Abstract
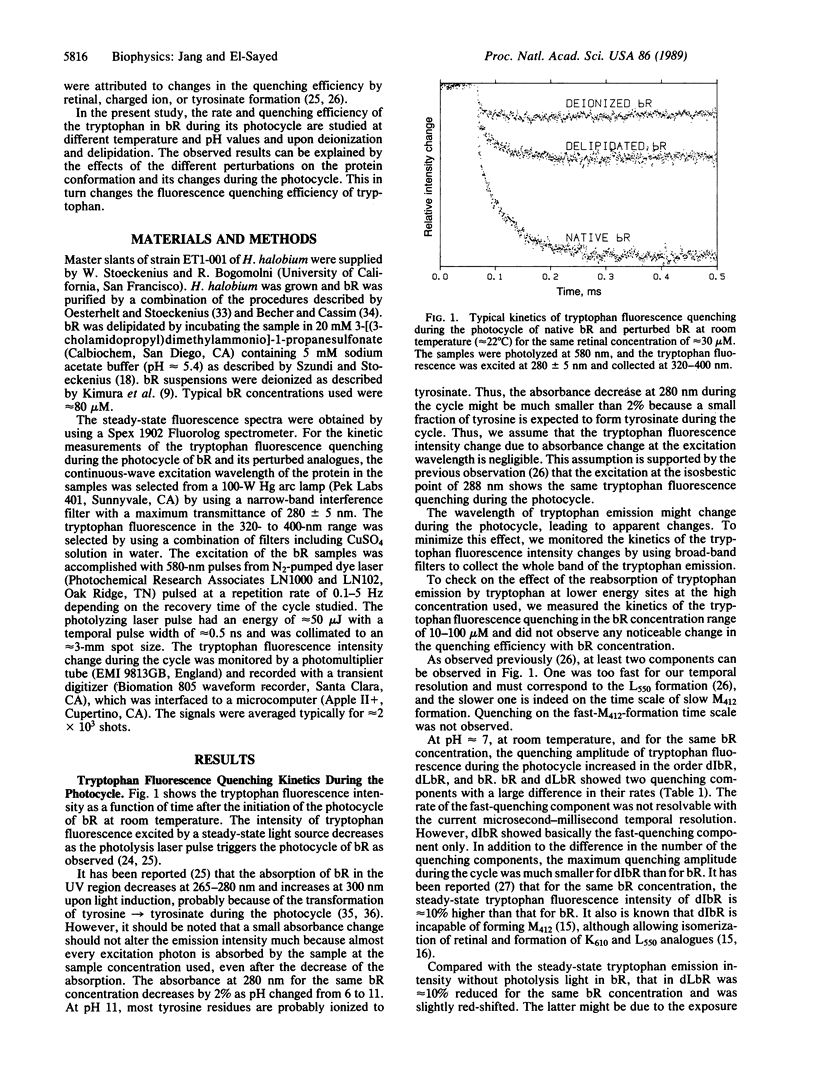
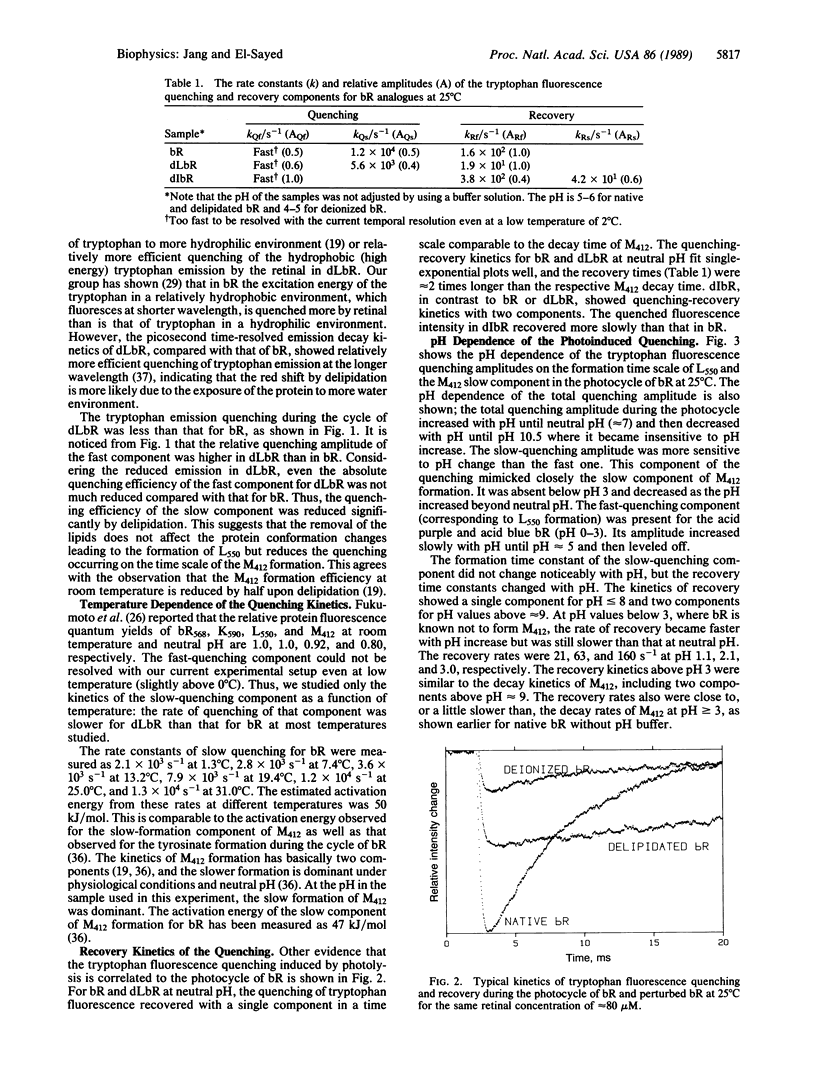
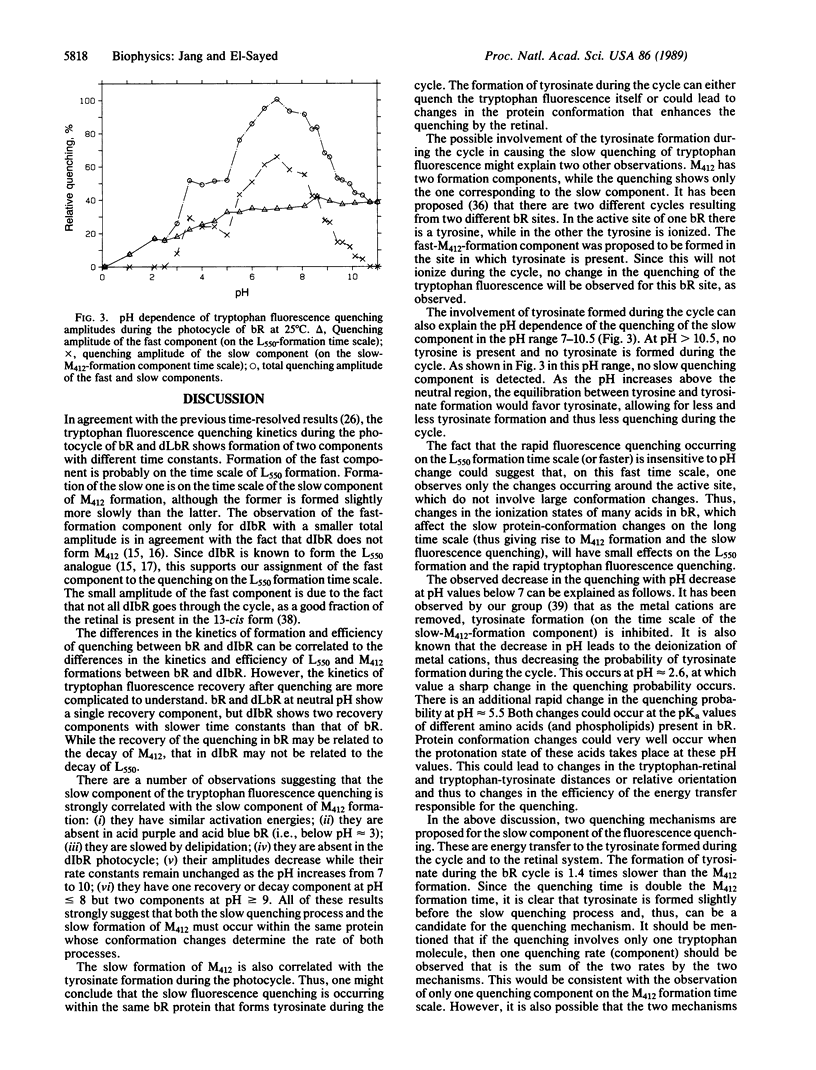
The rates of the quenching and recovery of tryptophan fluorescence are determined in the microsecond-millisecond time scale during the photocycle of bacteriorhodopsin under different perturbations. The kinetics suggest the presence of two quenching processes, a rapid one (on the time scale of photocycle intermediate L550 formation or faster) and a slow one (slightly slower than the slow component of intermediate M412 formation). The slow quenching process is found to respond to different perturbations in the same manner as the slow component of M412 formation. It has the same activation energy, it is inhibited if metal cations are removed, it is negligible at pH values greater than the pKa of tyrosine, and its rate is slowed down when 75% of the lipids are removed. These results, together with the observed value of the quenching activation energy, suggest that the rates of the tryptophan fluorescence quenching, like those of tyrosinate and M412 formations during the cycle, are all determined by the rates of the protein conformation changes. The pH studies of the slow quenching process show that the maximum quenching probability occurs at neutral pH. A rapid decrease in quenching occurs at lower pH (approximately 3 and approximately 5.5) and higher pH (approximately 9). Two quenching mechanisms involving energy transfer to either retinal or to tyrosinate are considered. Protein conformation changes resulting from a change in the ionization state of amino acids of different pKa values could change the tryptophan-retinal (or tryptophan-tyrosinate) coupling and thus the quenching efficiency.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ariki M., Lanyi J. K. Characterization of metal ion-binding sites in bacteriorhodopsin. J Biol Chem. 1986 Jun 25;261(18):8167–8174. [PubMed] [Google Scholar]
- Bayley H., Huang K. S., Radhakrishnan R., Ross A. H., Takagaki Y., Khorana H. G. Site of attachment of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2225–2229. doi: 10.1073/pnas.78.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B. M., Cassim J. Y. Improved isolation procedures for the purple membrane of Halobacterium halobium. Prep Biochem. 1975;5(2):161–178. doi: 10.1080/00327487508061568. [DOI] [PubMed] [Google Scholar]
- Bogomolni R. A., Stubbs L., Lanyi J. K. Illumination-dependent changes in the intrinsic fluorescence of bacteriorhodopsin. Biochemistry. 1978 Mar 21;17(6):1037–1041. doi: 10.1021/bi00599a015. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Stern L. J., Hackett N. R., Chao B. H., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: I. Tyrosine-185 protonates and deprotonates during the photocycle. Proteins. 1988;3(4):219–229. doi: 10.1002/prot.340030403. [DOI] [PubMed] [Google Scholar]
- Bridgen J., Walker I. D. Photoreceptor protein from the purple membrane of Halobacterium halobium. Molecular weight and retinal binding site. Biochemistry. 1976 Feb 24;15(4):792–798. doi: 10.1021/bi00649a010. [DOI] [PubMed] [Google Scholar]
- Chang C. H., Chen J. G., Govindjee R., Ebrey T. Cation binding by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1985 Jan;82(2):396–400. doi: 10.1073/pnas.82.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronister E. L., Corcoran T. C., Song L., El-Sayed M. A. On the molecular mechanisms of the Schiff base deprotonation during the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8580–8584. doi: 10.1073/pnas.83.22.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran T. C., Ismail K. Z., El-Sayed M. A. Evidence for the involvement of more than one metal cation in the Schiff base deprotonation process during the photocycle of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4094–4098. doi: 10.1073/pnas.84.12.4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencher N., Wilms M. Flash photometric experiments on the photochemical cycle of bacteriorhodopsin. Biophys Struct Mech. 1975 May 30;1(3):259–271. doi: 10.1007/BF00535760. [DOI] [PubMed] [Google Scholar]
- Dupuis P., Corcoran T. C., El-Sayed M. A. Importance of bound divalent cations to the tyrosine deprotonation during the photocycle of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3662–3664. doi: 10.1073/pnas.82.11.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis P., Corcoran T. C., El-Sayed M. A. Importance of bound divalent cations to the tyrosine deprotonation during the photocycle of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3662–3664. doi: 10.1073/pnas.82.11.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto J. M., Hopewell W. D., Karvaly B., El-Sayed M. A. Time-resolved protein fluorescence studies of intermediates in the photochemical cycle of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Jan;78(1):252–255. doi: 10.1073/pnas.78.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamoto J. H., Dupuis P., El-Sayed M. A. On the protein (tyrosine)-chromophore (protonated Schiff base) coupling in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7083–7087. doi: 10.1073/pnas.81.22.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang D. J., el-Sayed M. A. Deprotonation of lipid-depleted bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5918–5922. doi: 10.1073/pnas.85.16.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisky O., Feitelson J., Ottolenghi M. Photochemistry and fluorescence of bacteriorhodopsin excited in its 280-nm absorption band. Biochemistry. 1981 Jan 6;20(1):205–209. doi: 10.1021/bi00504a034. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Ikegami A., Stoeckenius W. Salt and pH-dependent changes of the purple membrane absorption spectrum. Photochem Photobiol. 1984 Nov;40(5):641–646. doi: 10.1111/j.1751-1097.1984.tb05353.x. [DOI] [PubMed] [Google Scholar]
- Mathies R. A., Brito Cruz C. H., Pollard W. T., Shank C. V. Direct observation of the femtosecond excited-state cis-trans isomerization in bacteriorhodopsin. Science. 1988 May 6;240(4853):777–779. doi: 10.1126/science.3363359. [DOI] [PubMed] [Google Scholar]
- Moore T. A., Edgerton M. E., Parr G., Greenwood C., Perham R. N. Studies of an acid-induced species of purple membrane from Halobacterium halobium. Biochem J. 1978 May 1;171(2):469–476. doi: 10.1042/bj1710469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowery P. C., Lozier R. H., Chae Q., Tseng Y. W., Taylor M., Stoeckenius W. Effect of acid pH on the absorption spectra and photoreactions of bacteriorhodopsin. Biochemistry. 1979 Sep 18;18(19):4100–4107. doi: 10.1021/bi00586a007. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Hess B. Reversible photolysis of the purple complex in the purple membrane of Halobacterium halobium. Eur J Biochem. 1973 Aug 17;37(2):316–326. doi: 10.1111/j.1432-1033.1973.tb02990.x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971 Sep 29;233(39):149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA Rhodopsin and bacteriorhodopsin: structure-function relationships. FEBS Lett. 1982 Nov 8;148(2):179–191. doi: 10.1016/0014-5793(82)80805-3. [DOI] [PubMed] [Google Scholar]
- Palmer P. L., Sherman W. V. Alkaline quenching of bacteriorhodopsin tryptophanyl fluorescence: evidence for aqueous accessibility or a hydrogen-bonded chain. Photochem Photobiol. 1985 Nov;42(5):541–547. doi: 10.1111/j.1751-1097.1985.tb01607.x. [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Roepe P., Ahl P. L., Earnest T. N., Bogomolni R. A., Das Gupta S. K., Mulliken C. M., Herzfeld J. Evidence for a tyrosine protonation change during the primary phototransition of bacteriorhodopsin at low temperature. Proc Natl Acad Sci U S A. 1986 Jan;83(2):347–351. doi: 10.1073/pnas.83.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., Mathies R. A. Resonance Raman spectra of the acidified and deionized forms of bacteriorhodopsin. Biophys J. 1985 Feb;47(2 Pt 1):251–254. doi: 10.1016/s0006-3495(85)83899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckenius W., Bogomolni R. A. Bacteriorhodopsin and related pigments of halobacteria. Annu Rev Biochem. 1982;51:587–616. doi: 10.1146/annurev.bi.51.070182.003103. [DOI] [PubMed] [Google Scholar]
- Szundi I., Stoeckenius W. Effect of lipid surface charges on the purple-to-blue transition of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3681–3684. doi: 10.1073/pnas.84.11.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szundi I., Stoeckenius W. Purple-to-blue transition of bacteriorhodopsin in a neutral lipid environment. Biophys J. 1988 Aug;54(2):227–232. doi: 10.1016/S0006-3495(88)82951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F., Ebrey T. The blue membrane: the 3-dehydroretinal-based artificial pigment of the purple membrane. Biochemistry. 1978 May 16;17(10):1915–1922. doi: 10.1021/bi00603a018. [DOI] [PubMed] [Google Scholar]


