2), which was annotated as an “opsin of unknown function,” associates with HmHtrM (htrM), and its function is yet to be determined. HmSRI (sop1) associates with reHmHtrI (htr1), and HmSRII (sop2) associates with HmHtrII (htr2) to mediate photoattractant and photorepellent responses, respectively. The blue box indicates the two rhodopsins that are in addition to the currently known four-rhodopsin system identified from H. salinarum.
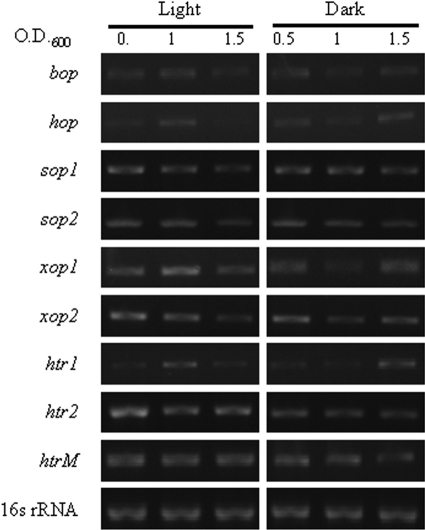
Experiments were performed to examine the biological expression of all six rhodopsins and the three proposed transducers, including the restored two-transmembrane htr for sop1 and the unusual short transducer for xop2 (Fig. 3), in H. marismortui. Total RNA was prepared from cells grown in the presence or absence of white-light illumination for reverse transcription-PCR, RT-PCR, and analysis, and despite minor differences in expression levels under different illumination conditions, mRNAs coding for all rhodopsins and transducers were found to be detectable throughout the early-log, mid-log, and stationary phases in both the presence and absence of white light (Fig. 3), showing that these genes are indeed expressed and physiologically active independent of environmental illumination, and these results offer further direct proof of the existence of a six-rhodopsin system in a single archaeon.
FIG. 3.
RT-PCR analysis of the rhodopsins and signal transducers. The mRNAs coding for all rhodopsins (bop, hop, sop1, sop2, xop1, and xop2) and transducers (htr1, htr2, and htrM) were detectable in the cDNA of H. marismortui grown with and without white-light illumination. The expression profiles of these genes in early-, mid-, and late-log-phase cultures (i.e., OD600s of 0.5, 1.0, and 1.5, respectively) were analyzed by RT-PCR with gene-specific primers. The 16S rRNA transcript was used as an internal reference for relative quantification (see Fig. SA1 in the supplemental material).
Cloning and purification of all six rhodopsins from genomic DNA.
Expression plasmids were prepared by obtaining each gene fragment from the genomic DNA of H. marismortui by PCR, and the gene fragments were further cloned into the pET-21b(+) vector before the vector was transformed into E. coli for expression as described in Materials and Methods to first observe the responding light wavelength for each protein encoded by the six opsin genes.
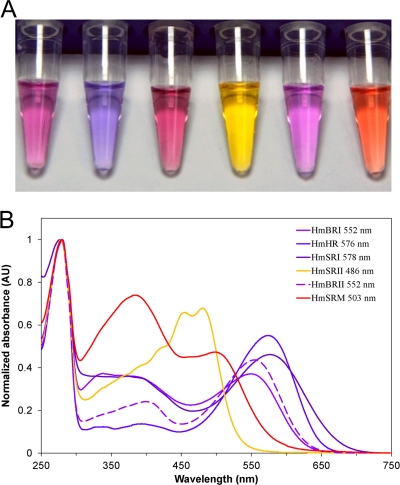
The purified proteins (Fig. 4A) from six genes all showed visually distinguishable colors, and their maximum absorbances were determined (Fig. 4B) with a UV/Vis spectrophotometer. All six proteins were found to absorb within the visible light range from 483 nm to 578 nm and were named (Fig. 2) according to their λmax, sequence analysis, and functional assays (described later).
FIG. 4.
Spectrum determination of the six individual purified rhodopsins from H. marismortui. (A) Group photo of all six rhodopsins purified from E. coli dissolved in MES buffer containing 0.05% n-dodecyl-β-d-maltoside, pH 5.8. From left to right, HmBRI, HmHR, HmSRI, HmSRII, HmBRII, and HmSRM. (B) Absorbance spectrum and absorbance peak of each rhodopsin. AU, arbitrary units.
HmSRII and HmSRM absorbed at 483 nm and 503 nm, respectively, and appeared to fall into the range of wavelengths known to trigger repellent responses based on the results of previous studies (32). Further toward the red, both of the products of two rhodopsin genes that were annotated as “bacteriorhodopsin” and “bacteriorhodopsin precursor” in the genome project (3) absorbed at the same wavelength of 552 nm and were named HmBRI and HmBRII, respectively. Toward the longer wavelengths, the protein absorbing at 578 nm was named HmSRI, while HmHR (explained later) was named for the protein maximally absorbed at 576 nm (32).
Flash-laser-induced photolysis measurement of the six rhodopsins.
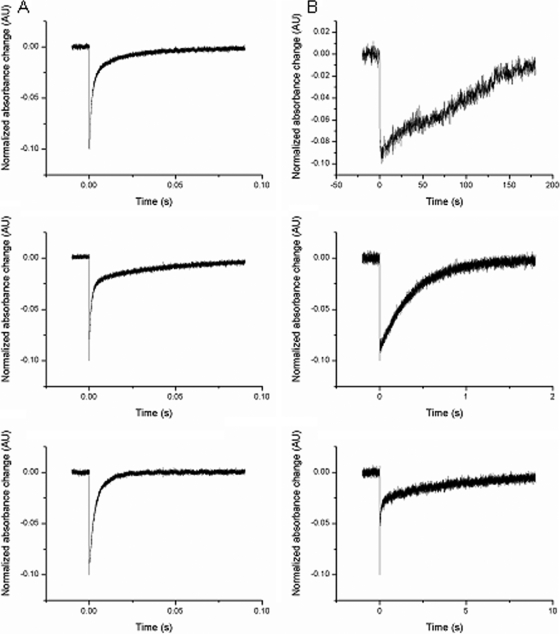
Photocycle time represents the basic definition for a light-inducible photoreceptor and the kinetic properties of its functionality (32). For each of those six rhodopsins (Fig. 5), the photocycle period, representing the decay and regeneration of the ground state, was measured and found to fall into two distinct groups; one, including HmBRI, HmBRII, and HmHR, was completed within ∼6 ms, while HmSRI, HmSRII, and HmSRM showed a slower recovery time of 4 to 6 s. Faster photocycle kinetics in the millisecond range has been proposed to indicate, but not determine, an ion transporter, while slower kinetics in the seconds range suggests sensory rhodopsin (7-8). HmSRM was thus classified as sensory-like based on the kinetics of the photocycle and the biologically expressed HmHtrM with which it associated.
FIG. 5.
Flash-induced absorption changes for the six purified H. marismortui rhodopsins. The purified proteins were dissolved in MES buffer containing 0.05% n-dodecyl-β-d-maltoside, pH 5.8, and transient absorbance changes were recorded at their corresponding ground state λmax. (A) Transients were measured for HmBRI at 550 nm (top), HmBRII at 550 nm (middle), and HmHR at 580 nm (bottom), and all photocycles were completed within a millisecond. (B) Transients were measured for HmSRI at 570 nm (top), HmSRII at 480 nm (middle), and HmSRM at 500 nm (bottom), and photocycles were completed with a second. The curves represent the change of absorbance corresponding to the λmax for each rhodopsin upon green-laser (532-nm) excitation.
Measurements of light-driven ion transportation.
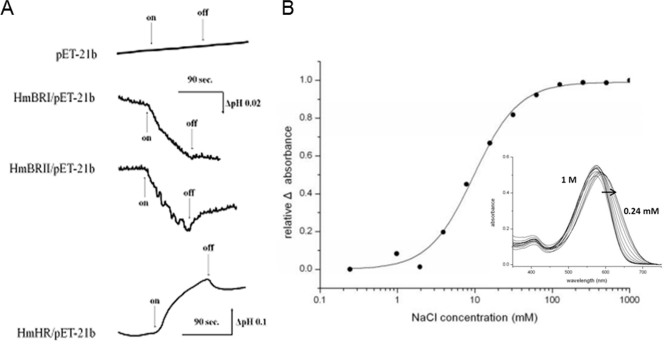
A unique light-driven proton transporter system composed of isochromatic rhodopsins HmBRI and HmBRII was proposed from the results of sequence alignments with bacteriorhodopsin, HmBRI and HmBRII having identical maximum absorbances at 552 nm and kinetics of photocycle in the millisecond range. To further confirm this speculation, the two genes were first expressed in E. coli cells and then suspended in nonbuffer as described in Materials and Methods for light-driven proton transportation measurements (Fig. 6A). Upon light activation, HmBRI and HmBRII were observed to indeed undergo light-dependent outward proton transportation, directly supporting the existence of a light-driven proton transportation system with two isochromatic rhodopsins.
FIG. 6.
Measurement of light-driven transport of ions. (A) A suspension of E. coli cells expressing HmBRI, HmBRII, or HmHR alone was used to measure light-induced pH changes. The beginning and end of illumination using a 10-mW, 532-nm continuous green laser are indicated by the arrows labeled “on” and “off,” respectively. (B) Chloride binding affinity determination for HmHR. The inset shows the spectral shifts of HmHR in different chloride concentrations. The direction of shift from high- to low-salt concentration is marked by an arrow.
HmHR was analyzed to mediate light-triggered inward chloride transportation and was examined through proton cotransportation associated with a chloride affinity assay (24). Passive proton transportation (Fig. 6A) showed that HmHR performed light-driven chloride transportation. The further experiment was performed to determine the chloride binding affinity (26). Within the chloride concentration decrease from 1 M to 0.24 mM (Fig. 6B), the spectrum exhibited a red-shift pattern and the determined Kd was 9.66 mM, a value close to those for both HsHR and Natronobacterium pharaonis HR (NpHR).
HmSRI and HmSRII mediated phototaxis for H. marismortui cells.
H. marismortui cells were cultured at 42°C for 2 days and then were observed under infrared light (>770 nm) under a phase-contrast light microscope. A motion observation and recording system with motion-analyzing software (17) developed in our laboratory were used to study the swimming responses to various wavelengths of light. When the swimming cells were illuminated with 465- ± 15-nm light, a rapid reverse swimming response was clearly observed (17), which led us to conclude that HmSRII mediated the photorepellent response. When light with a wavelength greater than 600 nm, designed to stimulate only HmSRI, was applied, the reverse-swimming rates decreased to a level below 50% of the control level, indicating that HmSRI is a photoattractant rhodopsin. HmSRM, on the other hand, showed no phototactic response upon light illumination (Fig. 7). The whole six-rhodopsin system is summarized in Fig. 2.
FIG. 7.
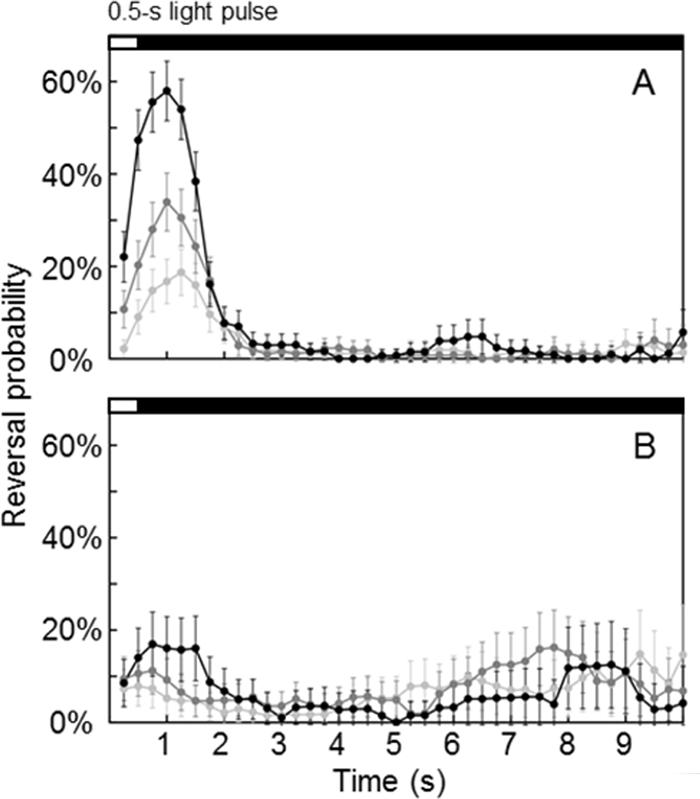
Reversal kinetics upon 0.5-s pulse stimulation of HmSRII (A) and HmSRM (B) with 465- ± 15-nm and 540- ± 10-nm lights, respectively. Light-gray, dark-gray, and black circles represent 25%, 50%, and 75% light intensity, respectively. The estimated 75% light intensities for the 465- ± 15-nm and 540- ± 10-nm lights were 3.7 × 106 and 2.9 × 106 photons·mm−2·s−1, respectively. Each panel was derived from 720 s of video from a single set of experiments. Error bars indicate 95% confidence intervals, assuming a binomial distribution; the number of trajectories ranged from 117 to 235.
DISCUSSION
In this study, a six-rhodopsin system containing the greatest number of rhodopsins so far identified in any single archaeon (3, 10, 32) was confirmed in H. marismortui. Three features not previously observed in any other archaea were unveiled, including the more diverse λmax distributions of the expressed rhodopsins, the light-driven proton transporter system with two isochromatic rhodopsins, and a new type of sensory rhodopsin-like protein with a transducer containing only the short cytoplasmic region without the usual histidine kinase binding domain.
First, H. marismortui has a more diverse photosensing system. Based on analysis of the λmax distribution pattern (Fig. SA2 in the supplemental material), it displayed a more finely tuned rhodopsin system than any currently known microbes with rhodopsins (32). In the known range of wavelength for sensory, i.e., 470 nm to 540 nm, the H. marismortui system has two dedicated rhodopsins, HmSRII (483 nm) and HmSRM (503 nm), instead of one, as do NpSRII (498 nm) and HsSRII (487 nm) for Natronobacterium pharaonis and H. salinarum, respectively (see Fig. SA2 in the supplemental material).
Second, in a previous study, the two isochromatic, light-driven proton transporter genes for both HmBRI and HmBRII were proposed to be two BR-like light-driven transporters with different λmaxs, extending the light-harvesting range (3). The results of this study revealed an unexpected isochromatic feature: both proteins absorb in the same range of light with the same λmax. In addition, functional measurements showed that both proteins transport protons out of the cell upon being illuminated, and these two proteins were also shown to be expressed under both dark and light conditions with similar photocycle kinetics. Further research into the differences between HmBRI and HmBRII will likely discover properties that have not yet been identified.
Third, the newly discovered type of sensory-like rhodopsin and transducer system, HmSRM-HmHtrM, was unexpected. The transducer associated with HmSRM, HmHtrM, appears to have an unusual short cytoplasmic region containing only a HAMP domain instead of only a HAMP domain and a histidine kinase binding domain, as have been found in all other known photosensory transducers. This configuration of HmHtrM therefore hints that direct controlling of flagellar proteins is unlikely. Our motion analysis experiments directly support this speculation, since cells under light designed to activate HmSRM showed no phototactic response.
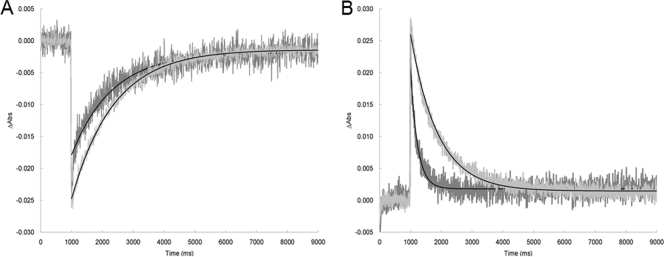
The existence of an HmSRM-HmHtrM system was supported by other results as well. In addition to (i) HmHtrM's lack of phototactic function, (ii) information from the htrM gene locus, (iii) translated-protein sequence analysis, and (iv) the fact it was shown to be biologically expressed, HmHtrM is yet to be directly shown to be a transducer for HmSRM. HmSRM-HmHtrM and HmSRM-HmHtrI fusion proteins were therefore constructed. The HmSRM-HmHtrM protein showed an increased duration time for M-intermediate formation upon activation with a 532-nm laser (Fig. 8). In contrast, the HmSRM-HmHtrI combination resulted in an extremely unstable protein complex which tended to denature immediately after purification and under any dim-light illumination (data not shown).
FIG. 8.
Laser flash-induced absorption changes for HmSRM and HmSRM-HmHtrM fusion proteins. (A) Dark-state depletion and recovery of HmSRM and the HmSRM-HmHtrM fusion protein. Laser flash photolysis (laser pulse time, 10 ms, 1 W) of HmSRM and the HmSRM-HmHtrM fusion protein in 50 mM Tris-HCl, 4 M NaCl, pH 6.8, 0.05% DDM at room temperature. The absorption change (ΔAbs) of HmSRM (dark gray) and the HmSRM-HmHtrM fusion (light gray) were monitored at 503 nm and 507 nm, respectively, and the flash was set to a time of 1 s. (B) Duration of the M-intermediate in HmSRM and the HmSRM-HmHtrM fusion protein. Laser-induced photolysis (laser pulse time, 10 ms, 1 W) of HmSRM and the HmSRM-HmHtrM fusion protein in 50 mM Tris-HCl, 4 M NaCl, pH 6.8, 0.05% DDM at room temperature. The absorption change of HmSRM (dark gray) and the HmSRM-HmHtrM fusion (light gray) were monitored at 378 nm and 379 nm, respectively, and the flash was set to a time of 1 s. Data fitting (black) based on a one-phase exponential-decay model and the duration of the signaling state, M-intermediate, was 164 ms in HmSRM and 622 ms in the HmSRM-HmHtrM fusion form.
The lack of a histidine kinase binding domain to directly contact flagellar proteins while still responding to a specific wavelength, 503 nm, of light suggests a plausible conclusion: the HmSRM-HmHtrM system must exert a function or mediate a mechanism which is new to our knowledge of the photosensory system. The possible physiological role of HmSRM-HmHtrM is still to be determined, but HmSRM-HmHtrM shows the appropriate configuration to mediate light-dependent interactions for other proteins. This speculation arises from the observation that HmHtrM features a unique short C terminus extruded into the cytosol with only one HAMP domain, which is known to initialize mainly protein-protein interactions (11), while lacking the whole histidine kinase binding domain that is universally observed in all other microbial transducers associated with sensory rhodopsins. Several possibilities can thus be proposed. Perhaps the HmSRM-HmHtrM complex modulates the availability of CheB or CheR to HmSRI or HmSRII by interacting with CheB or CheR and consequently regulates the sensitivity of HmSRI or HmSRII, or it may interact with other proteins for various illumination-related functions, like photoadaptation or circadian rhythm.
Two rhodopsin phototaxis receptors found in Chlamydomonas reinhardtii have the same function in vivo, namely, mediating light-gated cation channel activity (31). These receptors, called channelrhodopsins-1 and -2 differ by their absorption maxima, 460 nm versus 500 nm, respectively, as well as in their levels of light saturation, which extend the wavelength and intensity ranges for phototaxis by C. reinhardtii (30). Yet-undiscovered differences in properties of the two HmBR proteins are likely to provide an advantage to the organism, but our results rule out the possibility that they expand the wavelength range since they have identical absorption spectra. Another recently completed genomic project on another archaeon, Haloquadratum walsbyi, hinted at, but did not confirm, the presence of two annotated bacteriorhodopsin-like genes (5). Here, H. marismortui has two BRs simultaneously absorbing the same wavelength of light and both functioning as light-driven proton transporters. We further found that those two proteins exhibit different tolerances to pH changes, as the pKa- or pH-dependent λmax shift measurements showed HmBRII to leave unchanged the λmax in buffers with pHs of 3.0 to 9.0, while a >10-nm blue-shift for λmax in pH 9.0 was observed in HmBRI.
Retinal-binding proteins represent one of the most efficient signal-capturing and signal transduction systems that relay activation information to G-protein systems, histidine kinase systems, and channels (9). Exploring the physiological roles and significance of this six-rhodopsin system will surely expand our understanding of the already-complex rhodopsin system and will unveil more of the survival strategies which supported some life forms in the early global environment.
Supplementary Material
Acknowledgments
We thank John L. Spudich, Elena N. Spudich, Jun Sasaki, and Oleg A. Sineshchekov (Center for Membrane Biology, Department of Biochemistry and Molecular Biology, University of Texas Medical School) for providing the facilities for the laser flash photolysis study and discussion essential for this work. Special thanks go to Chin-Fa Shih (E-Hong Instruments Co., Ltd., Taipei, Taiwan) and Tzu-Ping Huang (National Synchrotron Radiation Research Center, Hsinchu, Taiwan) for their assistance in photocycle system construction. We are also grateful to Andrew H.-J. Wang (SINICA, Taiwan) for encouragement and discussion.
Footnotes
Published ahead of print on 27 August 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chiu, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14:2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogomolni, R. A., and J. L. Spudich. 1982. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc. Natl. Acad. Sci. U. S. A. 79:6250-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolhuis, H., P. Palm, A. Wende, M. Falb, M. Rampp, F. Rodriguez-Valera, F. Pfeiffer, and D. Oesterhelt. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., and J. L. Spudich. 2004. Five residues in the HtrI transducer membrane-proximal domain close the cytoplasmic proton-conducting channel of sensory rhodopsin I. J. Biol. Chem. 279:42964-42969. [DOI] [PubMed] [Google Scholar]
- 7.Chizhov, I., and M. Engelhard. 2001. Temperature and halide dependence of the photocycle of halorhodopsin from Natronobacterium pharaonis. Biophys. J. 81:1600-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chizhov, I., G. Schmies, R. Seidel, J. R. Sydor, B. Luttenberg, and M. Engelhard. 1998. The photophobic receptor from Natronobacterium pharaonis: temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fain, G. L., R. Hardie, and S. B. Laughlin. 2010. Phototransduction and the evolution of photoreceptors. Curr. Biol. 20:R114-R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falb, M., K. Muller, L. Konigsmaier, T. Oberwinkler, P. Horn, S. von Gronau, O. Gonzalez, F. Pfeiffer, E. Bornberg-Bauer, and D. Oesterhelt. 2008. Metabolism of halophilic archaea. Extremophiles 12:177-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrand, E., and N. Dencher. 1975. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature 257:46-48. [DOI] [PubMed] [Google Scholar]
- 14.Hohenfeld, I. P., A. A. Wegener, and M. Engelhard. 1999. Purification of histidine tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 442:198-202. [DOI] [PubMed] [Google Scholar]
- 15.Hou, S., A. Brooun, H. S. Yu, T. Freitas, and M. Alam. 1998. Sensory rhodopsin II transducer HtrII is also responsible for serine chemotaxis in the archaeon Halobacterium salinarum. J. Bacteriol. 180:1600-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, Y. C., H. Y. Fu, and C. S. Yang. 2010. Phototaxis of Haloarcula marismortui revealed through a novel microbial motion analysis algorithm. Photochem. Photobiol. 86:1084-1090. [DOI] [PubMed] [Google Scholar]
- 18.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 19.Matsuno-Yagi, A., and Y. Mukohata. 1980. ATP synthesis linked to light-dependent proton uptake in a rad mutant strain of Halobacterium lacking bacteriorhodopsin. Arch. Biochem. Biophys. 199:297-303. [DOI] [PubMed] [Google Scholar]
- 20.Mukohata, Y., K. Ihara, T. Tamura, and Y. Sugiyama. 1999. Halobacterial rhodopsins. J. Biochem. 125:649. [DOI] [PubMed] [Google Scholar]
- 21.Oesterhelt, D., and W. Stoeckenius. 1971. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233:149-152. [DOI] [PubMed] [Google Scholar]
- 22.Parkinson, J. S. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J. Bacteriol. 185:1492-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robb, F. T. 1995. Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Sasaki, J., L. S. Brown, Y. S. Chon, H. Kandori, A. Maeda, R. Needleman, and J. K. Lanyi. 1995. Conversion of bacteriorhodopsin into a chloride ion pump. Science 269:73-75. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki, J., and J. L. Spudich. 2008. Signal transfer in haloarchaeal sensory rhodopsin-transducer complexes. Photochem. Photobiol. 84:863-868. [DOI] [PubMed] [Google Scholar]
- 26.Scharf, B., and M. Engelhard. 1994. Blue halorhodopsin from Natronobacterium pharaonis: wavelength regulation by anions. Biochemistry 33:6387-6393. [DOI] [PubMed] [Google Scholar]
- 27.Schmies, G., I. Chizhov, and M. Engelhard. 2000. Functional expression of His-tagged sensory rhodopsin I in Escherichia coli. FEBS Lett. 466:67-69. [DOI] [PubMed] [Google Scholar]
- 28.Schobert, B., and J. K. Lanyi. 1982. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257:10306-10313. [PubMed] [Google Scholar]
- 29.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sineshchekov, O. A., E. G. Govorunova, and J. L. Spudich. 2009. Photosensory functions of channel rhodopsins in native algal cells. Photochem. Photobiol. 85:556-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sineshchekov, O. A., K. H. Jung, and J. L. Spudich. 2002. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 99:8689-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spudich, J. L. 2006. The multitalented microbial sensory rhodopsins. Trends Microbiol. 14:480-487. [DOI] [PubMed] [Google Scholar]
- 33.Spudich, J. L., and R. A. Bogomolni. 1984. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature 312:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich, J. L., C. S. Yang, K. H. Jung, and E. N. Spudich. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365-392. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, T., Y. Mochizuki, N. Kamo, and Y. Kobatake. 1985. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem. Biophys. Res. Commun. 127:99-105. [DOI] [PubMed] [Google Scholar]
- 36.Wang, W. W., O. A. Sineshchekov, E. N. Spudich, and J. L. Spudich. 2003. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 278:33985-33991. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, W., A. Brooun, M. M. Mueller, and M. Alam. 1996. The primary structures of the Archaeon Halobacterium salinarum blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc. Natl. Acad. Sci. U. S. A. 93:8230-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.