Abstract
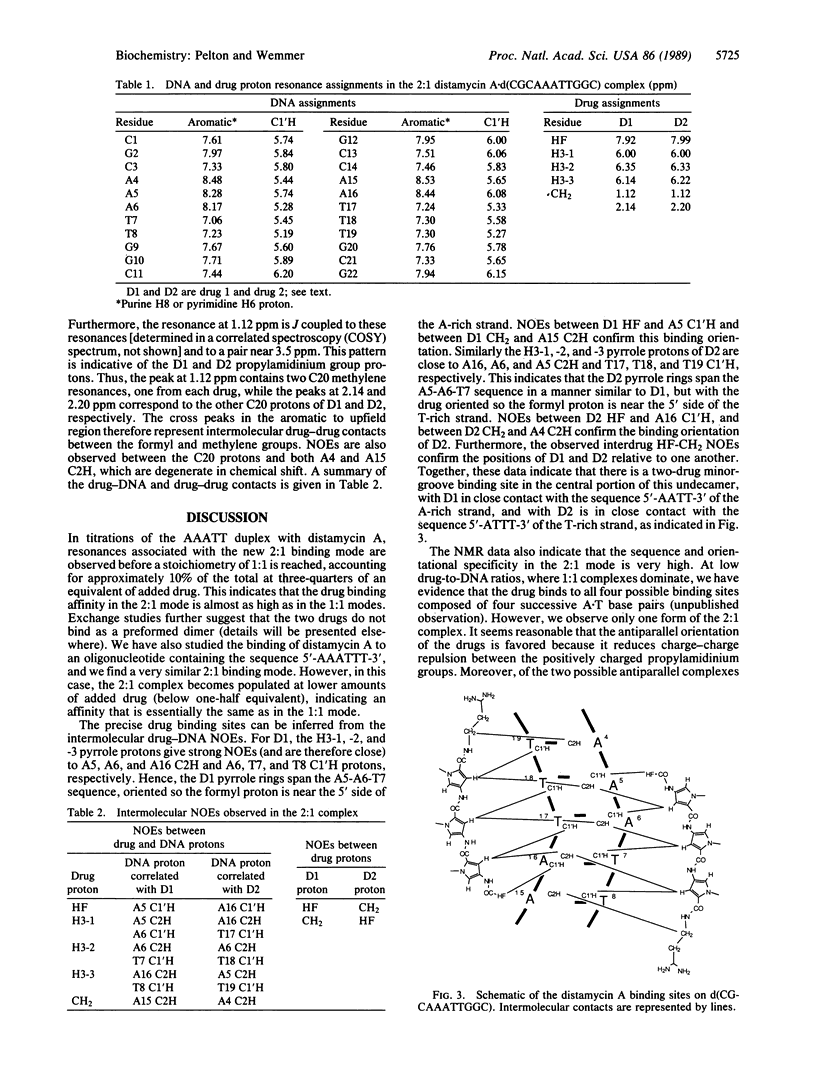
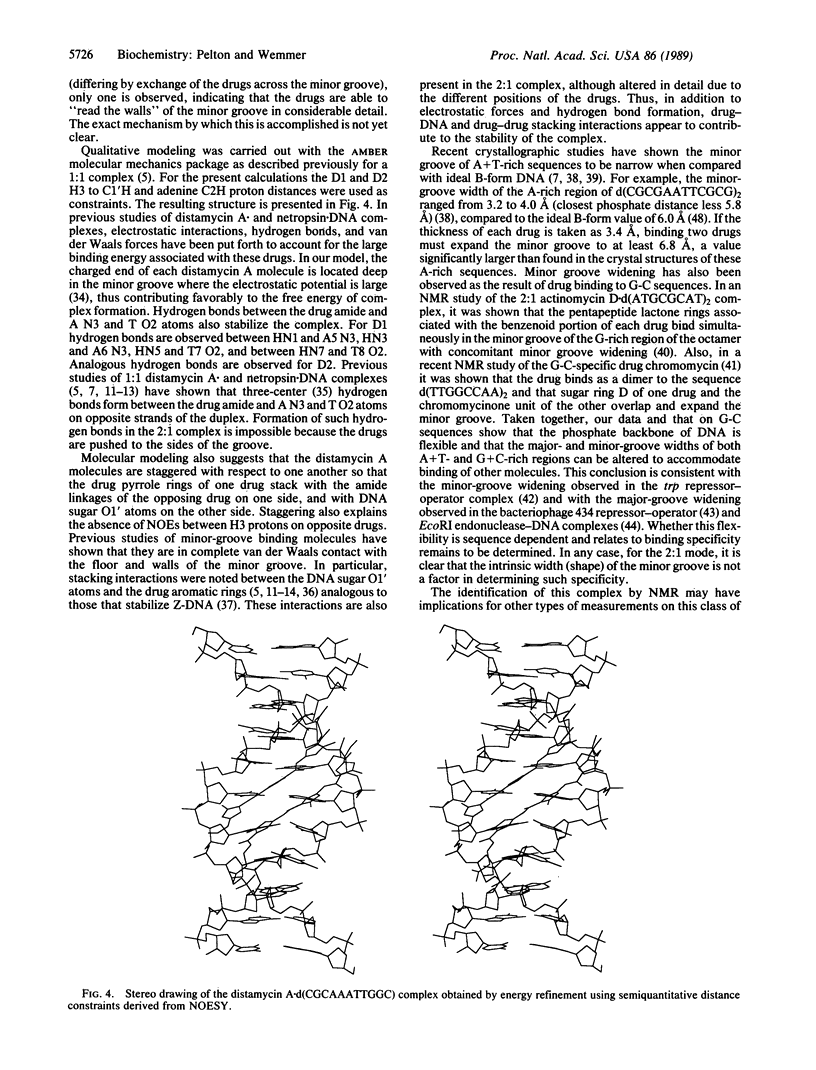
Two-dimensional NMR has been used to study the 2:1 distamycin A.d(CGCAAATTGGC).d(GCCAATTTGCG) complex. The nuclear Overhauser effect spectroscopy (NOESY) experiment was used to assign the aromatic and C1'H DNA protons and to identify drug-DNA contacts. These data indicate that two drug molecules bind simultaneously in the minor groove of the central 5'-AAATT-3' segment and are in close contact with both the DNA and one another. One drug binds with the formyl end close to the second adenine base of the A-rich strand, while the other drug binds with the formyl end close to the second adenine of the complementary strand. With this binding orientation, the positively charged propylamidinium groups are directed toward opposite ends of the helix. Molecular modeling shows that the minor groove must expand relative to the 1:1 complex to accommodate both drugs. Energy calculations suggest that electrostatic interactions, hydrogen bonds, and van der Waals forces contribute to the stability of the complex.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Behling R. W., Kearns D. R. 1H two-dimensional nuclear Overhauser effect and relaxation studies of poly(dA).poly(dT) Biochemistry. 1986 Jun 3;25(11):3335–3346. doi: 10.1021/bi00359a037. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Remeta D. P., Chou W. Y., Ferrante R., Curry J., Zaunczkowski D., Snyder J. G., Marky L. A. Enthalpy-entropy compensations in drug-DNA binding studies. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8922–8926. doi: 10.1073/pnas.84.24.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. H., Hare D. R., Wemmer D. E., Reid B. R. Sequence-specific recognition of deoxyribonucleic acid. Chemical synthesis and nuclear magnetic resonance assignment of the imino protons of lambda OR3 operator deoxyribonucleic acid. Biochemistry. 1983 Jun 21;22(13):3037–3041. doi: 10.1021/bi00282a002. [DOI] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish E. L., Lane M. J., Vournakis J. N. Determination of equilibrium binding affinity of distamycin and netropsin to the synthetic deoxyoligonucleotide sequence d(GGTATACC)2 by quantitative DNase I footprinting. Biochemistry. 1988 Aug 9;27(16):6026–6032. doi: 10.1021/bi00416a030. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Kintanar A., Klevit R. E., Reid B. R. Two-dimensional NMR investigation of a bent DNA fragment: assignment of the proton resonances and preliminary structure analysis. Nucleic Acids Res. 1987 Jul 24;15(14):5845–5862. doi: 10.1093/nar/15.14.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevit R. E., Wemmer D. E., Reid B. R. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986 Jun 3;25(11):3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Zakrzewska K., Pullman B. Binding of non-intercalating antibiotics to B-DNA: a theoretical study taking into account nucleic acid flexibility. J Biomol Struct Dyn. 1986 Jun;3(6):1155–1170. doi: 10.1080/07391102.1986.10508492. [DOI] [PubMed] [Google Scholar]
- Letsinger R. L., Lunsford W. B. Synthesis of thymidine oligonucleotides by phosphite triester intermediates. J Am Chem Soc. 1976 Jun 9;98(12):3655–3661. doi: 10.1021/ja00428a045. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Breslauer K. J. Calorimetric and spectroscopic investigation of drug-DNA interactions. I. The binding of netropsin to poly d(AT). Nucleic Acids Res. 1983 May 11;11(9):2857–2870. doi: 10.1093/nar/11.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of the imino protons of the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix in complexes with the antibiotics netropsin and/or actinomycin. Biochemistry. 1983 Mar 1;22(5):1107–1113. doi: 10.1021/bi00274a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Antibiotic-DNA interactions: intermolecular nuclear Overhauser effects in the netropsin-d(C-G-C-G-A-A-T-T-C-G-C-G) complex in solution. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6424–6428. doi: 10.1073/pnas.79.21.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Covalent carcinogenic O6-methylguanosine lesions in DNA. Structural studies of the O6 meG X A and O6meG X G interactions in dodecanucleotide duplexes. J Mol Biol. 1986 Apr 20;188(4):677–692. doi: 10.1016/s0022-2836(86)80014-6. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L. Molecular recognition in noncovalent antitumor agent-DNA complexes: NMR studies of the base and sequence dependent recognition of the DNA minor groove by netropsin. Biochimie. 1985 Jul-Aug;67(7-8):887–915. doi: 10.1016/s0300-9084(85)80181-4. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural modeling of the distamycin A-d(CGCGAATTCGCG)2 complex using 2D NMR and molecular mechanics. Biochemistry. 1988 Oct 18;27(21):8088–8096. doi: 10.1021/bi00421a018. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Gupta G., Sarma R. H. Netropsin specifically recognizes one of the two conformationally equivalent strands of poly(dA).poly(dT). One dimensional NMR study at 500 MHz involving NOE transfer between netropsin and DNA protons. J Biomol Struct Dyn. 1985 Jun;2(6):1085–1095. doi: 10.1080/07391102.1985.10507625. [DOI] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Schultz P. G., Dervan P. B. Distamycin and penta-N-methylpyrrolecarboxamide binding sites on native DNA. A comparison of methidiumpropyl-EDTA-Fe(II) footprinting and DNA affinity cleaving. J Biomol Struct Dyn. 1984 Mar;1(5):1133–1147. doi: 10.1080/07391102.1984.10507508. [DOI] [PubMed] [Google Scholar]
- Scott E. V., Zon G., Marzilli L. G., Wilson W. D. 2D NMR investigation of the binding of the anticancer drug actinomycin D to duplexed dATGCGCAT: conformational features of the unique 2:1 adduct. Biochemistry. 1988 Oct 4;27(20):7940–7951. doi: 10.1021/bi00420a053. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Determination of netropsin-DNA binding constants from footprinting data. Biochemistry. 1988 Feb 23;27(4):1198–1205. doi: 10.1021/bi00404a020. [DOI] [PubMed] [Google Scholar]
- Warshaw M. M., Cantor C. R. Oligonucleotide interactions. IV. Conformational differences between deoxy- and ribodinucleoside phosphates. Biopolymers. 1970;9(9):1079–1103. doi: 10.1002/bip.1970.360090910. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Patel D. J., Sauer R. T., Karplus M. 1H-NMR study of the lambda operator site OL1: assignment of the imino and adenine H2 resonances. Nucleic Acids Res. 1984 May 11;12(9):4035–4047. doi: 10.1093/nar/12.9.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]


