Abstract
Background
Malignant fibrous histiocytomas (MFHs), or undifferentiated pleomorphic sarcomas, are in general high-grade tumours with extensive chromosomal aberrations. In order to identify recurrent chromosomal regions of gain and loss, as well as novel gene targets of potential importance for MFH development and/or progression, we have analysed DNA copy number changes in 33 MFHs using microarray-based comparative genomic hybridisation (array CGH).
Principal findings
In general, the tumours showed numerous gains and losses of large chromosomal regions. The most frequent minimal recurrent regions of gain were 1p33-p32.3, 1p31.3-p31.2 and 1p21.3 (all gained in 58% of the samples), as well as 1q21.2-q21.3 and 20q13.2 (both 55%). The most frequent minimal recurrent regions of loss were 10q25.3-q26.11, 13q13.3-q14.2 and 13q14.3-q21.1 (all lost in 64% of the samples), as well as 2q36.3-q37.2 (61%), 1q41 (55%) and 16q12.1-q12.2 (52%). Statistical analyses revealed that gain of 1p33-p32.3 and 1p21.3 was significantly associated with better patient survival (P = 0.021 and 0.046, respectively). Comparison with similar array CGH data from 44 leiomyosarcomas identified seven chromosomal regions; 1p36.32-p35.2, 1p21.3-p21.1, 1q32.1-q42.13, 2q14.1-q22.2, 4q33-q34.3, 6p25.1-p21.32 and 7p22.3-p13, which were significantly different in copy number between the MFHs and leiomyosarcomas.
Conclusions
A number of recurrent regions of gain and loss have been identified, some of which were associated with better patient survival. Several specific chromosomal regions with significant differences in copy number between MFHs and leiomyosarcomas were identified, and these aberrations may be used as additional tools for the differential diagnosis of MFHs and leiomyosarcomas.
Introduction
The concept of malignant fibrous histiocytoma (MFH) has changed during the last decades, and is now used to describe a heterogeneous group of tumours without a specific known lineage of differentiation and with fibroblastic/myofibroblastic features. Tumours still classified as MFHs are also termed undifferentiated high grade pleomorphic sarcomas (UPSs) according to the latest World Health Organization (WHO) classification [1]. MFHs were initially assigned to several subgroups; pleomorphic, myxoid, giant cell and inflammatory, which are still used. However, several so-called giant cell MFHs are now reclassified as other giant cell sarcomas, and several so-called inflammatory MFHs are now recognized to be dedifferentiated liposarcomas [2]. The so-called myxoid MFHs are now termed myxofibrosarcomas [1], and this second common subtype have a better prognosis than the most common subtype, pleomorphic MFHs [3].
MFHs occur mainly late in life, between the age of 50 and 70 years, and the main locations are in the lower extremities followed by the upper extremities and the retroperitoneum [3]. The main exception is inflammatory MFHs, which are most often located in the retroperitoneum. MFHs were previously regarded as the most common soft tissue sarcoma of adults, and depending on the criteria used for classification, MFHs still account for a considerable portion of these tumours. Men are more frequently affected than women, about 2/3 of the tumours occur in men [3]. MFHs are in general high-grade tumours, and the 5-year survival is 65–70% [3].
Cytogenetic studies have revealed that most MFHs have complex karyotypes with numerous aberrations, both numerical and structural. Using chromosome-based comparative genomic hybridization (CGH), recurrent gains of regions in 1p, 1q, 5p, 17p, 17q and 20q have frequently been observed, as well as losses of regions in 2q, 9p, 10q, 11q and 13q [4], [5], [6], [7], [8], [9], [10], [11]. High-level amplification of the distal part of 13q has frequently been found [5], [6]. Gain of 17q has been associated with longer disease-free survival and low risk of developing distant metastasis [9], whereas gain of 7q32 has been associated with poor prognosis [5]. In addition, patients with gain of 1p31 showed a trend towards decreased overall survival [5].
More recently, microarray-based CGH (array CGH) has been used to analyse DNA copy number changes at higher resolution. In order to identify specific genomic events and candidate targets that may play a role in MFH development and/or progression, we have used array CGH to map the distribution and frequency of DNA copy number changes at high resolution in 33 MFH samples. Statistical analyses were performed in order to identify possible correlations between the experimental results and the clinical information. In addition, the results were compared to array CGH data from 44 leiomyosarcomas in order to identify chromosomal aberrations significantly different between the two tumour types, since their differential diagnosis may be difficult due to histological similarities.
Materials and Methods
Tumour samples
Thirty-one human sarcomas classified as MFHs were selected from a tumour collection at the Department of Tumour Biology at The Norwegian Radium Hospital. In addition, two samples initially diagnosed as leiomyosarcomas were included in the study after reclassification to MFH (MFH73x and MFH76x). All tumours were revised at the time of the study by an expert pathologist (B.B.) and diagnosed according to the current WHO classification [1].
Clinical samples were collected immediately after surgery, cut into small pieces, frozen in liquid nitrogen and stored at −70°C until use. Some of the samples were grown subcutaneously in immunodeficient mice as xenografts (suffix x). The clinical information was retrieved from the MEDinsight database at The Norwegian Radium Hospital. Clinical data for all samples are given in Table 1.
Table 1. Clinical data of malignant fibrous histiocytomas.
| Sample | Sample origin | Patient age/sex | Diagnosis | MFH Subtype | Grade1 | Primary tumour location | Size (cm)2 | Metastasis (months)3 | Status | Follow-up (months)4 |
| MFH1 | Rec | 87/M | MFH | Myxofibrosarcoma | 4 | Lower arm | 5 | 48 | DD | 50 |
| MFH2x | Rec | 71/M | MFH | Spindle and pleomorphic | 4 | Upper trunk | 19 | 4 | DD | 15 |
| MFH4 | Prim | 77/M | MFH | Spindle and pleomorphic | 4 | Upper arm | 15 | MD | DD | 3 |
| MFH7 | Prim | 68/M | MFH | Spindle and pleomorphic | 4 | Upper leg | 6 | NM | DOC | 74 |
| MFH8 | Met | 76/M | MFH | Spindle and pleomorphic | 4 | Knee5 | 6 | 15 | DD | 27 |
| MFH9 | Rec | 57/F | MFH | Spindle and pleomorphic | 4 | Upper leg | 3 | NM | DOC | 346 |
| MFH14 | Prim | 60/M | MFH | Spindle and pleomorphic | 4 | Knee | 5.5 | 38 | DD | 82 |
| MFH15 | Prim | 45/F | MFH | Pleomorphic with giant cells | 4 | Upper leg | 20 | 6 | DOC | 6 |
| MFH16 | Rec | 63/M | MFH | Myxofibrosarcoma | 4 | Upper trunk | NA | NM | DOC | 191 |
| MFH18 | Prim | 61/M | MFH | Spindle and pleomorphic | 4 | Lower leg | 18 | 16 | DD | 28 |
| MFH19 | Rec | 71/F | MFH | Spindle and pleomorphic | 4 | Upper leg | 5 | NM | NED | 168 |
| MFH20 | Met | 66/M | MFH | Spindle and pleomorphic | 4 | Upper leg6 | 18 | 17 | DD | 40 |
| MFH21 | Prim | 73/F | MFH | Spindle and pleomorphic | 4 | Upper trunk | 15 | 7 | DD | 21 |
| MFH24 | Prim | 92/F | MFH | Spindle and pleomorphic | 4 | Upper leg | 13 | NM | NED | 159 |
| MFH25 | Prim | 47/F | MFH | Myxofibrosarcoma | 4 | Upper leg | 20 | NM | NED | 182 |
| MFH27 | Rec | 60/M | MFH | Spindle and pleomorphic | 3 | Knee | 7.5 | 34 | DD | 181 |
| MFH30 | Prim | 56/M | MFH | Spindle and pleomorphic | 4 | Retroperitoneum | 15 | MD | DD | 3 |
| MFH34 | Rec | 79/F | MFH | Spindle and pleomorphic | 3 | Upper trunk | 1.7 | NM | DOC | 80 |
| MFH36 | Prim | 75/M | MFH | Myxofibrosarcoma | 4 | Shoulder | 20 | 7 | DD | 14 |
| MFH42 | Prim | 71/M | MFH | Pleomorphic with giant cells | 4 | Upper arm | 7.5 | 4 | DD | 22 |
| MFH44 | Rec | 80/F | MFH | Spindle and pleomorphic | 4 | Upper leg | 13 | NM | DOC | 15 |
| MFH45 | Prim | 69/F | MFH | Spindle and pleomorphic | 4 | Upper arm | 14 | NM | NED | 119 |
| MFH46 | Prim | 82/M | MFH | Spindle and pleomorphic | 4 | Upper leg | 11 | NM | NED | 86 |
| MFH47 | Prim | 56/M | MFH | Spindle and pleomorphic | 4 | Upper leg | 7 | 7 | DD | 19 |
| MFH48 | Prim | 63/M | MFH | Spindle and pleomorphic | 4 | Lower leg | 7 | MD | DD | 100 |
| MFH53 | Prim | 69/F | MFH | Pleomorphic with giant cells | 4 | Retroperitoneum | 18 | 5 | DD | 10 |
| MFH54 | Prim | 45/M | MFH | Myxofibrosarcoma | 4 | Upper leg | 20 | 6 | DD | 7 |
| MFH56 | Prim | 75/F | MFH | Spindle and pleomorphic | 4 | Upper arm | 10 | 9 | NED | 85 |
| MFH59 | Prim | 68/M | MFH | Spindle and pleomorphic | 4 | Upper trunk | 10.5 | MD | DD | 7 |
| MFH60 | Prim | 58/F | MFH | Pleomorphic | 4 | Upper arm | 11 | 9 | DD | 58 |
| MFH61 | Prim | 66/M | MFH | Myxofibrosarcoma | 4 | Upper leg | 9.5 | NM | NED | 74 |
| MFH73x | Rec | 82/F | MFH | Spindle cell | 4 | Lower arm | 7 | 14 | DOC | 59 |
| MFH76x | Prim | 69/F | MFH | Spindle and pleomorphic | 4 | Lower leg | 10 | 8 | DD | 18 |
Abbreviations: x, xenograft; Rec, recurrence; Prim, primary tumour; Met: metastasis; M, male; F, female; MFH, malignant fibrous histiocytoma; NA, not available; MD, metastasis at diagnosis; NM, no metastasis; DD, dead of disease; DOC, dead of other cause; NED, no evidence of disease;
Grading is based on a four-tiered system used in the Scandinavian Sarcoma Group (SSG);
Largest diameter of the tumour;
Time to first metastasis from diagnosis;
Time to last follow-up from diagnosis;
Metastasis located in the lung;
Metastasis located in the abdominal wall.
Ethics Statement
The information given to the patients, the written consent used, the collection of samples, and the research project were approved by the ethical committee of Southern Norway (Project S-06133). Human tumour samples were obtained with the corresponding written consent given by the patients. Animal care was in accordance with National and institutional guidelines and the project approved by the National Committee on Research on Animal Care (Project 1498).
Array CGH
he genomic microarray used contained 4,549 bacterial- and P1 artificial chromosome (BAC and PAC) clones representing the human genome at approximately 1 Mb resolution, as well as the minimal tiling-path between 1q12 and the beginning of 1q25. Detailed information on the construction and preparation of the microarray has been previously described [12]. The microarrays were provided by the Norwegian Microarray Consortium (www.microarray.no).
Array CGH was performed essentially as described previously [12]. In brief, approximately 500 ng of DpnII-digested total genomic DNA was labelled by random priming using BioPrime DNA Labeling System (Invitrogen, California, USA) and Cy3-dCTP (tumour) or Cy5-dCTP (reference) (PerkinElmer, Massachusetts, USA). Labelled tumour and reference DNA were combined together with 135 µg human Cot-1 DNA (Invitrogen). Hybridisation was performed using an automated hybridisation station, GeneTAC (Genomic Solutions/PerkinElmer), agitating the hybridisation solution for 42–46 hours at 37°C. The arrays were scanned using an Agilent G2565BA scanner (Agilent Technologies, California, USA), and the images were segmented using GenePix Pro 6.0 (Axon Laboratories, California, USA). Further data processing, including filtering and normalization, was performed using M-CGH as previously described [12], [13].
Array CGH data analysis
The complete array CGH dataset for the 33 MFHs can be viewed in the ArrayExpress microarray database (www.ebi.ac.uk/arrayexpress, accession number E-MEXP-1804). Clones belonging to chromosomes 1–22 with known unique chromosomal location in Ensembl (www.ensembl.org, v33, Sep 2005) were considered for analysis (3,351 clones). Due to experimental variation in normal control experiments, 22 clones (0.7%) were discarded as described previously [12]. In addition, clones with missing values in 10 or more of the 33 samples were discarded, leaving 3,144 clones for analysis. The remaining missing values were imputed via a K-Nearest Neighbour algorithm normalization using “Significance Analysis of Microarrays” (SAM) [14].
Clustering of all samples was performed using J-Express v. 2.7 [15], with average linkage (WPGMA) as the cluster method and Pearson correlation as the distance metric. In order to determine copy number changes, CGH-Explorer v. 3.1b was used [16]. “Analysis of Copy Errors” (ACE) was performed using a false discovery rate of 0.0001 and medium sensitivity. Chromosomal segments showing gains or losses in at least 10 of 33 MFHs (>30%) were used to identify minimal recurrent regions of alteration.
Statistical analysis
Statistical analyses were performed using SPSS 15.0. Samples were categorized based on the experimental results and compared with the clinical data (Table 1). Overall survival was analyzed using Kaplan-Meier survival curves and tested for significance using the Log Rank test for all clinical variables and minimal recurrent chromosomal regions altered. P values less than or equal to 0.05 were considered to be statistically significant. Multivariate analysis of experimental and clinical variables significantly associated with survival was performed using Cox regression.
In order to identify chromosomal regions with significantly different DNA copy number between two groups of samples, a two-class unpaired t-test was performed using SAM [14]. Using 100 permutations and a false discovery rate of <1%, a list of genomic clones showing significant copy number differences was generated. Chromosomal segments represented by multiple significant clones (at least five significant clones with less than 10 non-significant clones between two significant clones) were considered to be significantly different between the two groups.
Results
Recurrently altered chromosomal regions in malignant fibrous histiocytomas
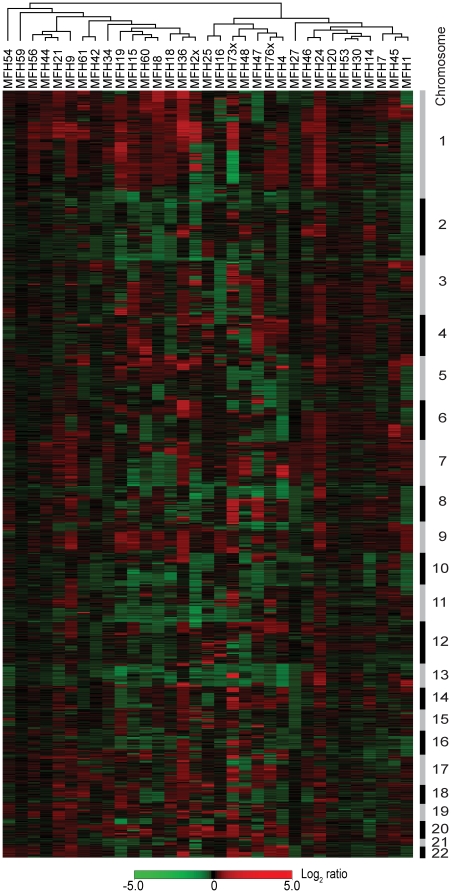
DNA copy number changes in a panel of 33 MFHs (Table 1) were analysed using a 1 Mb resolution BAC and PAC genomic microarray supplemented with the tiling-path between 1q12 and the beginning of 1q25. Hierarchical clustering of all samples based on the DNA copy number changes is shown in Figure 1. No associations between the clustering pattern and the clinical features were apparent. The panel consisted of mainly primary tumours and recurrences, and SAM was used to identify chromosomal regions with significantly different DNA copy number between the two subtypes, but no differences were found.
Figure 1. Hierarchical clustering of MFHs.
Hierarchical clustering of 33 MFHs using DNA copy number ratios relative to a pool of normal diploid DNA. A total of 3,144 unique genomic clones are shown in chromosomal order from 1ptel to 22qtel. Chromosomes are indicated with black and grey bars. Red, increases in DNA copy number; green, decreases in DNA copy number.
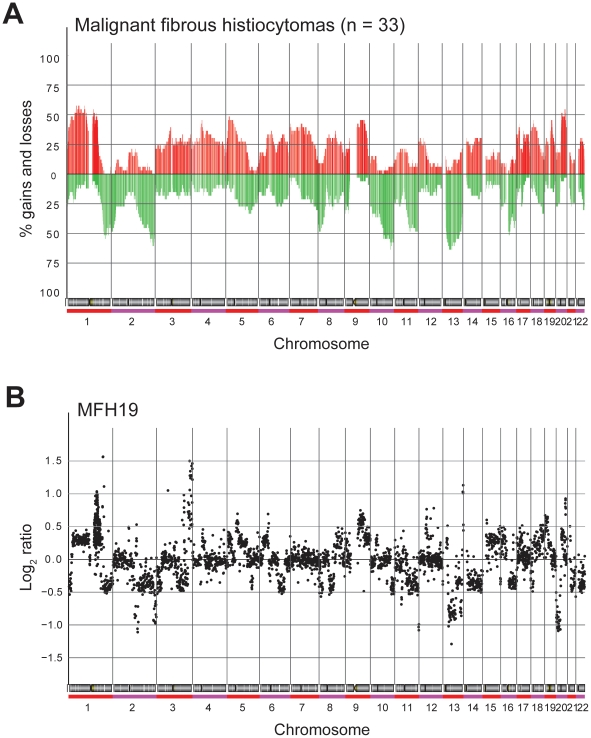
Regions with significant DNA copy number changes were identified in each sample using the ACE algorithm in CGH-Explorer. The resulting frequency plot of gains and losses is shown in Figure 2A, and a representative genome-wide ratio plot for this type of tumours is shown in Figure 2B. The ratio plots for all samples are shown in Figure S1. Minimal recurrent regions of alteration identified by ACE in at least 10/33 (>30%) samples are presented in Table 2. The complete list of data of all defined regions of gain and loss from the ACE analysis is presented in Table S1.
Figure 2. DNA copy number alterations.
(A) Genome-wide frequency plot of copy number alterations identified by ACE in 33 MFHs. Red, increases in DNA copy number; green, decreases in DNA copy number. (B) Whole genome DNA copy number profile of a representative MFH. Log2 ratio for each of the genomic clones is plotted according to chromosome position.
Table 2. Minimal recurrent regions altered in malignant fibrous histiocytomas (n = 33).
| Cytoband | Aberration | Start clone | End clone | Size (Mb) | Frequency |
| 1p33-p32.3 | Gain | RP1-86A18 | RP4-631H13 | 2.6 | 19/33 |
| 1p31.3-p31.2 | Gain | RP11-5P4 | RP5-1033K19 | 5.5 | 19/33 |
| 1p21.3 | Gain | RP11-17C2 | RP11-413P11 | 1.2 | 19/33 |
| 1q21.2-q21.3 | Gain | RP11-363I22 | RP11-316M1 | 0.5 | 18/33 |
| 1q41 | Loss | RP11-323K10 | RP11-241C9 | 2.3 | 18/33 |
| 2p25.3-p25.1 | Loss | RP11-352J11 | RP11-542B5 | 6.0 | 16/33 |
| 2q36.3-q37.2 | Loss | RP11-70L16 | RP11-84G18 | 6.9 | 20/33 |
| 3p12.1-p11.2 | Gain | RP11-382L10 | RP11-312H1 | 3.9 | 13/33 |
| 4q12 | Gain | RP11-738E22 | RP11-355L4 | 0.2 | 15/33 |
| 5p14.3 | Gain | RP11-28P24 | RP11-26L18 | 2.8 | 16/33 |
| 5q23.2-q31.3 | Loss | CTB-54G2 | RP11-515C16 | 13.2 | 11/33 |
| 6q12-q13 | Gain | RP1-160B9 | RP11-256L9 | 3.3 | 12/33 |
| 7p22.1 | Gain | RP11-172O13 | RP1-42M2 | 0.4 | 14/33 |
| 7q11.21-q11.22 | Gain | RP11-340I6 | RP11-471N21 | 8.5 | 14/33 |
| 7q36.1-q36.3 | Loss | RP11-24N19 | CTB-3K23 | 10.4 | 10/33 |
| 8p23.2-p22 | Loss | RP11-245H16 | RP11-44L18 | 9.9 | 16/33 |
| 8q22.1 | Gain | RP11-266D22 | RP11-3D19 | 2.6 | 12/33 |
| 9p24.2-p24.1 | Loss | RP11-509J21 | RP11-509D8 | 1.5 | 11/33 |
| 9q21.33-q31.3 | Gain | RP11-280P22 | RP11-470J20 | 24.1 | 15/33 |
| 10q25.3-q26.11 | Loss | RP11-96N16 | RP11-5G18 | 1.1 | 21/33 |
| 11q14.1 | Loss | RP11-118L16 | RP11-482L11 | 1.9 | 16/33 |
| 11q14.3-q21 | Loss | RP11-268B20 | RP11-83E23 | 2.2 | 16/33 |
| 11q23.3-q25 | Loss | CTD-3245B9 | RP11-469N6 | 16.0 | 16/33 |
| 12p13.31-p13.1 | Gain | RP11-277E18 | RP11-377D9 | 5.2 | 10/33 |
| 13q13.3-q14.2 | Loss | RP11-131F1 | RP11-174I10 | 9.5 | 21/33 |
| 13q14.3-q21.1 | Loss | RP11-384G23 | RP11-516G5 | 3.0 | 21/33 |
| 14q23.3-q31.3 | Gain | RP11-125H8 | RP11-203D9 | 17.3 | 10/33 |
| 14q32.13-q32.33 | Gain | RP11-371E8 | CTC-820M16 | 13.7 | 10/33 |
| 16p13.2 | Loss | RP11-114I12 | RP11-148F10 | 1.3 | 10/33 |
| 16q12.1-q12.2 | Loss | RP11-305A7 | RP11-467J12 | 3.5 | 17/33 |
| 17p12 | Gain | RP11-471L13 | RP11-488L1 | 1.1 | 13/33 |
| 17p12-p11.2 | Gain | RP11-459E6 | RP11-404D6 | 0.6 | 13/33 |
| 17p11.2 | Gain | RP11-189D22 | RP11-78O7 | 1.8 | 13/33 |
| 17q21.31-q21.32 | Gain | RP5-843B9 | RP11-510P20 | 0.9 | 12/33 |
| 18q11.2 | Gain | RP11-296E23 | RP11-17J14 | 2.1 | 14/33 |
| 18q22.1-q23 | Loss | RP11-21L20 | RP11-118I2 | 9.0 | 11/33 |
| 19q13.11-q13.2 | Gain | RP11-413M10 | CTB-186G2 | 4.7 | 15/33 |
| 20q13.2 | Gain | RP5-994O24 | RP11-6L15 | 3.1 | 18/33 |
| 22q12.2-q12.3 | Gain | RP1-76B20 | CTA-415G2 | 3.4 | 10/33 |
| 22q12.3-q13.1 | Gain | LL22NC01-132D12 | CTA-228A9 | 1.4 | 10/33 |
| 22q13.2-q13.33 | Loss | RP3-388M5 | CTA-722E9 | 5.8 | 10/33 |
In general, the samples showed numerous gains and losses of large chromosomal regions. Of the 41 minimal recurrent regions identified in the tumour samples, 24 represented gains and 17 losses of chromosomal segments. In at least 30% of the tumours, gain of regions in chromosome 1, 3–9, 12, 14, 17–20 and 22 was identified (Table 2). The most frequent gains observed were in chromosome 1, where minimal recurrent regions in 1p33-p32.3 (2.6 Mb), 1p31.3-p31.2 (5.5 Mb) and 1p21.3 (1.2 Mb) were gained in 19/33 (58%) samples, whereas 1q21.2-q21.3 (0.5 Mb) was gained in 18/33 (55%) samples. Gain of 20q13.2 (3.1 Mb) was identified in 18/33 (55%) samples. Scattered high-level amplification (log2 ratio>1) was observed in some of the tumours, but not consistently in any region (see Table S1).
Loss of regions in chromosome 1, 2, 5, 7–11, 13, 16, 18 and 22 was identified in at least 30% of the tumours (Table 2). The most frequent losses observed were in chromosome 10 and 13, where minimal recurrent regions in 10q25.3-q26.11 (1.1 Mb), 13q13.3-q14.2 (9.5 Mb) and 13q14.3-q21.1 (3.0 Mb) were lost in 21/33 (64%) samples. Loss of 2q36.3-q37.2 (6.9 Mb) was identified in 20/33 (61%) samples, whereas loss of 1q41 (2.3 Mb) was identified in 18/33 (55%) samples and 16q12.1-q12.2 (3.5 Mb) in 17/33 (52%) samples. Scattered homozygous deletion (log2 ratio <−1) was observed in some of the tumours, but not consistently in any region (see Table S1).
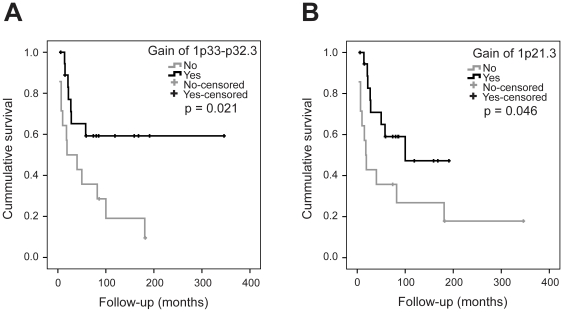
Clinical correlatesStatistical analyses were performed in order to identify possible correlations between the clinical information (Table 1) and the minimal recurrent chromosomal regions altered (Table 2). Survival analysis revealed that gain of 1p33-p32.3 and 1p21.3 was significantly associated with better patient survival (P = 0.021 and 0.046, respectively). Figure 3A and -B shows the corresponding Kaplan-Meier plots with overall survival curves. In contrast, male gender and metastasis at diagnosis were significantly associated with poor patient survival (P = 0.019 and 0.006, respectively). The corresponding Kaplan-Meier plots are shown in Figure S2.
Figure 3. Patient survival curves.
Kaplan-Meier plots with overall survival curves for (A) patients with gain of 1p33-p32.3 (n = 19) and patients with normal copy number or loss (n = 14) and (B) patients with gain of 1p21.3 (n = 19) and patients with normal copy number or loss (n = 14).
Multivariate analysis was performed in order to identify the most important prognostic factors. All experimental and clinical variables that were significantly associated with poor survival were tested using Cox regression. None of the variables were identified as independent prognostic factors, but metastasis at diagnosis showed almost significance as an independent prognostic factor (relative risk 4.0, P = 0.059).
Comparison with leiomyosarcomas
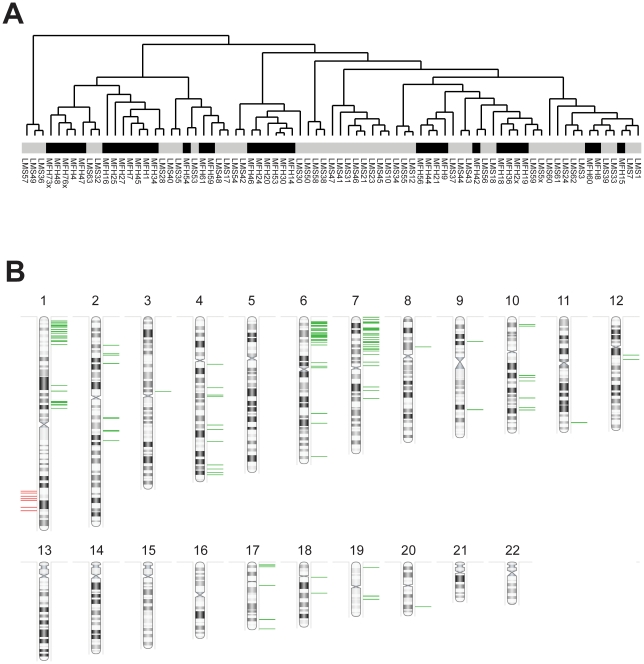
In order to investigate differences in DNA copy number aberrations between MFHs and leiomyosarcomas, a comparison with similar array CGH data from a panel of 44 leiomyosarcomas ([12] and Kresse et al., unpublished) was done. Figure 4A shows the hierarchical clustering dendogram of the 33 MFHs and 44 leiomyosarcomas. Although smaller groups of MFHs and leiomyosarcomas clustered separately, there were no overall significant differences in the clustering pattern between the two tumour types.
Figure 4. Comparison between MFHs and leiomyosarcomas.
(A) Hierarchical clustering dendogram of 33 MFHs and 44 leiomyosarcomas using DNA copy number ratios relative to a pool of normal diploid DNA. Black, MFHs; grey, leiomyosarcomas. (B) Graphical representation of genomic clones identified by SAM to be significantly different between the MFHs and leiomyosarcomas. The lines represent the chromosomal position of the 156 identified genomic clones. Green, higher copy number in MFHs compared to leiomyosarcomas; red, lower copy number.
SAM was used to identify chromosomal regions with significantly different DNA copy number between the MFHs and leiomyosarcomas. Using a false discovery rate of <1%, 156 genomic clones showing significant copy number differences between the two tumour types were identified. Figure 4B shows a graphical representation of the genomic clones, and the complete list is given in Table S2. Seven chromosomal segments represented by multiple significant clones were identified; 1p36.32-p35.2, 1p21.3-p21.1, 1q32.1-q42.13, 2q14.1-q22.2, 4q33-q34.3, 6p25.1-p21.32 and 7p22.3-p13. Of the 156 identified genomic clones, 104 mapped to these seven regions. The percentage of genomic clones in the identified regions showing significant differences varied between 25–100%, with the 1p21.3-p21.1 region showing the highest percentage of clones with significant differences (see Table S2). The 1q32.1-q42.13 region showed significantly lower copy number in MFHs compared to leiomyosarcomas, whereas all the other six regions showed significantly higher copy number.
Discussion
We have used array CGH to analyse DNA copy number changes in a panel of 33 MFHs, in order to identify recurrent copy number alterations at high-resolution and thus identify loci that may contain novel candidate oncogenes and/or tumour suppressor genes. The tumours were pathologically revised at the time of the study and classified according to the latest WHO classification [1]. The panel consisted mainly of the most common MFH subtype, spindle cell/pleomorphic MFHs, as well as some myxofibrosarcomas and pleomorphic MFHs with giant cells (Table 1). Hierarchical clustering of the samples based on the DNA copy number changes showed no associations between the clustering pattern and the tumour subtype or the other clinical features (Figure 1), and no chromosomal regions with significantly different DNA copy number between the primary tumours and recurrences were found.
The samples showed numerous gains and losses of large chromosomal regions. Forty-one minimal recurrent regions were identified as altered in at least 30% of the tumour samples, of which 24 showed increased and 17 decreased copy number (Table 2). The most common gains observed were in chromosome 1, where minimal recurrent regions in 1p33-p32.3, 1p31.3-p31.2 and 1p21.3 were gained in 58% of the samples, whereas 1q21.2-q21.3 was gained in 55% of the samples. Increased copy number of regions in chromosome 1 has been frequently observed in MFHs previously, in particular 1p31 and 1q21-q22 [5], [6], [8], [9]. High-level amplification of the 1q21-q22 region has been frequently found [6], and this is also a common finding in other types of sarcomas, like osteosarcomas [17], [18].
Although, gain of the 1p31 region has previously been associated with a trend to decreased overall patient survival in MFHs [5], no such association was seen in this tumour panel. On the contrary, gain of 1p33-p32.3 and 1p21.3 were significantly associated with better patient survival (P = 0.021 and 0.046, respectively) (Figure 3A and –B). However, these aberrations were not identified as independent prognostic factors in the multivariate analysis including all experimental and clinical variables significantly associated with survival. Notably, the other region in 1p frequently gained, 1p31.3-p31.2, was not associated with better (or worse) patient survival.
Gain of 20q13.2 was also identified in 55% of the samples, and increased copy number of regions in 20q has also been frequently observed in MFHs previously [4], [6], [8], [9]. Other regions of frequent gain including 5p14.3, 4q12, 9q21.33-q31.3 and 19q13.11-q13.2 (Table 2) have also been frequently observed in MFHs [4], [5], [6], [9], [10], [11], [19]. However, this higher resolution analysis also identified rather frequent regions of gain in MFHs (around 40%), not previously reported as such, including 7q11.21-q11.22, 18q11.2, 3p21.1-p11.2 and 6q12-q13.
The most frequent losses observed were in chromosome 10 and 13, where minimal recurrent regions in 10q25.3-q26.11, 13q13.3-q14.2 and 13q14.3-q21.1 were lost in 64% of the samples. Decreased copy number of regions in chromosome 13 has been frequently observed in MFHs previously [4], [5], [6], [8], [9], [10], [20]. The losses may involve the whole chromosome arm, or more specific regions like 13q14 and 13q21-q22. Loss of regions in chromosome 13 is a frequent finding in other types of sarcomas as well [7], [12], [21], [22], and the well-known tumour suppressor gene RB1 is considered to be the prime candidate target for the deletion involving the 13q14 region. The RB1 gene has also previously been suggested to be the target for loss of the 13q14-q21 region in MFHs [20].
Loss of regions of chromosome 10 is also a frequent finding in MFHs [4], [6], [8], as well as other types of sarcomas [12], [21], [22]. The minimal recurrent region identified here was 10q25.3-q26.11, and frequent loss of 10q25 has also been previously reported in MFHs [6]. Other recurrent regions of loss included 2q36.3-q37.2 (61% of the samples), 1q41 (55%), 16q12.1-q12.2 (52%), 2p25.3-p25.1, 11q14.1, 11q14.3-q21 and 11q23.3-q25 (all 48%), all previously reported as recurrent in MFHs [4], [6], [7], [8]. However, recurrent regions of loss not previously reported as frequent in MFHs were also identified, including 8p23.2-p22 (48% of the samples) and 5q23.2-q31.3 (33%).
Although several of the minimal recurrent regions identified were small in size, a number of genes are located in these segments, making it challenging to identify the target genes for the chromosomal aberrations based on these data only. Further analysis utilizing gene expression and functional data would be necessary for determining the most likely candidate target genes.
One of the main challenges in diagnosing MFHs is to distinguish them from other malignant tumours with a similar degree of cellular pleomorphism, like pleomorphic leiomyosarcomas, rhabdomyosarcomas and liposarcomas [3]. In order to investigate differences in DNA copy number aberrations between MFHs and leiomyosarcomas, a comparison with similar array CGH data from a panel of 44 leiomyosarcomas ([12] and Kresse et al., unpublished) was done. Hierarchical clustering of the samples based on the DNA copy number changes showed no major differences in the clustering pattern between the two tumour types (Figure 4A), similar to what has been reported by others [19], [23].
Seven chromosomal segments represented by multiple significant clones were identified as significantly different in copy number between the MFHs and leiomyosarcomas; 1p36.32-p35.2, 1p21.3-p21.1, 1q32.1-q42.13, 2q14.1-q22.2, 4q33-q34.3, 6p25.1-p21.32 and 7p22.3-p13 (Figure 4B). The 1q32.1-q42.13 region showed significantly lower copy number in MFHs compared to leiomyosarcomas, whereas all the other six regions showed significantly higher copy number. In a previous study comparing CGH data from 102 MFHs and 82 leiomyosarcomas, several chromosomal regions with differences in copy number between the two groups were identified [23]. Interestingly, the 1ptel-1p31 and 7p22-p15 regions were also shown to be more frequently gained in the MFHs, whereas the 1q32-qtel region was shown to be more frequently gained in the leiomyosarcomas, consistent with our findings. In addition, loss of the 6p region was more frequent in the leiomyosarcomas [23]. However, in another study comparing array CGH data from 31 MFHs/UPSs and 18 leiomyosarcomas, no chromosomal regions with significant differences in copy number were found using SAM, only a small set of single clones [19]. It is still uncertain whether MFHs represent a separate entity or if the majority correspond to highly pleomorphic leiomyosarcomas, but our work identified genomic differences between these two tumour groups and supports the existence of two separate entities.
Previously it was shown that a subset of MFHs could correspond to undifferentiated liposarcomas since these showed similar chromosomal aberrations, like high-level amplification of the 12q14-q15 region [24]. Further it was demonstrated that these MFH samples showed frequent co-amplification of either 1p32 or 6q23, which was not observed in the liposarcomas, suggesting that the lack of differentiation may be a consequence of amplification of target genes located in these regions, like the ASK1 (MAP3K5) gene in 6q23 [24], [25]. Increased copy number of regions in 12q was observed in this tumour panel as well, but not as frequent. Only two samples showed high-level amplification of parts of the region, but co-amplification of 1p32 or 6q23 was not identified in these samples (see Table S1), suggesting that this is not a general feature.
In summary, our array CGH analysis of a panel of MFHs identified a number of recurrent regions of gain and loss, some of which were associated with clinical features. Several of the regions have also been identified as frequently altered in previous CGH studies of MFHs, and may be characteristic for this type of tumours, although not necessarily specific. A comparison with a panel of leiomyosarcomas showed that the two tumour types could not be distinguished based on the overall DNA copy number profiles, but several specific chromosomal regions with significant differences in copy number were identified. If consistently found in larger panels of tumours, these aberrations may be used as additional tools for the differential diagnosis of MFHs and leiomyosarcomas.
Supporting Information
Genome-wide ratio plots of 33 MFHs. (PDF)
Kaplan-Meier plots with overall survival curves for A) female patients (n = 14) and male patients (n = 19) and B) patients with metastasis at diagnosis (n = 4) and patients without (n = 29). (TIF)
Identification of minimal recurrent regions in malignant fibrous histiocytomas using ACE (FDR = 0.0001 and medium sensitivity). Orange areas indicate increased DNA copy number detected by ACE and green areas decreased DNA copy number; Bold numbers indicate high-level amplification (log2>1) or homozygous deletion (<−1); Red triangles indicate missing values imputed via a K-Nearest Neighbor algorithm normalization using SAM; Recurrent regions are defined in light grey (≥30%) and grey (≥50%) and minimal recurrent regions in black frames. (XLS)
Identification of genomic clones significantly different in copy number between MFHs and leiomyosarcomas using SAM. Genomic areas significantly different are indicated in black frames. (XLS)
Acknowledgments
We thank Ana H. Barragan-Polania for excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Microarray analyses were supported by the FUnctional GEnomic (FUGE) Platform for Microarray Technology (Norwegian Microarray Consortium, www.microarray.no), funded by the Norwegian Research Council (www.forskningsradet.no). The present study was supported by The Norwegian Cancer Society (www.kreftforeningen.no). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fletcher C, van den Berg E, Molenaar W. Pleomorphic malignant fibrous histiocytoma/Undifferentiated high grade pleomorphic sarcoma. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2002. [Google Scholar]
- 2.Fletcher CD. The evolving classification of soft tissue tumours: an update based on the new WHO classification. Histopathology. 2006;48:3–12. doi: 10.1111/j.1365-2559.2005.02284.x. [DOI] [PubMed] [Google Scholar]
- 3.Weiss S, Goldblum J. Enzinger and Weiss's Soft Tissue Tumors: Mosby Elsevier 2008.
- 4.Idbaih A, Coindre JM, Derre J, Mariani O, Terrier P, et al. Myxoid malignant fibrous histiocytoma and pleomorphic liposarcoma share very similar genomic imbalances. Lab Invest. 2005;85:176–181. doi: 10.1038/labinvest.3700202. [DOI] [PubMed] [Google Scholar]
- 5.Larramendy ML, Tarkkanen M, Blomqvist C, Virolainen M, Wiklund T, et al. Comparative genomic hybridization of malignant fibrous histiocytoma reveals a novel prognostic marker. Am J Pathol. 1997;151:1153–1161. [PMC free article] [PubMed] [Google Scholar]
- 6.Mairal A, Terrier P, Chibon F, Sastre X, Lecesne A, et al. Loss of chromosome 13 is the most frequent genomic imbalance in malignant fibrous histiocytomas. A comparative genomic hybridization analysis of a series of 30 cases. Cancer Genet Cytogenet. 1999;111:134–138. doi: 10.1016/s0165-4608(98)00227-1. [DOI] [PubMed] [Google Scholar]
- 7.Parente F, Grosgeorge J, Coindre JM, Terrier P, Vilain O, et al. Comparative genomic hybridization reveals novel chromosome deletions in 90 primary soft tissue tumors. Cancer Genet Cytogenet. 1999;115:89–95. doi: 10.1016/s0165-4608(99)00082-5. [DOI] [PubMed] [Google Scholar]
- 8.Simons A, Schepens M, Jeuken J, Sprenger S, van de Zande G, et al. Frequent loss of 9p21 (p16(INK4A)) and other genomic imbalances in human malignant fibrous histiocytoma. Cancer Genet Cytogenet. 2000;118:89–98. doi: 10.1016/s0165-4608(99)00178-8. [DOI] [PubMed] [Google Scholar]
- 9.Weng WH, Ahlen J, Lui WO, Brosjo O, Pang ST, et al. Gain of 17q in malignant fibrous histiocytoma is associated with a longer disease-free survival and a low risk of developing distant metastasis. Br J Cancer. 2003;89:720–726. doi: 10.1038/sj.bjc.6601069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakabe T, Shinomiya T, Mori T, Ariyama Y, Fukuda Y, et al. Identification of a novel gene, MASL1, within an amplicon at 8p23.1 detected in malignant fibrous histiocytomas by comparative genomic hybridization. Cancer Res. 1999;59:511–515. [PubMed] [Google Scholar]
- 11.Hinze R, Schagdarsurengin U, Taubert H, Meye A, Wurl P, et al. Assessment of genomic imbalances in malignant fibrous histiocytomas by comparative genomic hybridization. Int J Mol Med. 1999;3:75–79. doi: 10.3892/ijmm.3.1.75. [DOI] [PubMed] [Google Scholar]
- 12.Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, Bjerkehagen B, Ohnstad HO, et al. Array comparative genomic hybridization reveals distinct DNA copy number differences between gastrointestinal stromal tumors and leiomyosarcomas. Cancer Res. 2006;66:8984–8993. doi: 10.1158/0008-5472.CAN-06-1972. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Meza-Zepeda LA, Kresse SH, Myklebost O. M-CGH: analysing microarray-based CGH experiments. BMC Bioinformatics. 2004;5:74. doi: 10.1186/1471-2105-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dysvik B, Jonassen I. J-Express: exploring gene expression data using Java. Bioinformatics. 2001;17:369–370. doi: 10.1093/bioinformatics/17.4.369. [DOI] [PubMed] [Google Scholar]
- 16.Lingjaerde OC, Baumbusch LO, Liestol K, Glad IK, Borresen-Dale AL. CGH-Explorer: a program for analysis of array-CGH data. Bioinformatics. 2005;21:821–822. doi: 10.1093/bioinformatics/bti113. [DOI] [PubMed] [Google Scholar]
- 17.Lau CC, Harris CP, Lu XY, Perlaky L, Gogineni S, et al. Frequent amplification and rearrangement of chromosomal bands 6p12-p21 and 17p11.2 in osteosarcoma. Genes Chromosomes Cancer. 2004;39:11–21. doi: 10.1002/gcc.10291. [DOI] [PubMed] [Google Scholar]
- 18.Ozaki T, Schaefer KL, Wai D, Buerger H, Flege S, et al. Genetic imbalances revealed by comparative genomic hybridization in osteosarcomas. Int J Cancer. 2002;102:355–365. doi: 10.1002/ijc.10709. [DOI] [PubMed] [Google Scholar]
- 19.Carneiro A, Francis P, Bendahl PO, Fernebro J, Akerman M, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest. 2009;89:668–675. doi: 10.1038/labinvest.2009.18. [DOI] [PubMed] [Google Scholar]
- 20.Chibon F, Mairal A, Freneaux P, Terrier P, Coindre JM, et al. The RB1 gene is the target of chromosome 13 deletions in malignant fibrous histiocytoma. Cancer Res. 2000;60:6339–6345. [PubMed] [Google Scholar]
- 21.Kresse SH, Ohnstad HO, Paulsen EB, Bjerkehagen B, Szuhai K, et al. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2009;48:679–693. doi: 10.1002/gcc.20675. [DOI] [PubMed] [Google Scholar]
- 22.Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 2010;456:201–217. doi: 10.1007/s00428-009-0853-4. [DOI] [PubMed] [Google Scholar]
- 23.Larramendy ML, Gentile M, Soloneski S, Knuutila S, Bohling T. Does comparative genomic hybridization reveal distinct differences in DNA copy number sequence patterns between leiomyosarcoma and malignant fibrous histiocytoma? Cancer Genet Cytogenet. 2008;187:1–11. doi: 10.1016/j.cancergencyto.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Chibon F, Mariani O, Derre J, Malinge S, Coindre JM, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet. 2002;139:24–29. doi: 10.1016/s0165-4608(02)00614-3. [DOI] [PubMed] [Google Scholar]
- 25.Chibon F, Mariani O, Derre J, Mairal A, Coindre JM, et al. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes Chromosomes Cancer. 2004;40:32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome-wide ratio plots of 33 MFHs. (PDF)
Kaplan-Meier plots with overall survival curves for A) female patients (n = 14) and male patients (n = 19) and B) patients with metastasis at diagnosis (n = 4) and patients without (n = 29). (TIF)
Identification of minimal recurrent regions in malignant fibrous histiocytomas using ACE (FDR = 0.0001 and medium sensitivity). Orange areas indicate increased DNA copy number detected by ACE and green areas decreased DNA copy number; Bold numbers indicate high-level amplification (log2>1) or homozygous deletion (<−1); Red triangles indicate missing values imputed via a K-Nearest Neighbor algorithm normalization using SAM; Recurrent regions are defined in light grey (≥30%) and grey (≥50%) and minimal recurrent regions in black frames. (XLS)
Identification of genomic clones significantly different in copy number between MFHs and leiomyosarcomas using SAM. Genomic areas significantly different are indicated in black frames. (XLS)