Abstract
Tissue-specific genetic variation in expression of the alcohol dehydrogenase, encoded by the Adh-1 gene, is found between C57BL/6J (B6) mice and B6.S congenic mice. B6.S mice contain a variant Adh-1 allele derived from a wild Danish strain in a B6 genetic background. B6 mice have nearly twice the alcohol dehydrogenase activity in liver but less than half the activity in kidney as B6.S mice. These tissue-specific genetic changes in alcohol dehydrogenase expression are manifest at the level of Adh-1-encoded mRNA. The regulatory site(s) involved act cis in both kidney and liver. These strains also differ in the extent to which androgen induces mRNA encoded by kidney Adh-1, with androgen increasing these levels 17-fold and 7.4-fold in the B6 and B6.S kidney, respectively. To identify the regulatory mechanism(s) underlying this strain variation in Adh-1 transcription in the B6 and B6.S kidney, liver, and androgen-induced kidney. For both uninduced and induced kidney, a difference in the transcription rate alone accounts for the strain difference in mRNA concentration. In contrast, because the Adh-1 transcription rate in liver does not differ significantly between B6 and B6.S mice, strain-specific variation in posttranscriptional regulation must be operative. Taken together these results indicate that the variation in Adh-1 expression between B6 and B6.S mice results from changes in both transcriptional and posttranscriptional control, and these controls are differentially operative in kidney and liver.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Back D. W., Wilson S. B., Morris S. M., Jr, Goodridge A. G. Hormonal regulation of lipogenic enzymes in chick embryo hepatocytes in culture. Thyroid hormone and glucagon regulate malic enzyme mRNA level at post-transcriptional steps. J Biol Chem. 1986 Sep 25;261(27):12555–12561. [PubMed] [Google Scholar]
- Balak K. J., Keith R. H., Felder M. R. Genetic and developmental regulation of mouse liver alcohol dehydrogenase. J Biol Chem. 1982 Dec 25;257(24):15000–15007. [PubMed] [Google Scholar]
- Belayew A., Tilghman S. M. Genetic analysis of alpha-fetoprotein synthesis in mice. Mol Cell Biol. 1982 Nov;2(11):1427–1435. doi: 10.1128/mcb.2.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F. G., Breen G. A., Paigen K. Genetic determination of the developmental program for mouse liver beta-galactosidase: involvement of sites proximate to and distant from the structural gene. Genetics. 1979 Aug;92(4):1187–1203. doi: 10.1093/genetics/92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsma D. J., Grichnik J. M., Gossett L. M., Schwartz R. J. Delimitation and characterization of cis-acting DNA sequences required for the regulated expression and transcriptional control of the chicken skeletal alpha-actin gene. Mol Cell Biol. 1986 Jul;6(7):2462–2475. doi: 10.1128/mcb.6.7.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Boulet A. M., Erwin C. R., Rutter W. J. Cell-specific enhancers in the rat exocrine pancreas. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3599–3603. doi: 10.1073/pnas.83.11.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M., Schibler U. Accumulation of rare and moderately abundant mRNAs in mouse L-cells is mainly post-transcriptionally regulated. J Mol Biol. 1984 Oct 5;178(4):869–880. doi: 10.1016/0022-2836(84)90316-4. [DOI] [PubMed] [Google Scholar]
- Ceci J. D., Lawther R., Duester G., Hatfield G. W., Smith M., O'Malley M. P., Felder M. R. Androgen induction of alcohol dehydrogenase in mouse kidney. Studies with a cDNA probe confirmed by nucleotide sequence analysis. Gene. 1986;41(2-3):217–224. doi: 10.1016/0378-1119(86)90101-0. [DOI] [PubMed] [Google Scholar]
- Ceci J. D., Zheng Y. W., Felder M. R. Molecular analysis of mouse alcohol dehydrogenase: nucleotide sequence of the Adh-1 gene and genetic mapping of a related nucleotide sequence to chromosome 3. Gene. 1987;59(2-3):171–182. doi: 10.1016/0378-1119(87)90325-8. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Stacy T. P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol Cell Biol. 1985 Aug;5(8):2104–2113. doi: 10.1128/mcb.5.8.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel W. L. Genetics of murine liver and kidney arysulfatase b. Genetics. 1976 Mar 25;82(3):477–491. doi: 10.1093/genetics/82.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Edlund T., Walker M. D., Barr P. J., Rutter W. J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5' flanking elements. Science. 1985 Nov 22;230(4728):912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- Felder M. R., Watson G., Huff M. O., Ceci J. D. Mechanism of induction of mouse kidney alcohol dehydrogenase by androgen. Androgen-induced stimulation of transcription of the Adh-1 gene. J Biol Chem. 1988 Oct 5;263(28):14531–14537. [PubMed] [Google Scholar]
- Felton J., Meisler M., Paigen K. A locus determining beta-galactosidase activity in the mouse. J Biol Chem. 1974 May 25;249(10):3267–3272. [PubMed] [Google Scholar]
- Fischer J. A., Maniatis T. Drosophila Adh: a promoter element expands the tissue specificity of an enhancer. Cell. 1988 May 6;53(3):451–461. doi: 10.1016/0092-8674(88)90165-1. [DOI] [PubMed] [Google Scholar]
- Foster J., Stafford J., Queen C. An immunoglobulin promoter displays cell-type specificity independently of the enhancer. 1985 May 30-Jun 5Nature. 315(6018):423–425. doi: 10.1038/315423a0. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Hoschek G., Vodkin L. O. An insertion sequence blocks the expression of a soybean lectin gene. Cell. 1983 Jun;33(2):465–475. doi: 10.1016/0092-8674(83)90428-2. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Hammer M. F., Wilson A. C. Regulatory and structural genes for lysozymes of mice. Genetics. 1987 Mar;115(3):521–533. doi: 10.1093/genetics/115.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K., Mullen R. J. Biochemical and genetic factors in the heat inactivation of murine beta-glucuronidase. Biochem Genet. 1977 Aug;15(7-8):641–653. doi: 10.1007/BF00484095. [DOI] [PubMed] [Google Scholar]
- Holmes R. S., Albanese R., Whitehead F. D., Duley J. A. Mouse alcohol dehydrogenase isozymes: products of closely localized duplicate genes exhibiting divergent kinetic properties. J Exp Zool. 1981 Aug;217(2):151–157. doi: 10.1002/jez.1402170202. [DOI] [PubMed] [Google Scholar]
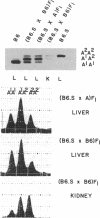
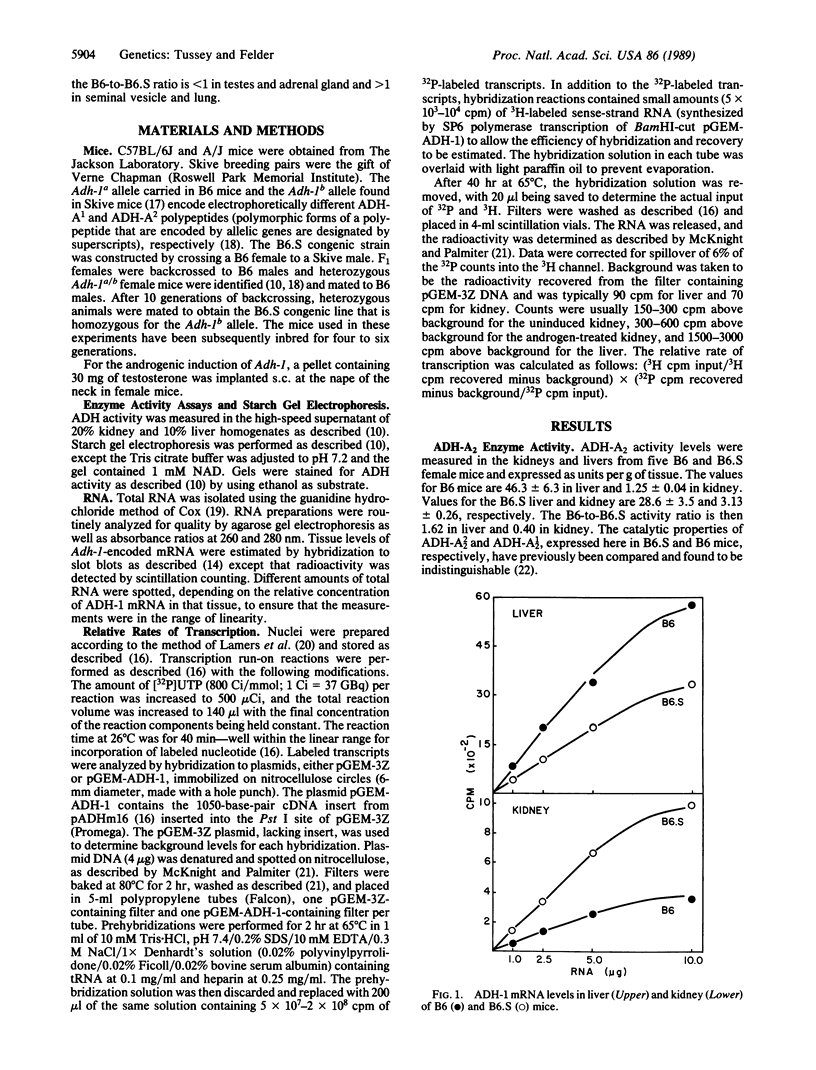
- Holmes R. S. Electrophoretic analyses of alcohol dehydrogenase, aldehyde dehydrogenase, aldehyde oxidase, sorbitol dehydrogenase and xanthine oxidase from mouse tissues. Comp Biochem Physiol B. 1978;61(3):339–346. doi: 10.1016/0305-0491(78)90134-7. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A. Growth-dependent expression of dihydrofolate reductase mRNA from modular cDNA genes. Mol Cell Biol. 1983 Sep;3(9):1598–1608. doi: 10.1128/mcb.3.9.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
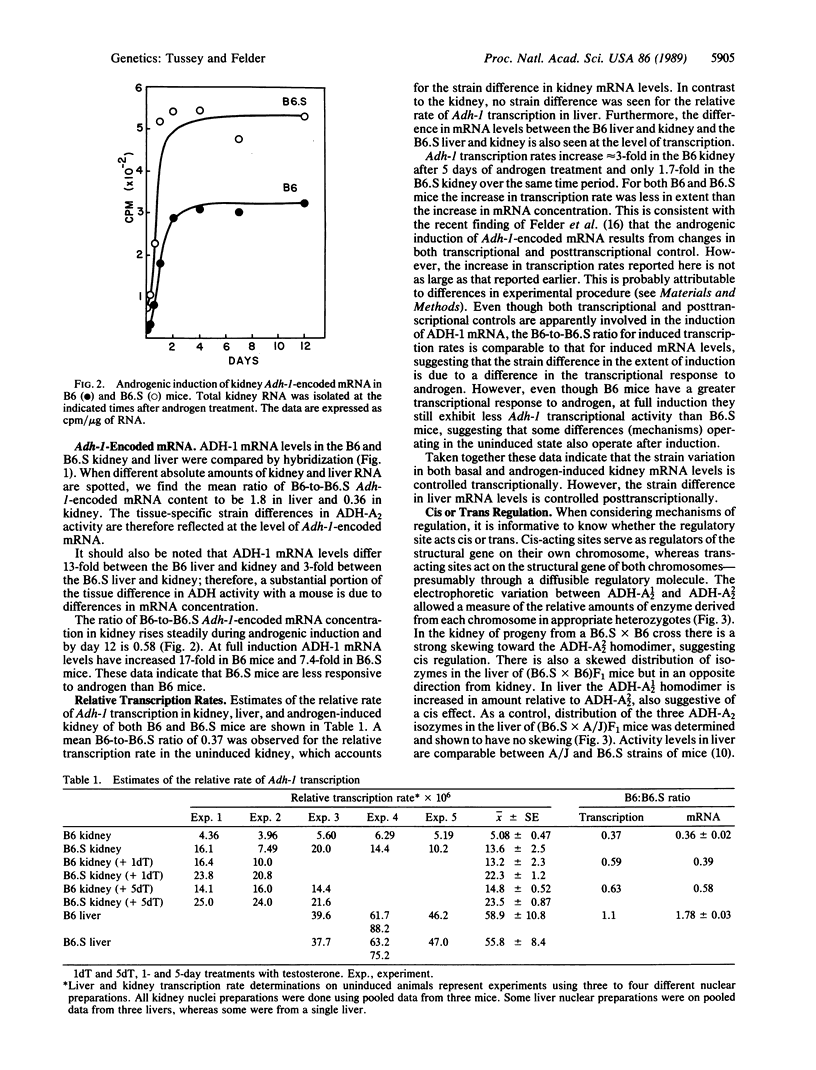
- Konieczny S. F., Emerson C. P., Jr Complex regulation of the muscle-specific contractile protein (troponin I) gene. Mol Cell Biol. 1987 Sep;7(9):3065–3075. doi: 10.1128/mcb.7.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak L. P., Ratner P. L. Genetic regulation of translatable mRNA levels for mouse sn-glycerol-3-phosphate dehydrogenase during development of the cerebellum. J Biol Chem. 1980 Aug 25;255(16):7589–7594. [PubMed] [Google Scholar]
- Lamers W. H., Hanson R. W., Meisner H. M. cAMP stimulates transcription of the gene for cytosolic phosphoenolpyruvate carboxykinase in rat liver nuclei. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5137–5141. doi: 10.1073/pnas.79.17.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis A. J., Paigen K. Genetic determination of the alpha-galactosidase developmental program in mice. Cell. 1975 Nov;6(3):371–378. doi: 10.1016/0092-8674(75)90186-5. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McKnight G. S., Palmiter R. D. Transcriptional regulation of the ovalbumin and conalbumin genes by steroid hormones in chick oviduct. J Biol Chem. 1979 Sep 25;254(18):9050–9058. [PubMed] [Google Scholar]
- Meredith S. A., Ganschow R. E. Apparent trans control of murine beta-glucuronidase synthesis by a temporal genetic element. Genetics. 1978 Dec;90(4):725–734. doi: 10.1093/genetics/90.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T., Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory roles in expression of the human alpha-cardiac actin gene. Mol Cell Biol. 1987 Aug;7(8):2803–2813. doi: 10.1128/mcb.7.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. D., Marks A. R., Buckley D. I., Kapler G., Payvar F., Goodman H. M. The first intron of the human growth hormone gene contains a binding site for glucocorticoid receptor. Proc Natl Acad Sci U S A. 1985 Feb;82(3):699–702. doi: 10.1073/pnas.82.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M., Lindahl G., Ruoslahti E. Genetic control of alpha-fetoprotein synthesis in the mouse. J Exp Med. 1977 Apr 1;145(4):819–827. doi: 10.1084/jem.145.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Stenius C., Christian L., Harris C., Ivey C. More about the testosterone induction of kidney alcohol dehydrogenase activity in the mouse. Biochem Genet. 1970 Oct;4(5):565–577. doi: 10.1007/BF00486095. [DOI] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex D. K., Bosron W. F., Dwulet F., Li T. K. Purification and characterization of the Danish (Skive) variant of mouse liver alcohol dehydrogenase. Biochem Genet. 1987 Feb;25(1-2):111–121. doi: 10.1007/BF00498955. [DOI] [PubMed] [Google Scholar]
- Rossi P., de Crombrugghe B. Identification of a cell-specific transcriptional enhancer in the first intron of the mouse alpha 2 (type I) collagen gene. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5590–5594. doi: 10.1073/pnas.84.16.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland L. J., Strommer J. N. Anaerobic treatment of maize roots affects transcription of Adh1 and transcript stability. Mol Cell Biol. 1986 Oct;6(10):3368–3372. doi: 10.1128/mcb.6.10.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland L. J., Strommer J. N. Insertion of an unstable element in an intervening sequence of maize Adh1 affects transcription but not processing. Proc Natl Acad Sci U S A. 1985 May;82(9):2875–2879. doi: 10.1073/pnas.82.9.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M., Alwine J. C. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984 Aug;4(8):1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampsell B. M., Held W. A. Variation in the major urinary protein multigene family in wild-derived mice. Genetics. 1985 Mar;109(3):549–568. doi: 10.1093/genetics/109.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Sordat B., Schibler U. Developmental coordination of alpha-amylase and psp gene expression during mouse parotid gland differentiation is controlled posttranscriptionally. Cell. 1986 Oct 10;47(1):107–112. doi: 10.1016/0092-8674(86)90371-5. [DOI] [PubMed] [Google Scholar]
- Sternberg E. A., Spizz G., Perry W. M., Vizard D., Weil T., Olson E. N. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988 Jul;8(7):2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Tsurushita N., Korn L. J. Effects of intron length on differential processing of mouse mu heavy-chain mRNA. Mol Cell Biol. 1987 Jul;7(7):2602–2605. doi: 10.1128/mcb.7.7.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widen S. G., Papaconstantinou J. Liver-specific expression of the mouse alpha-fetoprotein gene is mediated by cis-acting DNA elements. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8196–8200. doi: 10.1073/pnas.83.21.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3'-end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988 Apr 25;263(12):5780–5788. [PubMed] [Google Scholar]
- Zhang K., Bosron W. F., Edenberg H. J. Structure of the mouse Adh-1 gene and identification of a deletion in a long alternating purine-pyrimidine sequence in the first intron of strains expressing low alcohol dehydrogenase activity. Gene. 1987;57(1):27–36. doi: 10.1016/0378-1119(87)90173-9. [DOI] [PubMed] [Google Scholar]