Abstract
Lipid A coats the outer surface of the outer membrane of Gram-negative bacteria. In Francisella tularensis subspecies novicida lipid A is present either as the covalently attached anchor of lipopolysaccharide (LPS) or as free lipid A. The lipid A moiety of Francisella LPS is linked to the core domain by a single 2-keto-3-deoxy-D-manno-octulosonic acid (Kdo) residue. F. novicida KdtA is bifunctional, but F. novicida contains a membrane-bound Kdo hydrolase that removes the outer Kdo unit. The hydrolase consists of two proteins (KdoH1 and KdoH2), which are expressed from adjacent, co-transcribed genes. KdoH1 (related to sialidases) has a single predicted N-terminal transmembrane segment. KdoH2 contains 7 putative transmembrane sequences. Neither protein alone catalyzes Kdo cleavage when expressed in E. coli. Activity requires simultaneous expression of both proteins or mixing of membranes from strains expressing the individual proteins under in vitro assay conditions in the presence of non-ionic detergent. In E. coli expressing KdoH1 and KdoH2, hydrolase activity is localized in the inner membrane. WBB06, a heptose-deficient E. coli mutant that makes Kdo2-lipid A as its sole LPS, accumulates Kdo-lipid A when expressing the both hydrolase components, and 1-dephospho-Kdo-lipid A when expressing both the hydrolase and the Francisella lipid A 1-phosphatase (LpxE).
Introduction
Francisella tularensis, a highly infectious Gram-negative pathogen, is the causative agent of tularemia in mammals (Ellis et al., 2002, McLendon et al., 2006). As few as 10 bacteria of F. tularensis subsp. tularensis can cause fatal disease in humans, and F. tularensis is therefore considered a potential bio-weapon. The environmental isolate, F. tularensis subsp. novicida (F. novicida), is a mouse pathogen and is not infectious to humans (Rohmer et al., 2007), making it an excellent model system with which to study F. tularensis biochemistry. F. novicida, like other species of F. tularensis, is an intracellular pathogen that can survive and replicate within the mouse macrophage cytoplasm after its escape from phagolysosomes (McLendon et al., 2006, Checroun et al., 2006).
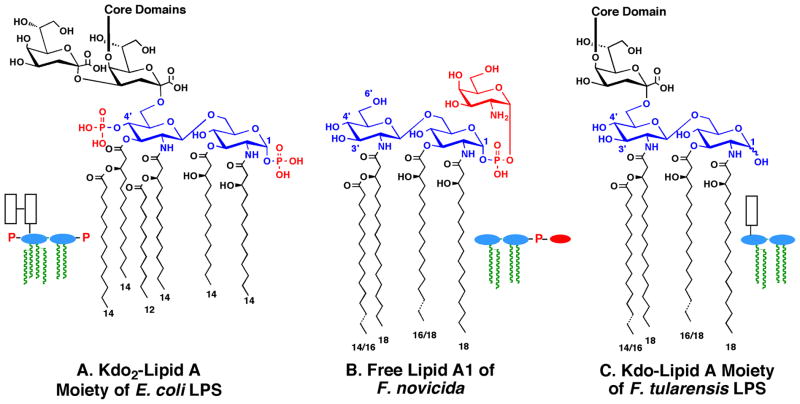
Lipid A, the hydrophobic moiety of lipopolysaccharide (LPS), is a major component of the outer leaflet of outer membranes of Gram-negative bacteria (Fig. 1A) (Raetz & Whitfield, 2002, Raetz et al., 2007). Although F. novicida and other strains of Francisella contain LPS (Vinogradov et al., 2002), over 70% of their lipid A is present in a “free” form (Fig. 1B), which is not covalently attached to the usual core and O-antigen sugars (Wang et al., 2006b). The two main species of free lipid A in F. novicida are designated A1 (Fig. 1B) and A2 (not shown) (Wang et al., 2006b). The minor component A2 differs from A1 in that it is further glycosylated at 6′-position with a glucose residue (Wang et al., 2006b). Both A1 and A2 lack the 4′-phosphate moiety and the 3′-hydroxyacyl chain present in E. coli lipid A (Wang et al., 2006b), and both are modified with a galactosamine residue on the 1-phosphate group (Wang et al., 2006b, Wang et al., 2009, Song et al., 2009) (Fig. 1B). Both A1 and A2 contain longer acyl chains when compared to E. coli lipid A (Figs. 1A and 1B), and Francisella lipid A displays much greater micro-heterogeneity in the lengths of its acyl chains (Shaffer et al., 2007).
Fig. 1. Structures of lipid A species found in E. coli and F. novicida.
Panel A. The Kdo2-lipid A moiety of E. coli LPS accumulates in heptose deficient mutants like WBB06 (Brabetz et al., 1997, Raetz et al., 2006). The lengths of the acyl chains are indicated by the numbers at the bottom. The 1 and 4′ positions are the sites of phosphate group attachment on the glucosamine disaccharide backbone. The core domain of E. coli LPS is attached to the inner Kdo residue, as indicated (Raetz & Whitfield, 2002). No free lipid A is present in wild-type E. coli. Panel B. Free lipid A makes up over 70% of the total lipid A of F. novicida. The A1 component of free lipid A is the predominant species (Wang et al., 2006b). The less abundant A2 component (not shown) contains an additional α-linked glucose residue at position 6′ (Wang et al., 2006b). Panel C. The Kdo-lipid A moiety of F. novicida LPS has a different structure than free lipid A (Wang et al., 2006b, Vinogradov et al., 2002). The LPS-bound lipid A of Francisella (less than 30% of the total lipid A) lacks the 1-phosphate group (Vinogradov et al., 2002), consistent with the presence of the LpxE 1-phosphatase (Wang et al., 2004) in Francisella. The rest of the core and O-antigen regions are not shown. Colors: black, Kdo; blue, glucosamine residues of lipid A; red, phosphate groups and galactosamine. The corresponding schematic structures are used throughout the paper: white boxes, Kdo; blue, glucosamine; red, phosphate groups or galactosamine; green, acyl chains.
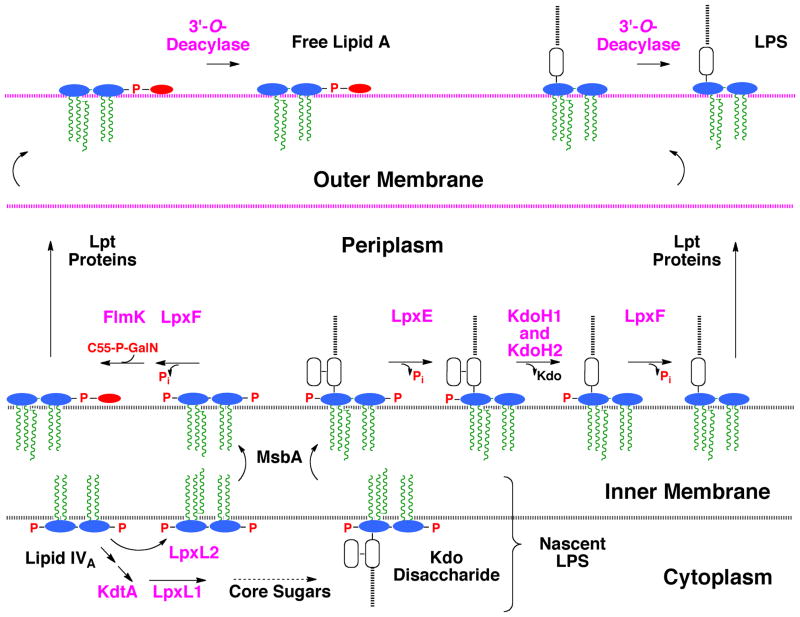
Despite these differences in lipid A structure, the F. novicida genome encodes orthologs of the key E. coli enzymes involved in Kdo2-lipid A biosynthesis, including the 4′-kinase LpxK and the Kdo transferase KdtA (Rohmer et al., 2007). The lack of a 4′-phosphate group is explained by the presence of a specific phosphatase in F. novicida, termed LpxF, which removes the 4′-phosphate moiety of both free lipid A and LPS on the outer surface of the inner membrane (Fig. 2) (Wang et al., 2006a). Mutants lacking LpxF (Wang et al., 2007) synthesize lipid A molecules that retain their 4′-phosphate group as well as their 3′-hydroxyacyl chain, suggesting an obligatory order of extra-cellular processing (Fig. 2). LpxF mutants of F. novicida are hypersensitive to cationic anti-microbial peptides and are highly attenuated in a mouse infection model (Wang et al., 2007).
Fig. 2. Topography of lipid A modification enzymes in the F. novicida cell envelope.
The differences and similarities in the processing of free lipid A and LPS-bound lipid A are compared to highlight the role of the Kdo hydrolase. The pathway diverges at the lipid IVA stage, which is acylated by LpxL2 and exported by MsbA in the case of free lipid A (Raetz et al., 2009), or is glycosylated by KdtA on the way to generating LPS. LpxE does not dephosphorylate free lipid A because it requires the Kdo moiety for activity (Wang et al., 2004). LpxE and LpxF are MsbA-dependent (Wang et al., 2004, Wang et al., 2006a), when expressed in E. coli, and therefore have active sites that face the periplasm. The order of LpxF action (if any) relative to FlmK (Wang et al., 2009) or KdoH1/H2 is not yet known. The gene encoding the F. novicida 3′-O-deacylase has not yet been identified. The enzyme probably resides in the outer membrane, based on studies with Salmonella LpxR (Rutten et al., 2009), which catalyzes a similar deacylation reaction. The schematic lipid A structures and the color schemes are the same as in Fig. 1.
The lipid A moiety of Francisella LPS shares many structural features with the free lipid A of Francisella, such as the absence of the 4′-phosphate group and the 3′-hydroxyacyl chain (Figs. 1B and C) (Vinogradov et al., 2002). Unlike the free lipid A (Fig. 1B) (Wang et al., 2006b), however, the lipid A component of Francisella LPS also lacks the 1-phosphate group and its attached GalN residue (Fig. 1C) (Vinogradov et al., 2002). The 1-phosphate group is removed on the outer surface of the inner membrane by a selective, Kdo-dependent phosphatase, designated LpxE (Fig. 2) (Wang et al., 2004). Furthermore, the LPS that is made by F. tularensis contains only one Kdo moiety in its core domain (Fig. 1C) (Vinogradov et al., 2002). The presence of one Kdo unit in the LPS core could be explained by a mono-functional Kdo-transferase KdtA, as seen in Haemophilus influenza (White et al., 1997, Chung & Raetz, 2010). However, we now demonstrate that F. novicida encodes a bifunctional Kdo-transferase. In addition, we have characterized a Kdo trimming activity in F. novicida membranes that removes the outer Kdo moiety from model substrates, such as E. coli Kdo2-lipid A (Fig. 1A) or its tetra-acylated precursor Kdo2-lipid IVA (Wang et al., 2004). We have identified two adjacent genes that encode the Francisella Kdo hydrolase, which consists of two inner membrane proteins designated KdoH1 and KdoH2. KdoH1 is the putative catalytic component, because it is distantly related to bacterial sialidases like NanI (Newstead et al., 2008), with which it appears to share several key active site residues. Extracts of F. novicida cells harboring mutations in either or both genes encoding the Kdo hydrolase lack Kdo trimming activity, but these deletions have no effect to the viability of F. novicida. Both proteins are likewise required for activity in vitro and in vivo when expressed in E. coli. A similar Kdo trimming activity is present in Helicobacter pylori (Stead et al., 2005). As shown in the accompanying manuscript (Stead et al., 2010), the H. pylori Kdo hydrolase also consists of two protein components. The availability of the genes encoding these Kdo hydrolases will be useful for re-engineering lipid A and LPS structures in diverse Gram-negative bacteria with possible applications in vaccine development.
Results
A bi-functional Kdo transferase in F. Novicida.
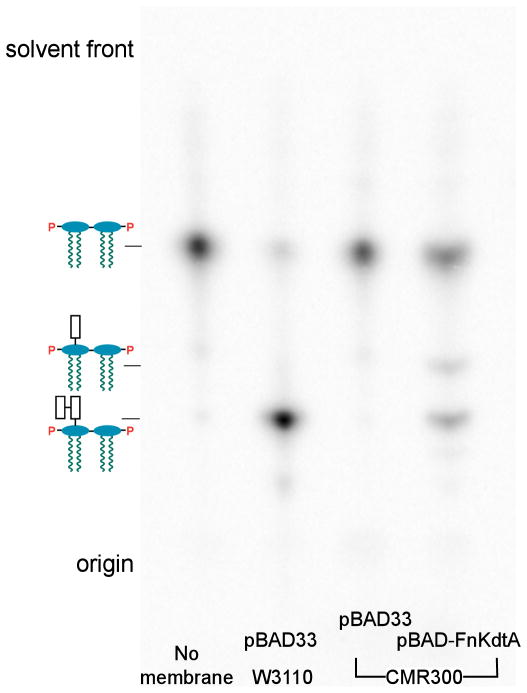
Kdo transferases (KdtAs) of Gram-negative bacteria can add one or more Kdo residues derived from the donor substrate CMP-Kdo to lipid IVA, a tetra-acylated precursor of lipid A (Supporting Fig. 1) (Raetz & Whitfield, 2002). KdtA of E. coli is a bi-functional enzyme (Belunis & Raetz, 1992) in that it incorporates both the inner and outer Kdo residues to generate the intermediate Kdo2-lipid IVA (Supporting Fig. 1). To determine whether F. novicida kdtA encodes a mono- or bi-functional Kdo transferase, the FnkdtA gene was cloned into pBAD33, generating pBAD-FnKdtA. Both the vector and pBAD-FnKdtA were then transformed into CMR300 (Reynolds & Raetz, 2009), an E. coli mutant with a deletion in its chromosomal kdtA gene. CMR300 harbors the covering plasmid pWMsbA that over-expresses MsbA (Reynolds & Raetz, 2009), an ABC transporter for core-lipid A (Zhou et al., 1998, Doerrler et al., 2001), enabling growth on rich medium at 37 °C with lipid IVA as the only remaining LPS substructure. Membranes were prepared from both constructs, as well as wild-type E. coli, and assayed for Kdo transferase activity (Fig. 3). The reactions were stopped after two hours at 30° C by spotting portions of the assay mixtures onto a TLC plate, which was developed with the solvent chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v). The membranes of the vector control CMR300/pBAD33 did not convert any [4′-32P]lipid IVA to more slowly migrating glycosylated substances. However, membranes of CMR300/pBAD-FnKdtA converted [4′-32P]lipid IVA to two products. The predominant slowly-migrating product corresponds to Kdo2- [4′-32P]lipid IVA, as shown by assaying membranes of wild-type E. coli in parallel (Fig. 3). The second product seen with the CMR300/pBAD-FnKdtA membranes corresponds to the Kdo-lipid IVA intermediate (Belunis & Raetz, 1992, White et al., 1997). The results show that FnKdtA is a bifunctional enzyme in vitro (Fig. 3).
Fig. 3. Kdo transferase assays of E. coli membranes and recombinant F. novicida KdtA.
Membranes (1 mg/mL) of E. coli W3110/pBAD33 or CMR300/pBAD-FnKdtA were assayed with 5 μM [4′-32P]lipid IVA (10,000 cpm/nmol) as the substrate. The first lane contains no membrane in the reaction mixture. The reactions were stopped after 2 hours at 30° C by spotting 4 μL onto a TLC plate, which was developed in the solvent chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v).
Mutants of F. novicida lacking Kdo hydrolase activity
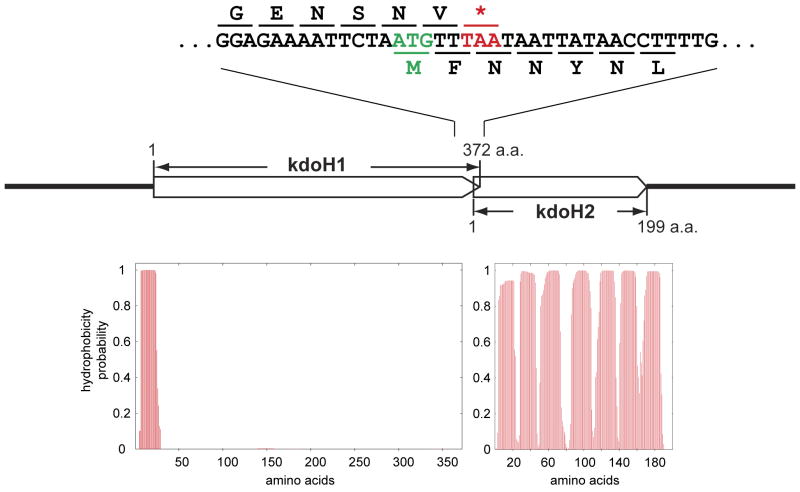
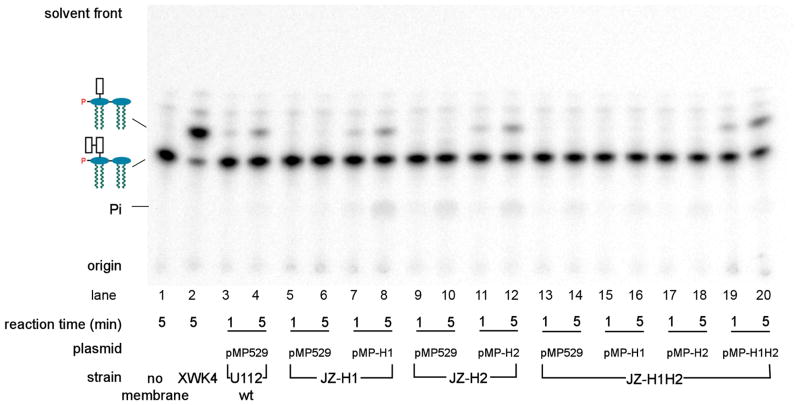
The Kdo hydrolase likely accounts for the presence of the single Kdo residue seen in the inner core of F. novicida LPS (Vinogradov et al., 2002, Wang et al., 2004). Although not present in E. coli, Kdo hydrolase activity has also been reported in Helicobacter pylori (Stead et al., 2005). The Helicobacter activity exhibits a strong dependency on the prior removal of the 1- phosphate group of Kdo2-lipid A (Stead et al., 2005). We postulated that the Kdo hydrolase sequences of F. novicida and H. pylori should be homologous to each other, but not to any proteins of E. coli. To identify potential Kdo hydrolase gene(s), we took advantage of the phylogenetic profiler service of the Integrated Microbial Genomes System (Markowitz et al., 2008). To include as many potential candidates as possible, the minimal percentage of identity at the protein level was set at 20%. In this manner, we identified 45 structural genes present in both F. novicida and H. pylori J99 that were absent in E. coli. The ones predicted to produce cytosolic proteins were ignored, because the Kdo hydrolase is membrane associated. Of the remaining 38 candidate genes, 35 were available with at least one transposon insertion mutation (Gallagher et al., 2007), which were obtained through the service available at www.francisella.org. Membranes were prepared from all these mutants and assayed for Kdo hydrolase activity. Under the conditions employed, the 4′-phosphatase activity of F. novicida, encoded by lpxF (Wang et al., 2006a), competes for the substrate (Supporting Fig. 1), generating 32Pi. In order to minimize the loss of label caused by LpxF, short reaction times (1 and 5 minutes) were used in these screening assays. Membranes of a mutant with a transposon insertion in the gene FTN_0495 (renamed kdoH1) (Fig. 4) did not show any Kdo trimming activity under standard assay conditions. To confirm this result, we constructed an in-frame deletion mutant, designated JZ-H1, in which the kdoH1 gene replaced by kanamycin resistant cassette, as described in the methods section. Again, no Kdo hydrolase activity was detected in membranes of this strain, either in the presence (Fig. 5, lanes 5/6) or absence (not shown) of the shuttle vector pMP529 (Maier et al., 2004).
Fig. 4. Overlapping genes encoding the F. novicida Kdo hydrolase components and hydropathy analysis of the predicted proteins.
Panel A. The organization of the F. novicida genes encoding the two protein components of the Kdo hydrolase (Rohmer et al., 2007). Panel B. The putative transmembrane segments of each Kdo hydrolase protein, as predicted by the TMHMM algorithm (Krogh et al., 2001).
Fig. 5. Absence of Kdo hydrolase activity in membranes of KdoH1 and KdoH2 mutants of F. novicida and its restoration by complementation.
The in vitro assays were carried out with 1-dephospho-Kdo-[4′-32P]lipid IVA as the substrate (Supporting Fig. 1). The membranes were from the F. novicida U112 wild-type, or from the mutants JZ-H1, JZ-H2 or JZ-H1H2, each harboring either the vector pMP529 (Maier et al., 2004) or the plasmids pMP-H1, pMP-H2, or pMP-H1H2 (Table 1), as indicated. The reactions were stopped at the indicated times at 30° C by spotting a portion of each reaction mixture onto a TLC plate. The products were separated using the solvent chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v). A negative control without added membranes and a positive control using membranes of the F. novicida 4′-phosphatase mutant XWK4 (Wang et al., 2007) are shown in the left two lanes.
A second gene (locus tag FTN_0494), transcribed in the same direction, is located next to kdoH1 on the F. novicida chromosome with 8 overlapping base pairs (Fig. 4). An in-frame deletion mutation in this downstream gene was constructed as for kdoH1, which likewise eliminated Kdo trimming activity when membranes were assayed in vitro (Fig. 5, lanes 9/10). This gene was therefore designated as kdoH2 (Fig. 4), and the correspondingly mutated strain as JZ-H2. Because of the overlap of these two genes, the kanamycin cassette that was used to replace kdoH2 was preceded by a ribosome binding sequence, located immediately upstream of kdoH2 and downstream of kdoH1. The mutant, JZ-H1H2, harboring an in-frame deletion of both kdoH1 and kdoH2 with replacement by a kanamycin resistance cassette, likewise lost its Kdo hydrolase activity (Fig. 5, lanes 13/14).
Phenotypes of mutants and hydropathy analysis of KdoH1 and KdoH2
The doubling time of the kdtA deletion mutant of F. novicida was 56 minutes when grown at 37° C, while that of wild-type cells was 40 minutes. LPS was absent in these mutants (Supporting Fig. 2A), but the levels of free lipid A were slightly higher than wild-type (Supporting Fig. 2B). Deletion mutants in either or both protein components of the Kdo hydrolase displayed normal growth rates under laboratory conditions. Their LPS profiles were normal, as judged by gel electrophoresis (Supporting Fig. 2A), and the amount of free lipid A was unchanged (Supporting Fig. 2B). The Kdo hydrolase mutants were not hypersensitive to polymyxin.
Based on a PSI-BLAST analysis (Altschul et al., 1997), the C-terminal portion of the protein expressed from the kdoH1 gene shows distant homology to the soluble sialidase NanI of Clostridium perfringens (Supporting Fig. 3A), which is secreted into the growth medium (Newstead et al., 2008). However, KdoH1 is predicted to contain one N-terminal transmembrane helix, while KdoH2 is predicted to have 7 transmembrane segments without any large connecting loops or other unusual structural features, as judged by the TMHMM algorithm (Krogh et al., 2001) (Fig. 4). A significant homolog of KdoH1 is present in H. pylori (see accompanying manuscript and Supporting Fig. 3B) (Stead et al., 2010). No obvious homologues of KdoH2 were detected in any genomes other than related strains of Francisella. However, as in F. novicida, a second gene encoding a protein with 6 predicted transmembrane helices follows the kdoH1 homolog of H. pylori with 3 base pairs of overlap (Stead et al., 2010). This downstream H. pylori gene product likewise displays no homology to any known protein sequences, but its appears to have a similar function to F. novicida kdoH2 (see accompanying manuscript) (Stead et al., 2010).
Complementation of F. novicida mutants defective in the Kdo hydrolase
A shuttle vector for E. coli and Francisella, pMP529, kindly provided by Dr. T. Zahrt of The Medical College of Wisconsin (Maier et al., 2004), carries a hygromycin resistance cassette under the control of the F. novicida groESL promoter. A unique MluI restriction enzyme site was used for inserting kdoH1 and/or kdoH2. Three plasmids were constructed: pMP-H1, pMP-H2, and pMP-H1H2, which express KdoH1, KdoH2 or both. In each case, the 300-base pair sequence immediately upstream of kdoH1 in F. novicida was used as the promoter and ribosomal binding site. These plasmids and the control vector were transformed into our three F. novicida mutants, JZ-H1, JZ-H2 and JZ-H1H2 (Fig. 5). Membranes derived from the three Kdo hydrolase mutants carrying the pMP529 vector displayed no Kdo trimming activity in vitro (Fig. 5, lanes 5/6, 9/10, and 13/14). The Kdo hydrolase activity of mutants JZ-H1 and JZ-H2 could be reconstituted by plasmids pMP-H1 and pMP-H2, respectively (Fig. 5, lanes 7/8 and 11/12), but they could not rescue the enzyme defect in the double deletion mutant JZ-H1H2 (Fig. 5, lanes 15/16 and 17/18). Only plasmid pMP-H1H2 expressing both proteins restored Kdo hydrolase activity to membranes of JZ-H1H2 (Fig. 5, lanes 19/20).
Reconstitution of hydrolase activity by mixing of membranes or co-expression in E. coli
Membranes from the F. novicida strains JZ-H1 and JZ-H2, harboring the single protein mutations, showed no Kdo hydrolase activity under standard assay conditions in the presence of 0.1 % Triton X-100 (Fig. 5). To demonstrate that hydrolase activity could be reconstituted, the membranes from the two mutant strains were solubilized by diluting them into 1% Triton X-100 in phosphate buffered saline (PBS), pH 7.4, containing 250mM NaCl, at a final protein concentration of 1 mg/mL. After 30 minutes at 4° C and removal of insoluble materials by centrifugation at 200,000 × g for 1 h, the supernatants were added to a Kdo hydrolase assay mixture at a final concentration of 0.1 mg/mL each. The reactions were incubated at 30° C for 25 hours. This resulted in the cleavage of about 30 % of the substrate when the mixed membranes were used, but not with the individual membranes (Supporting Fig. 4).
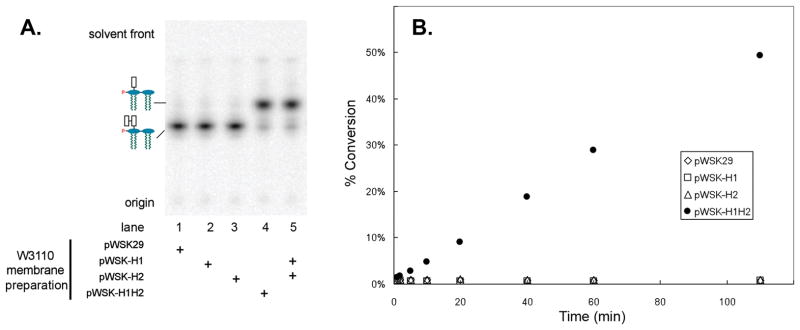
The kdoH1 and kdoH2 genes, or both, were cloned into the vector pWSK29 (Wang & Kushner, 1991) with a ribosome binding site derived from pET21b. The resulting constructs were designated pWSK-H1, pWSK-H2 and pWSK-H1H2. These plasmids and the vector control pWSK29 were transformed into wild-type E. coli W3110 (Table 1). Membranes were prepared and used in the standard Kdo hydrolase in vitro assay at 0.1 mg/mL in the presence of 0.1 % Triton X-100. The reactions were stopped after 30 minutes at 30° C by spotting onto a TLC plate that was developed in chloroform, methanol, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v). Membranes from cells carrying the vector pWSK29, or the plasmids pWSK-H1 or pWSK-H2 expressing the single proteins, showed no Kdo trimming activity (Fig. 6A, lanes 1–3). However, membranes from W3110/pWSK-H1H2 displayed robust activity (Fig. 6A, lane 4). Alternatively, hydrolase activity could be reconstituted by mixing solubilized membranes from W3110 cells expressing the individual proteins derived from pWSK-H1 or pWSK-H2 (Fig. 6A, lane 5).
Table 1.
Bacterial strains and plasmids.
| Strain or plasmids | Description | Source |
|---|---|---|
| Strains | ||
| W3110 | Wild-type E. coli, F−, λ− | E. coli Gentic Stock Center, Yale University |
| WBB06 | W3110 (waaC-waaF::tet6) | (Brabetz et al., 1997, Raetz et al., 2006) |
| CMR300 | W3110 (kdtA::kan) pWMsbA | (Reynolds & Raetz, 2009) |
| U112 | Wild-type F. novicida | Dr. F. Nano, University of Victoria, Canada |
| XWK4 | U112 (lpxF::kan) | (Wang et al., 2007) |
| JZ-H1 | U112 (kdoH1::kan) | this work |
| JZ-H2 | U112 (kdoH2::kan) | this work |
| JZ-H1H2 | U112 (kdoH1-kdoH2::kan) | this work |
| Plasmids | ||
| pACYC184 | Medium copy vector, TetR, CamR | New England Biolabs |
| pWSK29 | Low copy vector, lac promoter, AmpR | (Wang & Kushner, 1991) |
| pWMsbA | pWSK29 harboring E. coli msbA | (Reynolds & Raetz, 2009) |
| pWSK-H1 | pWSK29 harboring F. novicida kdoH1 | this work |
| pWSK-H2 | pWSK29 harboring F. novicida kdoH2 | this work |
| pWSK-H1H2 | pWSK29 harboring F. novicida kdoH1 and kdoH2 | this work |
| pBAD33 | Medium copy vector, CamR | (Guzman et al., 1995) |
| pBAD-FnLpxE | pBAD33 harboring F. novicida lpxE (Previously designated pBAD33-lpxE) | (Ma et al., 2008) |
| pBAD-FnKdtA | pBAD33 harboring F. novicida kdtA | this work |
| pMP529 | Shuttle vector for F. novicida and E. coli | (Maier et al., 2004) |
| pMP-H1 | pMP529 harboring F. novicida kdoH1 | this work |
| pMP-H2 | pMP529 harboring F. novicida kdoH2 | this work |
| pMP-H1H2 | pMP529 harboring F. novicida kdoH1 and kdoH2 | this work |
Fig. 6. Hydrolase activity in membranes of E. coli cells expressing both KdoH1 and KdoH2 or by mixing membranes containing the individual protein components.
Panel A. Membranes, prepared from E. coli W3110 harboring pWSK29, pWSK-H1, pWSK-H2 or pWSK-H1H2, were used in the Kdo hydrolase assays at 0.1 mg/mL each with 1-dephospho-Kdo-[4′-32P]lipid IVA as the substrate (Supporting Fig. 1). The reactions were stopped after 30 min at 30° C, and the products were separated as in Fig. 5. Panel B. Time course of product formation at membrane concentrations of 0.005 mg/mL. The reactions at 30° C were stopped at the indicated times by spotting a portion of each reaction mixture onto a TLC plate, as above. Product formation was quantified by PhosphorImager analysis.
These results were confirmed with a quantitative time course assay shown in Fig. 6B, which was carried out at 0.005 mg/mL of each membrane protein preparation. Membranes expressing the individual proteins had no detectable hydrolase activity (Fig. 6B). Taken together, these results strongly support the idea that the kdoH1 and kdoH2 are structural genes encoding a novel two-component Kdo trimming enzyme.
Substrate selectivity of the Kdo hydrolase
As demonstrated previously, membranes of wild-type F. novicida catalyze the cleavage of the outer Kdo unit from Kdo2-lipid IVA or Kdo2-lipid A (Wang et al., 2004). Prior removal of the phosphate group from the 1-position of lipid A by LpxE greatly accelerates Kdo trimming activity (Wang et al., 2004). Although tetra-acylated 1-dephospho-Kdo2-lipid IVA was used as the substrate in our initial experiments (Figs. 5 - 8), the hydrolase was ~5 fold more active with hexa-acylated 1-dephospho-Kdo2-lipid A as the substrate under our standard assay conditions. With membranes from E. coli cells expressing both KdoH1 and KdoH2, the specific activity of Kdo hydrolysis was 0.022 nmol/min/mg with 5 μM Kdo2-lipid IVA, 6.6 nmol/min/mg with 5 μM 1-dephospho,Kdo2-lipid IVA, and 33.8 nmol/min/mg with 5 μM 1-dephospho-Kdo2-lipid A.
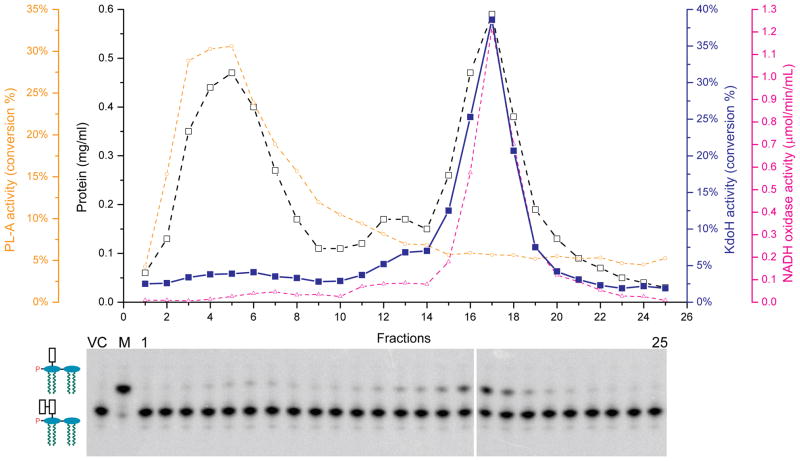
Fig. 8. Inner membrane localization of the recombinant Kdo hydrolase in E. coli.
Kdo hydrolase was expressed from the plasmid pWSK-H1H2 in E. coli W3110. The inner and outer membranes were separated by sucrose gradient centrifugation (Osborn & Munson, 1974). Each fraction from was assayed for protein concentration using the bicinchoninic acid Protein Assay Kit (Pierce, Rockford, IL) (Smith et al., 1985) (black open squares), the outer membrane marker phospholipase A (yellow open circles), the inner membrane marker NADH oxidase (magenta open triangles) (Zhou et al., 1998), and the Kdo hydrolase (blue solid squares and lower panel). VC, vector control; M, total membranes; 1 and 25, the first and last fractions of the gradient.
Dependence of the recombinant Kdo hydrolase on pH and detergent
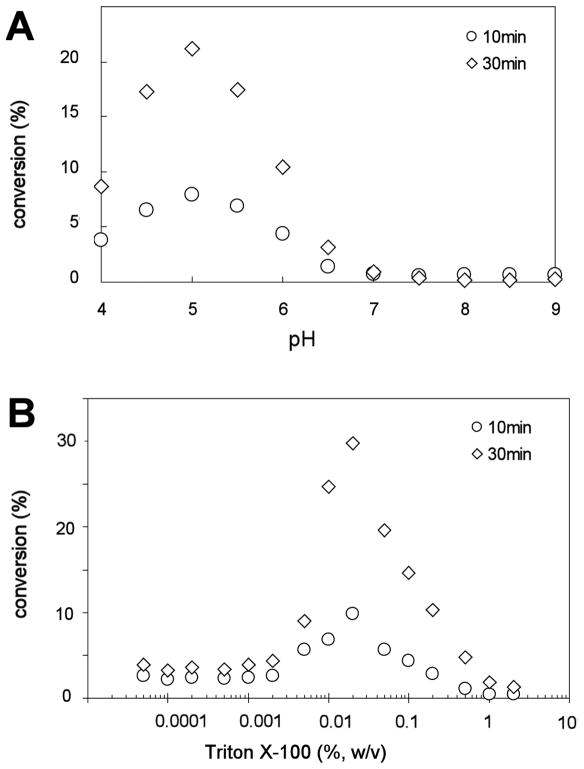
Membranes of E. coli cells expressing both Kdo hydrolase components were assayed over the pH range of 4.0 to 9.0, using a triple buffer system consisting of 100 mM sodium acetate, 50 mM bis(2-hydroxyethyl)iminotris(hydroxymethyl)hexane, and 50 mM Tris (McClerren et al., 2005). The optimal pH for Kdo hydrolase activity was 5.0 under these conditions, and product formation was proportional to time (Fig. 7A). In a phosphate buffered assay system, the pH optimum was around 6.0 (not shown). Although the pH optima were slightly different in the two systems, the Kdo hydrolase activity was always higher under mildly acidic conditions.
Fig. 7. pH and Triton X-100 dependence of the recombinant Kdo hydrolase.
Panel A. The pH dependence of the Kdo hydrolase activity was determined in a triple buffer system (McClerren et al., 2005). Panel B. The Triton X-100 dependency was analyzed in the range from 0.00005% to 2% w/v. The residual activity was the same in the absence of Triton X-100 as at 0.00005%.
To study the detergent dependence of the hydrolase, both the non-radioactive carrier and the radiolabeled 1-dephospho-Kdo2-lipid IVA substrates (Supporting Fig. 1) were initially suspended in 25 mM Tris-HCl, pH 7.8, containing 1 mM EDTA and EGTA, in the absence of Triton X-100. The assay concentrations of Triton X-100 could then be varied from 0% to 2% in w/v in the assay system by dilution from a 10% stock (Fig. 7B). Again, 0.005 mg/mL of total membrane protein from cells expressing both KdoH1 and KdoH2 was used as the enzyme source. The reactions were stopped after 10 min and 30 min at 30 ° C, and the maximal conversion of substrate to product was always less than 30%. Over the range of Triton X-100 concentrations tested, Kdo hydrolase activity was highest at 0.02%, but was strongly inhibited by 2% (Fig. 7B). The activity was not completely abolished at very low detergent concentrations (Fig. 7B) or in the absence of Triton X-100 (data not shown).
Membrane localization of the F. novicida Kdo hydrolase expressed in E. coli
Kdo hydrolase activity is associated with membranes whether expressed in F. novicida or E. coli. The cytosolic fraction contained little or no activity. To determine whether the hydrolase is localized in outer or inner membranes when expressed in E. coli, wild-type cells of strain W3110, harboring the plasmid pWSK-H1H2 (Table 1), were grown in LB medium and induced with 1 mM IPTG. The inner and outer membranes were separated by a minor modification of the sucrose gradient centrifugation method described previously (Osborn & Munson, 1974, Trent et al., 2001). Twenty five fractions were collected. Analysis of the protein concentration revealed two major peaks (Fig. 8), which were confirmed to be inner and outer membranes by assaying for the marker enzymes NADH oxidase and phospholipase A, respectively (Zhou et al., 1998, Trent et al., 2001) (Fig. 8). Kdo hydrolase assays were performed using equal volumes of each fraction. The highest protein concentration in the assay was 0.1 mg/mL for fraction 12. The reactions were stopped after 90 seconds at 30° C, and Kdo hydrolysis was analyzed by TLC (Fig. 8, lower panel). The Kdo hydrolase activity coincided with the inner membrane marker, strongly suggesting that the KdoH1 and KdoH2 are localized to the inner membrane when expressed in E. coli, consistent with their hydropathy profiles (Fig. 4).
Identification of the in vitro reaction products of the recombinant Kdo hydrolase
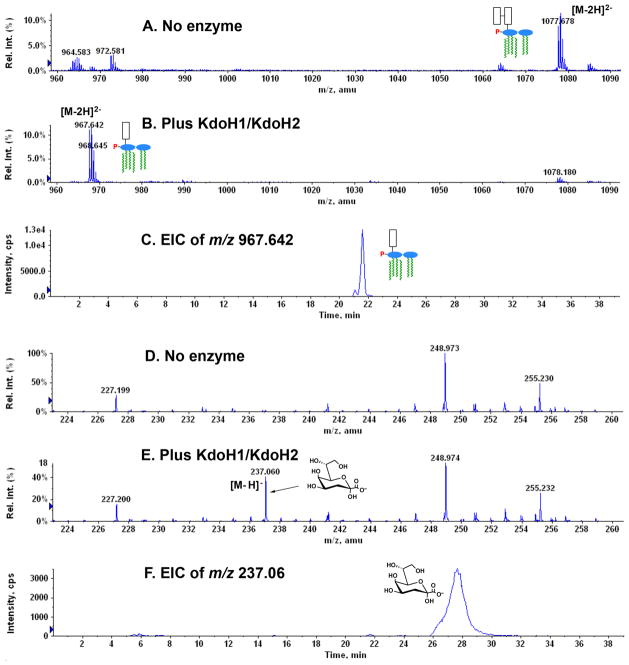
LC-ESI/MS in the negative ion mode (Fig. 9) was used to confirm the proposed structures of the reaction products generated in vitro by the recombinant Kdo hydrolase. In this experiment 100 μM 1-dephospho-Kdo2-lipid A was used as the substrate (Fig. 9A), which was isolated from E. coli WBB06 (Brabetz et al., 1997) expressing LpxE (Wang et al., 2004). Analysis of the chloroform-soluble lipid product in the acidic Bligh-Dyer lower phase (Bligh & Dyer, 1959, Nishijima & Raetz, 1979) of a complete reaction mixture containing 0.5 mg/ml of recombinant KdoH1 and KdoH2 showed a strong peak at m/z 967.642 (Fig. 9B), interpreted as the [M-2H]2− ion of 1-dephospho-Kdo-lipid A (predicted [M-2H]2− at m/z 967.648). The compound migrated as a discrete peak between minutes 21 and 22, as judged by analysis of the extracted ion current (EIC) (Fig. 9C), which is diagnostic for the presence of 1-dephospho-Kdo-lipid A in the column elution profile. The no enzyme control reaction (Fig. 9A) contained very little 1-dephospho-Kdo-lipid A, but showed the appropriate substrate peak at m/z 1077.678, as expected for the [M-2H]2− ion of 1-dephospho-Kdo2-lipid A (predicted [M-2H]2− at m/z 1077.677).
Fig. 9. Identification of the in vitro reaction products generated by the recombinant Kdo hydrolase by normal phase LC/ESI/MS.
For this purpose, 100 μM hexa-acylated 1-dephospho-Kdo2-lipid A was used as the substrate together with 0.5 mg/mL of membranes from E. coli expressing both proteins. Panels A, B and C. Analysis of the chloroform-soluble (lower phase) substrate and product before or after incubation with KdoH1/KdoH2. Panels D, E and F. Analysis of the water-soluble (upper phase) product. The reaction mixtures were extracted with a two phase Bligh-Dyer system (Bligh & Dyer, 1959), and the compounds in the lower and upper phases were subjected to normal phase LC/ESI/MS analysis in the negative ion mode. EIC, extracted ion current.
Analysis of the water-soluble product(s) (Figs. 9D, 9E and 9F) revealed a strong peak at m/z 237.060 in the Kdo hydrolase sample (Fig. 9E) but not in the no enzyme control (Fig. 9D), consistent with [M-H] − ion of the released Kdo (predicted [M-H] − at m/z 237.061). The Kdo product migrated as a broad peak between minutes 26 and 29, as judged by analysis of the EIC (Fig. 9F).
Modification of Kdo2-lipid A in E. coli by the combined action of the lipid A 1-phosphatase and the Kdo hydrolase
Expression of the F. novicida phosphatase LpxE in the heptose deficient E. coli mutant WBB06, which makes Kdo2-lipid A without any other core sugars (Brabetz et al., 1997, Kanipes et al., 2001), removes the phosphate group from the 1-position of Kdo2-lipid A, resulting in the accumulation of 1-dephospho-Kdo2-lipid A in living cells (Wang et al., 2004). When both LpxE and KdoH1/KdoH2 were co-expressed in WBB06, they modified Kdo2-lipid A synergistically. In this case, a version of WBB06 was constructed that carries both the pBAD-FnLpxE and the pWSK-H1H2 plasmids (Fig. 10A). In general, 0.2% arabinose and 1mM IPTG were added as protein expression inducers when cell cultures reached density of A600 ~0.5. Addition of the inducers to WBB06 expressing both LpxE and KdoH1/KdoH2 at the time of inoculation was toxic (data not shown).
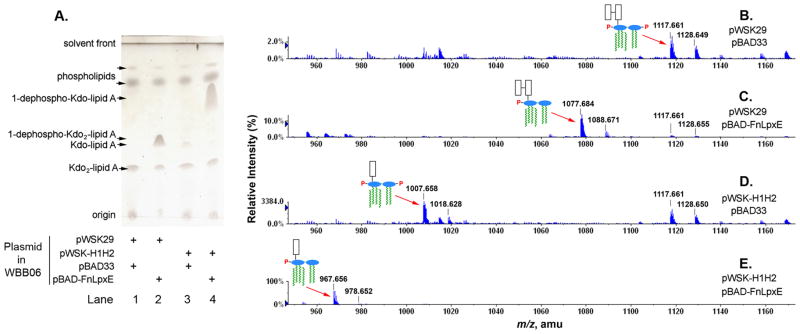
Fig. 10. Tandem modification of Kdo2-lipid A by LpxE and KdoH1/KdoH2 in E. coli.
Panel A. The heptose deficient strain, WBB06 (Brabetz et al., 1997), was used as the host to express LpxE, KdoH1/KdoH2, or both. When expressed individually, a new lipid A species appears in each strain, but migrating with differently. When both LpxE and KdoH1/KdoH2 are expressed together, a rapidly migrating species accumulates, indicating the combined effect of both enzymes on Kdo2-lipid A. Panel B. The ESI/MS analysis in the negative ion mode of the total lipids confirms that some Kdo2-lipid A (Raetz et al., 2006) is still present in all the strains, which was used to calibrate the spectra. Expression of LpxE causes accumulation of 1-dephospho- Kdo2-lipid A, whereas Kdo hydrolase expressed from pWSK-H1H2 removes one Kdo residue from Kdo2-lipid A. When both KdoH1/KdoH2 and LpxE are expressed, the combination generates the novel species 1-dephospho-Kdo-lipid A.
The lipids were extracted by the acidic Bligh-Dyer method (Nishijima & Raetz, 1979) and subjected to TLC analysis in chloroform, methanol, acetic acid, water (25:15:4:4, v/v/v/v) (Fig. 10A). In WBB06 cells containing only the vectors pWSK29 and pBAD33, Kdo2-lipid A was found as the major lipid A species (Fig. 10A, lane 1). WBB06/pWSK29/pBAD-FnLpxE, which expresses LpxE, produces a new LPS species, previously identified as 1-dephospho-Kdo2- lipid A (Fig. 10A, lane 2). The strain WBB06/pWSK-H1H2/pBAD33, which expresses KdoH1/KdoH2, produces some Kdo-lipid A, which migrates differently than the 1-dephospho-Kdo2-lipid A (Fig. 10A, lane 3). When both modification enzymes were expressed simultaneously in WBB06/pWSK-H1H2/pBAD-FnLpxE, a different lipid A species was produced, which migrated faster than others, as expected for 1-dephospho-Kdo-lipid A (Fig. 10A, lane 4).
The compositions of the predicted lipid A species were confirmed by ESI/MS analysis (Figs. 10B to 10E). In the strain containing the vectors only, the [M-2H]2− ion at m/z 1117.661, which arises from E. coli Kdo2-lipid A and was used to calibrate the spectrum, was predominant, followed by its sodium adduct ion [M+Na-3H]2− (predicted m/z 1128.651) at m/z 1128.649 (Fig. 10B). When LpxE was expressed from the plasmid pBAD-FnLpxE in WBB06, a major peak consistent with the [M-2H]2− ion of 1-dephospho-Kdo2-lipid A (predicted m/z 1077.677) was observed at m/z 1077.684 (Fig. 10C). Similarly, the [M-2H]2− ion for Kdo-lipid A (predicted m/z 1007.631) was observed at m/z 1007.658 in WBB06 expressing KdoH1/KdoH2 (Fig. 10D). Lastly, a large peak at m/z 967.656, consistent with the [M-2H]2− of 1-dephospho-Kdo-lipid A (predicted m/z 967.648), was observed in WBB06 expressing both KdoH1/KdoH2 and LpxE (Fig. 10E). In each case, lower levels of sodium adduct ions were also detected.
In summary, when KdoH1 and KdoH2 are expressed in an E. coli mutant lacking heptose, significant amounts of Kdo-lipid A accumulate in living cells (Fig. 10D). When KdoH1 and KdoH2 are expressed together with the 1-phosphatase LpxE, the cells produce even larger amounts of 1-dephospho-Kdo-lipid A (Fig. 10E). These findings, together with the in vitro assays described above, provide definitive evidence that kdoH1 and kdoH2 are the structural genes encoding the Kdo hydrolase.
Discussion
Most Gram-negative bacteria synthesize LPS molecules containing two or three Kdo residues within their inner core domains (Raetz & Whitfield, 2002, Belunis et al., 1992), but strains of Haemophilus influenzae (Zamze et al., 1987), Bordetella pertussis (Isobe et al., 1999), Helicobacter pylori (Stead et al., 2005), and Francisella tularensis (Vinogradov et al., 2002), among others, synthesize LPS with only a single Kdo unit attached to their lipid A. In Haemophilus and Bordetella, the Kdo transferase (KdtA) is monofunctional (White et al., 1997, Isobe et al., 1999), whereas in E. coli and most other organisms KdtA is bifunctional (Belunis & Raetz, 1992, Raetz & Whitfield, 2002). Bacteria with a monofunctional Kdo transferase usually contain a Kdo kinase (White et al., 1997, White et al., 1999), which adds a phosphate group to the Kdo 4-position that is usually occupied by the outer Kdo residue. There is no homolog of Kdo kinase in either F. novicida or H. pylori. However, the F. novicida and H. pylori Kdo transferases are bifunctional. The single Kdo unit found in their LPS arises as a consequence of a novel Kdo trimming reaction, catalyzed by an unusual hetero-dimeric Kdo hydrolase. This enzyme is associated with the inner bacterial membrane. It consists of a putative catalytic protein that is oriented towards the periplasm (see accompanying manuscript) (Stead et al., 2010) and a polytopic inner membrane protein that probably serves as part of the membrane anchor or is needed to deliver the nascent LPS substrate to the catalytic domain. Both proteins are absolutely required for function (Figs. 5 and 6), as judged by genetic analysis and reconstitution of enzymatic activity in E. coli.
The genes encoding the enzymes that catalyze Kdo cleavage have not been reported previously. The Francisella enzyme that we have investigated removes only the outer Kdo residue. It would be interesting to explore the possibility that it could remove more than one Kdo moiety from LPS cores of organisms like Chlamydia that contain three or four Kdo units (Brade, 1999). The putative catalytic component of KdoH1 is distantly related to secreted Gram-positive bacterial sialidases, some of which have been studied by x-ray crystallography (Newstead et al., 2008). Interestingly, the two essential arginine residues that interact with the carboxylic acid moiety of sialic acid substrate in the sialidase are also conserved in KdoH1 (Newstead et al., 2008) (Supporting Fig. 3A). Bacterial sialidases cleave terminal sialic acid residues from the oligosaccharides of glycoproteins and glycolipids on eukaryotic cells (Corfield, 1992). The physiological significance of the sialidase activity has been linked with the pathogenicity and nutrition of some bacterial strains (Corfield, 1992).
The function of the Kdo hydrolase is not yet known. The pH dependence profile of KdoH1/KdoH2 shows that a slight acidic condition is preferred (Fig. 7). Other F. novicida lipid A modification enzymes, such as LpxE, LpxF and FlmK, also prefer a pH of around 6.0 (Wang et al., 2004, Wang et al., 2006a, Wang et al., 2009). This observation may reflect the low pH environment within macrophages in which F. novicida cells replicate (McLendon et al., 2006). Francisella mutants lacking the hydrolase continue to make LPS (Supporting Fig. 2A), albeit with two Kdo moieties (J. Zhao and C. R. H. Raetz, in preparation), and they grow normally under laboratory conditions. Their virulence in animal models remains to be examined. As shown in the accompanying manuscript (Stead et al., 2010), H. pylori mutants lacking the hydrolase are hypersensitive to cationic anti-microbial peptides because their LpxF phosphatase cannot efficiently remove the 4′-phosphate group of their lipid A, implying an obligatory order of lipid A processing by the various extracellular modification enzymes of that organism.
A unique feature of the Francisella system is the relative abundance of free lipid A (Wang et al., 2006b), which is apparently exported to the outer membrane without prior attachment of Kdo (Fig. 2). Free lipid A does not arise by cleavage of the inner Kdo moiety, which is not a substrate for KdoH1/KdoH2. Deletion of kdtA in F. novicida results in the complete loss of LPS (Supporting Fig. 2A) with a modest (~25%) increase in free lipid A (Supporting Fig. 2B) and a reduction in growth rate. In wild-type Francisella, free lipid A is more abundant than LPS-bound lipid A (Wang et al., 2006b), which makes up less than 30% of the total.
The core region of Francisella lacks heptose, which is replaced by mannose (Vinogradov et al., 2002), a structurally similar sugar that is also found in Rhizobium etli (Kadrmas et al., 1998, Forsberg & Carlson, 1998). The Rhizobium mannosyl transferase, encoded by the lpcC gene (Kadrmas et al., 1998), requires a Kdo disaccharide for activity (Kanipes et al., 2003), and it is an intracellular enzyme. LpcC is inactive with substrates containing a single Kdo unit (Kanipes et al., 2003). If Francisella incorporated only a single Kdo moiety into its nascent LPS, it would presumably be unable to assemble its core and O-antigen domains. To test this idea one could replace the bifunctional Kdo transferase of Francisella with the monofunctional Kdo transferase of H. influenzae and analyze the resulting LPS and free lipid A structures.
A unique feature of the F. novicida and H. pylori Kdo hydrolases is the requirement for two distinct proteins. So far, we have been unable to complement our Francisella mutants with the Helicobacter genes encoding KdoH1 and KdoH2 (Stead et al., 2010). Given that reconstitution by mixing of membranes expressing the individual protein components is quite efficient (Fig. 6), it should be possible to examine the kinetics and affinity of these two components for each other in a purified system. So far, it has been difficult to over-express both proteins at high levels in E. coli. Use of codon optimized genes or expression in Francisella itself, which contains some unusual membrane phospholipids (Wang et al., 2004), may be necessary to achieve high levels of protein expression for structural studies.
As shown in previous work, mutation of lipid A or LPS modification enzymes can have profound effects on pathogenesis, often resulting in attenuation (Gunn, 2001, Wang et al., 2007, Raetz et al., 2007). Characterization of the pathogenesis phenotypes of the Kdo hydrolase mutants therefore requires further investigation. Expression of modification enzymes in foreign hosts, as shown for KdoH1/KdoH2 in the case of E. coli (Fig. 10), permits unusual structural modifications of LPS that likewise may have profound effects on outer membrane assembly and pathogenesis. KdoH1/KdoH2 can modify lipid A in E. coli synergistically with LpxE, the lipid A 1-phosphatase. The product of both KdoH1/KdoH2 and LpxE, 1-dephospho-Kdo-lipid A, accumulates to high levels in the E. coli strains expressing both enzymes (Fig. 10). The loss of the 1-phosphate group and the outer Kdo residue greatly reduces the charge of the LPS molecules and likely alters the electrostatic interactions between them. The stability of the outer membrane may be compromised when 1-dephospho-Kdo-lipid A is the major component of outer leaflet of outer membrane. Combining heterologous modification enzymes in diverse pathogens that normally do not contain them could lead to the development of novel attenuated strains with utility as vaccines.
Experimental procedures
Materials and reagents
Trypticase soy broth, Mueller-Hinton broth, yeast extract, tryptone and Bacto agar were purchased from Difco, Detroit, MI. Glass-backed silica gel 60 thin layer chromatography (TLC) plates were from E. Merck, Darmstadt, Germany. Chloroform, pyridine, methanol, acetic acid and 88% formic acid were from EMD Science, Gibbstown, NJ. [γ -32P]-ATP was purchased from PerkinElmer, Waltham, MA.
Bacterial growth and membrane preparation
E. coli strains were usually grown in LB broth (1% tryptone, 0.5% yeast extract and 1% NaCl) at 37° C. F. novicida U112 (Rohmer et al., 2007) and derived strains were grown in 3% trypticase soy broth supplemented with 0.1% cysteine (w/v). When necessary, ampicillin, chloramphenicol, kanamycin, hygromycin or tetracycline was added at 100, 25, 30, 200 or 12.5 μg/mL, respectively for E. coli strains, while kanamycin, hygromycin or tetracycline was added at 15, 200 or 12.5 μg/mL respectively for F. novicida strains.
To prepare membranes, the bacteria were typically grown in 100 mL of medium at 37° C and were harvested when A600 reached 1.0. The inducers, IPTG or L-arabinose, were added when necessary as indicated. Cells were harvested by low-speed centrifugation and washed twice with 50 mL PBS, pH7.4. The cells were then re-suspended in 2 mL PBS and passed through French pressure cell once at 16,000 psi. The unbroken cells and large debris were removed by centrifugation at 10,000 × g for 15 minutes at 4 ° C. The supernatant was subjected to ultracentrifugation at 200,000 × g for 1 hour at 4° C. The membrane-containing pellets were resuspended in 0.4 mL PBS, pH7.4, and stored at −80° C in aliquots. The protein concentration of the membrane preparations was determined with the bicinchoninic acid assay (Pierce, St. Louis, MO) (Smith et al., 1985).
Expression cloning of the F. novicida kdtA, lpxE and Kdo hydrolase genes
The F. novicida kdtA (FnkdtA) was amplified by PCR and cloned into the pBAD33 vector at the SacI and PstI restriction sites (Table 1). The forward primer nKdtA_for and the reverse primer nKdtA_rev used for this purpose are shown in Supporting Table 1. The PCR was performed with Pfu turbo polymerase (Invitrogen, Carlsbad, CA) and F. novicida genomic DNA as template, prepared with the Easy-DNA kit (Invitrogen). The 50-μL reaction mixture contained 100 ng template, 200 ng of each primers, and 2.5 units of Pfu polymerase. The reaction was started at 95° C for 1 min, followed by 25 cycles of denaturation (1 min at 95° C), annealing (1 min at 55° C) and extension (3 min at 72° C). A 10-min extension at 72° C was used at the end. The DNA product was located on 0.7% Agarose gel, and the desired band was excised and purified with Gel Extraction Kit (Qiagen, Valencia, CA). The PCR product was digested with SacI and PstI, and ligated into similarly digested and Antarctic Phosphatase treated pBAD33 vector. The ligation mixture was transformed into E. coli DH5α and a portion of the cells were spread onto a LB agar plate, containing chloramphenicol. The insertion of FnkdtA into pBAD33 was confirmed by DNA sequencing, and the desired plasmid was designated as pBAD-FnKdtA. All restriction enzymes, T4 DNA ligase and phosphatase were purchased from New England BioLabs (Ipswich, MA). The plasmid pBAD-FnLpxE was constructed similarly by cloning the F. novicida lpxE gene into the pBAD33 vector, and was kindly provided by Dr. B. Ma (Ma et al., 2008).
The individual protein components of the F. novicida Kdo hydrolase, or both, were cloned into the E. coli vector pWSK29 (Wang & Kushner, 1991) or into pMP529 (Maier et al., 2004), a shuttle vector for E. coli and F. novicida. To clone kdoH1 or both kdoH1 and kdoH2 into pMP529, the same forward primer nMpH1_for was used for PCR (Supporting Table 1). For cloning kdoH1 alone, the reverse primer nMpH1_for was employed (Supporting Table 1). The reverse primer nMpH2_rev (Supporting Table 1) was used for cloning both kdoH1 and kdoH2. The PCR products were digested with MluI, purified and ligated to pMP529 at the MluI restriction site, using similar procedure previously described for pBAD-FnKdtA. The resulting plasmids carrying kdoH1 and kdoH1-kdoH2 are designated as pMP-H1 and pMP-H1H2, respectively. The shuttle vector and its derivatives were transformed to F. novicida by electroporation (LoVullo et al., 2006).
The kdoH2 gene was cloned into pMP529 preceded by the promoter region upstream of kdoH1 to allow expression in trans. A two-step PCR protocol was applied for plasmid construction. First, the promoter and the kdoH2 coding region were amplified separately. The region about 300bp immediately upstream of kdoH1 contains the promoter and ribosomal binding site. To amplify the promoter region, an additional sequence matching the beginning of kdoH2 was incorporated into the reverse primer. This region was required for the fusion PCR in the second step to anneal to the kdoH2 coding sequence and generate the final DNA product containing the promoter, ribosomal binding site and kdoH2. The promoter region was amplified by PCR using the forward primer nMpH1_for (as above) about 300bp upstream of kdoH1 and a reverse primer nPromH2_rev (Supporting Table 1). The kdoH2 coding region was amplified with the forward primer nMpH2_for (Supporting Table 1); the reverse primer nMpH2_rev (Supporting Table 1) was the same as used for cloning kdoH1-kdoH2. Both above PCR reactions used F. novicida genomic DNA as template. In the second PCR step, 50 ng each of the purified PCR products of promoter region and the kdoH2 coding sequence were used as templates. About 200 ng of the common forward primer for the promoter region and the reverse primer for kdoH2 (Supporting Table 1) were used in a reaction that included 1 unit of KOD Hot Start DNA polymerase (Novagen). The reaction started with 2 min incubation at 95° C, followed by 30 cycles of 30 seconds denaturation at 95° C, 30 seconds annealing at 50° C and 1 min extension at 70 ° C. A 10-min extension at 70° C was added in the end. The PCR product was purified, digested with appropriate restriction enzyme, and ligated into pMP529 at the MluI site. The resulting plasmid carrying kdoH2 is designated as pMP-H2.
To clone kdoH1 into pWSK29, the gene was amplified with the forward primer nWskH1_for and the reverse primer nWskH1_rev (Supporting Table 1). The kdoH2 gene was amplified with forward primer nWskH2_for and the reverse primer nWskH2_rev (Supporting Table 1). To clone both kdoH1 and kdoH2, the forward primer for kdoH1 and the reverse primer for kdoH2 were used in the PCR reaction (Supporting Table 1). The PCR products were purified, digested with XbaI and BamHI, and ligated into pWSK29. The plasmid constructs harboring kdoH1, kdoH2 or kdoH1 and kdoH2 were designated pWSK-H1, pWSK-H2 or pWSK-H1H2, respectively.
Construction of F. novicida mutations with deletions in kdoH1 and kdoH2
The kanamycin cassette was amplified from the pET28b vector with the forward primer nKan_for and the reverse primer nKan_rev (Supporting Table 1).
To construct F. novicida mutants with deletions in kdoH1, kdoH2, or both, the following primers were used. To amplify the 2kb region upstream of kdoH1, forward primer nHmut_for was used together with reverse primer nH1Kan_rev (Supporting Table 1). To amplify the 2kb region downstream of kdoH1, reverse primer nHmut_rev was used in conjunction with forward primer nH1Kan_for (Supporting Table 1). Both PCR reactions used F. novicida genomic DNA as the template.
The two PCR products and the kanamycin cassette were then used as templates in a final PCR that was done with forward primer nHmut_for and reverse primer nHmut_rev (Supporting Table 1). The desired DNA species was purified and transformed to wild type F. novicida that had been rendered competent by chemical treatment (Anthony et al., 1991). The mutants were selected on 3% trypticase soy broth agar plates supplemented with kanamycin and 0.1% cysteine.
To construct the kdoH2 deletion mutant, the 2kb upstream sequence of kdoH1 was amplified with forward primer nHmut_for and reverse primer nH2Kan_rev (Supporting Table 1). The downstream 2kb sequence of kdoH2 was amplified with reverse nHmut_rev and forward primer nH2Kan_for (Supporting Table 1). Finally, a fusion PCR reaction was set up as described above for kdoH1, and the PCR product was transformed into wild type F. novicida followed by selection on kanamycin plates.
The DNA for constructing the kdoH1-kdoH2 deletion mutant was prepared in a fusion PCR reaction using the 2kb upstream sequence for kdoH1, the 2kb downstream sequence for kdoH2 and the kanamycin cassette as templates with forward primer nHmut_for and reverse primer nHmut_rev. The linear DNA product was transformed into wild type F. novicida, and the kdoH1-kdoH2 mutant was selected as above. The deletions in kdoH1, kdoH2, or kdoH1-kdoH2 were confirmed by DNA sequencing, and the mutated strains were designated JZ-H1, JZ-H2 and JZ-H1H2, respectively.
Preparation of 1-dephospho-Kdo2-[4′-32P]lipid IVA
The tetraacyl-disaccharide 1-phosphate precursor of lipid A was converted to [4′-32P]lipid IVA using [γ -32P]ATP and BLR(DE3)/pLysS/pJK2 membranes containing the overexpressed 4′-kinase LpxK (Garrett et al., 1997). Purified E. coli Kdo transferase (KdtA) (Basu et al., 1999), was used to convert [4′-32P]lipid IVA to Kdo2-[4′-32P]lipid IVA (Supporting Fig. 1). The 1- dephospho-Kdo2-[4′-32P]lipid IVA was generated from Kdo2-[4′-32P]lipid IVA with E. coli membranes containing recombinant F. novicida 1-phosphatase LpxE (Supporting Fig. 1) (Wang et al., 2004). The 1-dephospho-Kdo2-[4′-32P]lipid IVA was purified by preparative TLC (Basu et al., 1999).
In vitro assay for the F. novicida Kdo hydrolase
Kdo hydrolase assays were usually done in a 15 μL reaction mixture at 30° C, containing 50 mM potassium phosphate buffer, pH 6.0, 0.1% Trition X-100 and 5 μM 1-dephospho-Kdo2-[4′-32P]lipid IVA (100,000 cpm/nmol). The reactions were terminated by spotting 4 μL samples onto a TLC plate. The reaction times were varied as indicated. The plate was dried under a cool air stream and developed in the solvent chloroform, pyridine, 88% formic acid, water (30:70:16:10, v/v/v/v). The plate was dried with hot air stream and exposed to a PhosphorImager screen. Reactants and products were quantified using a Molecular Dynamics PhosphorImager equipped with ImageQuant software. Specific activity was calculated from the amount of product in nmol generated at a given time in the assay volume, divided by the incubation time in minutes and the total amount of protein in the given assay volume in mg.
Lipid preparation
Unless otherwise indicated, lipids were extracted using the Bligh-Dyer system (Bligh & Dyer, 1959). Typically, 100 mL cultures were grown to A600 of 1.0. The cells were harvested by centrifugation and washed twice with 50 mL PBS, pH7.4. The cells were resuspended in 8 mL PBS (for the neutral condition) or 0.1N HCl (for the acidic condition), and then 10 mL chloroform and 20 mL methanol were added to form a single phase mixture (chloroform, methanol and aqueous ratio of 1:2:0.8, v/v/v). After incubation at room temperature for 60 min, the insoluble debris was removed by centrifugation at 3000 × g for 30 min. The supernatant was transferred to clean solvent-resistant bottles, and then 10 mL chloroform and 10 mL PBS (for the neutral extraction) or 0.1N HCl (for the acidic extraction) were added to generate a two-phase system. After thorough mixing, the samples were centrifuged at 3000 × g for 15 min, and the lower phase was collected; for the acidic extraction, several drops of pyridine were added. The sample was dried under a stream of nitrogen.
ESI/MS analysis
Lipid samples were dissolved in chloroform and methanol (2:1, v/v) containing 1% piperidine and immediately subjected to ESI/MS analysis in negative ion mode (Raetz et al., 2006, Wang et al., 2007) via direct infusion, as previously described. All mass spectra were collected on a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (ABI/MDS-Sciex) equipped with an ESI source. Data acquisition and analysis were performed with Analyst QS software supplied by manufacturer.
LC-ESI/MS analysis
Normal phase LC-ESI/MS of lipids was performed using an Agilent 1200 Quaternary LC system coupled to a QSTAR XL quadrupole time-of-flighttandem mass spectrometer (as above). An AscentisR Si HPLC column (5 μm, 25 cm × 2.1 mm) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform/methanol/water/ aqueous ammonium hydroxide (600:340:50:5, v/v/v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v/v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min, and finally returned to 100% A over 0.5 min and held at 100% A for 5 min. The total LC flow rate was 300 μl/min. The post-column splitter diverted ~10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer. Nitrogen was used as the collision gas for MS/MS experiments.
Supplementary Material
Acknowledgments
This research was supported by NIH Grant GM-51796 to C. R. H. Raetz. The mass spectrometry facility in the Department of Biochemistry of the Duke University Medical Center is supported by the LIPID MAPS Large Scale Collaborative Grant number GM-069338 from NIH. The authors are especially grateful to Drs. Hak Suk Chung and Ziqiang Guan for their critical reading of the manuscript and for Dr. Guan’s help with the mass spectrometry.
This research was supported by NIH Grant GM-51796 to C. R. H. Raetz.
Abbreviations
- EIC
extracted ion current
- ESI-MS
electrospray ionization mass spectrometry
- IPTG
isopropyl-β-D-thiogalactoside
- Kdo
2-keto-3-deoxy-D-manno-octulosonic acid
- LC
liquid chromatography
- LPS
lipopolysaccharide
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LS, Gu MZ, Cowley SC, Leung WW, Nano FE. Transformation and allelic replacement in Francisella spp. J Gen Microbiol. 1991;137:2697–2703. doi: 10.1099/00221287-137-12-2697. [DOI] [PubMed] [Google Scholar]
- Basu SS, York JD, Raetz CRH. A phosphotransferase that generates phosphatidylinositol 4-phosphate (PtdIns-4-P) from phosphatidylinositol and lipid A in Rhizobium leguminosarum. A membrane-bound enzyme linking lipid A and PtdIns-4-P biosynthesis. J Biol Chem. 1999;274:11139–11149. doi: 10.1074/jbc.274.16.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belunis CJ, Mdluli KE, Raetz CRH, Nano FE. A novel 3-deoxy-D-manno-octulosonic acid transferase from Chlamydia trachomatis required for expression of the genus-specific epitope. J Biol Chem. 1992;267:18702–18707. [PubMed] [Google Scholar]
- Belunis CJ, Raetz CRH. Biosynthesis of endotoxins: purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid transferase from Escherichia coli. J Biol Chem. 1992;267:9988–9997. [PubMed] [Google Scholar]
- Bligh EG, Dyer JJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brabetz W, Muller-Loennies S, Holst O, Brade H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur J Biochem. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- Brade H. Chlamydial lipopolysaccharide. In: Brade H, Opal SM, Vogel SN, Morrison DC, editors. Endotoxin in Health and Disease. New York: Marcel Dekker, Inc; 1999. pp. 229–242. [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Raetz CR. Interchangeable Domains in the Kdo Transferases of Escherichia coli and Haemophilus influenzae. Biochemistry. 2010;49:824–832. doi: 10.1021/bi100343e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield T. Bacterial sialidases--roles in pathogenicity and nutrition. Glycobiology. 1992;2:509–521. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- Doerrler WT, Reedy MC, Raetz CRH. An Escherichia coli mutant defective in lipid export. J Biol Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Carlson RW. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J Biol Chem. 1998;273:2747–2757. doi: 10.1074/jbc.273.5.2747. [DOI] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TA, Kadrmas JL, Raetz CRH. Identification of the gene encoding the Escherichia coli lipid A 4'-kinase. Facile synthesis of endotoxin analogs with recombinant LpxK. J Biol Chem. 1997;272:21855–21864. doi: 10.1074/jbc.272.35.21855. [DOI] [PubMed] [Google Scholar]
- Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe T, White KA, Allen AG, Peacock M, Raetz CRH, Maskell DJ. Bordetella pertussis waaA encodes a monofunctional 2-keto-3-deoxy-D-manno-octulosonic acid transferase that can complement an Escherichia coli waaA mutation. J Bacteriol. 1999;181:2648–2651. doi: 10.1128/jb.181.8.2648-2651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Allaway D, Studholme RE, Sullivan JT, Ronson CW, Poole PS, Raetz CRH. Cloning and overexpression of glycosyltransferases that generate the lipopolysaccharide core of Rhizobium leguminosarum. J Biol Chem. 1998;273:26432–26440. doi: 10.1074/jbc.273.41.26432. [DOI] [PubMed] [Google Scholar]
- Kanipes MI, Lin S, Cotter RJ, Raetz CRH. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J Biol Chem. 2001;276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- Kanipes MI, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. A mannosyl transferase required for lipopolysaccharide inner core assembly in Rhizobium leguminosarum. Purification, substrate specificity and expression in Salmonella waaC mutants. J Biol Chem. 2003;278:16356–16364. doi: 10.1074/jbc.M301255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting trans-membrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- LoVullo ED, Sherrill LA, Perez LL, Pavelka MS., Jr Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- Ma B, Reynolds CM, Raetz CRH. Periplasmic orientation of nascent lipid A in the inner membrane of an Escherichia coli LptA mutant. Proc Natl Acad Sci USA. 2008;105:13823–13828. doi: 10.1073/pnas.0807028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Havig A, Casey M, Nano FE, Frank DW, Zahrt TC. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IM, Dubchak I, Anderson I, Lykidis A, Mavromatis K, Ivanova NN, Kyrpides NC. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36:D528–533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerren AL, Zhou P, Guan Z, Raetz CRH, Rudolph J. Kinetic analysis of the zinc-dependent deacetylase in the lipid A biosynthetic pathway. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- McLendon MK, Apicella MA, Allen LA. Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead SL, Potter JA, Wilson JC, Xu G, Chien CH, Watts AG, Withers SG, Taylor GL. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J Biol Chem. 2008;283:9080–9088. doi: 10.1074/jbc.M710247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima M, Raetz CRH. Membrane lipid biogenesis in Escherichia coli: identification of genetic loci for phosphatidylglycerophosphate synthetase and construction of mutants lacking phosphatidylglycerol. J Biol Chem. 1979;254:7837–7844. [PubMed] [Google Scholar]
- Osborn MJ, Munson R. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. (Kdo)2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Guan ZQ, Ingram BO, Six DA, Song F, Wang XY, Zhao JS. Discovery of new biosynthetic pathways: the lipid A story. Journal of Lipid Research. 2009;50:S103–S108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CM, Raetz CRH. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry. 2009;48:9627–9640. doi: 10.1021/bi901391g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, Gallagher LA, Manoil C, Ernst RK, Drees B, Buckley D, Haugen E, Bovee D, Zhou Y, Chang J, Levy R, Lim R, Gillett W, Guenthener D, Kang A, Shaffer SA, Taylor G, Chen J, Gallis B, D'Argenio DA, Forsman M, Olson MV, Goodlett DR, Kaul R, Miller SI, Brittnacher MJ. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten L, Mannie JP, Stead CM, Raetz CRH, Reynolds CM, Bonvin AM, Tommassen JP, Egmond MR, Trent MS, Gros P. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium. Proc Natl Acad Sci U S A. 2009;106:1960–1964. doi: 10.1073/pnas.0813064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1080–1092. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Song F, Guan Z, Raetz CRH. Biosynthesis of undecaprenyl phosphate-galactosamine and undecaprenyl phosphate-glucose in Francisella novicida. Biochemistry. 2009 doi: 10.1021/bi802212t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead C, Tran A, Ferguson D, Jr, McGrath S, Cotter R, Trent S. A novel 3-deoxy-D-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J Bacteriol. 2005;187:3374–3383. doi: 10.1128/JB.187.10.3374-3383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead CM, Zhao J, Raetz CRH, Trent MS. Removal of the outer Kdo residue from Helicobacter pylori lipopolysaccharide and its impact on the bacterial surface. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07304.x. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Pabich W, Raetz CRH, Miller SI. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4'-phosphatase LpxF. J Biol Chem. 2006a;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CRH. Attenuated virulence of a Francisella mutant lacking the lipid A 4'-phosphatase. Proc Natl Acad Sci U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, McGrath S, Cotter R, Raetz CRH. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006b;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, Raetz CRH. Identification of undecaprenyl phosphate-β-D-galactosamine in Francisella novicida and its function in lipid A modification. Biochemistry. 2009;48:1162–1172. doi: 10.1021/bi802211k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, I, Kaltashov A, Cotter RJ, Raetz CRH. A mono-functional 3-deoxy-D-manno-octulosonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J Biol Chem. 1997;272:16555–16563. doi: 10.1074/jbc.272.26.16555. [DOI] [PubMed] [Google Scholar]
- White KA, Lin S, Cotter RJ, Raetz CR. A Haemophilus influenzae gene that encodes a membrane bound 3-deoxy-D-manno-octulosonic acid (Kdo) kinase. Possible involvement of kdo phosphorylation in bacterial virulence. J Biol Chem. 1999;274:31391–31400. doi: 10.1074/jbc.274.44.31391. [DOI] [PubMed] [Google Scholar]
- Zamze SE, Ferguson MAJ, Moxon ER, Dwek RA, Rademacher TW. Identification of phosphorylated 3-deoxy-manno-octulosonic acid as a component of Haemophilus influenzae lipopolysaccharide. Biochem J. 1987;245:583–587. doi: 10.1042/bj2450583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CRH. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.