Abstract
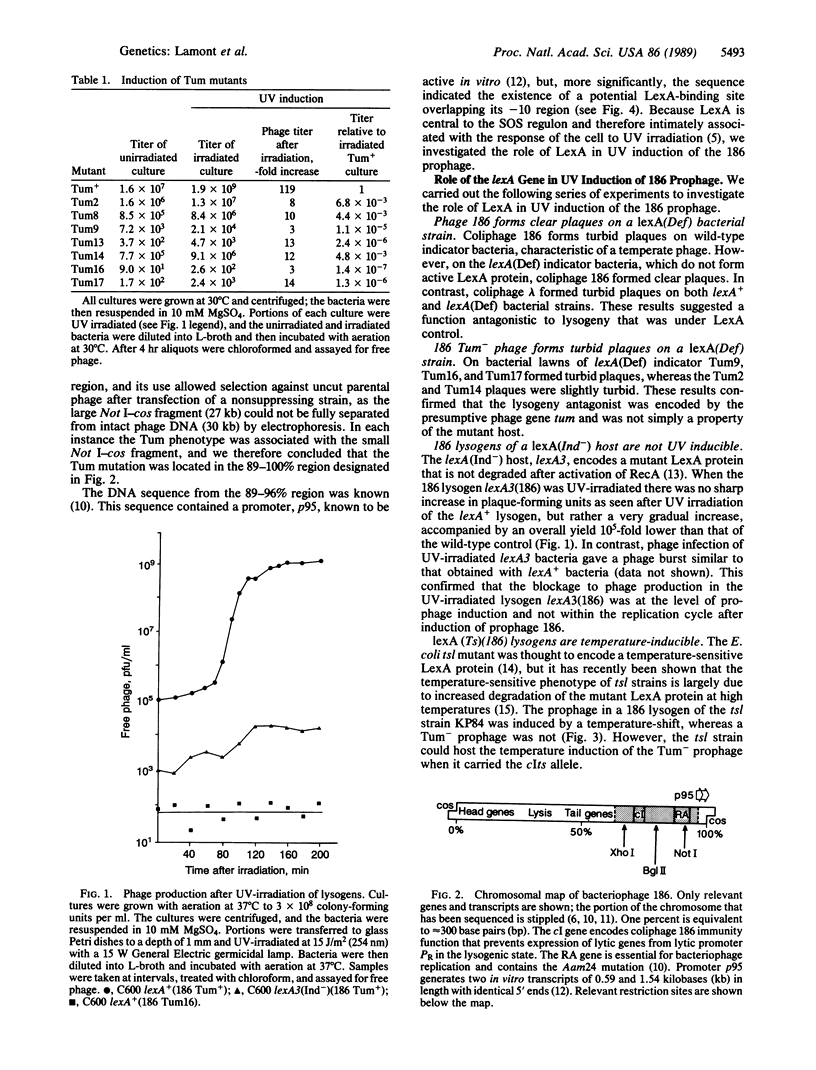
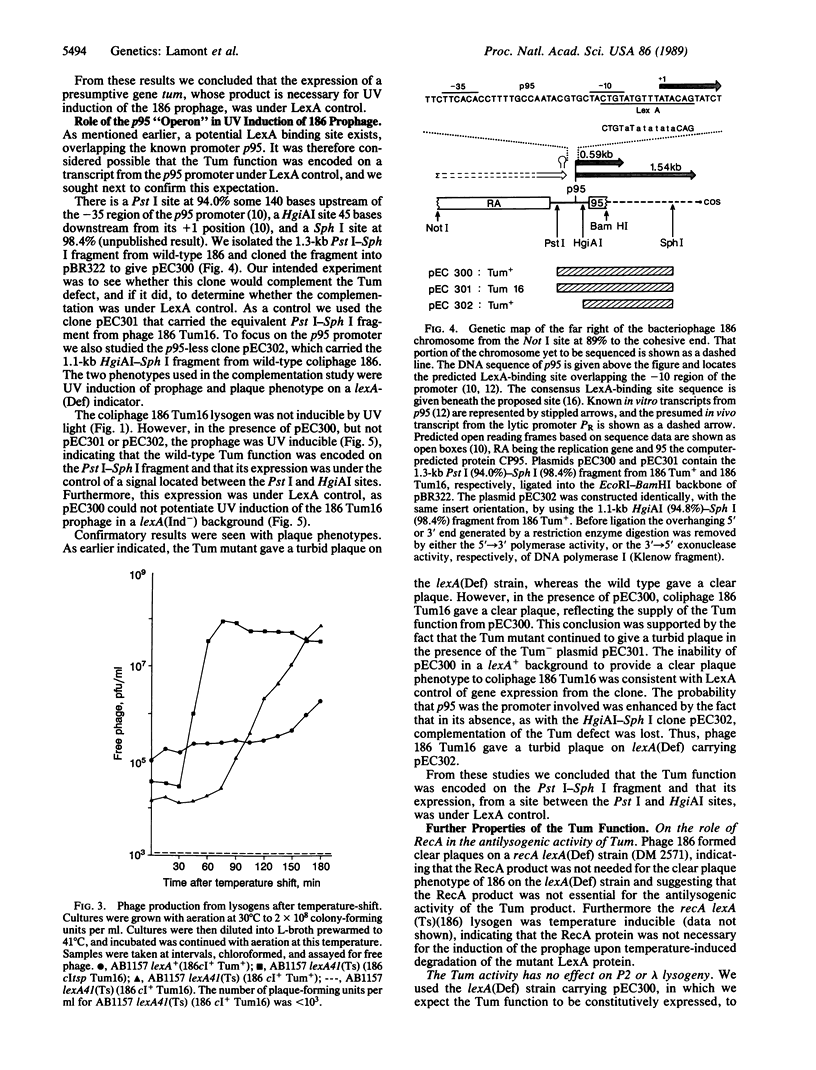
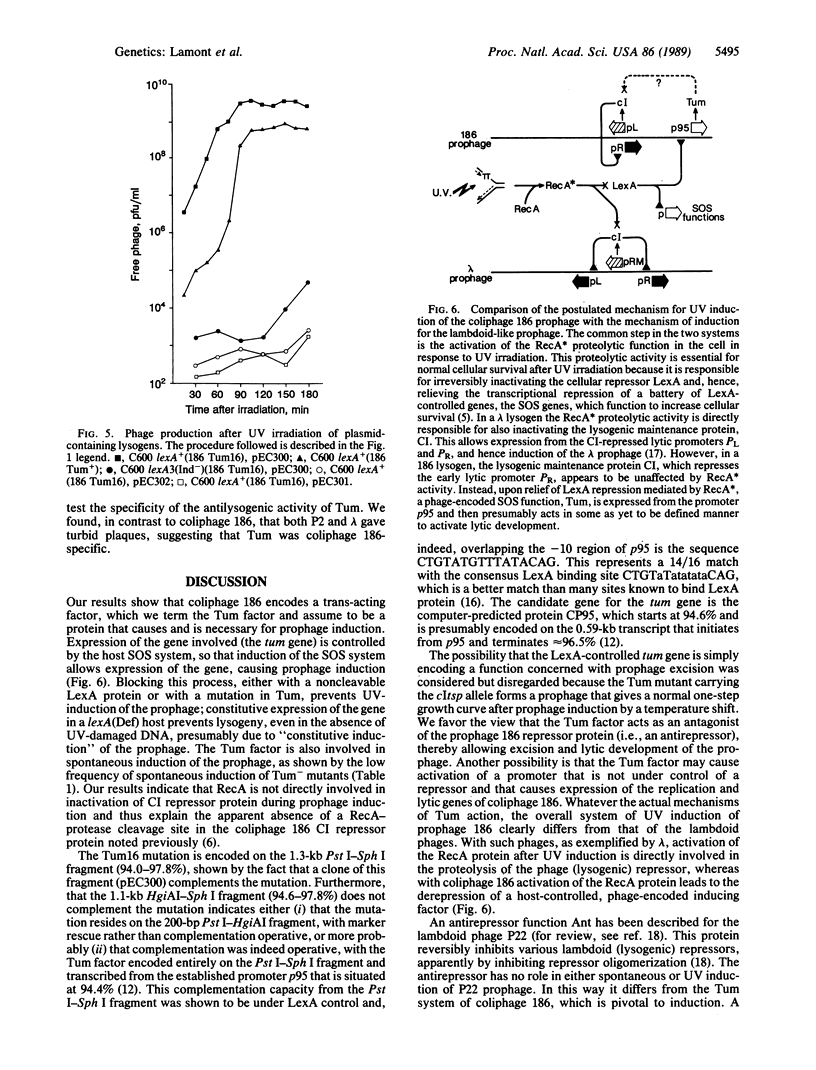
Our results show that UV induction of the 186 prophage depends upon the phage function Tum, with the mutant phenotype of turbid plaques on mitomycin plates and the expression of which is controlled by the host LexA protein. Tum function, encoded near the right-hand end of the coliphage 186 chromosome, is under the control of promoter p95. This promoter is overlapped by a sequence closely related to the consensus sequence of the LexA-binding site. It is proposed that inactivation of LexA after UV irradiation (or by genetic means) leads to prophage induction by permitting expression of Tum which, by unknown means, induces prophage. This mechanism is basically different from that seen with the UV-inducible lambdoid coliphages, which are not regulated by LexA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley C., Ling O. P., Egan J. B. Isolation of phage P2-186 intervarietal hybrids and 186 insertion mutants. Mol Gen Genet. 1975 Sep 29;140(2):123–135. doi: 10.1007/BF00329780. [DOI] [PubMed] [Google Scholar]
- Hooper I., Woods W. H., Egan B. Coliphage 186 Replication is delayed when the host cell is UV irradiated before infection. J Virol. 1981 Nov;40(2):341–349. doi: 10.1128/jvi.40.2.341-349.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalionis B., Dodd I. B., Egan J. B. Control of gene expression in the P2-related template coliphages. III. DNA sequence of the major control region of phage 186. J Mol Biol. 1986 Sep 20;191(2):199–209. doi: 10.1016/0022-2836(86)90257-3. [DOI] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987 Nov;1(9):1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- Little J. W., Edmiston S. H., Pacelli L. Z., Mount D. W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Low K. B., Edmiston S. J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J Bacteriol. 1972 Nov;112(2):886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. R., Mount D. W. Differential repression of SOS genes by unstable lexA41 (tsl-1) protein causes a "split-phenotype" in Escherichia coli K-12. J Mol Biol. 1987 Jan 5;193(1):27–40. doi: 10.1016/0022-2836(87)90623-1. [DOI] [PubMed] [Google Scholar]
- Pritchard M., Egan J. B. Control of gene expression in P2-related coliphages: the in vitro transcription pattern of coliphage 186. EMBO J. 1985 Dec 16;4(13A):3599–3604. doi: 10.1002/j.1460-2075.1985.tb04123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillardet P., Moreau P. L., Ginsburg H., Mount D. W., Devoret R. Cell survival, UV-reactivation and induction of prophage lambda in Escherichia coli K12 overproducing RecA protein. Mol Gen Genet. 1982;188(1):37–43. doi: 10.1007/BF00332993. [DOI] [PubMed] [Google Scholar]
- Richardson H., Puspurs A., Egan J. B. Control of gene expression in the P2-related temperate coliphage 186. VI. Sequence analysis of the early lytic region. J Mol Biol. 1989 Mar 5;206(1):251–255. doi: 10.1016/0022-2836(89)90539-1. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W., Craig N. L. Escherichia coli recA gene product inactivates phage lambda repressor. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka S. A., Hanson P. Comparisons of the distribution of nucleotides and common sequences in deoxyribonucleic acid from selected bacteriophages. J Virol. 1972 Apr;9(4):583–593. doi: 10.1128/jvi.9.4.583-593.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprizhitsky YuA, Kopylov V. M. The SOS system of Escherichia coli in the regulation of bacteriophage lambda development. FEBS Lett. 1983 Aug 22;160(1-2):7–10. doi: 10.1016/0014-5793(83)80925-9. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hoess R. The molecular genetics of bacteriophage P1. Annu Rev Genet. 1983;17:123–154. doi: 10.1146/annurev.ge.17.120183.001011. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman K. F., Mount D. W. Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J Bacteriol. 1985 Jul;163(1):376–384. doi: 10.1128/jb.163.1.376-384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods W. H., Egan J. B. Prophage induction of noninducible coliphage 186. J Virol. 1974 Dec;14(6):1349–1356. doi: 10.1128/jvi.14.6.1349-1356.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younghusband H. B., Egan J. B., Inman R. B. Characterization of the DNA from bacteriophage P2-186 hybrids and physical mapping of the 186 chromosome. Mol Gen Genet. 1975 Sep 29;140(2):101–110. doi: 10.1007/BF00329778. [DOI] [PubMed] [Google Scholar]


