Abstract
Current efforts to identify protein biomarkers of disease use mainly mass spectrometry (MS) to analyze tissue and blood specimens. The low-molecular-weight “peptidome” is an attractive information archive because of the facile nature by which the low-molecular-weight information freely crosses the endothelial cell barrier of the vasculature, which provides opportunity to measure disease microenvironment-associated protein analytes secreted or shed into the extracellular interstitium and from there into the circulation. However, identifying useful protein biomarkers (peptidomic or not) which could be useful to detect early detection/monitoring of disease, toxicity, doping, or drug abuse has been severely hampered because even the most sophisticated, high-resolution MS technologies have lower sensitivities than those of the immunoassays technologies now routinely used in clinical practice. Identification of novel low abundance biomarkers that are indicative of early-stage events that likely exist in the sub-nanogram per milliliter concentration range of known markers, such as prostate-specific antigen, cannot be readily detected by current MS technologies. We have developed a new nanoparticle technology that can, in one step, capture, concentrate, and separate the peptidome from high-abundance blood proteins. Herein, we describe an initial pilot study whereby the peptidome content of ovarian and prostate cancer patients is investigated with this method. Differentially abundant candidate peptidome biomarkers that appear to be specific for early-stage ovarian and prostate cancer have been identified and reveal the potential utility for this new methodology
Key words: Peptidome, Cancer, Biomarker, Nanoparticle, Mass spectrometry
INTRODUCTION
Prostate and ovarian cancers pose a great health burden on world population principally due to the very high incidence and variability in the clinical course of the former and overall poor prognosis of the latter. Prostate cancer (CaP) is an age-related tumor as it accounts for about 25% of all the newly diagnosed cancers in American men and caused more than 28,000 deaths in 2008. As the population ages, the number of new cases per year is expected to increase by >60% and reach 300,000 by 2015 (1). This high incidence, coupled with the clinical course variability of the disease, makes CaP a particularly appropriate candidate for prevention and early intervention strategies.
Since the FDA approval and introduction into the clinic in the mid-1980s, the prostate-specific antigen (PSA) levels combined with digital rectal examination are the standard screening tests for the early detection of prostate cancer (2). The European Randomized Study of Screening for Prostate Cancer, initiated in the early 1990s and ended in 2006 to evaluate the effect of screening with PSA, concluded that PSA-based screening reduced the rate of death from prostate cancer by 20%, but is associated with a high risk of overdiagnosis (3). Actually, PSA is secreted from benign as well as cancerous cells of the prostate, and it has been demonstrated to be mostly a marker of prostate volume than of malignant conditions (4). An increase in prostate volume, naturally growing with age, can occur for benign as well as for pathologic conditions, and thus, serum PSA values correlate closely with both benign prostatic hyperplasia and CaP (4). Moreover, while it appears that some levels of CaP biopsies are carried out without actual clinical necessity (with the associated economic and social consequences), 15% of patients with levels of PSA lower than the recognized 4 ng/ml disease diagnosis threshold have cancer, with some of these being very lethal aggressive forms (5). Indeed, PSA is unable to predict with certainty the biological aggressiveness of the disease, so prostate cancer screening results in the detection of disease that in many patients is not clinically significant.
Ovarian cancer (OC), according to the late onset of symptoms and the subsequent poor prognosis by the time of diagnosis, has come to be known by the haunting definition of “the silent killer.”OC is the fourth most frequent cause of cancer death and the most lethal of all gynecologic tumors in women from North America and northern and western Europe. Each year, approximately 20,000 American women are diagnosed with ovarian cancer and about 15,000 women die from the disease. The median age at diagnosis is 63. In 2009, the National Cancer Institute estimated that 21,650 women in the USA would be diagnosed with ovarian cancer and 15,520 women would die from the disease (6). Despite its incidence being ten times lower than breast cancer, it is threefold more lethal and is usually not diagnosed until its advanced stages. The prognosis for survival from ovarian cancer largely depends on the extent of disease at diagnosis: while the overall 5-year relative survival rate for all women with ovarian cancer is 46%, this rate improves greatly to 93% if the cancer is diagnosed at an early stage before it has spread beyond the ovaries (7,8). Accordingly, detection of early stage disease offers an opportunity to reduce mortality. Unfortunately, to date, no screening protocol for ovarian cancer has been shown to achieve this aim, and only 19% of ovarian cancer cases are diagnosed at local stage (9,10)
Despite advances in molecular biology, surgery, and chemotherapy, survival rates have changed very little in the last three decades. The glycoprotein CA-125 is the most thoroughly investigated biomarker for ovarian cancer screening and is currently the only marker used in combination with transvaginal ultrasound to detect a significant proportion of ovarian cancer (8,11–13). The serum CA 125 assay has been evaluated for ovarian cancer screening as a tool to differentiate benign from malignant ovarian masses and as an indicator of tumor status during and after chemotherapy. Unfortunately, CA-125 has not been found to be sufficiently specific or sensitive for screening the general population. It is believed that up to 20% of ovarian cancers do not express CA-125, suggesting a maximum theoretical sensitivity of 80% for this marker, and for early-stage disease, this theoretical sensitivity is likely to be lower (7,8,13).
Consequently, ovarian and prostate cancers share unsolved diagnostic and therapeutic dilemmas, underpinning the need for new reliable biomarkers for early detection and better prognostic classification. Blood-borne biomarkers currently available for these two cancers do not fulfill the requirements for implementing reliable and acceptable screening tests on the general population currently performing a tissue biopsy represents the only definitive way to achieve diagnosis. Thus, the search for new, highly sensitive and specific markers, and combinations or signatures of multiple markers, has been intensive. The circulatory proteome has become one of the most promising molecular archives for the discovery of biomarkers of human diseases (14–17). Even as some investigators have turned their attention to urine and abdominal fluid as a source of molecular diagnostic information for prostate and ovarian cancers, among all physiological fluids, human plasma and serum have become the most intensively investigated biological samples. Blood, since it flows through the entire body every second, can be considered to most accurately reflect ongoing physiology and represents a protein-rich information reservoir, containing the traces of what has been encountered during constant perfusion of tissues (18).
Due to the ability of the technique to investigate and profile the complex proteomes of biological specimens in an unbiased fashion, mass spectrometry (MS)-based proteomic technologies are currently at the forefront for the discovery of blood-borne biomarkers. Of special recent interest in biomarker discovery efforts, the peptidome, or low-molecular-weight (LMW) proteome, may likely contain many of the most clinically important undiscovered markers since this repository contains analytes that are able to passively diffuse though the endothelial cell barrier of the vasculature, a barrier that effectively prevents the passive perfusion of molecules above 60 kDa. The peptidome contains protein fragments and small protein molecules generated within the tissue microenvironment that could reflect early-stage pathophysiological changes (19).
However, despite the recent technological advances in mass spectrometry and sample fractionation methods, analytical and physiological barriers hinder the deep exploration of circulatory peptidome and biological marker discovery. Firstly, most biomarkers that will have any clinical impact on early cancer detection will be found in very low abundances, with concentrations likely in the sub-nanogram per milliliter range, well below the detection limits of even the most sophisticated MS instrumentation. A second major problem is the apparently insurmountable abundance of resident proteins such as albumin and immunoglobulins, which hinder the MS identification of new candidate biomarkers, that may be one billion or more times less abundant than these endogenous proteins. Moreover, most of the time, low abundance biomarkers are non-covalently and endogenously associated with resident proteins that account for >95% of circulating proteins, confounding the isolation of rare LMW biomarkers (19–25). As a consequence, many researchers endeavor in the extremely complicated and time-consuming task to enrich the biospecimen for targeted analytes, attempting to avoid loss of the LMW fraction bound to high-molecular-weight proteins.
To that end, herein we present a novel method for body fluid-based biomarker discovery in which a novel nanoparticle-based biomarker capture technology is applied to amplify, fractionate, and enrich the LMW peptidome archive providing a powerful pre-analytical (MS) concentration and fractionation method for mass spectrometry-based biomarker discovery. We have developed these core–shell hydrogel nanoparticles to perform, in solution and in one step, the rapid (within 30 min) complete sequestration of the peptidome simultaneously performing size sieving enrichments and dramatic concentration of the LMW analytes (26,28). The bait within the particles, comprising a high-affinity dye molecule, competes with carrier proteins such as albumin for the binding of LMW cationic proteins and peptides that are thus trapped in the particle. The shell acts as a sieve, excluding high molecular weight, high abundance proteins. Feasibility of this workflow is presented whereby LMW candidate biomarkers have been identified using well-controlled study sets of serum samples from patients with and without CaP or OC that are processed though nanoparticle enrichment followed by high-resolution MS to illustrate the potential of this new workflow.
MATERIALS AND METHODS
Synthesis and Characterization of Hydrogel NIPAm/AAc Core–Shell Particles
Particles were synthesized as previously described (26–28) using NIPAm (Sigma-Aldrich) and BIS (Sigma-Aldrich) by precipitation polymerization. Acrylic acid (AAc; Sigma-Aldrich) was incorporated into NIPAm particle to provide a charge-based affinity moiety bait for affinity capture of peptides and small molecules
Sera Sample Collection and Storage
Urologic Patients
A unique study set of 30 patient-matched serum samples were obtained from 15 patients suffering from prostate cancer under full IRB approval and patient consent. The pretreatment serum was collected before radical prostatectomy and the post-surgery samples obtained 6 weeks to 3 months afterward. Patient epidemiologic and clinical information is shown in Table I.
Table I.
Clinical Features of Prostate Cancer Patients Included in the Study
| Age | Pre-surgical PSA (ng/ml) | c-stage | Gleason score biopsy | Bx | Lymph node status |
|---|---|---|---|---|---|
| 48 | 8.1 | T1c | 3 + 3 = 6 | Left | No |
| 63 | 5.3 | T2b | 3 + 4 = 7 | Bilateral | Yes |
| 66 | 4.9 | T1c | 3 + 3 = 6 | Bilateral | Yes |
| 67 | 6.3 | T1c | 3 + 3 = 6 | Left | No |
| 69 | 7 | T1c | 3 + 4 = 7 | Left | No |
| 62 | 5.7 | T1c | 4 + 4 = 8 | Right | Yes |
| 72 | 8.6 | T1c | 3 + 3 = 6 | Right | No |
| 67 | 3.1 | T1c | 3 + 3 = 6 | Right | No |
| 70 | 5.1 | T1c | 3 + 3 = 6 | Right | No |
| 68 | 8.3 | T1c | 3 + 3 = 6 | Right | No |
| 55 | 7.3 | T1c | 3 + 4 = 7 | Bilateral | Yes |
| 70 | 10.7 | T2a | 3 + 4 = 7 | Bilateral | Yes |
| 55 | 16.3 | T1c | 3 + 3 = 6 | Right | Yes |
| 63 | 5.7 | T1c | 3 + 3 = 6 | Right | No |
| 78 | 4.1 | T1c | 3 + 3 = 6 | No |
Gynecological Patients
A study set of serum samples from 20 early-stage epithelial ovarian cancer patients and 20 patients affected by benign gynecologic disease requiring surgical intervention (controls) was collected at the Department of Obstetrics and Gynaecology, University of Brescia, Italy, between 2004 and 2006 under IRB approval and patient consent. Clinical and epidemiologic information is shown in Tables II and III. All women in the study population underwent preoperative clinical examination, ultrasound morphologic scoring (in all cases performed as a transvaginal examination, the abdominal approach was used when indicated), and color Doppler imaging. Samples were obtained from fasting patients either the day prior to, or the day of, surgical intervention before induction of anesthesia for treatment.
Table II.
Clinical Features of Ovarian Cancer Patients
| Age | Menopausal status | Ultrasound risk | Stage | Pathology |
|---|---|---|---|---|
| 64 | Post- | High | II | S. Bord. |
| 63 | Post- | High | IC | M. bord. |
| 56 | Post- | Unknown | IIA | Cl. C. Ca. |
| 57 | Post- | High | IA | Cl. C. Ca. |
| 48 | Post- | Intermediate | IA | M. ACa. |
| 42 | Pre- | High | IA | S. Bord. |
| 24 | Pre- | High | I | M. Bord. |
| 24 | Pre- | Intermediate | IA | S. Ca. |
| 19 | Pre- | Intermediate | I | M. ACa. |
| 54 | Post- | High | IIA | E. ACa. |
| 58 | Post- | High | IA | Cl. C. Ca. |
| 28 | Pre- | Intermediate | IA | M. Bord. |
| 59 | Post- | Intermediate | I | M. Bord. |
| 58 | Post- | Unknown | IC | S. ACa. |
| 68 | Post- | Low | IC | S. ACa. |
| 36 | Pre- | Low | IC | Unknown |
| 33 | Pre- | Unknown | IA | ACa. |
| 53 | Pre- | Unknown | IA | E. ACa. |
| 59 | Post- | High | IC | S. ACa. |
| 29 | Pre- | Low | IC | S. ACa. |
Cl. C. Ca. clear cell carcinoma, ACa. adenocarcinoma, S. serous, E. ACa. endometrioid adenocarcinoma, M. mucinous
Table III.
Clinical Information of Patient Affected by Benign Gynecological Disease
| Age | Menopausal status | Ultrasound risk | Pathology |
|---|---|---|---|
| 25 | Pre | Intermediate | M.C.T. |
| 21 | Pre- | Intermediate | M.C.T. |
| 39 | Pre- | Intermediate | C. L C. |
| 39 | Pre- | Intermediate | E. C. |
| 36 | Pre- | Intermediate | F. C. |
| 68 | Post- | Intermediate | S. CAd |
| 50 | Pre- | High | F. C. |
| 55 | Post- | Intermediate | S. Cad. |
| 41 | Pre- | Intermediate | Unknown |
| 52 | Pre- | Intermediate | M.C.T. |
| 58 | Post- | Intermediate | S. Cad. |
| 52 | Pre- | Intermediate | S. CAF. |
| 65 | Post- | Intermediate | M.C.T. |
| 49 | Pre- | Unknown | F. C. |
| 28 | Pre- | Unknown | D. C. |
| 58 | Post- | Unknown | S. CAF. |
| 74 | Post- | Unknown | M. CAd. |
| 53 | Unknown | Unknown | S. CAd. |
| 48 | Pre- | Low | S-M. Cad. |
| 50 | Post- | Low | S. Cad |
M.C.T. mature cystic teratoma, S. CAF. serous cystoadenofibroma, F. C. follicular cyst, D. C. dermoid cyst, S. CAd. serous cystoadenoma, C. L C. corpus luteum cyst, M. C. mucinous cystoadenoma, E. C. endometriosic cyst, S-M. CAd. serous–mucinous cystoadenoma
Peptidome Capture and Isolation
Serum samples were centrifuged (7 min, 4°C, 16,100 rcf) and diluted 1:2 with 50 mM Tris–HCl, pH 7.0. Four hundred microliters of sera from ovarian cancer and benign controls was used per patient, and 200 μL of sera for pre- and post-prostatectomy specimens was used for biomarker discovery. Two chemokines, CCL28, known also as mucose-associated epithelial chemokine (MEC)m with a predicted MW 14 kDa (Antigenix America), and CXCL12 (Antigenix America), also known as SDF-1β with a predicted MW of 11 kDa, were added to the serum samples as internal process control standards. Pre-prostatectomy samples (200 μL aliquots) were spiked with CXCL12/SDF-1β and of CCL28/MEC at the concentration of 0.5 and 0.05 ng/µL, respectively, while 0.5 ng/µL of MEC and 0.05 ng/µL SDF-1β were added to the post-prostatectomy sera. Ovarian cancer patient samples were spiked with 0.5 ng/µL MEC and 0.05 ng/µL SDF-1β, as opposed to benign pelvic disease controls which were spiked with 0.05 ng/µL MEC and 0.5 ng/µL SDF-1β Subsequently, nanoparticles were incubated with each diluted serum sample for 15 min at room temperature under slow rotation. After incubation and peptidome capture, samples were centrifuged (15 min, 25°C, 16,100 rcf), the supernatant was discarded, and the particles were washed two times by means of resuspending the pellets in 1 mL of the washing buffer (20% acetonitrile/0.5× PBS) and centrifuging (4 min, 25°C, 16,100 rcf). The pellet of washed particles, obtained as described above, was vortexed with 100 μL of elution buffer (60% acetonitrile/2% acetic acid) at room temperature. The elution mix was sonicated three times for 10 s each. The eluate was saved. The elution step was repeated twice and the eluates pooled together. The eluates were then lyophilized until dry. The dried-up samples were stored at −20°C until use. Eluted proteins from the nanoparticles were reduced by dithiothreitol, alkylated by iodoacetamide, and digested by trypsin overnight at 37°C as described (27).
SDS-PAGE Analysis
The incubation of serum with particles has been performed as previously described (27). Serum, supernatant, and eluate derived from particle incubation were loaded on 4–20% Tris-Glycine gel (Invitrogen Corporation). Proteins were detected by Comassie staining.
Mass Spectrometry Analysis
The peptides from each sample were analyzed by reversed-phase liquid chromatography/electrospray tandem mass spectrometry (MS/MS) using a linear trap quadrupole (LTQ)-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with an in-line autosampler. The reverse-phase C18 column (0.5-mm i.d. × 50-mm length) was from Michrom BioResources. The column was washed for 2 min with mobile phase A (0.1% formic acid), and peptides were eluted using a linear gradient of 0% mobile phase B (0.1% formic acid, 80% acetonitrile) to 50% mobile phase B in 50 min at 2 μL/min, then to 100% B for an additional 2 min.
The mass spectrometer was operated in a data-dependent mode in which each full MS scan was followed by five MS/MS scans in which the five most abundant molecular ions were dynamically selected and fragmented by collision-induced dissociation using a normalized collision energy of 30%. To decrease carryover, one blank sample was analyzed using a 30-min, double high-performance liquid chromatograph (HPLC) gradient program after analyzing one sample. To verify HPLC reproducibility, two standard samples (yeast enolase digest, from Michrom BioResources) were analyzed using a 30-min HPLC program after analyzing 15 serum samples.
Bioinformatic Analysis
Protein identification was performed with SEQUEST. The data were searched against a fully tryptic indexed, human protein database maintained by the National Center for Biotechnology Information with a variable oxidized methionine and a static carboxyamidomethylated cysteine modification. Search results were filtered with the following criteria: minimum XCorr = 2.2 (+2), 3.5 (+3), minimum dCn = 0.1, and a maximum precursor ion mass deviation of 15 ppm. An MS1-based comparative data analysis was accomplished using BioSieve (Thermo and Vast Scientific). Spectral counting (MS2-based) analysis was accomplished using Scaffold (Proteome Software, Inc. Portland, OR). Candidate differentially abundant peptides were evaluated by manual inspection of the raw data to confirm the peptide identifications, and differences in peptide relative abundances were determined using a two-tailed t test applied to the spectral counts with the assumption of normally distributed data.
RESULTS
Particles Perform Size Exclusion and Concentration of Serum Proteins
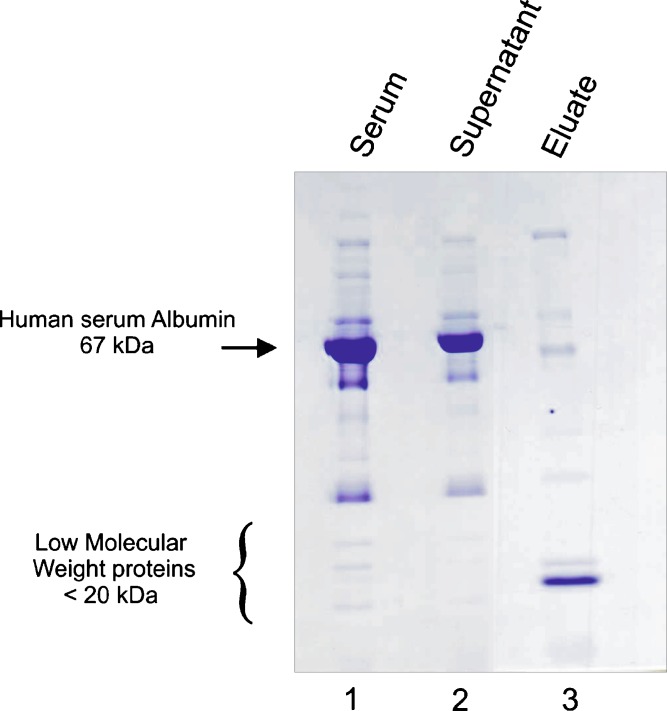
In order to evaluate, in a qualitative manner, the efficiency of the nanoparticle capture methodology as a pre-processing step to eliminate the excess of high abundant proteins and concentrate the low-molecular-weight protein archive, a serum sample was processed and the starting sample, the post-particle supernatant, and eluate from particles were analyzed by SDS-PAGE. As shown in Fig. 1, in the eluate from particle (lane 3), the amount of albumin, and in general serum high abundant proteins, is tremendously reduced compared to the remaining albumin in the supernatant and starting raw sample. Conversely, the nanoparticle captured archive within the eluate reveals a number of highly concentrated and enriched proteins, which demonstrates the ability of particles to concentrate proteins based on the nature of the chemical bait used. Since the incubation of the sample was in pH values >3.5, the acrylic acid bait is deprotonated and thus carries a negative charge, so the affinity of this particle type is most specific for positively charged polypeptides and proteins. Consequently, proteins/peptides that are negatively charged (such as those remaining in the supernatant), will not be captured by this specific nanoparticle.
Fig. 1.
SDS-PAGE analysis of human serum sample pre- and post-incubation with NIPAm/AAc nanoparticles. Lane 1 Serum from healthy donor before incubation with particles. Lane 2 Supernatant obtained after centrifugation of the mix of incubation containing serum and nanoparticles. Lane 3 Eluate from nanoparticles: the samples are enriched in low-molecular-weight proteins, while albumin is mostly excluded
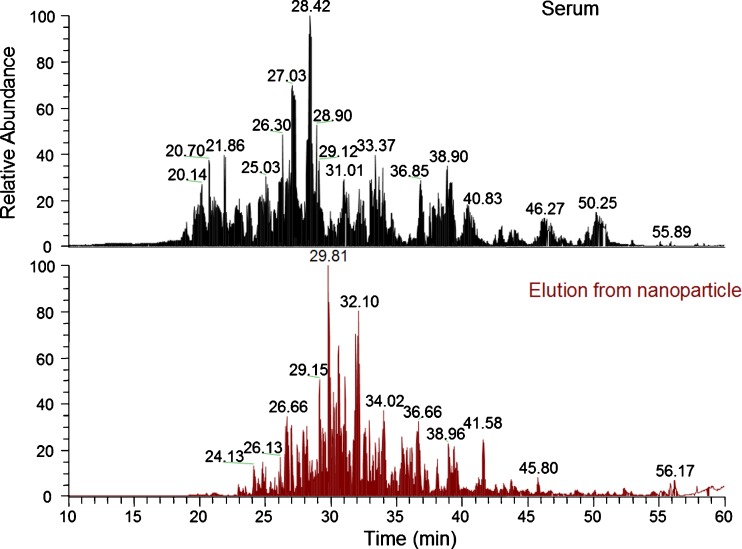
The nanoparticles’ capacity to reduce albumin can be also seen by a qualitative analysis of a mass spectrometry chromatogram of raw serum and the nanoparticle eluate. As can be observed in Fig. 2 the peak intensities of several tryptic peptides of albumin, high in raw serum, are notably reduced in the elution from nanoparticle sample.
Fig. 2.
Comparison of the LC-MS chromatography of serum and elution from nanoparticle. The abundant base peaks are labeled with their retention time. The peak intensities of albumin tryptic peptides such as AAFTECCQAADK (retention time 21.86 min), FQNALLVR (retention time 27.03 min), LVNEVTEFAK (retention time 28.42 min), KVPQVSTPTLVEVSR (retention time 28.90 min), RPCFSALEVDETYVPK (retention time 31.01 min), and AVMDDFAAFVEK (33.37 min) are high in the serum sample and are significantly low in the elution from nanoparticle sample
Evaluation of the Nanoparticle Capture–MS Workflow Performance
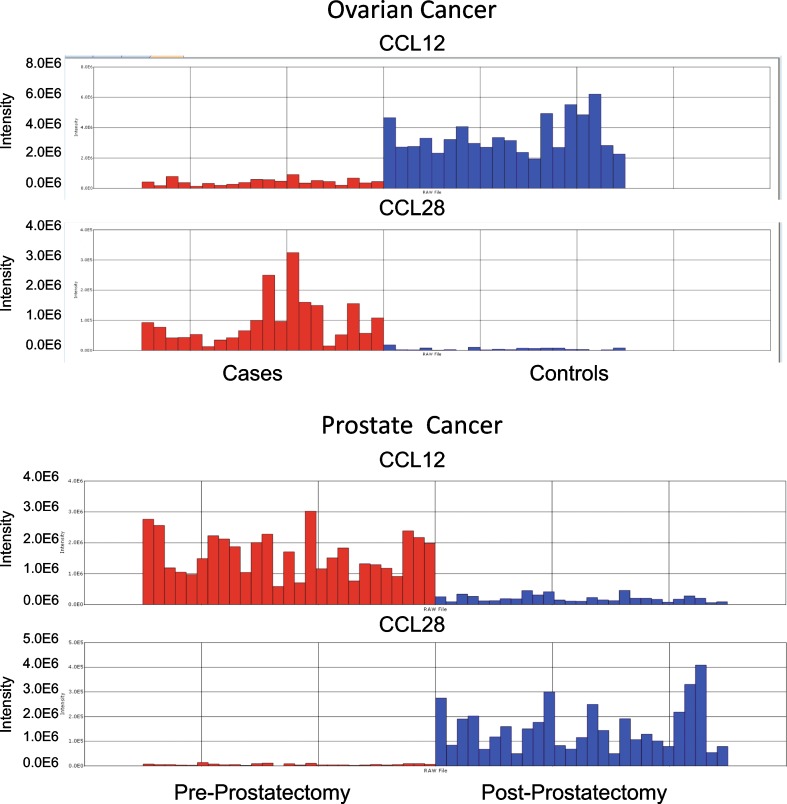
In order to evaluate the nanoparticle capture workflow’s process performance and the reproducibility of the data, two chemokines, stromal cell-derived factor 1β (SDF-1β, also known as CCL12) and mucosae epithelial chemokine (MEC, known as CCL28), were spiked into the serum specimens before the nanoparticle processing step as internal standards. The two chemokines were chosen because of their small size (in the MW range of the peptidomic archive under evaluation for this study) and their physicochemical properties (volability and ionizability) for MS suitability, and the affinity for these two analytes for the specific nanoparticle bait molecule utilized has been previously demonstrated (27,28). SDF-1β was added in a ratio of 10:1 in the pre-prostatectomy vs. post-prostatectomy samples and 1:10 in the cases vs. controls of the prostate cancer study set, while MEC was spiked in a ratio of 1:10 in the pre-prostatectomy vs. post-prostatectomy samples set and 10:1 in the ovarian cancer sera vs. controls samples. The chemokines were found to be differentially abundant with the expected ratio by BioSieve analysis (Fig. 3) in both study sets, and the MS data analysis confirmed the expected recovery of the internal standard controls .Since the levels of the endogenous, native CCL28 and CCL12 within each patient sample are unknown and will vary from patient to patient, we can estimate the approximate reproducibility of the overall process by an analysis of the expected and actual ratios produced by the spiked-in chemokines over the population of samples rather than any one specific sample. The ratio of CCL12 intensity in the cases/controls of ovarian cancer is predicted to be 0.1 since a 1:10 amount was spiked in respectively and the experimentally derived value was 0.106. For CCL28, which was spiked in at 10:1 case vs. control, the expected value of 10 compares to the actual value obtained of 10.052. For the prostate study, the ratio of CCL12 on the pre/post-prostatectomy samples resulted 8.592 compared to a theoretical value of 10, and for CCL28, the experimental value of 0.063 was determined compared to the theoretical value of 0.1 These results indicate excellent process reproducibility and variance across the entire set and provide increased confidence in the experimental results obtained for newly identified markers.
Fig. 3.
Verification of nanoparticle capture workflow. The chemokines CCL12 and CCL28 were spiked, as internal standard process controls, in 1:10 and 10:1 ratios in ovarian cancer case and benign control sera, respectively (top), while in pre- and post-prostatectomy sera in the ratios of 10:1 and 1:10 (bottom). Each vertical bar in the histogram represents one serum sample from a cancer patient (red) or a control subject (blue), and the bar height corresponds to the peptide ion abundance measured
Identification of Candidate CaP and OC LMW/Peptidome Biomarker Candidates Using Nanoparticle Capture–MS
In order to identify LMW protein/peptide serum biomarker candidates for CaP, 30 sera from 15 patients suffering from CaP, collected before radical prostatectomy and 6 weeks to 3 months following surgery (Table I), and 40 sera from gynecologic patients (20 samples from women affected by ovarian cancer and 20 samples from controls; Tables II and III) were subjected to our novel nanoparticle capture workflow followed by LC-MS/MS analysis (Fig. 4). The label-free spectral counting Scaffold analyses of MS/MS data from the nanoparticle-captured protein analytes yielded the determination of 14 proteins with higher abundance in the pre-prostatectomy samples compared to the post-prostatectomy samples (Table IV) and 59 proteins with higher abundance in the early-stage OC sample compared to the age-matched benign gynecologic controls (Table V). Only analytes whose spectral counts achieved at least a 50% differential amount were included in these tables. Highlighted in red are the spiked-in internal standard proteins that, as expected, were found by both BioSieve and Scaffold (shown) to be differentially abundant. This result reveals that the entire process produces the expected results, thus providing increased confidence in the new experimentally determined candidate biomarker proteins. To provide a better sense of the complexity of the samples after preprocessing with nanoparticles, the total number of proteins and peptides that have been identified on average for the group of case/controls in the ovarian cancer or pre/post-prostatectomy in the prostate cancer study are shown in Table VI.
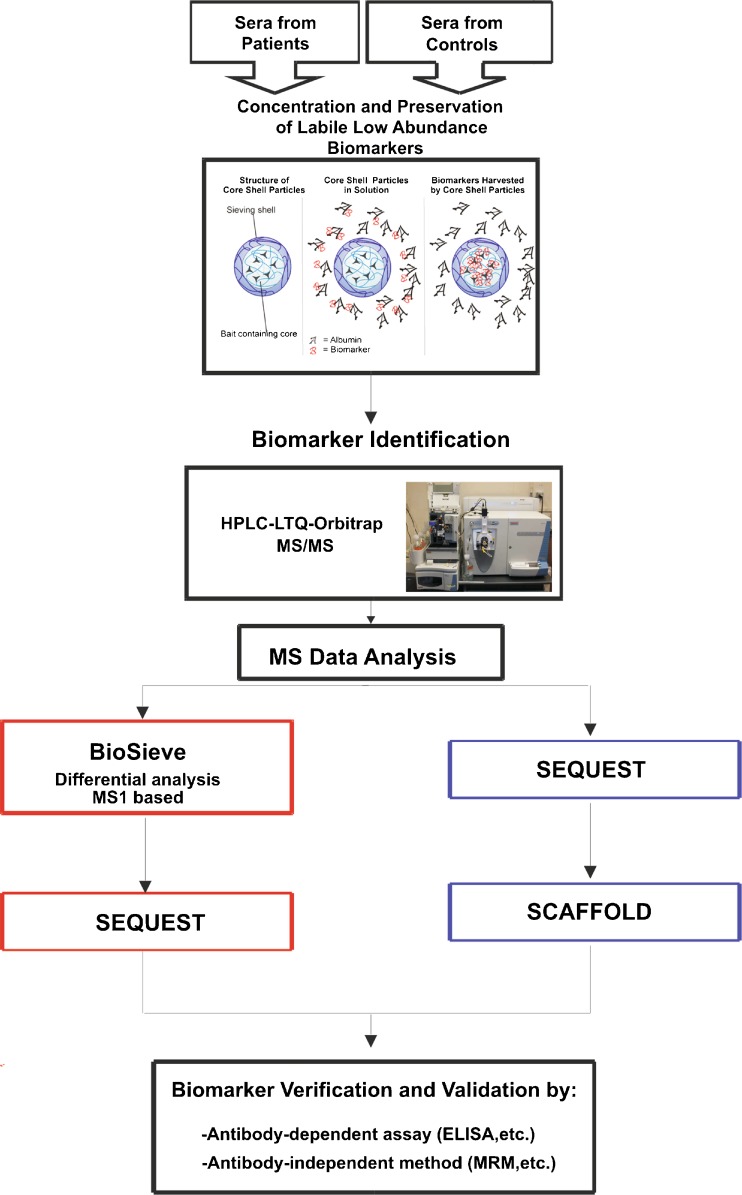
Fig. 4.
Nanoparticle capture–mass spectrometry workflow. Sera from patients and controls are incubated with NIPAm/AAc particles, eluates from particles are analyzed by HPLC-LTQ-Orbitrap mass spectrometry, and the data analysis is performed using BioSieve and Scaffold software
Table IV.
Scaffold Spectral Count Analysis of Differentially Abundant Analytes Elevated in the Serum of Pre-prostatectomy Patients Compared to Patient Matched Post-prostatectomy Patients
| Protein ID | Description | Prea | Postb | Relative difference (%)c |
|---|---|---|---|---|
| gi|55956899|ref|NP_000217.2| | Keratin 9 | 32 | 19 | 51.0 |
| gi|4502153|ref|NP_000375.1| | Apolipoprotein B precursor | 55 | 32 | 52.9 |
| gi|50659080|ref|NP_001076.2| | Serpin peptidase inhibitor, clade A, member 3 precursor | 28 | 15 | 60.5 |
| gi|88853069|ref|NP_000629.3| | Vitronectin precursor | 4 | 2 | 66.7 |
| gi|4557287|ref|NP_000020.1| | Angiotensinogen preproprotein | 11 | 5 | 75.0 |
| gi|4504315|ref|NP_003536.1| | H4 histone family, member J | 10 | 4 | 85.7 |
| gi|4504489|ref|NP_000403.1| | Histidine-rich glycoprotein precursor | 5 | 2 | 85.7 |
| gi|4503745|ref|NP_001447.1| | Filamin 1 (actin-binding protein-280) | 10 | 3 | 107.7 |
| gi|76563933|ref|NP_001029058.1| | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) isoform gamma | 224 | 62 | 113.3 |
| gi|47132551|ref|NP_997640.1| | Fibronectin 1 isoform 2 preproprotein | 4 | 1 | 120.0 |
| gi|9257232|ref|NP_000598.1| | Orosomucoid 1 precursor | 9 | 2 | 127.3 |
| gi|4502511|ref|NP_001728.1| | Complement component 9 | 15 | 3 | 133.3 |
| gi|16753233|ref|NP_006280.2| | Talin 1 | 10 | 2 | 133.3 |
| gi|4757960|ref|NP_004351.1| | Cadherin 1, type 1 preproprotein | 5 | 0 | 200.0 |
aSum of spectra found in all the pre-prostatectomy samples
bSum of spectra founded in all the post-prostatectomy serum samples
cRelative difference = ((Pre − Post)/Average of number of spectra for the Pre and Post)) × 100
Table V.
Scaffold Spectral Count Analysis of Differentially Abundant Analytes Elevated in the Serum of Early Stage Ovarian Cancer Patients Compared to Patients with Benign Gynecologic Conditions
| Protein ID | Description | Casea | Controlsb | Relative difference (%)c |
|---|---|---|---|---|
| gi|22538811|ref|NP_683513.1| | Chemokine (C-C motif) ligand 28 precursor | 29 | 0 | 200.0 |
| gi|88758615|ref|NP_000410.2| | Integrin alpha 2b preproprotein | 28 | 0 | 200.0 |
| gi|13562114|ref|NP_110400.1| | Beta tubulin 1, class VI | 17 | 0 | 200.0 |
| gi|41281905|ref|NP_848537.1| | UNC-112 related protein 2 long form | 14 | 0 | 200.0 |
| gi|7669550|ref|NP_054706.1| | Vinculin isoform meta-VCL | 13 | 0 | 200.0 |
| gi|4502153|ref|NP_000375.1| | Apolipoprotein B precursor | 358 | 3 | 196.7 |
| gi|47132551|ref|NP_997640.1| | Fibronectin 1 isoform 2 preproprotein | 226 | 3 | 194.8 |
| gi|16753233|ref|NP_006280.2| | Talin 1 | 73 | 2 | 189.3 |
| gi|4503745|ref|NP_001447.1| | Filamin 1 (actin-binding protein-280) | 65 | 4 | 176.8 |
| gi|31652249|ref|NP_004130.2| | Lipopolysaccharide-binding protein precursor | 28 | 2 | 173.3 |
| gi|4503643|ref|NP_000121.1| | Coagulation factor V precursor | 20 | 2 | 163.6 |
| gi|4501891|ref|NP_001093.1| | Actinin, alpha 1 | 16 | 2 | 155.6 |
| gi|17921989|ref|NP_005991.1| | Tubulin, alpha 1 | 21 | 4 | 136.0 |
| gi|4557287|ref|NP_000020.1| | Angiotensinogen preproprotein | 15 | 3 | 133.3 |
| gi|4557485|ref|NP_000087.1| | Ceruloplasmin (ferroxidase) | 75 | 16 | 129.7 |
| gi|4502067|ref|NP_001624.1| | Alpha-1-microglobulin/bikunin precursor | 9 | 2 | 127.3 |
| gi|4502511|ref|NP_001728.1| | Complement component 9 | 62 | 15 | 122.1 |
| gi|21735614|ref|NP_003652.2| | Apolipoprotein L1 isoform a precursor | 16 | 4 | 120.0 |
| gi|73858568|ref|NP_000053.2| | Complement component 1 inhibitor precursor | 104 | 27 | 117.6 |
| gi|55770842|ref|NP_000558.2| | C-reactive protein, pentraxin-related | 15 | 4 | 115.8 |
| gi|16418467|ref|NP_443204.1| | Leucine-rich alpha-2-glycoprotein 1 | 9 | 3 | 100.0 |
| gi|55956899|ref|NP_000217.2| | Keratin 9 | 112 | 38 | 98.7 |
| gi|4501887|ref|NP_001605.1| | Actin, gamma 1 propeptide | 72 | 30 | 82.4 |
| gi|4504781|ref|NP_002206.1| | Inter-alpha (globulin) inhibitor H1 | 9 | 4 | 76.9 |
| gi|4502161|ref|NP_001637.1| | Apolipoprotein C-IV | 10 | 5 | 66.7 |
| gi|40317626|ref|NP_003237.2| | Thrombospondin 1 precursor | 157 | 83 | 61.7 |
| gi|4505981|ref|NP_002695.1| | Pro-platelet basic protein precursor | 67 | 39 | 52.8 |
| gi|62912466|ref|NP_001017366.1| | Complement component 4 binding protein, beta chain isoform 2 precursor | 22 | 13 | 51.4 |
| gi|50659080|ref|NP_001076.2| | Serpin peptidase inhibitor, clade A, member 3 precursor | 70 | 42 | 50.0 |
| gi|8922987|ref|NP_060856.1| | PCI domain containing 2 | 3 | 0 | 200.0 |
| gi|19923106|ref|NP_000437.3| | Paraoxonase 1 | 3 | 0 | 200.0 |
| gi|21361302|ref|NP_006206.2| | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 | 3 | 0 | 200.0 |
| gi|4507021|ref|NP_000333.1| | Solute carrier family 4, anion exchanger, member 1 | 3 | 0 | 200.0 |
| gi|89027630|ref|XP_941134.1| | PREDICTED: similar to actin-related protein 3-beta isoform 1 isoform 3 | 3 | 0 | 200.0 |
| gi|4505879|ref|NP_002655.1| | Pleckstrin | 3 | 0 | 200.0 |
| gi|22035665|ref|NP_055874.1| | Talin 2 | 3 | 0 | 200.0 |
| gi|70906435|ref|NP_005132.2| | Fibrinogen, beta chain preproprotein | 3 | 0 | 200.0 |
| gi|34577083|ref|NP_689937.2| | Ras suppressor protein 1 isoform 2 | 4 | 0 | 200.0 |
| gi|113420458|ref|XP_941447.2| | PREDICTED: similar to Protein FAM82B | 4 | 0 | 200.0 |
| gi|47419932|ref|NP_000164.3| | Platelet glycoprotein Ib alpha polypeptide precursor | 5 | 0 | 200.0 |
| gi|54112429|ref|NP_212132.2| | Dedicator of cytokinesis 7 | 5 | 0 | 200.0 |
| gi|4503631|ref|NP_000120.1| | Coagulation factor XIII A1 subunit precursor | 6 | 0 | 200.0 |
| gi|4557443|ref|NP_000069.1| | Cholesteryl ester transfer protein, plasma precursor | 7 | 0 | 200.0 |
| gi|5031635|ref|NP_005498.1| | Cofilin 1 (non-muscle) | 8 | 1 | 155.6 |
| gi|73858566|ref|NP_000176.2| | Heparin cofactor II precursor | 6 | 1 | 142.9 |
| gi|34335256|ref|NP_003808.1|-R | A disintegrin and metalloproteinase domain 7 | 4 | 1 | 120.0 |
| gi|45580723|ref|NP_066275.3| | Haptoglobin-related protein | 4 | 1 | 120.0 |
| gi|4502261|ref|NP_000479.1| | Serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | 3 | 1 | 100.0 |
| gi|70778918|ref|NP_002207.2| | Inter-alpha globulin inhibitor H2 polypeptide | 3 | 1 | 100.0 |
| gi|31982945|ref|NP_689564.2| | Solute carrier family 5 (sodium/glucose cotransporter), member 10 | 3 | 1 | 100.0 |
| gi|4557391|ref|NP_000057.1| | Complement component 8, beta polypeptide preproprotein | 3 | 1 | 100.0 |
| gi|94721347|ref|NP_006656.2| | Heparanase | 3 | 1 | 100.0 |
| gi|4507357|ref|NP_003555.1| | Transgelin 2 | 3 | 1 | 100.0 |
| gi|21361820|ref|NP_064538.2| | Hypothetical protein LOC56912 | 4 | 2 | 66.7 |
| gi|14917119|ref|NP_066293.2| | Keratin 34 | 4 | 2 | 66.7 |
| gi|20336724|ref|NP_115809.1|-R | Zinc finger protein 333 | 6 | 3 | 66.7 |
| gi|31559819|ref|NP_853515.1| | Keratin 25C | 6 | 3 | 66.7 |
| gi|21735625|ref|NP_663723.1| | Tyrosine 3/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 5 | 3 | 50.0 |
| gi|14917115|ref|NP_002268.2| | Keratin 31 | 5 | 3 | 50.0 |
aCase = sum of spectra founded in all the case samples
bControl = sum of spectra founded in all the Control samples
cRelative difference = ((Case − Control)/Average of number of spectra for the Case and the Control)) × 100
Table VI.
Total Peptide and Protein Count for Ovarian and Prostate Sample Set Obtained by Scaffold
| Peptides | Proteins | ||
|---|---|---|---|
| Ovarian cancer | Case | 569 | 126 |
| Control | 513 | 118 | |
| Prostate cancer | Pre-prostatectomy | 271 | 36 |
| Post-prostatectomy | 266 | 35 | |
An average number of protein and peptides for all the case, controls, and pre- and post-prostatectomy have been calculated
Moreover, a list of the most abundant proteins identified in the prostate and ovarian sets samples are reported (Tables VII and VIII). Proteins such as albumin and complement component 3 are still the most abundant proteins regardless of upfront preprocessing. It is important to consider that a nanoparticle-based sample preparation is not a depletion-based approach but is an enrich-based strategy to “amplify” and concentrate low molecular analytes; thus, the spectral counts of the albumin and complement component 3 peptides are dramatically reduced and the ratios of the endogenous high abundance analytes to the newly discovered analytes is lower. Due to the extraordinarily high abundance of proteins such albumin that exist in one to ten million-fold molar excess to low abundance proteins, it would be impossible to completely eliminate them from downstream analysis. Since mass spectrometry works in a dynamic range of four to five orders of magnitude, the reduction of a consistent part of the most abundant protein (as showed in the gel) still provides a great advantage in this analysis, allowing the identification of the less abundant peptides/proteins that usually are masked.
Table VII.
Protein Identified as Most Abundant in Prostate Cancer Pre-prostatectomy and Post-prostatectomy Samples
| Protein identify as most abundant in prostate cancer study | MW |
|---|---|
| Albumin precursor (Homo sapiens) | 69 |
| Complement component 3 precursor (Homo sapiens) | 187 |
| Apolipoprotein A-I preproprotein (Homo sapiens) | 31 |
| Apolipoprotein A-IV precursor (Homo sapiens) | 45 |
| Transferrin (Homo sapiens) | 77 |
| Complement component 4A preproprotein (Homo sapiens) | 193 |
| Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (Homo sapiens) | 47 |
| Apolipoprotein E precursor (Homo sapiens) | 36 |
| Platelet factor 4 (chemokine (C-X-C motif) ligand 4; Homo sapiens) | 11 |
| Complement component 1, r subcomponent (Homo sapiens) | 80 |
Table VIII.
Protein Identified as Most Abundant in Ovarian Cancer and Controls Samples
| Protein identify as most abundant in ovarian cancer study | MW |
|---|---|
| Albumin precursor (Homo sapiens) | 69 |
| Complement component 3 precursor (Homo sapiens) | 187 |
| Complement component 4A preproprotein (Homo sapiens) | 193 |
| Apolipoprotein A-I preproprotein (Homo sapiens) | 31 |
| Transferrin (Homo sapiens) | 77 |
| Complement component 1, r subcomponent (Homo sapiens) | 80 |
| Platelet factor 4 (chemokine (C-X-C motif) ligand 4; Homo sapiens) | 11 |
| Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (Homo sapiens) | 47 |
| Apolipoprotein A-II preproprotein (Homo sapiens) | 11 |
| Apolipoprotein C-III precursor (Homo sapiens) | 11 |
In order to determine, within the proteins identified, candidate molecules that could be more specific for prostate cancer and ovarian cancer and thereby produce more specific markers for further analysis and validation, the candidates obtained from the two study sets were compared. The results of the analysis (Fig. 5 and Table IX) revealed a series of proteins whose increased relative abundance, as determined by label-free spectral counting methodology, was either CaP-specific or OC-specific. Comparison of the list of ovarian cancer candidates to biomarker candidates discovered using the identical workflow applied to serum sets from patients with prostate cancer revealed only a marginal overlap, indicating potential for specificity in the candidates uncovered to date.
Fig. 5.
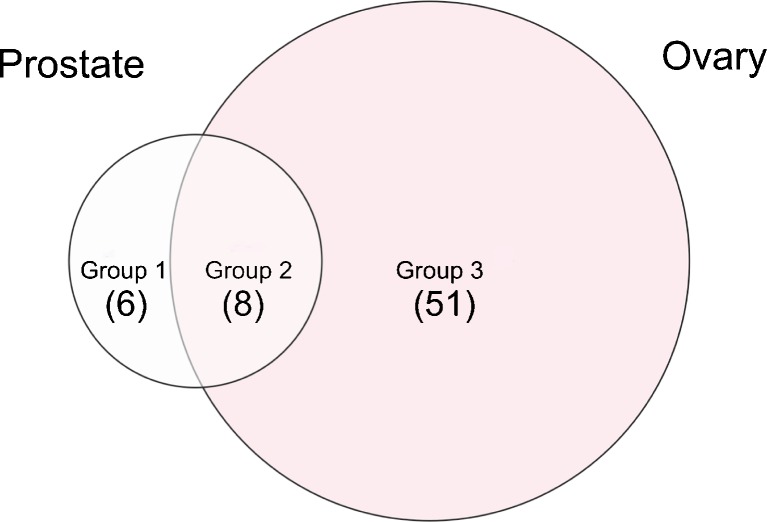
Venn diagram visualization of group 1 (CaP-specific proteins; increased spectral counts in pre- vs. post-prostatectomy serum) not found upregulated in OC; group 3 (OC-specific proteins; increased spectral counts in OC vs. benign control) not found upregulated in CaP; and group 2 commonly upregulated proteins in both CaP and OC
Table IX.
Summary List of Nanoparticle-Captured Analytes
| Group 1 (prostate)a | Group 2 (common)b | Group 3 (ovary)c |
|---|---|---|
| Cadherin 1, type 1 preproprotein | Angiotensinogen preproprotein | Actin, gamma 1 propeptide |
| H4 histone family, member J | Apolipoprotein B precursor | Actinin, alpha 1 |
| Histidine-rich glycoprotein precursor | Complement component 9 | Alpha-1-microglobulin/bikunin precursor |
| Orosomucoid 1 precursor | Fibronectin 1 isoform 2 preproprotein | Apolipoprotein C-IV |
| Vitronectin precursor | Filamin 1 (actin-binding protein-280) | Apolipoprotein L1 isoform a precursor |
| Chemokine (C-X-C motif) ligand 12 | Keratin 9 | Beta tubulin 1, class VI |
| Serpin peptidase inhibitor, clade A, member 3 precursor | Ceruloplasmin (ferroxidase) | |
| Talin 1 | Cholesteryl ester transfer protein, plasma precursor | |
| Chemokine (C-C motif) ligand 28 precursor | ||
| Coagulation factor V precursor | ||
| Coagulation factor XIII A1 subunit precursor | ||
| Cofilin 1 (non-muscle) | ||
| Complement component 1 inhibitor precursor | ||
| Complement component 4 binding protein, beta chain isoform 2 precursor | ||
| C-reactive protein, pentraxin-related | ||
| Dedicator of cytokinesis 7 | ||
| Fibrinogen, beta chain preproprotein | ||
| Integrin alpha 2b preproprotein | ||
| Inter-alpha (globulin) inhibitor H1 | ||
| Leucine-rich alpha-2-glycoprotein 1 | ||
| Lipopolysaccharide-binding protein precursor | ||
| Paraoxonase 1 | ||
| PCI domain containing 2 | ||
| Platelet glycoprotein Ib alpha Polypeptide precursor | ||
| Pleckstrin | ||
| PREDICTED: similar to actin-related protein 3-beta isoform 1 isoform 3 | ||
| PREDICTED: similar to Protein FAM82B | ||
| Pro-platelet basic protein precursor | ||
| Ras suppressor protein 1 isoform 2 | ||
| Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 | ||
| Solute carrier family 4, anion exchanger, member 1 | ||
| Talin 2 | ||
| Thrombospondin 1 precursor | ||
| Tubulin, alpha 1 | ||
| UNC-112 related protein 2 long form | ||
| Vinculin isoform meta-VCL | ||
| Heparin cofactor II precursor | ||
| A disintegrin and metalloproteinase domain 7 | ||
| Haptoglobin-related protein | ||
| Serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | ||
| Inter-alpha globulin inhibitor H2 polypeptide | ||
| Solute carrier family 5 (sodium/glucose cotransporter), member 10 | ||
| Complement component 8, beta polypeptide preproprotein | ||
| Heparanase | ||
| Transgelin 2 | ||
| Hypothetical protein LOC56912 | ||
| Keratin 34 | ||
| Zinc finger protein 333 | ||
| Keratin 25C | ||
| Tyrosine 3/tryptophan 5-monooxygenase activation protein, zeta polypeptide | ||
| Keratin 31 |
aGroup 1: CaP-specific (elevated in CaP, not found in OC)
bGroup 2: elevated in both CaP and OC
cGroup 3: OC-specific (elevated in OC, not found in CaP). Only those analytes whose total spectral counts were different by >50% relative to the matched control group are shown (pre- vs. post-prostatectomy for CaP and cancer vs. benign for OC)
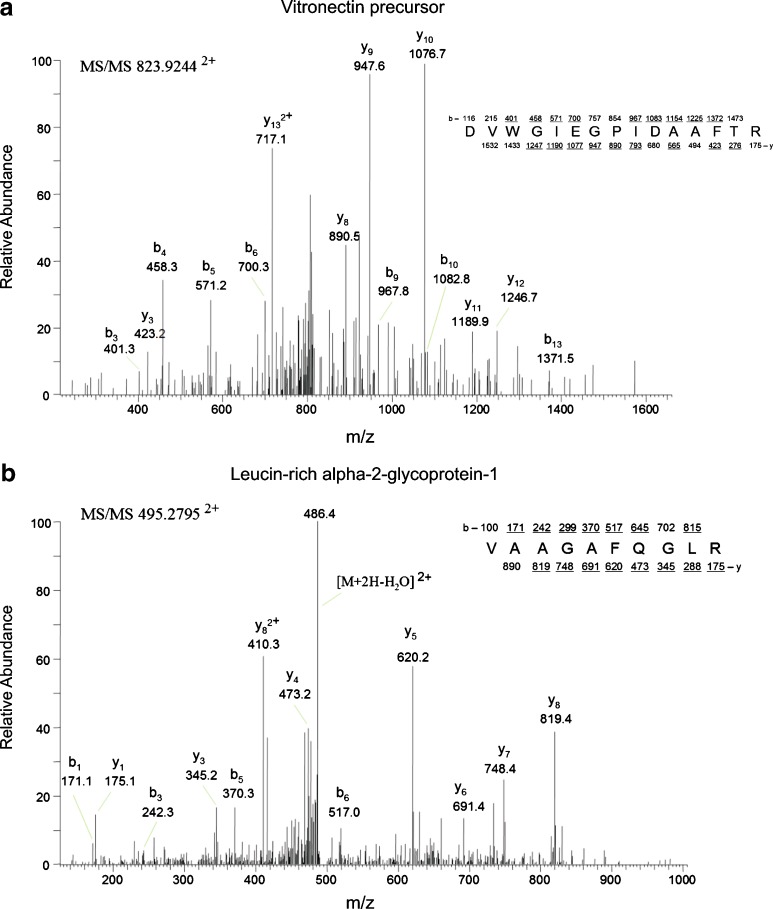
Comparing the results from different neoplastic processes could be a useful means to identify proteins involved more specifically in the neoplastic process of the specific type of tumor (i.e., ovarian cancer vs. prostate cancer) vs. those that may be more reflective of any neoplastic growth or inflammatory process. Manual verification vitronectin precursor and leucin-rich alpha-2-glycoprotein 1, taken as a representative example of differentially abundant analytes for CaP (Fig. 6a) and OC (Fig. 6b), are shown and reveal that the MS/MS analysis was correct for these example candidate biomarkers, providing increased confidence in the Scaffold results.
Fig. 6.
MS/MS spectra for peptide analytes with statistically significant elevations. The precursor b and y ion masses are shown, as well as the matched amino acid sequence (top right for each panel) for MS/MS spectra corresponding to a a CaP-specific peptide from vitronectin precursor and b an OC-specific peptide from leucine-rich alpha 2 glycoprotein 1
DISCUSSION
The search for novel clinically relevant disease-specific protein biomarkers that can be measured in an easily accessible body fluid such as the blood remains a huge challenge for researches because of the complexity of the circulatory proteome and the likely extremely low concentration of any candidate biomarkers coupled with the lability of the markers in vivo and ex vivo. The intensity of biomarker discovery efforts has lead to the increased investigation of the peptidome/LMW proteome because of the facile nature of this archive to easily pass between the tissue and bloodstream. This information archive may be especially a rich source of disease-specific information for the early detection of cancer, toxicity, and drug abuse/doping whereby complex changes within the tissue microenvironment may be reflected in altered pathophysiology of the affected organ(s).
To that end, we developed a novel workflow for LMW proteome/peptidome biomarker research of low-molecular-weight proteins/peptides by the novel application of hydrogel nanoparticle technology as a tool to rapidly capture and concentrate analytes in solution coupled with high-sensitivity mass spectrometry analysis (Fig. 4). The novel aspect of this new biomarker discovery method lies within the capacity of the core–shell nanoparticles (described in (26–28)) to perform both an affinity capture step and size exclusion chromatography in solution in just one step. In this instance, we used an acrylic acid affinity capture reagent that is immobilized within the core region of the nanoparticle, which is then covered with a shell matrix with tuned porosity to allow in analytes <30 kDa.
Consequently, for this feasibility study, the particles will bind only protein analytes that are net positively charged at the buffered pH of 7.0. Other marker capture nanoparticles with different affinity “baits” can be easily constructed with other types of affinity dye reagents, for example Cibacron blue dye which we showed could effectively capture 100% of hGH in urine (27). The ability of the particles to perform two-dimensional separation in just one step, whereby subsets of the peptidome (in this case the positively charged archive) are captured from hundreds of microliters of serum along with excluding all of the high abundance resident proteins such as albumin and immunoglobulins, provides a concentration step that is equivalent to injecting in 400 μL (for ovarian cancer study) and 200 μL (for prostate cancer study) into the Orbitrap at once, without the need for any of the types of pre-fractionation step normally required to analyze blood proteins by mass spectrometry (29). Most of the procedures commonly used can result in the loss of part of important information, either because many LMW serum proteins are bound to high-molecular-weight carrier proteins or because of lengthy sample handling procedures. Moreover, mass spectrometry normally works in a dynamic range of three to four orders of magnitude, meaning that highly abundant proteins such as albumin mask most of the lower abundance proteins.
We applied the nanoparticle-based biomarker capture technique for the determination of differentially abundant peptidomes from two important human cancers as a feasibility case study and not a definitive biomarker discovery effort. We utilized a unique set of pretreatment serum samples taken from CaP patients with organ-confined disease who were undergoing radical prostatectomy whereby patient-matched pre- and post-prostatectomy samples were obtained. This type of serum collection represents a powerful opportunity to understand the changes in protein expression from the same individual with and without organ-confined cancer. We utilized a second study set serum obtained from women with early-stage ovarian cancer and women with benign gynecologic conditions as an age-matched control group. The use of differentially spiked-in cytokine standards into the starting serum sample that were then analyzed at the end of the entire process by MS/MS allowed us to evaluate the robustness of the overall process, with all samples showing the expected 10:1 or 1:10 ratios as seen by BioSieve analysis (Fig. 3). While each individual independently run sample did not show the expected ion abundance ratios for peptides corresponding to the two spiked proteins, the average of the case and control sample ratios was approximately equal to the expected ratios and indicated overall process fidelity. Based on this result, we have higher confidence in the biomarker candidate information generated by MS/MS and differential spectral counting methods within the cancer study sets utilized. Furthermore, to evaluate the specificity of the candidate markers found by nanoparticle harvesting, we compared the resultant candidates within the two cancers to each other. We used the results of both Scaffold and BioSieve software, the former to analyze the MS2 data and the other to analyze the MS1 data and integrate the information from both methods. An interesting aspect of a MS1-based approach is that the comparison is not related to an MS2 identification, and as long as the independent MS chromatograms align well, the software performs a statistically driven analysis on any reproducibly seen peak without regard to whether or not an MS2 spectrum is not available, and thus, lower abundant analytes are also analyzed. Peptides that are found to be potentially differentially abundant can then be identified searching the obtained MS2 data against a human protein database (NCBI) with SEQUEST. An MS targeting experiment could be performed when the MS2 spectra were not obtained from the first analysis and differentially abundant peptides taken forward for further validation by mass spectrometry-based methods such as multiple reaction monitoring (MRM).
We chose a label-free spectral counting approach since this method has been found to provide a cost-efficient reproducible means of obtaining accurate differences in relative peptide abundances between different input samples based on the differences in the number of times a specific tryptic fragment is selected for MS/MS within a data-dependent experiment (30).
An overview of the proteins identified for both OC and CaP, found by either Scaffold (results shown in Tables IV and V) or Biosieve (not shown) and in aggregate summary of both (Table IX), reveals low-molecular-weight protein such as Profilin 1, Ras suppressor protein 1, S100A7, Ribonuclease RNAse Family, Platelet factor 4, and enzymes. The nanoparticles captured LMW fragments of high-molecular-weight proteins (e.g., ceruloplasmin, fibronectin 1, Talin 1, actinin alpha 1, apolipoprotein B) that were found to be differentially abundant in cancer vs. control sera as defined by spectral counting differences. The inferred proteins, corresponding to identified peptides, included serum plasma proteins such as serpin peptidase inhibitor, clade A, member 3 precursor, angiotensinogen, and orosomucoid 1. The identified tryptic fragments originate from both cytoplasmic and nuclear proteins which reinforce the value of the LMW proteome/peptidome as being populated by cellular information, reflecting ongoing tissue pathophysiology. The presence in the serum of peptide fragments of intracellular proteins may yield valuable information about the processes and signaling networks operating within the tumor cells. Integrins, LRG, and LPB have been identified in this pilot study as proteins differentially abundant uniquely in ovarian cancer. Integrins constitute a family of transmembrane receptor proteins that mediate the extracellular matrix influence on cell growth and differentiation (31–34). Decreased or increased integrin expression has been shown to be associated with cytoskeletal changes coincident with tumor transformation, and a role for integrins in tumor growth and metastasis has repeatedly been proposed (35–38).
If these results are extensively validated in larger independent study sets, development of the expression profiles of LMW peptide fragments of integrin might be useful as tumor progression markers for prognostic and for diagnostic purposes. Moreover, for individual tumors, this may have further potential in identifying a cell surface signature for a specific tumor type and/or stage. (39). Leucine-rich alpha-2-glycoprotein-1 (LRG) is a serum glycoprotein of unknown function that has shown promise based on qualitative assessments as a biomarker for certain diseases, including microbial infections and cancer (40–42).
Increased serum LRG was observed in patients with several types of cancer, including pancreatic (43), liver (44), lung (45), and epithelial ovarian cancers (46,47). However, the lack of a quantitative assay for LRG has so far limited its application. (48). Lipopolysaccharide-binding protein gene has been found upregulated in clear cell ovarian cancer compared to other major histological types (49).
Differentially abundant peptides/LMW fragments found in the prostate cancer study set include proteins already correlated with carcinogenesis or prostate cancer. Vitronectin and cadherin 1 are cell adhesion proteins whose involvement in cancer progression and cellular spreading has been investigated and demonstrated (50,51). Vitronectin is recognized by certain members of the integrin family and acts as cell-to-substrate adhesion molecule, while cadherin 1 is involved in mechanisms regulating cell–cell adhesions, mobility, and proliferation of epithelial cells (52,53). The presence of vitronectin and cadherin 1 in the circulation could indicate the local loss of diffusion barriers, such as cell junction and the basement membrane. (54). Vitronectin has recently been identified as an extrinsic inducer of stem cell differentiation through an integrin alpha(v)beta(3)-dependent mechanism (55) and may be important in mediating bone metastasis in CaP (56). Moreover, high expression levels of cadherin 1 fragment in serum have been observed in advanced metastatic prostate cancer (57,58).
The spiked-in internal standard standards, MEC and SDF-1, were both found to be differentially abundant in the correct expected pattern by both Scaffold (shown) and BioSieve (not shown), which provides increased confidence to the validity of the newly identified LMW candidates found. Of course, further analysis will be required for full validation. Future work will encompass antibody-based (e.g., ELISA) and antibody-independent (MRM-MS) verification and validation of differential expression in independent blinded serum study sets, including benign and inflammatory diseases as well as other cancers, to understand the specificity and sensitivity of each candidate alone and in combination with each other.
Overall, we identified more differentially abundant proteins in the OC study sets than with CaP samples, and this is very likely due to the fact that we used a smaller input starting volume of serum for CaP studies (due to limitations in sample available) compared to OC samples. This is in keeping with expected results from nanoparticle capture since the entire contents are analyzed from the particle eluates: more input sample, more LMW peptidome analyzed/MS run. Consequently, it is highly likely that the number of CaP-specific candidates would increase if input starting sample volume increased. Moreover, this then highlights the potential for this workflow to enable detection of lower level endogenous proteins and protein fragments as larger volumes of starting sample are used. To that end, among other investigations, we are evaluating experimental results whereby very large volumes (e.g., 8 cc) of body fluids such as serum are used as the input into the nanoparticle capture workflow. The results described in this study are not meant in any way to reflect a comprehensive analysis of the serum peptidome or the entirety of the prostate and ovarian cancer LMW biomarker candidates that exist. Indeed, the identified analyte content is entirely dependent on the bait chemistry used within the nanoparticle. We used an acrylic acid capture chemistry that will selectively bind only analytes with a net positive charge at pH 7.0. Of course, the charge structure of the LMW analyte is dependent on the amino acid composition of the peptide/LMW protein. In the future, one can imagine the use of cocktails of particles with differential binding chemistries for positively charged proteins, negatively charged proteins, phosphoproteins, glycoproteins, etc. that will provide larger screening capabilities and wider coverage of the peptidome for biomarker discovery.
CONCLUSIONS
With this work, we have begun to test a novel workflow for body fluid-based biomarker discovery based on the application of a novel nanoparticle-based biomarker capture and concentration technology to amplify, fractionate, and enrich the peptidome/LMW analytes in a single simple step that in just a few minutes can harvest candidate biomarker analytes from raw unprocessed serum. This workflow provides subsequent high-resolution RPLC-MS/MS analysis with a concentrated and fractionated sample enriched for peptidome content. Using this approach, we have identified an initial set of OC and CaP-specific candidate peptides/LMW proteins from what appears to be a rich source of information. These candidates will require further extensive verification and validation in order to understand clinical utility either alone or in combination.
Acknowledgments
The authors appreciate the generous support of Dr. Vikas Chandhoke and the Department of Life Sciences at George Mason University. This work was partly supported by the Italian Istituto Superiore di Sanita` in the framework Italy/USA cooperation agreement between the U.S. Department of Health and Human Services, George Mason University, and the Italian Ministry of Public Health. This work was partially supported by the U.S. Department of Energy grant no. 201270.
Footnotes
Claudia Fredolini and Francesco Meani equally contributed to this work.
References
- 1.Crawford ED. Understanding the epidemiology, natural history, and key pathways involved in prostate cancer. Urology. 2009;73(Suppl 5):4–10. doi: 10.1016/j.urology.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med. 2001;344:1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 3.Schröder FH, Hugosson J, Roobol MJ, ERSPC Investigators et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Roehrborn CG. The utility of serum prostatic-specific antigen in the management of men with benign prostatic hyperplasia. Int J Impot Res. 2008;20(Suppl 3):S19–S26. doi: 10.1038/ijir.2008.53. [DOI] [PubMed] [Google Scholar]
- 5.Thompson IA, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level of ≤4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 6.Horner MJ, Ries LAG, Krapcho M, et al., editors. SEER Cancer Stat Fact Sheets. 2009, Bethesda, MD. National Cancer Institute. http://seer.cancer.gov/statfacts/html/ovary.html, accessed December 2, 2009.
- 7.Rosenthal AN, Menon U, Jacobs IJ. Screening for ovarian cancer. Clinical Obstetrics and Gynecology. 2006;49:433–447. doi: 10.1097/00003081-200609000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Duffy MJ, Bonfrer JM, Kulpa J, et al. CA125 in ovarian cancer. European Group on Tumor Markers guidelines for clinical use. Int J Gynecol Cancer. 2005;15:679–691. doi: 10.1111/j.1525-1438.2005.00130.x. [DOI] [PubMed] [Google Scholar]
- 9.Bast RC., Jr Status of tumor markers in ovarian cancer screening. J Clin Oncol. 2003;21(Suppl 10):200s–205s. doi: 10.1200/JCO.2003.01.068. [DOI] [PubMed] [Google Scholar]
- 10.Azad NS, Rasool N, Annunziata CM, Minasian L, Whiteley G, Kohn EC. Proteomics in clinical trials and practice: present uses and future promise. Mol Cell Proteomics. 2006;5:1819–1829. doi: 10.1074/mcp.R600008-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Buys SS, Partridge E, Greene MH, et al. Ovarian cancer screening in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial: findings from the initial screen of a randomized trial. Am J Obstet Gynecol. 2005;193:1630–1639. doi: 10.1016/j.ajog.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Skates SJ, Menon U, MacDonald N, et al. Calculation of the risk of ovarian cancer from serial CA-125 values for preclinical detection in postmenopausal women. J Clin Oncol. 2003;21(Suppl 10):206–210s. doi: 10.1200/JCO.2003.02.955. [DOI] [PubMed] [Google Scholar]
- 13.Menon U, Skates SJ, Lewis S, et al. Prospective study using the risk of ovarian cancer algorithm to screen for ovarian cancer. J Clin Oncol. 2005;23:7919–7926. doi: 10.1200/JCO.2005.01.6642. [DOI] [PubMed] [Google Scholar]
- 14.Berven FS, Flikka K, Berle M, et al. Proteomic-based biomarker discovery with emphasis on cerebrospinal fluid and multiple sclerosis. Curr Pharm Biotechnol. 2006;7:147–158. doi: 10.2174/138920106777549713. [DOI] [PubMed] [Google Scholar]
- 15.Farina A, Dumonceau JM, Lescuyer P. Proteomic analysis of human bile and potential applications for cancer diagnosis. Expert Rev Proteomics. 2009;6:285–301. doi: 10.1586/epr.09.12. [DOI] [PubMed] [Google Scholar]
- 16.Jamaspishvili T, Kral M, Khomeriki I, et al. Urine markers in monitoring for prostate cancer. Prostate Cancer Prostatic Dis. 2010;13:12–19. doi: 10.1038/pcan.2009.31. [DOI] [PubMed] [Google Scholar]
- 17.Drake RR, White KY, Fuller TW, Igwe E, Clements MA, Nyalwidhe JO, et al. Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease. J Proteomics. 2009;72:907–917. doi: 10.1016/j.jprot.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chem Rev. 2007;107:3601–3620. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 19.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, et al. An investigation into the human serum “interactome”. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 20.Wang YY, Cheng P, Chan DW. A simple affinity spin tube filter method for removing high-abundant common proteins or enriching low-abundant biomarkers for serum proteomic analysis. Proteomics. 2003;3:243–248. doi: 10.1002/pmic.200390036. [DOI] [PubMed] [Google Scholar]
- 21.Gundry RL, Van Eyk JE. Unraveling the complexity of circulating forms of brain natriuretic peptide. Clin Chem. 2007;53:1181–1182. doi: 10.1373/clinchem.2007.086611. [DOI] [PubMed] [Google Scholar]
- 22.Lowenthal MS, Mehta AI, Frogale K, Bandle RW, Araujo RP, Hood BL, et al. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- 23.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 25.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 26.Luchini A, Geho DH, Bishop B, et al. Smart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradation. Nano Lett. 2008;8:350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredolini C, Meani F, Alex Reeder K, et al. Concentration and preservation of very low abundance biomarkers in urine, such as human growth hormone (hGH), by Cibacron Blue F3G-A loaded hydrogel particles. Nano Res. 2008;1:502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longo C, Patanarut A, George T, Bishop B, Zhou W, Fredolini C, et al. Core–shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS One. 2009;4(3):e4763. doi: 10.1371/journal.pone.0004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed FE. Sample preparation and fractionation for proteome analysis and cancer biomarker discovery by mass spectrometry. J Sep Sci. 2009;32:771–779. doi: 10.1002/jssc.200900014. [DOI] [PubMed] [Google Scholar]
- 30.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 31.Streuli CH. Integrins and cell-fate determination. J Cell Sci. 2009;122:171–177. doi: 10.1242/jcs.018945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eliceiri BP. Integrin and growth factor receptor crosstalk. Circ Res. 2001;89:1104–1110. doi: 10.1161/hh2401.101084. [DOI] [PubMed] [Google Scholar]
- 33.Danen EH, Yamada KM. Fibronectin, integrins, and growth control. J Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- 34.Howe A, Aplin AE, Alahari SK, Juliano RL. Integrin signaling and cell growth control. Curr Opin Cell Biol. 1998;10:220–231. doi: 10.1016/S0955-0674(98)80144-0. [DOI] [PubMed] [Google Scholar]
- 35.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 36.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goel HL, Li J, Kogan S, Languino LR. Integrins in prostate cancer progression. Endocr Relat Cancer. 2008;15:657–664. doi: 10.1677/ERC-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monniaux D, Huet-Calderwood C, Le Bellego F, Fabre S, Monget P, Calderwood DA. Integrins in the ovary. Semin Reprod Med. 2006;24:251–261. doi: 10.1055/s-2006-948554. [DOI] [PubMed] [Google Scholar]
- 39.Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. doi: 10.1046/j.1525-1373.1999.d01-122.x. [DOI] [PubMed] [Google Scholar]
- 40.Weivoda S, Andersen JD, Skogen A, Schlievert PM, Fontana D, Schacker T, et al. ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J Immunol Methods. 2008;336:22–29. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382:776–779. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen JD, Boylan KL, Xue FS, Anderson LB, Witthuhn BA, Markowski TW, et al. Identification of candidate biomarkers in ovarian cancer serum by depletion of highly abundant proteins and differential in-gel electrophoresis. Electrophoresis. 2010;31:599–610. doi: 10.1002/elps.200900441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakisaka T, Kondo T, Okano T, Fujii K, Honda K, Endo M, et al. Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2007;852:257–267. doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawakami T, Hoshida Y, Kanai F, Tanaka Y, Tateishi K, Ikenoue T, et al. Proteomic analysis of sera from hepatocellular carcinoma patients after radiofrequency ablation treatment. Proteomics. 2005;5:4287–4295. doi: 10.1002/pmic.200401287. [DOI] [PubMed] [Google Scholar]
- 45.Okano T, Kondo T, Kakisaka T, Fujii K, Yamada M, Kato H, et al. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006;6:3938–3948. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Lim BK, Hashim OH. Different altered stage correlative expression of high abundance acute-phase proteins in sera of patients with epithelial ovarian carcinoma. J Hematol Oncol. 2009;2:37. doi: 10.1186/1756-8722-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Lim BK, Peh SC, Abdul-Rahman PS. Hashim OH profiling of serum and tissue high abundance acute-phase proteins of patients with epithelial and germ line ovarian carcinoma. Proteome Sci. 2008;6:20. doi: 10.1186/1477-5956-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weivoda S, Andersen JD, Skogen A, Schlievert PM, Fontana D, et al. ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J Immunol Methods. 2008;336:22–29. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 50.Paschos KA, Canovas D, Bird NC. The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis. Cell Signal. 2009;21:665–674. doi: 10.1016/j.cellsig.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Heyman L, Leroy-Dudal J, Fernandes J, Seyer D, Dutoit S, Carreiras F. Mesothelial vitronectin stimulates migration of ovarian cancer cells. Cell Biol Int. 2010;34:493–502. doi: 10.1042/CBI20090331. [DOI] [PubMed] [Google Scholar]
- 52.Goodwin M, Yap AS. Classical cadherin adhesion molecules: coordinating cell adhesion, signaling and the cytoskeleton. J Mol Histol. 2004;35:839–844. doi: 10.1007/s10735-004-1833-2. [DOI] [PubMed] [Google Scholar]
- 53.Felding-Habermann B, Cheresh DA. Vitronectin and its receptors. Curr Opin Cell Biol. 1993;5:864–868. doi: 10.1016/0955-0674(93)90036-P. [DOI] [PubMed] [Google Scholar]
- 54.De Wever O, Derycke L, Hendrix A, De Meerleer G, Godeau F, Depypere H, et al. Soluble cadherins as cancer biomarkers. Clin Exp Metastasis. 2007;24:685–697. doi: 10.1007/s10585-007-9104-8. [DOI] [PubMed] [Google Scholar]
- 55.Hurt EM, Chan K, Serrat MA, Thomas SB, Veenstra TD, Farrar WL. Identification of vitronectin as an extrinsic inducer of cancer stem cell differentiation and tumor formation. Stem Cells. 2010;28:390–398. doi: 10.1002/stem.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper CR, Chay CH, Pienta KJ. The role of alpha(v)beta(3) in prostate cancer progression. Cooper Neoplasia. 2002;4:191–194. doi: 10.1038/sj.neo.7900224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuefer R, Hofer MD, Gschwend JE, et al. The role of an 80 kDa fragment of E-cadherin in the metastatic progression of prostate cancer. Clin Cancer Res. 2003;9:6447–6452. [PubMed] [Google Scholar]
- 58.Kuefer R, Hofer MD, Zorn CS, Engel O, Volkmer BG, Juarez-Brito MA, et al. Assessment of a fragment of e-cadherin as a serum biomarker with predictive value for prostate cancer. Br J Cancer. 2005;92:2018–2023. doi: 10.1038/sj.bjc.6602599. [DOI] [PMC free article] [PubMed] [Google Scholar]