Abstract
Helicobacter pylori produces a unique surface lipopolysaccharide (LPS) characterized by strikingly low endotoxicity that is thought to aid the organism in evading the host immune response. This reduction in endotoxicity is predicted to arise from the modification of the Kdo-lipid A domain of Helicobacter LPS by a series of membrane bound enzymes including a Kdo (3-deoxy-D-manno-octulosonic acid) hydrolase responsible for the modification of the core-oligosaccharide. Here we report that Kdo hydrolase activity is dependent upon a putative two-protein complex composed of proteins Hp0579 and Hp0580. Inactivation of Kdo hydrolase activity produced two phenotypes associated with cationic antimicrobial peptide (CAMP) resistance and O-antigen expression. Kdo hydrolase mutants were highly sensitive to polymyxin B, which could be attributed to a defect in downstream modifications to the lipid A 4′-phosphate group. Production of a fully extended O-antigen was also diminished in a Kdo hydrolase mutant, with a consequent increase in core-lipid A. Finally, expression of O-antigen Lewis X and Y epitopes, known to mimic glycoconjugates found on human tissues, was also affected. Taken together, we have demonstrated that loss of Kdo hydrolase activity affects all three domains of H. pylori LPS, thus highlighting it’s role in the maintenance of the bacterial surface.
Introduction
Helicobacter pylori is a Gram-negative bacterium with only one well defined niche, the human stomach. After colonizing the stomach H. pylori can persist for several years without manifesting any symptoms, although over time serious sequelae can appear, including peptic ulcer disease and gastric cancer (Blaser, 1996, Parsonnet et al., 1991). Like the majority of Gram-negative bacteria the outer surface of H. pylori is composed primarily of lipopolysaccharide (LPS), consisting of three domains known as lipid A, core and O-antigen.
The core and O-antigen represent the polysaccharide component of LPS. The core sugars are generally well conserved within a bacterial species; however, the O-antigen can show great diversity. H. pylori O-antigen expression is highly variable and generally mimics human blood group antigens (Appelmelk et al., 1998, Wang et al., 2000). This molecular mimicry is thought to contribute to the chronic nature of H. pylori infections by making the bacterium appear like its host and preventing detection by the immune system (Wirth et al., 1997, Appelmelk et al., 2000). H. pylori O-antigen has also been implicated in other aspects of pathogenesis, including adhesion to gastric epithelial cells and immune system modulation, mediated by interaction with the C-type lectin DC-SIGN (Bergman et al., 2004).
The lipid A domain of LPS acts as a hydrophobic anchor holding the LPS molecule in the outer membrane. Lipid A is also responsible for the endotoxic properties associated with LPS. Biosynthesis of lipid A is well conserved throughout Gram-negative bacteria and proceeds via a nine-step enzymatic pathway (Raetz & Whitfield, 2002). Despite its conserved synthesis, a great deal of variation is seen when comparing the lipid A structures of various Gram-negative bacterial species (Raetz et al., 2007). This variation is generated primarily by the action of lipid A modification enzymes and, with human pathogens, may well be a consequence of evolutionary pressure applied by the human innate immune system. For example, the modification of phosphate groups present on the lipid A disaccharide backbone provides resistance to cationic anti-microbial peptides (CAMPs). CAMPs are small positively charged peptides that bind to negatively charged structural motifs (e.g. lipid A) present on the surface of bacteria leading to eventual cell lysis and death (Diamond et al., 2009). Gram-negative bacteria resist the action of CAMPs by adding positively charged substituents, such as phosphoethanolamine or L-4-aminoarabinose, to the negatively charged phosphate groups of lipid A or by removing the phosphate groups of lipid A (Tran et al., 2006, Gunn et al., 1998).
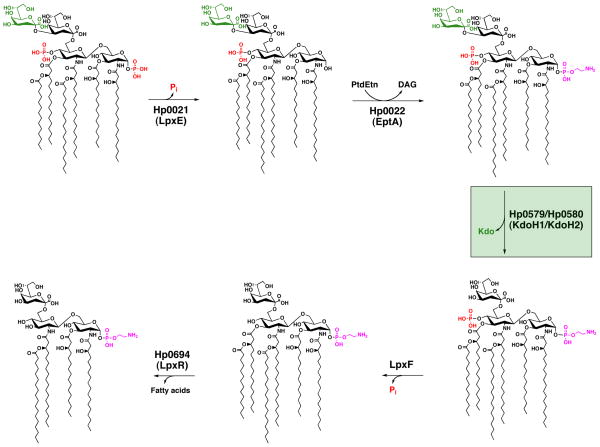
In a number of organisms lipid A modifications are regulated and only occur after specific environmental cues are detected; however, it appears that in the case of H. pylori, lipid A modifications are constitutive. H. pylori lipid A modification is a complex process that occurs via a five step enzymatic pathway (Fig. 1) and in the laboratory a single distinct lipid A species is produced. A lack of regulation could be explained by the fact that H. pylori has only one known reservoir, the human stomach, thus rendering adaptation unnecessary. However, it is not possible to rule out whether or not the organism is capable of changing its lipid A domain within its human host.
Fig. 1. Modification pathway of H. pylori Kdo-lipid A.
H. pylori produces a highly modified lipid A species via a five step enzymatic pathway. The 1-phosphate is first cleaved by Hp0021 (LpxE), leaving a free hydroxyl group, followed by addition of a phosphoethanolamine by Hp0022 (EptA). Presumably, phosphatidylethanolmaine (PtdEtN) serves as the donor for the latter. Next Hp0579 (KdoH1) and Hp0580 (KdoH2) work in concert to remove the terminal Kdo sugar. The 4′-phosphate group is removed by an as yet unidentified phosphatase; however, the remaining hydroxyl group is not further modified as is the case at the 1 position. The final step involves the removal of the 3′-O-linked acyl chains by Hp0694 (LpxR), resulting in a tetra-acylated lipid A.
Several of the H. pylori lipid A modification enzymes have already been characterized by previous studies from our laboratory, including the identification of a novel 3-deoxy-D-manno-octulosonic acid-hydrolase (Kdo-hydrolase) enzymatic activity present in H. pylori membranes (Stead et al., 2005). The Kdo-hydrolase removes the outer Kdo sugar from the core region, with optimal activity occurring after the prior removal of the 1-phosphate group by a dedicated inner membrane phosphatase encoded by hp0021. The presence of a single Kdo sugar had previously been attributed to a mono-functional Kdo transferase (KdtA). Investigations into H. pylori KdtA functionality proved it to be bi-functional, indicating that it was no longer possible to predict how many Kdo sugars a particular KdtA may transfer based upon characterization of the Kdo-lipid A species alone. A similar situation was thought to exist in Francisella novicida, which has also been reported to produce a Kdo-hydrolase (Wang et al., 2004).
Although the Kdo-hydrolase enzymatic activity is well characterized, the gene responsible for encoding the enzyme has yet to be determined. Here we report the identification of the Kdo-hydrolase of Helicobacter pylori and show that Kdo-hydrolase activity is, in fact, dependent upon two gene products. We also demonstrate that Kdo-hydrolase activity is necessary for subsequent lipid A modifications (Fig. 1), resistance to CAMPs, as well as efficient O-antigen expression, indicating a key role in H. pylori pathogenesis.
Results
The H. pylori Kdo-hydrolase is encoded by hp0579 & hp0580
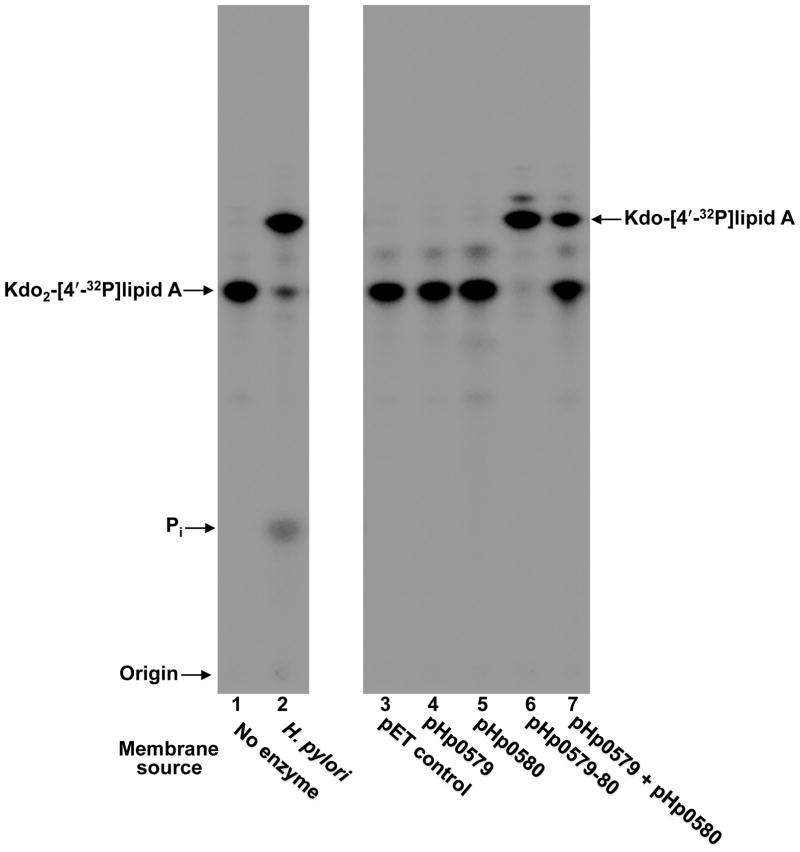
Given that membranes isolated from both Helicobacter pylori and Francisella novicida had been shown to catalyze the removal of the outer Kdo sugar from Kdo2-lipid A substrates (Stead et al., 2005, Wang et al., 2004), a genomic comparison of the two organisms was performed to identify a possible gene encoding for the Kdo hydrolase (see accompanying manuscript by Raetz and Zhao). The genomic comparison yielded a candidate gene annotated as FTN_0495 in Francisella and hp0580 in Helicobacter strain 26695 (Fig. S1). PSI-BLAST (Altschul et al., 1997) analysis shows that the soluble C-terminal domain of both Hp0580 (Fig. S2) and its counterpart in Francisella show distant homology to the sialidase NanI (Newstead et al., 2008) produced by Clostridium perfringens (see Fig. S3 of the accompanying manuscript, Zhao and Raetz). Sialidases function to bind and hydrolyze a terminal monosaccharide from glycoconjugates, lending support to the prediction of Hp0580 functioning in the cleavage of the outer Kdo sugar. Hp0580 was cloned into pET21a, heterologously expressed in E. coli HMS174 and membranes were isolated for use in an in vitro assay. The in vitro assay system makes use of radiolabeled lipid A substrates, which are incubated with an enzyme source followed by separation of the reaction products via thin layer chromatography (TLC). The solvent used for TLC separates species according to hydrophobicity, such that more hydrophobic products migrate faster. In this case we utilized Kdo2-[4′-32P]lipid A as the substrate and membranes isolated from HMS174/pHp0580 as the enzyme source. After analysis of the reactant products by TLC, we were surprised to see no removal of a Kdo sugar from the starting substrate (Fig. 2, lane 5). Membranes from H. pylori showed Kdo hydrolase activity (lane 2) and served as the positive control.
Fig. 2. In vitro assay of Kdo hydrolase candidate proteins, Hp0579 and Hp0580, heterologously expressed in E. coli.
Hp0579, Hp0580 and Hp0579-Hp0580 were overexpresed in E. coli HMS174 and membranes were isolated for use as the enzyme source in an in vitro Kdo-lipid A modification assay, using Kdo2-[4′-32P]lipid A as the substrate. H. pylori membranes isolated from strain Hp7-91 harboring a mutation in hp0021 (1-phosphatase) acted as a positive control and the empty pET21a vector acted as a negative control, demonstrating that E. coli has no endogenous Kdo hydrolase activity. When assayed individually, Hp0579 and Hp0580 were devoid of Kdo hydrolase activity; however, after co-expression a robust Kdo hydrolase activity could be detected by the appearance of a faster migrating product, which co-migrated with the positive control. Mixing membranes from individually expressed Hp0579 and Hp0580 could also generate Kdo hydrolase activity, reinforcing the necessity for a two protein complex. The minor product spots present above both the starting substrate Kdo2-[4′-32P]lipid A and the major product Kdo-[4′-32P]lipid A can be attributed to endogenous E. coli PagP activity, an outer membrane lipid A acyl transferase (Bishop et al., 2000).
This anomalous result prompted closer inspection of the FTN_0495 and hp0580 genomic regions. In the case of Francisella it appeared that FTN_0495 could be transcribed as part of an operon, which contained only one other gene, annotated as FTN_0494, with no predicted function. The genome of H. pylori showed a similar organization, with the gene encoding the putative sialidase (hp0580) possibly organized in an operon with hp0579 (Fig. S1). Unlike hp0580, hp0579 is not homologous to its Francisella counterpart FTN_0494. However, hydropathy analysis of hp0579 and FTN_0494 indicated that both proteins contain multiple transmembrane domains (6 or 7) absent of additional notable structural features (Fig. S2). Therefore, we speculated that hp0579 might also be required for Kdo hydrolase activity in Helicobacter. Both hp0579 and hp0580 were cloned into pET21a and overexpressed for use in an in vitro assay. When using Kdo2-[4′-32P]lipid A as the substrate and membranes isolated from HMS174/pHp0579-80 as the enzyme source, we observed a robust Kdo hydrolase activity (Fig. 2, lane 6), suggesting that both Hp0579 and Hp0580 were required for Kdo hydrolase activity. Confirmation that the product spot was in fact generated by a Kdo hydrolase was provided after performing mass spectrometry on the products from a large-scale Kdo hydrolase reaction (Fig S3). To demonstrate that Hp0579 alone wasn’t responsible for the Kdo hydrolase activity, hp0579 was cloned into pET21a and assayed using Kdo2-[4′-32P]lipid A as the substrate and, as expected, no Kdo hydrolase activity was apparent (Fig. 2, lane 4). Kdo hydrolase activity could also be generated by mixing membranes isolated from HMS174/pHp0579 with membranes isolated from HMS174/pHp0580 in the presence of Kdo2-[4′-32P]lipid A substrate (Fig. 2, lane 7), further supporting evidence for a two-protein enzymatic complex.
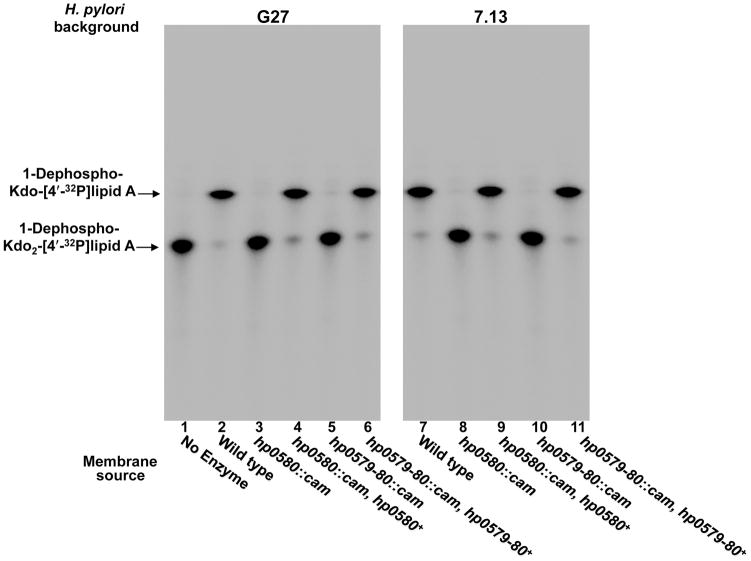
The results obtained with heterologous expression of hp0579 and hp0580 prompted us to generate mutations in H. pylori. Two H. pylori strains were chosen for this study: strain G27 for its ability to be readily transformed and the gerbil adapted strain 7.13 for its potential in animal studies (Censini et al., 1996, Franco et al., 2005). Furthermore, it is important to confirm results in a second strain due to the high degree of phase variation found with H. pylori, which can lead to strain specific phenotypes (Tran et al., 2006). Mutations were made in the catalytic subunit (hp0580) along with a double mutation, which removed both hp0579 and hp0580, as described in experimental procedures. Each of these mutations were also complemented chromosomally by insertion of the relevant gene(s) into the rdxA locus. Although G27 is now sequenced, this was not the case when these mutants were constructed and consequently complemented, which necessitated the use of strain 26695 genome sequence (Baltrus et al., 2009). The G27 homolog of Hp0580 shows 93% identity (E-value of 0.0 with 96% coverage) and the Hp0579 homolog shows 96% identity (E-value of 1×10−82 with 97% coverage) to those of H. pylori 26695. Membranes were isolated from each of the mutants and complemented mutants for use in an in vitro assay, using 1-dephosphorylated-Kdo2-[4′-32P]lipid A as the substrate. The assay clearly showed complete loss of Kdo hydrolase function in both strains G27 and 7.13 for the hp0580 single mutant (Fig. 3, lanes 3 & 8) and the hp0579-hp0580 double mutant (Fig. 3, lanes 5 & 10). Kdo hydrolase activity was fully restored in each of the complemented mutants for both strains (Fig. 3, lanes 4, 6, 9 & 11), ruling out any polar effects. This data, along with the heterologous expression data, demonstrates that Hp0579 and Hp0580 are responsible for removal of the Kdo sugar and that both proteins are required for activity.
Fig. 3. In vitro assay of H. pylori Kdo hydrolase mutants.
Membranes from both Hp0580 and Hp0579-Hp0580 mutants in strains G27 and 7.13 were assayed using 1-dephosphorylated-Kdo2-[4′-32P]lipid A as the substrate. Each of the mutants showed a complete loss of Kdo hydrolase activity, which was fully restored in complemented strains.
Demonstration of Kdo hydrolase activity in vivo
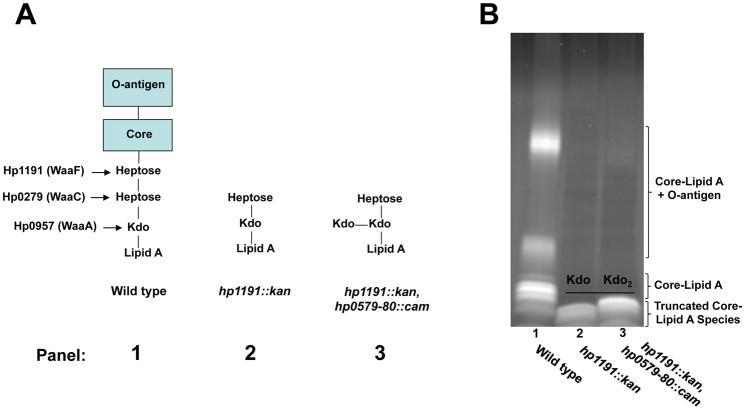
Analysis of Kdo hydrolase activity in vivo is complicated by the fact that standard lipid A analyses, such as radiolabeling or mass spectrometry, rely upon removal of the polysaccharide portion of LPS for lipid A isolation. The polysaccharides are removed by mild acid hydrolysis at 100°C, which cleaves the bond between the Kdo sugar and the lipid A backbone, preventing any further analysis of the Kdo sugars. To circumvent this problem, we made use of a previously well characterized H. pylori heptosyl transferase mutant (26695 hp1191::kan), which produces a severely truncated LPS with only a Kdo and heptose remaining in the core region (Fig. 4A) (Chandan et al., 2007). This truncation allows accurate analysis of LPS via SDS-PAGE, using a 16% tricine gel, to show differences in mass corresponding to one sugar (Lesse et al., 1990). We proceeded to make a mutation in the hp0579 and hp0580 genes of strain 26695 hp1193::kan to produce the double mutant. One would expect to see a slower migrating LPS species in the heptosyl transferase/Kdo hydrolase double mutant as compared to the heptosyl transferase mutant, because the number of sugar moieties present in the core region should increase from 2 to 3 (Fig. 4A). SDS-PAGE analysis of the LPS from the two mutants displays the expected phenotype (Fig. 4B), confirming the function of Hp0579 and Hp0580, using an in vivo system.
Fig. 4. SDS-PAGE analysis of H. pylori 26695 hp1191::kan and hp1191::kan, hp0579-80::cam LPS profiles.
A. Hp1191 is a heptosyl transferase that is responsible for transferring the second heptose sugar found in the core region of H. pylori 26695 LPS (panel 1). An Hp1191 mutant produces a severely truncated LPS with only two sugars, heptose and Kdo, extending past the lipid A domain (panel 2). The in vitro assays depicted in Figs. 2 and 3 would predict that an hp1191, hp0579-hp0580 mutant would produce an LPS species with 3 sugars extended past the lipid A domain (panel 3). B. The predicted difference in LPS composition, between the hp1191 and hp1191, hp0579-hp0580 mutants was evaluated by SDS-PAGE. As expected the hp1191, hp0579-hp0580 mutant produced a higher mass LPS (lane 3) as compared to the hp1191 mutant (lane 2), demonstrating an in vivo activity for the Kdo hydrolase.
Hp0580 has a periplasmic active site
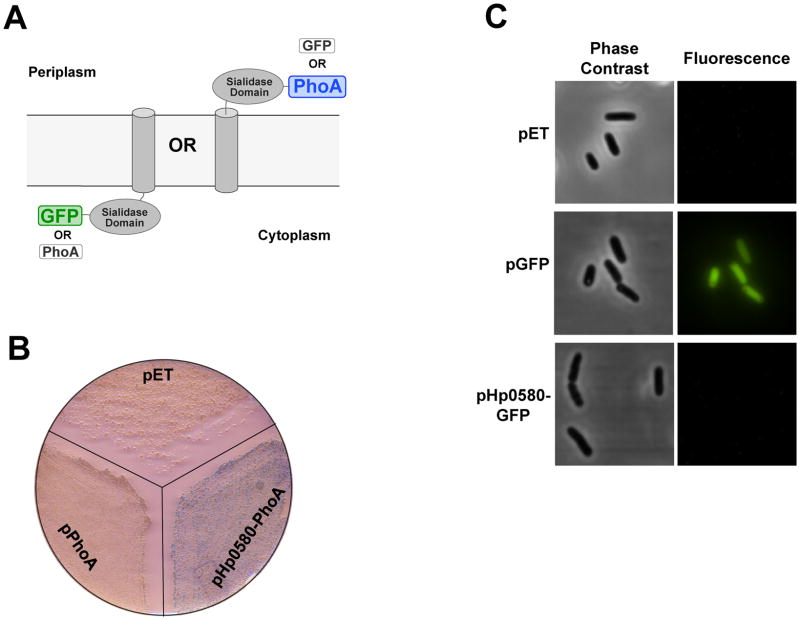
Our previous studies characterizing the Kdo hydrolase activity present in H. pylori membranes indicated that the enzyme utilized a 1-dephosphorylated lipid A substrate when working in the linear range of activity (Stead et al., 2005). The enzyme responsible for removing the 1-phosphate group of H. pylori lipid A, Hp0021 (LpxE) (Fig. 1), has been shown to have a periplasmic active site (Tran et al., 2004). This would suggest that Hp0580 also has a periplasmic active site, given its dependence on the prior removal of the 1-phosphate group. Furthermore, one would predict that it would be energetically unfavorable to remove the outer Kdo sugar in the cytoplasm given that both Kdo sugars are assembled into the core oligosaccharide in the same cytoplasmic compartment by the Kdo transferase, WaaA (Brozek et al., 1989). Therefore, we sought to confirm the location of the Hp0580 active site by expressing C-terminal PhoA (alkaline phosphatase) and GFP (green fluorescent protein) fusion proteins (Fig. 5A).
Fig. 5. Hp0580 topology studies.
A. Hp0580 topology was investigated using PhoA and GFP C-terminal fusion proteins. PhoA is only active when located in the periplasm, whilst GFP will only fluoresce when folded in the cytoplasm (see text for details). B. W3110 (DE3) PhoA-expressing hp0580-phoA was grown on LB agar in the presence of X-phosphate substrate. The appearance of blue colonies indicates a periplasmic orientation for the Hp0580 soluble domain. W3110 (DE3) PhoA− plus empty vector and W3110 (DE3) PhoA- expressing phoA minus a signal peptide acted as negative controls. C. W3110 (DE3) expressing hp0580-GFP or GFP alone was visualized under a fluorescent microscope. Only the strain expressing GFP alone fluoresced, indicating a periplasmic orientation for the Hp0580 soluble domain.
PhoA is only active if present in the periplasm (Hoffman & Wright, 1985) and therefore a C-terminal Hp0580-PhoA fusion protein would display phosphatase activity only if the Hp0580 soluble sialidase domain was periplasmic. Phosphatase activity can be determined by growing the bacteria on solid agar medium containing the colorimetric substrate X-phosphate, which produces a blue colony phenotype after cleavage by a phosphatase. E. coli PhoA mutants expressing either the Hp0580-PhoA fusion or PhoA minus its signal peptide, and the vector control were grown in the presence of X-phosphate. Only the strain expressing the Hp0580-PhoA fusion protein produced blue colonies, indicating that the Hp0580 active site is in fact periplasmic (Fig. 5B). This finding was corroborated with an Hp0580-GFP fusion protein. The inverse scenario to PhoA is true with GFP, in that GFP will fluoresce only if it is folded in the cytoplasm, meaning that the Hp0580-GFP fusion protein will not fluoresce if the Hp0580 active site is periplasmic (Feilmeier et al., 2000). Fluorescence microscopy of E. coli expressing the Hp0580-GFP fusion protein revealed no fluorescent bacteria, while the positive control strain expressing cytoplasmic GFP only, produced fluorescent bacteria (Fig. 5C), confirming the periplasmic orientation of the Hp0580 active site.
Inactivation of the Kdo hydrolase leads to an increase in polymyxin B sensitivity
CAMPs primary mode of action is to bind to the negative charges present on the LPS molecule, prior to exerting their bactericidal activity (Diamond et al., 2009). Kdo sugars contain a carboxylic acid moiety, which may be negatively charged at neutral pH. Therefore, the Kdo hydrolase mutant could increase the net negative charge present at the bacterial surface, thereby increasing sensitivity to CAMPs. We tested this hypothesis using polymyxin B, a common experimental substitute for CAMPs, which has a similar mechanism of action (Gutsmann et al., 2005). Minimum inhibitory concentrations (MICs) were determined for the hp0579-hp0580 mutants and hp0579-hp0580 complemented mutants in both strains G27 and 7.13, using Etest® strips, as described in experimental procedures. The G27 hp0579-hp0580 mutant showed over a 250-fold decrease in its polymyxin B MIC (Table 1) and the 7.13 hp0579-hp0580 mutant showed over a 180-fold decrease in its polymyxin B MIC (Table 1) as compared to wild type levels. The complemented Hp0579-Hp0580 mutants of each strain regained wild type levels of polymyxin B resistance, eliminating the possibility of any downstream polar effects.
Table 1.
Polymyxin minimal inhibitory concentration (MIC) of H. pylori strains
| Strain | Kdo Hydrolase Activity | Polymyxin B MIC (μg/ml) |
|---|---|---|
| 7.13 | + | 256.0 ± 0.0 |
| 7.13/hp0580::cam | − | 1.0 ± 0.0 |
| 7.13/hp0580::cam, hp0580+ | + | 234.7 ± 37.0 |
| 7.13/hp0579-80::cam | − | 1.0 ± 0.0 |
| 7.13/hp0579-80::cam, hp0579-80+ | + | 256.0 ± 0.0 |
| G27 | + | 128.0 ± 0.0 |
| G27/hp0580::cam | − | 0.7 ± 0.1 |
| G27/hp0580::cam, hp0580+ | + | 117.3 ± 18.5 |
| G27/hp0579-80::cam | − | 0.5 ± 0.0 |
| G27/hp0579-80::cam, hp0579-80+ | + | 128.0 ± 0.0 |
This initial experiment utilized the Kdo hydrolase double mutant, which is deficient in both Hp0579 and Hp0580. Hp0580 contains homology to a sialidase and, therefore, is likely responsible for the actual cleavage of the Kdo-Kdo linkage within the LPS core oligosaccharide. Hp0579, however, has no predicted function and, therefore, could also be involved in other distinct processes. To confirm that the increase in polymyxin B sensitivity is a result of the action of the Kdo hydrolase and not a secondary function attributable to Hp0579 we repeated the experiment using the Hp0580 single mutant and Hp0580 complemented mutant. We saw identical results as compared to the Hp0579-Hp0580 mutant and Hp0579-Hp0580 complemented mutant in both strains, indicating that the increase in sensitivity is attributable to the action of the Kdo hydrolase (Table 1).
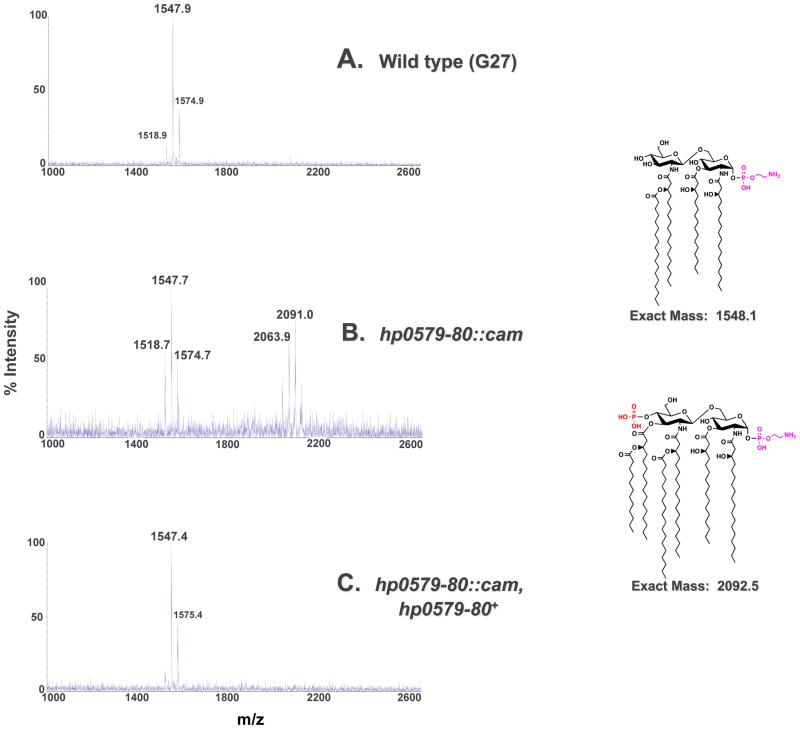
The dramatic increase in polymyxin B sensitivity of the Kdo hydrolase mutants is comparable to that of the increase in polymyxin B sensitivity observed when lipid A phosphate modifications are disrupted (Tran et al., 2006). This was a surprising result given that a phosphate group carries more negative charges as compared to a carboxylic acid. To explain this phenomenon we hypothesized that loss of Kdo hydrolase function could affect other downstream lipid A modifications, such as removal of the 4′-phosphate group (Fig. 1). The requirement of H. pylori lipid A modification enzymes for a specific substrate has been shown previously by our group, lending credence to our hypothesis (Tran et al., 2004, Stead et al., 2005). We isolated lipid A from G27 wild type, the G27 Hp0579-Hp0580 mutant and the G27 Hp0579-Hp0580 complemented mutant and subjected each lipid A species to analysis by MALDI-TOF (matrix-assisted laser desorption/ionization-time of flight) mass spectrometry in the negative ion mode. The wild type spectrum showed a predominant peak at m/z 1547.9 consistent with the [M-H]− ion of the wild type structure of H. pylori lipid A (predicted [M-H]− at m/z 1548.1), which is tetra-acylated without a phosphate group at the 4′-position and a phosphoethanolamine residue at the 1-position (Fig. 6). The Hp0579-Hp0580 mutant spectrum showed two predominant peaks at m/z 1547.7 and m/z 2091.0 (Fig. 6). The peak at m/z 1547.7 is consistent with the [M-H]− ion of the wild type structure; however, the peak at m/z 2091.0 is consistent with the [M-H]− of a lipid A species that is hexa-acylated with a phosphate group at the 4′-position and a phosphoethanolamine at the 1-position (predicted [M-H]− at m/z 2092.5). This indicates that the 4′-phosphatase and 3′-O-deacylase are no longer 100% efficient, suggesting an ordered lipid A modification pathway, determined by enzyme substrate specificity. The Hp0579-Hp0580 complemented mutant displayed a predominant peak at m/z 1547.4, consistent with a wild type lipid A species. These same analyses were completed in strain 7.13 harboring the hp0579-hp0580 mutation (Fig. S4) and in strains harboring the single hp0580 mutation (data not shown). In all cases the results were identical to those shown in Fig. 6.
Fig. 6. Mass spectrometry of the G27 Kdo hydrolase mutant.
Lipid A was isolated from G27, G27 hp0579-80::cam and G27 hp0579-80::cam, hp0579-80+ and analyzed by MALDI-TOF mass spectrometry in the negative-ion mode. G27 produced a peak at 1547.9 m/z corresponding to the published wild type H. pylori lipid A mass (A). In addition to the wild type peak at m/z 1547.7 G27 hp0579-80::cam also displayed a peak at m/z 2091.0 (B). 2091.0 corresponds to a hexa-acylated lipid A species, with the 4′-phosphate still present. G27 hp0579-80::cam, hp0579-80+ showed complete reversion to the wild type phenotype, with a single peak present at a m/z of 1547.4 (C).
The presence of a lipid A sub-population still possessing a 4′-phosphate group may explain the increase in polymyxin B sensitivity seen with the Kdo hydrolase mutants, although it does not rule out a possible direct contribution to polymyxin B sensitivity brought about by the presence of a second Kdo sugar and its associated carboxylic acid moiety. Francisella mutants lacking a 4′-phosphatase also show an increase in polymyxin B sensitivity, which correlates to an attenuation of virulence (Wang et al., 2007), highlighting the importance of such modifications for bacterial survival.
Kdo hydrolase activity modulates O-antigen expression
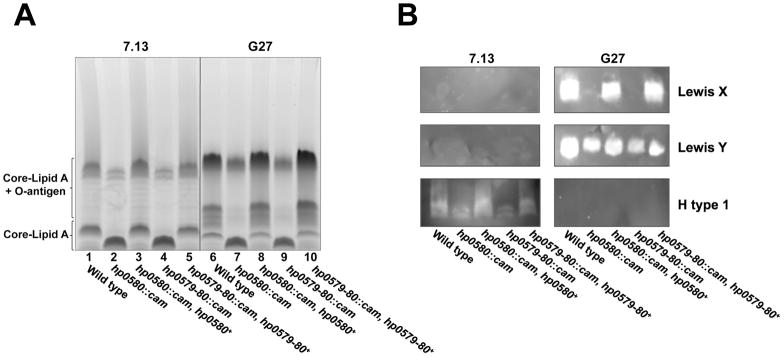
Addition of core sugars to the Kdo-lipid A domain occurs at the cytoplasmic membrane, prior to transport of core-lipid A across the inner membrane by MsbA (Raetz & Whitfield, 2002). O-antigen is synthesized, transported across the inner membrane and ligated to the core-lipid A, by the ligase WaaL, to produce the finished LPS molecule (Raetz & Whitfield, 2002). To determine if the presence of a second Kdo sugar, present in a Kdo hydrolase mutant, had any effect on the LPS synthesis pathway we analyzed the LPS profile of each Kdo hydrolase mutant by SDS-PAGE. The resultant gel shows that in both strains G27 and 7.13 the Hp0580 mutant (Fig. 7A, lanes 2 & 7) and Hp0579-Hp0580 mutant (Fig. 7A, lanes 4 & 9) have a reduced amount of fully extended LPS and, inversely, the amount of core-lipid A increases as compared to wild type bacteria (Fig. 7A, lanes 1 & 6). Complementation of the Hp0580 and Hp0579-Hp0580 mutants fully restores the wild type phenotype (Fig. 7A, lanes 3, 5, 8 & 10), ruling out any polar effects.
Fig. 7. SDS-PAGE analysis of Kdo hydrolase mutants and complements.
Proteinase K treated whole cell lysates from Hp0580, Hp0579-Hp0580 mutants and Hp0580, Hp0579-Hp0580 complemented mutants, in strains G27 and 7.13, were separated by SDS-PAGE and stained with Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (A) or blotted to a nitrocellulose membrane and probed with antibodies raised against various human blood group antigens (B). The Kdo hydrolase mutants in both strains displayed a reduced amount of fully extended O-antigen and an increase in core-lipid A, as compared to wild type. A wild type phenotype was fully restored in each of the complemented Kdo hydrolase mutants. Strain 7.13 only expressed the H type 1 blood group antigen, which was reduced in the Kdo hydrolase mutants. Strain G27 expressed both Lewis X and Lewis Y blood group antigens. Lewis Y expression was unaffected in the Kdo hydrolase mutants; however, Lewis X expression was absent in each of the Kdo hydrolase mutants.
H. pylori O-antigen usually mimics human blood group antigens, although the specific epitope seen can be highly variable (Appelmelk et al., 1998, Wang et al., 2000). Despite the possibility for variation, Lewis X and Lewis Y blood group antigens are the most prevalent epitopes found on the bacterial cell surface (Simoons-Smit et al., 1996). The Lewis X motif is composed of a galactose and N-acetylglucosamine backbone decorated with fucose on the N-acetylglucosamine sugar by the α(1,3)-fucosyl transferase FutA (Moran, 2008). Lewis Y is a terminal motif with a galactose and N-acetylglucosamine backbone decorated with a fucose on both sugars by the α(1,3)-fucosyl transferase FutA and the α(1,2)-fucosyl transferase FutC (Moran, 2008). The presence of poly-C tracts in the fucosyl transferase genes allows the proteins to effectively be ‘on’ or ‘off’, providing the mechanism of variation in O-antigen expression (Appelmelk et al., 1999, Wang et al., 1999b).
After observing a reduction in fully extended O-antigen with the Kdo hydrolase mutants, it was logical to see if this reduction also corresponded with a change in blood group antigen expression. To achieve this aim LPS SDS-PAGE gels were blotted to a nitrocellulose membrane and the O-antigen profile was determined using specific antibodies, as described in the experimental procedures. Wild type G27 expressed both Lewis X & Y antigens (Fig. 7B). Interestingly this was not the case for the Kdo hydrolase mutants. Both the Hp0580 and Hp0579-Hp0580 mutants no longer expressed the Lewis X antigen; however, expression of the Lewis Y antigen was unaffected (Fig. 7B). The wild type phenotype was fully restored in the Hp0580 and Hp0579-Hp0580 complemented mutants (Fig. 7B). To prove that this phenotype was not a consequence of variation in the levels of Lewis X and Y undecaprenyl-linked precursors, each sample was subjected to mild acid hydrolysis followed by SDS-PAGE and immunoblot analysis. Mild acid hydrolysis cleaves undecaprenyl-linked sugars while leaving LPS intact. Figure S5 shows no changes in the Kdo hydrolase mutants after mild acid hydrolysis, indicating that a variation in Lewis antigen expression is not responsible for the observed phenotype. Each strain was also probed with anti-H type 1 (Fig. 7B), anti-Lewis A (data not shown) and anti-Lewis B (data not shown) antibodies and shown not to express any of those epitopes. Not only does the Kdo hydrolase have an effect on the amount of fully extended O-antigen produced, it is also capable of modulating which blood group antigen is presented at the bacterial cell surface.
Strain 7.13 did not have the same phenotype as strain G27 because it did not express the Lewis X or Lewis Y antigens (Fig. 7B). Blots probing with anti-Lewis A and anti-Lewis B antibodies also produced negative results (data not shown). However, we were able to show that wild type strain 7.13 expressed the H type 1 epitope (Fig. 7B). Although the production of an H type 1 epitope did not disappear in the 7.13 Hp0580 and Hp0579-Hp0580 mutants, it was reduced (Fig. 7B), mirroring the results seen in the LPS gel (Fig. 7A).
Discussion
Modification of the Kdo-Lipid A domain of LPS is a common theme amongst Gram-negative pathogens, which invariably provide a direct benefit to the invading microbe. H. pylori is no exception to this rule and produces a highly modified Kdo-lipid A, which provides resistance to the human innate immune system in an in vitro setting (Tran et al., 2006). One step of the H. pylori Kdo-lipid A modification pathway involves the removal of a Kdo sugar, a trait known to be shared with only one other bacterium – Francisella tularensis (Stead et al., 2005, Wang et al., 2004). Although other bacteria can produce lipid A species with only one Kdo sugar, this arrangement is generated by a mono-functional Kdo transferase and the Kdo sugar is consequently modified by the addition of a phosphate containing group (Hankins & Trent, 2009, Isobe et al., 1999, White et al., 1997).
After previously characterizing the H. pylori Kdo hydrolase we have now shown that the Kdo hydrolase enzymatic activity is dependent on the presence of two proteins, Hp0579 and Hp0580, in vitro. Hp0580 has homology to a sialidase, a family of sugar cleaving enzymes, indicating that Hp0580 is likely responsible for the actual cleavage of the Kdo-lipid A linkage. However, Hp0580 cannot function without the co-expression of Hp0579, suggesting that both proteins work in concert to remove the outer Kdo sugar. This raises questions as to the exact role played by Hp0579 during the cleavage process. Hp0579 has numerous alpha-helical trans-membrane domains (Fig. S2), a feature that is often found in inner membrane transport proteins. Perhaps Hp0580 actually transfers the Kdo sugar to Hp0579, which then shuttles the Kdo sugar across the inner membrane, back to the cytoplasm, for recycling. Another intriguing possibility is that Hp0579 acts as a ‘docking station’, presenting the Kdo2-lipid A substrate to Hp0580 in a specific confirmation, making it amenable for cleavage. Although easy to speculate, determining the exact function of Hp0579 will undoubtedly be a challenging project for future research.
What makes the Kdo hydrolase stand out from other H. pylori Kdo2-lipid A modification enzymes is the downstream effect it has on H. pylori CAMP resistance and O-antigen expression. In an H. pylori Kdo hydrolase mutant two lipid A species are present, one representing a lipid A species typically seen in wild type H. pylori the other a hexa-acylated lipid A species, with an intact 4′-phosphate group. The appearance of a 4′-phosphorylated lipid A sub-population in the Kdo hydrolase mutant goes a long way towards explaining the increase in polymyxin B sensitivity. It is well documented that removal or ‘masking’ of lipid A phosphate groups is the primary mechanism involved in CAMP resistance (Trent et al., 2006). The reason for inefficient removal of the 4′-phosphate group could be explained by substrate specificity, a mechanism previously shown to be important for Kdo hydrolase function and the Helicobacter lipid A 1-phosphatase, Hp0021 (LpxE) (Stead et al., 2005, Tran et al., 2004). Thus, it appears that the enzymatic machinery responsible for these modifications has evolved into a highly ordered pathway producing a lipid A that is perhaps more suited for the unique lifestyle of H. pylori.
It has been documented that in Salmonella species, ligation of the O-antigen to core-lipid A is specific for a particular core structure (Kaniuk et al., 2004). This specificity was shown not to be imparted by the WaaL ligase itself, but rather by an unknown accessory molecule. H. pylori Kdo hydrolase mutants also showed differences in O-antigen expression after alteration to the core region, in the form of an extra Kdo sugar. In both strains G27 and 7.13, production of a fully extended LPS was diminished with the Kdo hydrolase mutants, suggesting that WaaL could no longer function at optimal efficiency. The reduction in fully extended LPS correlated to an increase in core-lipid A, as would be expected. Given that H. pylori O-antigen has been well characterized as a virulence factor, it will be interesting to see if merely a reduction in surface exposed O-antigen, as seen with the Kdo hydrolase mutant, has an any impact on pathogenesis, or if a complete truncation is required to see attenuation. However, these results could be complicated by the increased sensitivity of the Kdo hydrolase mutant to polymyxin B.
The G27 Kdo hydrolase mutant also possessed a unique O-antigen phenotype, related to the expression of specific blood group antigens. Wild type G27 expresses both Lewis X and Lewis Y blood group antigens, as shown in Fig. 7B. However, after the introduction of a Kdo hydrolase mutation, only Lewis X expression was affected. Because FutA activity is required for the generation of both epitopes and the fucosyl transferases do not have overlapping activity (Wang et al., 1999a, Ge et al., 1997), this phenomenon cannot be explained by a FutA transcriptional effect. Perhaps strain G27 encodes two WaaL accessory molecules, one specific for Lewis X and the other for Lewis Y, and only the Lewis X accessory molecule is sensitive to the presence of a second Kdo sugar. Recent studies directed towards H. pylori WaaL showed that the enzyme was capable of ligating diverse substrates in an in vitro assay (Hug et al., 2010); however, because the ligation assay used purified WaaL it is possible that the WaaL accessory molecules were not present in the reaction mixture.
Variation in H. pylori O-antigen expression is a well studied process, which has been shown to occur at the level of fucosyl transferase transcription (Appelmelk et al., 1999, Wang et al., 1999b). The fucosyl transferases decorate the O-antigen backbone with fucose sugars to produce the blood group antigen epitopes. Each fucosyl transferase gene contains a poly-C tract, which makes transcription prone to mistakes, leading to production of non-functional proteins. Inactivation of a fucosyl transferase produces a distinct clonal population of bacteria, which under favorable conditions can outgrow the progenitor strain and become predominant. Although this system can generate changes in O-antigen expression, it is not capable of producing a rapid response to sudden environmental changes. It is tempting to speculate that the Kdo hydrolase gene is actually regulated and could turn off unnecessary modifications in an entire bacterial population.
A definitive role for the Kdo hydrolase enzyme has yet to be determined in relationship to H. pylori pathogenesis, although we have clearly demonstrated its importance for CAMP resistance and O-antigen expression, two well characterized virulence determinants. Thus, the periplasmic modification of the Kdo domain is critical for maintenance of the H. pylori outer surface. We have also demonstrated previously a Kdo hydrolase activity in several H. pylori clinical strains (Stead et al., 2005), supporting the importance for a functional Kdo hydrolase enzyme in the H. pylori life cycle.
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are summarized in Table 2. Primary plate cultures of H. pylori were grown from methyl cellulose stocks on blood agar medium at 37 °C for 24–48 h in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2). The resultant colonies were inoculated into Brucella broth supplemented with 7% fetal bovine serum (Hyclone) and vancomycin (10 μg/ml). Cells were grown to an A600 of ~ 1.0 at 37 °C under microaerobic conditions for 24–48 h. Prior to every experiment, confirmation of H. pylori was performed by both Gram stain and urease test (Jerris, 1995). E. coli were typically grown at 37 °C in LB broth(Miller, 1972). When required for selection of plasmids, cells were grown in the presence of ampicillin (100 μg/ml). Antibiotics ampicillin (100 μg/ml), kanamycin (8 μg/ml), chloramphenicol (8 μg/ml) and metronidazole (12 μg/ml) were used where appropriate.
Table 2.
Bacterial strains and plasmids used in this study
| Strain or Plasmid | Genotype or Description | Source or Reference |
|---|---|---|
| Strains | ||
| Helicobacter pylori | ||
| 26695 | Wild type laboratory strain | ATCC 700392 |
| 7.13 | Gerbil adapted B128 | Boneca, I.G. |
| G27 | Wild type laboratory strain | Salama, N.R. |
| J99 | Wild type laboratory strain | ATCC 700824 |
| Hp7-91 hp0021::cam | Hp7-91 with chloramphenicol resistance cassette in hp0021 (lpxE) | (Tran et al., 2006) |
| 26695/hp1191::kan | 26695 with kanamycin resistance cassette in hp1191 (waaF) | (Chandan et al., 2007) |
| 26695/hp1191::kan/hp0579-80::cam | 26695 with kanamycin resistance cassette in hp1191 (waaF) & chloramphenicol resistance cassette in hp0579 & hp0580 | This work |
| 7.13/hp0580::cam | 7.13 with chloramphenicol resistance cassette in hp0580 | This work |
| 7.13/hp0580::cam, hp0580+ | 7.13/hp0580::cam, rdxA::hp0580 | This work |
| 7.13/hp0579-80::cam | 7.13 with chloramphenicol resistance cassette in hp0579 & hp0580 | This work |
| 7.13/hp0579-80::cam, hp0579-80+ | 7.13/hp0579-80::cam, rdxA::hp0579-80 | This work |
| G27/hp0580::cam | G27 with chloramphenicol resistance cassette in hp0580 | This work |
| G27/hp0580::cam, hp0580+ | G27/hp0580::cam, rdxA::hp0580 | This work |
| G27/hp0579-80::cam | G27 with chloramphenicol resistance cassette in hp0579 & hp0580 | This work |
| G27/hp0579-80::cam, hp0579-80+ | G27/hp0579-80::cam, rdxA::hp0579-80 | This work |
| Escherichia coli | ||
| XL-1 Blue | recA1 endA1 gyrA96thi-1 hsdR17 supE44 relA1 lac [F′proAB lacIqZΔM15::Tn10 (TetR)] | Stratagene |
| HMS174 (DE3) | F− recA1 hsdR (rK12− mK12+) (DE3) RifR | Novagen |
| W3110 (DE3) | W3110, (DE3- novagen), TetR | (Cullen & Trent, 2010) |
| W3110 (DE3) phoA− | W3110, (DE3- novagen), phoA::kan, TetR | This work |
| JW0374 | BW25113 phoA::kan (Keio Collection) | (Baba et al., 2006) |
| Plasmids | ||
| pBluescript II SK (+) | Cloning vector; AmpR, lac promoter (lacZ), f1, ColE1 | Stratagene |
| pET21a | Vector containing a T7 promotor, Ampr | Novagen |
| pGFPuv | Vector containing the “cycle 3” variant of GFP, Ampr | Clontech |
| pBAD24 | Bacterial expression vector, Ampr | (Guzman et al., 1995) |
| pHp0579 | pET21a containing hp0579 | This work |
| pHp0580 | pET21a containing hp0580 | This work |
| pHp0579-80 | pET21a containing hp0579-80 | This work |
| pBS580KO | pBluescript II SK (+) containing hp0580 flanking regions with a chloramphenicol resistance cassette insert | This work |
| pBS579-80KO | pBluescript II SK (+) containing hp0579-80 flanking regions with a chloramphenicol resistance cassette insert | This work |
| pET634comp | jhp0634 complemrnt plasmid | (Stead et al., 2008) |
| pET580comp | pEThp0954 with hp0580 inserted in hp0954 | This work |
| pET579-80comp | pEThp0954 with hp0579-80 inserted in hp0954 | This work |
| pPhoA | pET21a containing phoA | This work |
| pBAD580 | pBAD24 containing hp0580 | This work |
| pBAD580phoA | pBAD580 containing phoA | This work |
| pHp0580-PhoA | pET21a containing hp0580 with an C-terminal phoA fusion | This work |
| pGFP | pET21a containing gfp | This work |
| pGFPuv580 | pGFPuv containing hp0580 | This work |
| pHp0580-GFP | pET21a containing hp0580 with an C-terminal gfp fusion | This work |
Recombinant DNA techniques
Plasmids were isolated using the QIAprep Spin Minirep Kit (Qiagen). Custom primers were obtained from Integrated DNA Technologies (Table S1). PCR reagents were purchased from Stratagene and PCR products were isolated using Qiaquick PCR Purification Kit (Qiagen). DNA fragments were isolated from agarose gels using the Qiaquick Gel Extraction Kit (Qiagen). All other modifying enzymes were purchased from New England Biolabs and were used according to the manufacturers’ instructions.
Overexpression of the Kdo hydrolase proteins behind a T7lac promotor
Hp0579 and hp0580 were both separately and jointly subcloned into pET21a behind the T7lac promoter, using the following primer sets: hp0579 – Fhp0579 & Rhp0579, hp0580 – Fhp0580 & Rhp0580, hp0579-hp0580 – Fhp0580 & Rhp0579 (Table S1). The generated plasmids pHp0579, pHp0580, and pHp0579-80 were transformed into HMS174 (DE3) for over-expression of the protein.
Construction of H. pylori Kdo hydrolase defective non-polar mutants
Hp0580 and hp0579-hp0580 knock out vectors were constructed as described previously (Chalker et al., 2001). Briefly an upstream (primers Hp0580P1 and Hp0580P2 for both hp0580 and hp0579-hp0580) and downstream (primers Hp0580P3 and Hp0580P4 for hp0580 and primers Hp0579P3 and Hp0579P4 for hp0579-hp0580) region of each gene was PCR amplified from H. pylori strain 26695 genomic DNA. Each P2 and P3 primer also incorporated an overhang complimentary to the chloramphenicol resistance (Cam) cassette from the plasmid pHel2. The Cam cassette was PCR amplified from pHel2 (primers CamF and CamR) using a reverse primer, which incorporated a ribosomal binding sequence and start codon to enable generation of a non-polar mutant. The upstream and downstream amplicons for each individual gene were then combined with the Cam amplicon, plus the corresponding P1 and P4 primers, in a PCR reaction to generate the knockout inserts. The inserts were then cloned into pBluescript SK II + to yield the completed knockout vectors. The knockout vectors were transformed into H. pylori by natural transformation and resistant colonies selected on blood agar plates containing 8 μg/ml of chloramphenicol. Resistant colonies were re-purified on chloramphenicol containing plates and the successful insertion of the resistance cassette was verified by PCR of genomic DNA.
Chromosomal complementation of Kdo hydrolase mutants
RdxA (hp0954) was chosen as the site for chromosomal complementation. RdxA is a nitroreductase, which is capable of converting metronidazole from an inactive pro-drug, into its active form (Gerrits et al., 2004). The disruption and consequent inactivation of rdxA, by insertion of hp0580 or hp0579-hp0580, will render the bacteria resistant to metronidazole. The hp0580 and hp0579-hp0580 complementation vectors were constructed using the previously made vector pET634comp (Stead et al., 2008). Plasmid pET634comp was digested with BamHI and EcoRI to remove the jhp0634 insert and gel purified, leaving the rdxA flanking regions intact. Hp0580 (primers F580comp and R580comp) and hp0579-hp0580 (primers F580comp and R579comp) plus approximately 100 base pairs of upstream sequence were PCR amplified from H. pylori strain 26695 using primers, which incorporated BamHI and EcoRI restriction sites. Each amplicon was then ligated between the rdxA flanking regions of the gel-isolated vector to generate the complementation vectors. The complementation vectors were transformed into H. pylori by natural transformation and resistant colonies selected on blood agar plates containing 12 μg/ml of metronidazole. Resistant colonies were re-purified on metronidazole containing plates and the successful insertion of the complementation cassette was verified by PCR of genomic DNA.
Preparation of cell-free extracts, double-spun cytosol, and washed membrane
Typically, 200 ml of culture was grown to an A600 of ~ 1.0 at 37 °C and harvested by centrifugation at 10,000 × g for 10 min. All samples were prepared at 4 °C. Cell-free extract, membrane-free cytosol, and washed membranes were prepared as previously described (Tran et al., 2004) and were stored in aliquots at −20 °C. Protein concentration was determined by the bicinchoninic acid method, using bovine serum albumin as the standard.
Preparation of radiolabeled substrates
Kdo2-[4′-32P]lipid A and 1-dephosphorylated Kdo2-[4′-32P]lipid A were prepared as previously described (Stead et al., 2005).
Assay of Kdo hydrolase activity
Kdo hydrolase activity was assayed under optimized conditions in a 10-μl reaction mixture containing 50 mM Hepes, pH 8, 0.1% Triton X-100 and 2.5 μM lipid A substrate (Kdo2-[4′-32P]lipid A or 1-dephosphorylated Kdo2-[4′-32P]lipid A at ~5000 cpm/nmol). Washed membranes at 0.5 mg/ml for E. coli over expressed samples and 1 mg/ml for H. pylori samples were employed as the enzyme source, as indicated. Enzymatic reactions were incubated at 30 °C for the indicated times and terminated by spotting 4.5 μl portions of the mixtures onto silica gel 60 TLC plates. The reaction products were separated using the following solvent system: chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v). TLC plates were exposed overnight to a PhosphorImager Screen and product formation detected and analyzed using a Bio-Rad Molecular Imager PhosphorImager equipped with Quantity One Software.
Large-scale isolation of lipid A and mass spectrometry analysis
Typically, 25 ml cultures of each strain were grown at 37 °C until each culture reached an A600 of <1.0. Lipid A was released from cells and purified as described previously (Tran et al., 2006). The lipid A species were analyzed in the UT-Austin Analytical Instrumentation Facility Core using a MALDI-TOF (ABI Voyager-DE PRO) mass spectrometer equipped with a N2 laser (337 nm) using a 20 Hz firing rate. The spectra were acquired in negative ion reflectron mode. The matrix used was a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 5% tribasic ammonium citrate (20:1, v/v). 0.6μl of matrix solution was deposited on the sample plate, followed by 0.4μl of sample dissolved in chloroform-methanol (4:1, v/v).
Construction of hp0580-phoA fusion vectors
Hp0580 minus the stop codon was PCR amplified from H. pylori strain 26695 genomic DNA using primers F580phoA and R580phoA, which introduced XbaI and PstI restriction sites, respectively. R580phoA also incorporated an AvrII restriction site to facilitate insertion of phoA. Hp0580 was inserted between the XbaI and PstI restriction sites of pBAD24 to create the vector pBAD580. PhoA was PCR amplified from E. coli W3110 genomic DNA using primers FphoAfus and RphoAfus, which introduced AvrII and HindIII restriction sites, respectively. FphoAfus was designed to remove the first 13 codons from phoA, thereby disrupting the signal peptide (Hoffman & Wright, 1985). The phoA amplicon was inserted into pBAD580 between the AvrII and HindIII restriction sites to produce the vector pBAD580phoA. The completed hp0580-phoA fusion insert was PCR amplified from pBAD580phoA using the primers F580phoApet and RphoAfus, which introduced NdeI and HindIII restriction sites, respectively. The hp0580-phoA amplicon was inserted between the NdeI and HindIII restriction sites of pET21a to produce the vector pHp0580-PhoA. PhoA minus the first 13 codons was cloned into pET21a using the primer set FphoA and RphoAfus, to act as a negative control. The FphoA primer incorporated a start codon to allow transcription of the gene. Both pET580phoA and pPhoA were transformed into W3110 (DE3) phoA::kan for use in consequent assays.
Construction of hp0580-gfp fusion vectors
Hp0580 minus the stop codon was PCR amplified from H. pylori strain 26695 genomic DNA using primers F580gfp and R580gfp, which introduced HindIII and XbaI restriction sites, respectively. F580gfp also incorporated an NheI restriction site to facilitate insertion of hp0580-gfp into pET21a. Hp0580 was inserted between the HindIII and XbaI restriction sites of pGFPuv to create the vector pGFPuv580. Hp0580-gfp was excised from pGFPuv using NheI and EcoRI restriction enzymes, followed by insertion into pET21a, to produce the vector pHp0580-GFP. Gfp was PCR amplified from pGFPuv using primers Fgfp and Rgfp, then cloned into pET21a, to act as a positive control. Both pHp0580-GFP and pGFP were transformed into W3110 (DE3)for use in consequent assays.
Construction of E. coli W3110 (DE3) phoA defective mutant
P1 vir phage transduction was used to move the selectable phoA::kan mutation from E. coli strain JW0374 (Keio collection) to strain W3110 (DE3) as previously described (Sambrook & Russell, 2001).
Alkaline phosphatase assay
The alkaline phosphatase activity of the Hp0580-PhoA fusion protein was determined qualitatively by growth of W3110 (DE3) phoA::kan/pHp0580-PhoA on LB agar plates containing 100μg/ml ampicillin, 0.05mM IPTG and 40μg/ml 5-bromo-4-chloro-3-indoyl phosphate (X-phosphate, Sigma). X-phosphate is a colorimetric substrate, which turns blue in the presence of a phosphatase, leading to the production of blue colonies on the agar plate.
Fluorescence Microscopy
Cultures were grown to an A600 of ~ 0.7, then induced with 1mM IPTG and allowed to grow for a further 2 hours. 20 μl of a 1:1 culture dilution was added to a poly-L-lysine coated slide (Electron Microscopy Sciences) and a coverslip added. Bacteria were viewed at a magnification of 1000× with a Nikon Eclipse 80i microscope equipped with a 100× 1.4NA PLAN APO lens, a GFP band pass emission filter set with a 480 ± 15 nm excitation range and a 535 ± 20 nm emission range and a Photometrics Cool SNAP HQ2 camera. NIS-Elements AR 3.0 software was used to capture the images.
Determination of polymyxin B minimum inhibitory concentrations (MIC)
MICs were determined using Polymyxin B Etest® strips (Biomérieux). 200μl of culture at an A600 of 0.1 was spread evenly onto a blood agar plate and allowed to dry completely, followed by addition of the Etest® strip to the center of the plate. The plates were incubated at 37°C in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) for 24 hours before reading. Each experiment was repeated in triplicate, the results averaged and a standard deviation calculated.
H. pylori LPS analyses
Cultures were grown to an A600 of ~ 1, then the equivalent to 1 ml of culture at an A600 of 1 was harvested at 16,000 × g in a microcentrifuge and washed once with 1× PBS. Cell pellets were resuspended in 100μl of 1× LDS sample buffer (Invitrogen) + 4% BME. Cell suspensions were boiled for 10 minutes to lyse the cells and allowed to cool. Proteinase K (New England Biolabs) was added to a concentration of 125 ng/μl and the mixture was incubated at 55°C for 16 hours. The proteinase K was heat inactivated at 100°C for 5 minutes. The proteinase K treated whole cell lysates were separated by SDS polyacrylamide gel electrophoresis using a 4–12% bis-tris gradient gel (Invitrogen). The gels were either stained with Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (Molecular Probes) or blotted to a nitrocellulose membrane (Invitrogen) for immunodetection. The blots were probed with the following primary mouse monoclonal antibodies (Covance): anti-BG-4 (H type 1 chain) clone 17–206, anti-BG-5 (Lewis A) clone T174, anti-BG-6 (Lewis B) clone T218, anti-BG-7 (Lewis X) clone P12, anti-BG-8 (Lewis Y) clone F3. The secondary antibody was goat anti-mouse conjugated to peroxidase (MP Biomedicals, Cappel). Bands were detected using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants AI064184 and AI76322 to M.S.T, and Grant GM51796 to C. R. H. Raetz. We thank others members of the Trent laboratory for critical reading of the manuscript. We thank E. Altman for the 26695 hp1191::kan strain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelmelk BJ, Martin SL, Monteiro MA, Clayton CA, McColm AA, Zheng P, Verboom T, Maaskant JJ, van den Eijnden DH, Hokke CH, Perry MB, Vandenbroucke-Grauls CM, Kusters JG. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in alpha3-fucosyltransferase genes. Infect Immun. 1999;67:5361–5366. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelmelk BJ, Monteiro MA, Martin SL, Moran AP, Vandenbroucke-Grauls CM. Why Helicobacter pylori has Lewis antigens. Trends in microbiology. 2000;8:565–570. doi: 10.1016/s0966-842x(00)01875-8. [DOI] [PubMed] [Google Scholar]
- Appelmelk BJ, Shiberu B, Trinks C, Tapsi N, Zheng PY, Verboom T, Maaskant J, Hokke CH, Schiphorst WE, Blanchard D, Simoons-Smit IM, van den Eijnden DH, Vandenbroucke-Grauls CM. Phase variation in Helicobacter pylori lipopolysaccharide. Infection and immunity. 1998;66:70–76. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. Journal of bacteriology. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman MP, Engering A, Smits HH, van Vliet SJ, van Bodegraven AA, Wirth HP, Kapsenberg ML, Vandenbroucke-Grauls CM, van Kooyk Y, Appelmelk BJ. Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. The Journal of experimental medicine. 2004;200:979–990. doi: 10.1084/jem.20041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. Embo J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. The bacteria behind ulcers. Sci Am. 1996;274:104–107. doi: 10.1038/scientificamerican0296-104. [DOI] [PubMed] [Google Scholar]
- Brozek KA, Hosaka K, Robertson AD, Raetz CR. Biosynthesis of lipopolysaccharide in Escherichia coli. Cytoplasmic enzymes that attach 3-deoxy-D-manno-octulosonic acid to lipid A. J Biol Chem. 1989;264:6956–6966. [PubMed] [Google Scholar]
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, Warren PV, Lupas A, Stanhope MJ, Brown JR, Hoffman PS. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. Journal of bacteriology. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandan V, Logan SM, Harrison BA, Vinogradov E, Aubry A, Stupak J, Li J, Altman E. Characterization of a waaF mutant of Helicobacter pylori strain 26695 provides evidence that an extended lipopolysaccharide structure has a limited role in the invasion of gastric cancer cells. Biochem Cell Biol. 2007;85:582–590. doi: 10.1139/o07-056. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0913451107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Current pharmaceutical design. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. Journal of bacteriology. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L, Perez-Perez GI, Hatakeyama M, Whitehead R, Gaus K, O’Brien DP, Romero-Gallo J, Peek RM., Jr Activation of beta-catenin by carcinogenic Helicobacter pylori. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Chan NW, Palcic MM, Taylor DE. Cloning and heterologous expression of an alpha1,3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. The Journal of biological chemistry. 1997;272:21357–21363. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- Gerrits MM, van der Wouden EJ, Bax DA, van Zwet AA, van Vliet AH, de Jong A, Kusters JG, Thijs JC, Kuipers EJ. Role of the rdxA and frxA genes in oxygen-dependent metronidazole resistance of Helicobacter pylori. J Med Microbiol. 2004;53:1123–1128. doi: 10.1099/jmm.0.45701-0. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- Gutsmann T, Hagge SO, David A, Roes S, Bohling A, Hammer MU, Seydel U. Lipid-mediated resistance of Gram-negative bacteria against various pore-forming antimicrobial peptides. Journal of endotoxin research. 2005;11:167–173. doi: 10.1179/096805105X37330. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins JV, Trent MS. Secondary acylation of Vibrio cholerae lipopolysaccharide requires phosphorylation of Kdo. The Journal of biological chemistry. 2009;284:25804–5812. doi: 10.1074/jbc.M109.022772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:5107–111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Couturier MR, Rooker MM, Taylor DE, Stein M, Feldman MF. Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS pathogens. 2010;6:e1000819. doi: 10.1371/journal.ppat.1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe T, White KA, Allen AG, Peacock M, Raetz CR, Maskell DJ. Bordetella pertussis waaA encodes a monofunctional 2-keto-3-deoxy-D-manno-octulosonic acid transferase that can complement an Escherichia coli waaA mutation. J Bacteriol. 1999;181:2648–651. doi: 10.1128/jb.181.8.2648-2651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerris RC. Helicobacter. In: Murray PR, editor. Manual of Clinical Microbiology. Washington, D. C: American Society of Microbiology; 1995. pp. 492–498. [Google Scholar]
- Kaniuk NA, Vinogradov E, Whitfield C. Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus Salmonella: WaaL “ligase” is not the sole determinant of acceptor specificity. The Journal of biological chemistry. 2004;279:36470–36480. doi: 10.1074/jbc.M401366200. [DOI] [PubMed] [Google Scholar]
- Lesse AJ, Campagnari AA, Bittner WE, Apicella MA. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990;126:109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- Miller JR. Experiments in Molecular Genetics. Cold Springs Harbor Laboratory; Cold Springs Harbor, NY: 1972. [Google Scholar]
- Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydrate research. 2008;343:1952–1965. doi: 10.1016/j.carres.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Newstead SL, Potter JA, Wilson JC, Xu G, Chien CH, Watts AG, Withers SG, Taylor GL. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J Biol Chem. 2008;283:9080–9088. doi: 10.1074/jbc.M710247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A Modification Systems in Gram-Negative Bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Simoons-Smit IM, Appelmelk BJ, Verboom T, Negrini R, Penner JL, Aspinall GO, Moran AP, Fei SF, Shi BS, Rudnica W, Savio A, de Graaff J. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. Journal of clinical microbiology. 1996;34:2196–2200. doi: 10.1128/jcm.34.9.2196-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead C, Tran A, Ferguson D, Jr, McGrath S, Cotter R, Trent S. A novel 3-deoxy-D-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J Bacteriol. 2005;187:3374–3383. doi: 10.1128/JB.187.10.3374-3383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead CM, Beasley A, Cotter RJ, Trent MS. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J Bacteriol. 2008;190:7012–7021. doi: 10.1128/JB.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Karbarz MJ, Wang X, Raetz CR, McGrath SC, Cotter RJ, Trent MS. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol. 2006;188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- Wang G, Boulton PG, Chan NW, Palcic MM, Taylor DE. Microbiology (Reading, England) Pt 11. Vol. 145. 1999a. Novel Helicobacter pylori alpha1,2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens; pp. 3245–3253. [DOI] [PubMed] [Google Scholar]
- Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Molecular microbiology. 2000;36:1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Rasko DA, Sherburne R, Taylor DE. Molecular genetic basis for the variable expression of Lewis Y antigen in Helicobacter pylori: analysis of the alpha (1,2) fucosyltransferase gene. Molecular microbiology. 1999b;31:1265–1274. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CR. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of francisella novicida LpxE expressed in Escherichia coli. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CR. Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, I, Kaltashov A, Cotter RJ, Raetz CR. A mono-functional 3-deoxy-D-manno-octulosonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J Biol Chem. 1997;272:16555–16563. doi: 10.1074/jbc.272.26.16555. [DOI] [PubMed] [Google Scholar]
- Wirth HP, Yang M, Peek RM, Jr, Tham KT, Blaser MJ. Helicobacter pylori Lewis expression is related to the host Lewis phenotype. Gastroenterology. 1997;113:1091–1098. doi: 10.1053/gast.1997.v113.pm9322503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.