Summary
How bacteria respond to chromosome replication stress has been traditionally studied using temperature-sensitive mutants and chemical inhibitors. These methods inevitably arrest all replication and lead to induction of transcriptional responses and inhibition of cell division. Here, we used repressor proteins bound to operator arrays to generate a single stalled replication fork. These replication roadblocks impeded replisome progression on one arm, leaving replication of the other arm and re-initiation unaffected. Remarkably, despite robust generation of RecA-GFP filaments and a strong block to cell division during the roadblock, patterns of gene expression were not significantly altered. Consistent with these findings, division inhibition was not mediated by the SOS-induced regulator YneA nor by RecA-independent repression of ftsL. In support of the idea that nucleoid occlusion prevents inappropriate cell division during fork arrest, immature FtsZ-rings formed adjacent to the DNA mass but rarely on top of it. Furthermore, mild alterations in chromosome compaction resulted in cell division that guillotined the DNA. Strikingly, the nucleoid occlusion protein Noc had no discernable role in division inhibition. Our data indicate that Noc-independent nucleoid occlusion prevents inappropriate cell division during replication fork arrest. They further suggest that Bacillus subtilis normally manages replication stress rather than inducing a stress-response.
Keywords: DNA replication, SOS response, nucleoid occlusion, Noc, RecA
Introduction
Eukaryotes and prokaryotes have evolved a diverse collection of mechanisms for monitoring and responding to the integrity of their genomes. For example, when replication does not proceed normally, mechanisms are triggered to repair the replication defect and to delay the cell division cycle until the problem has been resolved (Zhou & Elledge, 2000). In bacteria, protective responses to replication stress are mediated, in part, by surveillance mechanisms that trigger new patterns of gene expression and, in part, by factors that normally function to coordinate cell division with chromosome segregation in every cell cycle. Here, we are concerned with factors that inhibit cell division in response to replication fork arrest.
In most bacteria, cell division initiates with the polymerization of the tubulin homolog FtsZ into a ring-like structure (the Z-ring) at mid cell (Errington et al., 2003). The Z-ring serves as a scaffold for the assembly of additional septal ring components into a division machine, which is capable of coordinated constriction of the cell envelope. In rod-shaped bacteria like E. coli and B. subtilis, spatial control over Z-ring assembly is largely achieved through the combined action of two negative regulatory systems: the Min system and nucleoid occlusion (Adams & Errington, 2009, Harry et al., 2006, Barak & Wilkinson, 2007). The Min system prevents FtsZ-ring assembly at or near the cell poles and nucleoid occlusion prevents its assembly in the vicinity of the chromosome mass (referred to as the nucleoid). As the replicated chromosomes are segregated, nucleoid occlusion is relieved between them allowing division in the mid-cell region. In B. subtilis, the protein that mediates nucleoid occlusion is called Noc (Wu & Errington, 2004); in E. coli it is named SlmA (Bernhardt & de Boer, 2005). These factors perform similar functions but are not homologous. Both associate with the chromosome and act (directly or indirectly) to inhibit Z-ring assembly on top of the DNA. Noc was recently shown to bind to specific sequence elements that are largely excluded from the terminus region of the chromosome (Wu et al., 2009). These results imply that Noc-dependent cell division inhibition is relieved as replication nears completion and the terminus region occupies the mid-cell position. Accordingly, Noc (and likely SlmA (Den Blaauwen et al., 1999, Bernhardt & de Boer, 2005)) not only functions as a spatial regulator of cell division but also acts to couple cell division to chromosome replication and segregation.
In addition to their roles as spatial and temporal regulators of cell division, Noc and SlmA function to inhibit cell division during replication arrest. When replication was blocked using conditional alleles of genes required for replication initiation or chemical inhibitors of replication elongation, cells lacking Noc or SlmA divided on top of the DNA mass more frequently than cells containing these nucleoid occlusion factors (Bernhardt & de Boer, 2005, Wu & Errington, 2004). These inappropriate divisions led to bisection of the chromosome and loss of genomic integrity and/or cell death. Thus, these topological regulators of cell division are thought to comprise one arm of the protective response to replication fork arrest.
The other arm of this protective response involves surveillance mechanisms that respond to replication stress by altering gene expression. The best-characterized and principal pathway is the SOS response, which involves two regulatory factors RecA and LexA (Freidberg et al., 1995). Cells normally have limited amounts of single stranded DNA (ssDNA) at the replication forks, but during replication fork arrest the amount of ssDNA increases. RecA binds to ssDNA forming a nucleoprotein filament. In addition to their role in recombination, repair, and protection of the ssDNA from degradation, these filaments stimulate the autocleavage and subsequent degradation of the transcriptional repressor LexA. Cleavage of LexA results in induction of the SOS regulon. This regulon includes genes involved in DNA recombination, repair and cell division inhibition (Fernandez De Henestrosa et al., 2000, Au et al., 2005). The B. subtilis SOS-induced cell division inhibitor is called YneA (Kawai et al., 2003). The E. coli inhibitor is named SulA (Huisman & D’Ari, 1981). These proteins perform similar functions but are not homologous. SulA directly binds FtsZ preventing polymerization (Mukherjee et al., 1998). The mechanism by which YneA inhibits septal ring assembly remains unknown but does not appear to involve direct inhibtion of FtsZ (Mo & Burkholder, Kawai & Ogasawara, 2006). In addition to the SOS-response, a RecA-independent response to replication arrest has been described in B. subtilis (Goranov et al., 2005, Yasbin et al., 1991). This response is mediated, in part, by the replication initiator protein DnaA. This pathway also contributes to cell division inhibition by down-regulating the expression of the cell division gene ftsL (Goranov et al., 2005). FtsL is an unstable protein (Daniel & Errington, 2000, Bramkamp et al., 2006, Robson et al., 2002) and repression of ftsL transcription prevents cell division during replication inhibition.
In virtually all studies characterizing the response to replication stress, replication was arrested using temperature-sensitive mutations in replication genes or chemical inhibitors that directly or indirectly block replication elongation. Here, we investigate the response of B. subtilis to a single arrested replication fork, a condition most likely encountered in vivo. We did this using repressor proteins bound to an array of operators to generate a “replication roadblock” (Possoz et al., 2006). We show that virtually all cells experiencing these roadblocks generated RecA-GFP filaments and were inhibited for cell division. Remarkably, SOS was not significantly induced during these roadblocks. Moreover, patterns of gene expression were not significantly altered. Furthermore, we show that the block to cell division was not mediated by any of the characterized pathways for division inhibition during replication stress. Specifically, division inhibition in response to fork arrest was not mediated by up-regulation of YneA or down-regulation of ftsL nor was it due to Noc. Our data are most consistent with a model in which Noc-independent nucleoid occlusion prevents inappropriate cell division during fork arrest. We hypothesize that the increase in DNA mass from ongoing replication on the unblocked arm and new rounds of initiation play a vital role in preventing cell division during fork arrest. Our data further suggest that B. subtilis normally manages replication stress rather than inducing a stress-response. We hypothesize that avoidance of mutagenic repair processes associated with these responses is a more common strategy than previously appreciated.
Results
Repressors bound to an operator array block DNA replication and inhibits cell division
In the course of generating new fluorescent repressor protein-operator pairs to visualize chromosomal loci in B. subtilis (Wang et al., 2005, Webb et al., 1997, Viollier et al., 2004), we discovered that the TetR-GFP fusion bound to a large array of tet operators (tetO) caused growth arrest. This discovery arose from our inability to construct strains containing (tetO)120 that constitutively expressed TetR-GFP. Based on the reported replication roadblocks caused by repressor binding to operator arrays in E. coli (Possoz et al., 2006), we repeated our strain constructions in the presence of the inducer anhydrotetracycline (aTC), which binds TetR and reduces its affinity for tetO (Lau et al., 2003). Under these conditions, we successfully generate the desired strains. Interestingly, when we streaked them on LB agar plates lacking aTC, we observed a high rate of spontaneous suppression, resulting in 5–10 colonies in the primary streak of cells (data not shown). In all cases analyzed, the arrays had shrunken to ~24 or fewer copies of tetO (see Experimental Procedures). Finally, the growth arrest and spontaneous suppression were observed regardless of the genomic location of the array insertion. These results are consistent with the findings of Possoz and Sherratt in E. coli, that tight binding of TetR-GFP to an array of tetO operators impairs replication fork movement (Possoz et al., 2006) (Figure 1A).
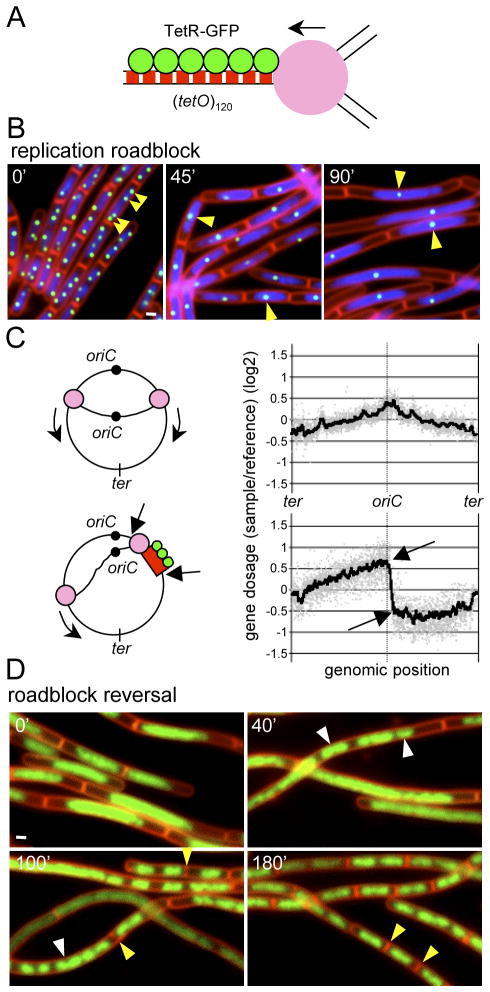
Figure 1.
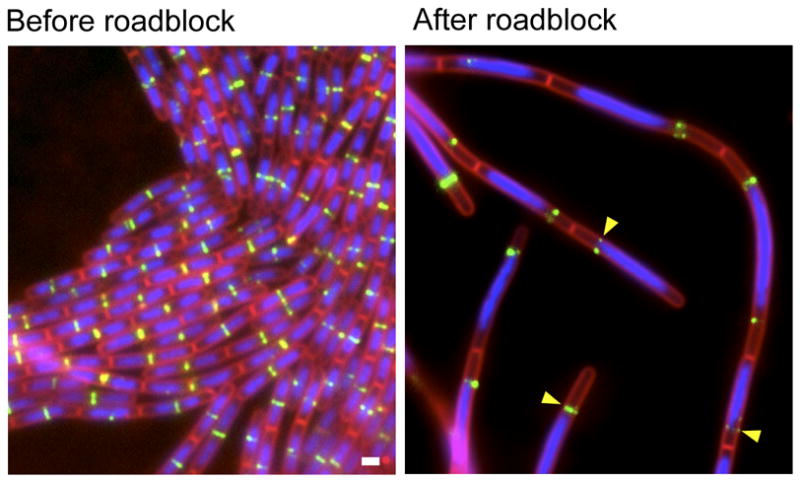
TetR bound to tetO arrays causes a reversible replication block. (A) Schematic representation of a stalled replication fork generated by TetR-GFP (green circles) bound to an array of tet operators (red boxes). The repressor proteins block replisome (pink circle) progression. (B) Images before and after induction of a replication roadblock at +7° (81.7 kb from the origin of replication) in strain BRB63. Time (in min) after removal of the inducer aTC is indicated. Images show membranes stained with FM4-64 (red), DAPI-stained DNA (blue), and TetR-GFP (green) bound to (tetO)120. The reduction in TetR-GFP foci (and therefore the number of +7° loci per cell) following the removal of aTC is highlighed (yellow carets). White bar is 1 μm. (C) Genomic microarray analysis of strain BRB63 before (upper panel) and after (lower panel) the replication roadblock. Gene dosage (log2) relative to a reference DNA is on the y-axis. All the probed genes in the B. subtilis chromosome arranged from −188° to +172° (ter-oriC-ter) are shown (grey dots). The smooth line was generated by plotting the average gene dosage of the 25 genes before and 25 genes after each gene probed. Arrows indicate a position before (+6°) and after (+8°) the site of insertion of the (tetO)120 array (+7°). Schematic representations of the two conditions are shown to the left of the graphs. (D) Chromosome segregation and cell division upon release of fork arrest. The replication roadblock was induced for 90 min in strain BRB63. aTC was added to the culture to release the replication fork and cells were visualized by fluorescence microscopy at the times indicated. Images show membranes stained with FM4-64 (red) and DAPI-stained DNA (false-colored green). New cell division events (yellow carets) and resolution and segregation of the DNA mass into distinct nucleoids (white carets) are highlighted.
To investigate whether a similar replication roadblock was occurring in B. subtilis, we visualized TetR-GFP by fluorescence microscopy in a strain (BRB63) harboring (tetO)120 inserted 81.7 kb (+7°) from the origin of replication. The cells were pre-cultured in rich medium containing aTC to prevent growth arrest. At mid-exponential growth (OD600=0.5), the cells were washed in rich medium lacking inducer, diluted 10-fold in the same medium, and visualized by fluorescence microscopy in a time course (see Experimental Procedures). At early time points following the removal of aTC, the cells appeared similar to wild-type, containing segregated and compacts nucleoids and an average cell length of 3.5 ± 1 μm (compared to 3.2 ± 1 μm in wild-type) (Figures 1B and Sl). As expected, TetR-GFP formed discrete foci (2 to 4 per cell) that co-localized with the DNA (Figures 1B and S1). The number of foci per cell was consistent with the number of origins expected for cells growing in rich medium undergoing multi-fork replication (Wang & Levin, 2009, Yoshikawa et al., 1964). In support of the idea that TetR-GFP bound to the operator prevents replication fork progression, 45 min after removal of aTC, the number of TetR-GFP foci decreased to one per cell in virtually all (>95%) cells in the field (Figures 1B and S1). Moreover, cell division was inhibited resulting in an increased average cell length (4.7 ± 1 μm). By 90 min after the removal of inducer, most cells (~95%) maintained a single TetR-GFP focus and the cells were filamentous with an average cell length of 8.8 ± 1 μm. In addition, the DNA mass appeared unstructured, extending along much of the cell filament (Figures 1B and S1). Finally, after 140 min, cell lysis was observed (Figure S1), consistent with the inability to form colonies on agar plates.
To investigate whether replication of the left chromosomal arm was impaired by a roadblock on the right arm, we inserted a small array of lac operators at −7° (in the yycR gene) on the left arm. LacI-CFP bound to this lacO array did not impair cell growth (data not shown). Examination of LacI-CFP foci bound at −7° during a roadblock at +7° revealed that the left arm was efficiently replicated (Figure S2). Moreover and interestingly, the replicated loci at −7° were distributed along the entire DNA mass (Figure S2). Altogether, these data suggest that TetR-GFP bound to a tetO array in B. subtilis blocks replication fork progression at a single site (the operator insertion site), leading to inhibition of cell division and ultimately cell death.
To more rigorously test whether TetR-GFP bound to (tetO)120 serves as a replication roadblock, we directly monitored DNA content after removal of aTC using DNA microarrays. Genomic DNA was isolated from strain BRB63 that was grown for 45 min in the presence or absence of inducer. After cleavage by restriction endonucleases, the DNA was used as a template to generate Cy5-labeled probes that were hybridized to a B. subtilis oligo array. Genomic DNA from a control strain in which replication initiation was blocked and all ongoing replication had been completed was used as a template to generate Cy3-labeled probes. The ratio of replication origins to replication termini (oriC/ter) of this control was defined as 1.0 and this DNA was used in all microarray experiments as a reference sample (see Experimental Procedures). The upper panel in Figure 1C shows the data from a representative experiment in which BRB63 was grown in the presence of inducer resulting in unimpeded bidirectional replication. In these plots, the fluorescence intensity of each locus is compared to the intensity of the same locus from the reference DNA and the ratio is plotted relative to genomic position. The results were typical for a fast-growing asynchronous population of cells, with an oriC/ter ratio averaging 2.0 (Figures 1C and S3) (Wang et al., 2007). By contrast, in cells in which aTC was removed and TetR-GFP was able to bind to the array of operators inserted at +7°, there was a striking discontinuity in DNA content at the +7° position (Figure 1C, lower panel). Specifically, the DNA content of loci terminus-proximal to the tetO array was significantly reduced relative to loci that were origin-proximal to the array. Moreover, and consistent with the experiments described above, DNA content at the same position on the left arm of the chromosome (−7°) lacked this discontinuity and appeared similar to the strain grown in the presence of aTC. The difference in DNA content on either side of the roadblock was even more pronounced in cells grown for 90 min lacking aTC (Figure S3). Furthermore, similar results were obtained with a strain in which the tetO array was inserted at +130° (Figure S3). These data strongly support the idea that TetR-GFP bound to tetO functions as a replication roadblock that impedes or halts replication progression and does so at a single site. As would be expected for a roadblock, all loci on the right arm of the chromosome downstream of the tetO array had reduced DNA content. Interestingly, sites near the replication terminus had higher DNA content compared to sites closer to the roadblock (Figures 1C and S3). We interpret this to mean that replisomes engaged in replicating the left arm will sometimes replicate beyond the terminus and continue back toward the origin. The small increase in DNA content suggests that replication beyond the termination (ter) sites (Smith & Wake, 1992) is a relatively rare event and/or that replication in this direction is slower due to collisions with RNA polymerase (Wang et al., 2007, Pomerantz & O’Donnell). Replication beyond the terminus during similar roadblocks was also observed in E. coli (Possoz et al., 2006). Altogether, our results indicate that TetR-GFP bound to a tetO array blocks the progression of replication on one arm of the chromosome while leaving the other unimpeded. Furthermore, our data show that this roadblock strongly inhibits cell division.
Efficient chromosome segregation upon release of the replication roadblock
We have shown that during the replication roadblock, DNA content increases and the nucleoid loses its characteristic structure and fills the elongating cell (Figures 1B and 1D). Moreover, an origin-proximal locus (−7°) on the unimpeded arm is actively replicated and these replicated loci appear to be distributed throughout the DNA mass (Figure S2). Although the machinery that governs bacterial chromosome segregation remains ill-defined, we wondered whether this machinery would be capable of segregating the chromosomes into distinct nucleoids, if the replication roadblock was relieved. To address this, we induced the roadblock at +7° for 90 minutes, generating filamentous cells (average cell length of 10 ± 1 μm) that possessed an extended and unstructured DNA mass (Figure 1D, 0 min). aTC was then added to the culture to relieve the block and the cells were monitored by fluorescence microscopy. 40 minutes after replication release, an increase in the size of the DNA mass was readily apparent suggesting active replication of the blocked arm. Visualization of an origin proximal locus on the left arm (−7°) using LacI-CFP bound to a small array of lac operators revealed that the origins had taken on a more segregated appearance (Figure S4). Moreover, we observed some re-organization of the DNA mass into distinct nucleoids (Figures 1D and S4). Strikingly, by 100 minutes, much of the DNA was resolved into discrete and compact nucleoids (Figure 1D). Moreover, incipient septa were visible in regions between some of the nucleoids. 180 min after releasing the replication block, the cells appeared similar to vegetatively growing wild-type cells, with regularly spaced septa between segregated nucleoids (Figure 1D). These results indicate that the segregation machinery is surprisingly robust and can efficiently segregate chromosomes in which replication of the left and right arms are not fully coordinated. Finally, the reversibility of cell division inhibition and the efficient and rapid return to vegetative growth, raised the possibility that the replication roadblock induces a checkpoint-like response that serves a protective function for the cell.
The replication roadblock does not significantly induce the SOS response
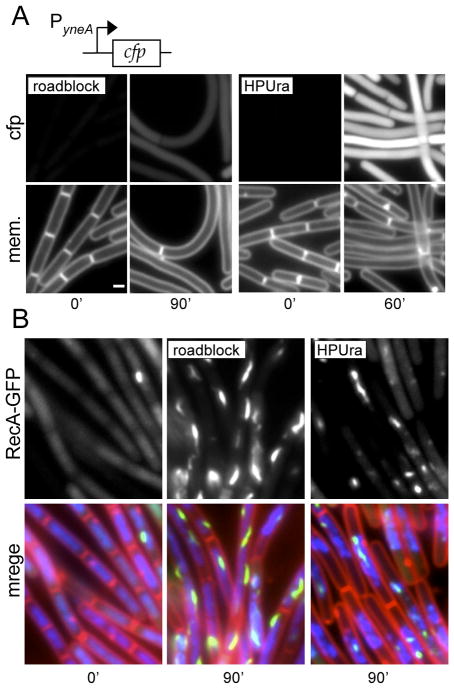
In response to DNA damage and chemical inhibition of DNA replication elongation, B. subtilis induces the SOS response (Au et al., 2005, Goranov et al., 2006). This response involves the RecA-dependent autocleavage of the LexA transcriptional repressor and the subsequent up-regulation of genes involved in DNA repair, DNA recombination and cell division inhibition (Au et al., 2005, Goranov et al., 2006, Friedberg et al., 2006, Kawai et al., 2003). Cell division inhibition is mediated by a protein called YneA that interferes with divisome assembly (Kawai & Ogasawara, 2006, Mo & Burkholder). To investigate whether the replication roadblock induces this checkpoint-like response, we constructed an SOS reporter by fusing a strongly induced SOS-responsive promoter (PyneA) (Au et al., 2005) to the gene encoding the cyan fluorescent protein (cfp). The promoter fusion was then inserted into a nonessential locus in the B. subtilis chromosome. Cells containing the reporter were strongly induced following incubation with classical chemical inhibitors (HPUra and Mytomycin C) that directly or indirectly block replication elongation and are known to induce SOS (Goranov et al., 2006) (Figure 2A and data not shown). Importantly, the expression of cfp was entirely dependent on the SOS response since CFP fluorescence was undetectable in a strain harbouring a LexA mutant (LexAind-) that was unable to undergo autocleavage (data not shown). To determine whether the SOS-response was induced during the replication roadblock, we introduced the SOS reporter into a strain that constitutively expresses TetR-YFP and harbours (tetO)120. Surprisingly, 90 min after the replication roadblock was induced and cells were filamentous, only low levels of CFP were detected (Figure 2A). The fluorescence intensity was >3-fold lower than cells treated with HPUra and >8-fold lower than cells treated with Mytomycin C. Similar results were obtained using a second SOS-responsive promoter fusion (PtagC-cfp) (data not shown).
Figure 2.
The replication roadblock generates RecA-GFP foci and filaments but does not significantly induce the SOS response. (A) Visualization of an SOS-reporter (PyneA-cfp) in response to a replication roadblock (left panel; strain BRB190) or following HPUra treatment (right panel; strain BRB175). Membranes (mem.) were visualized with FM4-64. Exposure times for the fluorescent reporter were identical in all strains and the fluorescent intensities were scaled identically. White bar is 1 μm. A TetR-YFP fusion was used instead of TetR-GFP to generate the roadblock to ensure the emission spectra of the SOS reporter and fluorescent repressor protein were distinct. TetR-YFP was as efficient as TetR-GFP in blocking replication fork progression and inhibiting cell division (data not shown). (B) Localization of RecA-GFP in cells before and after replication arrest in response to a replication roadblock (BRB636, middle panel) or to HPUra (BDR2429, right panel). The left panel shows exponentially growing cells from BDR2429 before addition of HPUra. A time course of RecA-GFP foci/filament formation is shown in Figure S6B. Images show membranes stained with FM4-64 (red), DAPI-stained DNA (blue) and RecA-GFP (green). A TetR-CFP fusion was used instead of TetR-GFP to generate the replication roadblock. TetR-CFP was slightly less efficient at blocking replication fork progression as TetR-GFP (data not shown).
In E. coli and B. subtilis, autocleavage of LexA and subsequent induction of the SOS regulon is stimulated by nucleoprotein filaments of RecA bound to single-stranded DNA (Courcelle & Hanawalt, 2003, Miller et al., 1996). In B. subtilis, these filaments appear to be necessary but not sufficient to trigger SOS-induction (Simmons et al., 2009). To investigate whether or not RecA was engaged in maintaining the stalled forks, we took advantage of a functional RecA-GFP fusion that forms fluorescent foci and thread-like structures (thought to be nucleoprotein filaments) in response to chemical inhibition of replication and DNA damage (Kidane & Graumann, 2005, Renzette et al., 2005, Simmons et al., 2007) (Figure 2B). In support of the idea that RecA helps maintain the stalled fork, 45 minutes after induction of the replication roadblock (using a TetR-CFP fusion), ~60% of the cells had RecA-GFP foci. Strikingly, by 90 minutes, ~80% of the cells had one or more RecA-GFP foci or thread-like structure (Figures 2B and S5). The RecA-GFP foci and filaments generated during the replication roadblock were similar to those observed during chemical inhibition of replication elongation using HPUra (Figures 2B and S5). These results strongly suggest that RecA helps maintain the stalled forks during the replication roadblock. Furthermore, they reinforce the observations of Simmons and colleagues that B. subtilis does not readily induce SOS even in the presence of RecA filaments generated by double strand breaks (Simmons et al., 2009).
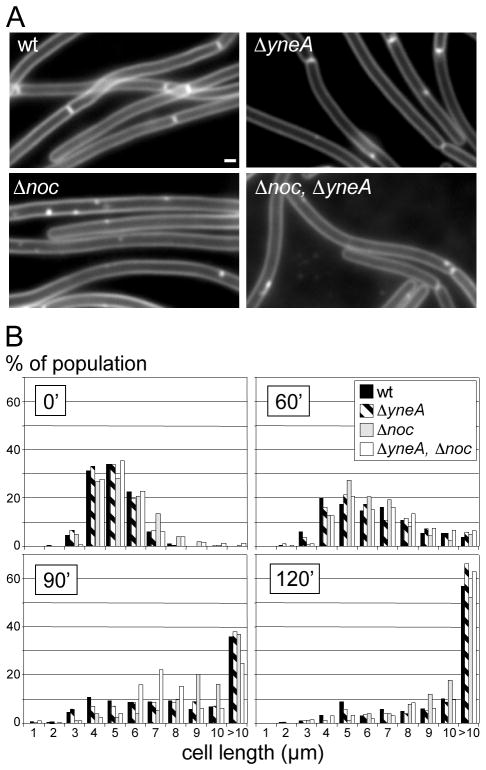
The lack of a significant SOS response during the replication roadblock suggested that YneA is not responsible for the observed inhibition of cell division. To rigorously test this, we monitored the increase in cell length during the replication roadblock in a strain lacking YneA. As expected, the YneA mutant cells filamented in response to the roadblock and did so in a manner that was indistinguishable from wild-type (Figure 3). Similar results were obtained when the replication roadblock was induced in a LexA mutant (LexAind-) that was unable to undergo autocleavage and therefore was unable to induce the entire SOS regulon (Figure S6) (Goranov et al., 2006, Fabret et al., 2002).
Figure 3.
Cell division inhibition during the replication roadblock is independent of YneA and Noc. (A) Representative images of cell filaments after 120′ of replication fork arrest in wild-type (BRB1), or strains lacking yneA (ΔyneA; BRB35), noc (Δnoc; BRB117), or both (Δnoc, ΔyneA; BRB117). White bar is 1 μm. (B) Histograms show the length distribution of the indicated strains during fork arrest. Time (in min) after induction of the roadblock is indicated. >1000 cells (from 3 independent experiments) were measured for each strain and time point.
A RecA-independent response to replication stress also exists in B. subtilis (Goranov et al., 2005). This response is mediated, in part, by the replication initiator protein DnaA. One arm of this response is the inhibition of cell division through transcriptional repression of the cell division gene ftsL. B. subtilis FtsL is an unstable protein (Daniel & Errington, 2000, Bramkamp et al., 2006, Robson et al., 2002) and the down-regulation of ftsL transcription, which is thought to occur by direct binding of DnaA to the ftsL promoter, results in a reduction in cell division (Goranov et al., 2005). To assess whether repression of ftsL was contributing to cell division inhibition during the replication roadblock, we used a strain harboring an IPTG-inducible promoter fusion to ftsL. Using this same fusion, constitutive expression of ftsL in the presence of IPTG was previously shown to suppress division inhibition during replication stress (Goranov et al., 2005). By contrast, constitutive expression of ftsL did not significantly impact the degree of filamentation during the replication roadblock (Figure S7).
Altogether, these results indicate that the RecA-dependent SOS response is not significantly activated during the replication roadblock. These results further show that the strong inhibition of cell division observed during the roadblock is not due to YneA or any other SOS-induced gene, nor is it due to RecA-independent repression of ftsL.
The replication roadblock does not significantly alter gene expression
Since the replication roadblock did not activate SOS, we wondered whether a different checkpoint-like response was induced to inhibit cell division and possibly to help maintain the stalled replication forks. To investigate this, we performed transcriptional profiling on cells undergoing the replication roadblock. We used BRB63 to induce a roadblock at +7° and a reference strain (BRB95) that contained the same tetO array at +7° but lacked TetR-GFP and therefore retained bidirectional replication. Accordingly, the two strains could be treated identically. Both strains were pre-cultured in the presence of aTC and then 45 minutes after removal of the inducer, total RNA was isolated. cDNAs were used as templates to generate Cy3- and Cy5-labelled probes and hybridized to a B. subtilis oligo array (see Experimental Procedures). Surprisingly, we observed very little difference in gene expression in the presence or absence of the roadblock; out of 4200 genes, only 42 were induced and 14 repressed (P < 0.01). The induction ratios measured for these genes were weak; most were less than 2-fold (Table S1). Most of the repression ratios were less than 3-fold; the highest was 4.6-fold. Furthermore, accounting for gene dosage downstream of the roadblock, the induction and repression ratios were even smaller. The low induction ratios and, in some cases, the absence of induction/repression of adjacent genes in the same operon suggest that many of the changes in gene expression we observe are probably not significant. In accordance with the experiments described above using SOS reporters, the expression of only 2 out of ~63 genes in SOS regulon (Au et al., 2005) (ruvA and uvrB) were induced during the replication roadblock and both were induced less than 2-fold. No other genes known to be involved in DNA repair or replication elongation were significantly affected. In addition, expression of all known cell division genes and cell division inhibitors were not measurably altered. Altogether these results suggest that the replication roadblock does not trigger a specific transcriptional response. Thus, these results imply that vegetatively growing B. subtilis can tolerate and manage acute and chronic fork arrest without significantly altering gene expression.
The nucleoid occlusion protein Noc is not required for cell division inhibition during the replication roadblock
Our data suggesting that there is no specific transcriptional response to the replication roadblock left open the question of what prevents cell division during fork arrest. Since the DNA mass fills much of the cell filament during the roadblock, an obvious candidate was the nucleoid occlusion protein Noc (Wu & Errington, 2004). Noc is a site-specific DNA binding protein that inhibits cell division over unsegregated DNA (Wu et al., 2009). To investigate whether Noc was responsible for inhibiting cell division during fork arrest, we monitored the cell length in its absence. Remarkably, the increase in cell length during the roadblock was indistinguishable from wild-type (Figure 3). Moreover, there was no significant septation on top of the DNA mass in the Noc mutant (see below). Finally, cells lacking both YneA and Noc exhibited a similar filamentation phenotype to the single mutants and to wild-type (Figure 3). Thus, although Noc functions to prevent cell division on top of DNA, these results suggest that it plays little role in preventing cell division during replication roadblocks.
YneA and Noc inhibit cell division when replication elongation is blocked by HPUra
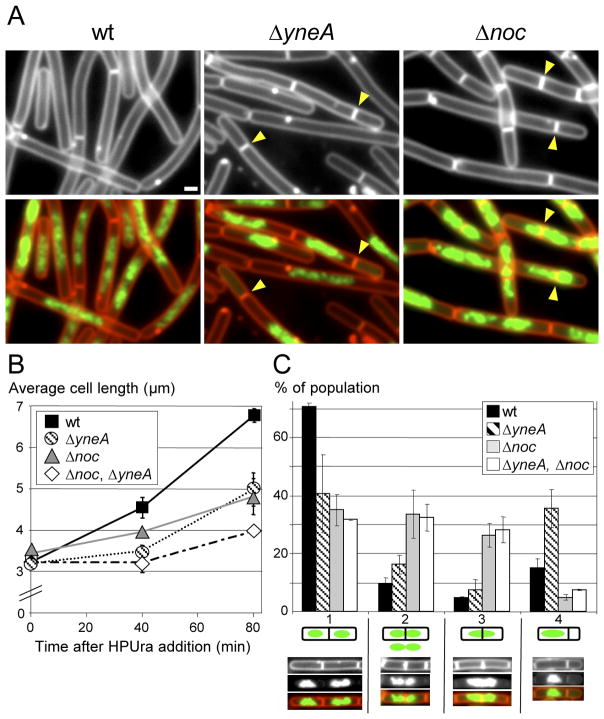
Previous studies using chemical inhibitors of replication suggest that both YneA and Noc function in cell division inhibition when replication elongation is blocked (Moriya et al., 2010, Wu & Errington, 2004, Biller & Burkholder, 2009). Yet neither one appears to act during replication roadblocks. To confirm that these proteins inhibit cell division under our assay conditions, we grew wild-type and cells lacking Noc, YneA, or both in the same rich medium used for the roadblock experiments and treated them with the DNA polymerase III inhibitor HPUra (Brown 1970, Cozzarelli & Low, 1973, Gass et al., 1973). We then monitored the increase in cell length (Figures 4A and B) and the frequency of divisions on top of or adjacent to the nucleoid (Figure 4C). As previously reported, HPUra efficiently inhibited cell division in wild-type cells resulting in filamentation (Biller & Burkholder, 2009, Goranov et al., 2005, Wu & Errington, 2004). The average cell length increased from 3.2 ± 0.1 μm to 6.8 ± 0.2 μm after 80 minutes of the drug treatment (Figure 4B). Since all replication elongation is blocked in the presence of HPUra, DNA content did not increase, however, the nucleoid became elongated and somewhat unstructured (Figure 4A). As expected, cells lacking YneA or Noc continued to undergo some divisions during treatment with HPUra resulting in shorter cells (Figure 4B); their increase in average cell length after 80 min of drug treatment was 5 ± 0.4 μm and 4.8 ± 0.4 μm, respectively. Even more divisions occurred in the double mutant leading to even shorter cells (4 ± 0.1 μm). Next, we examined the frequency of septation on top of or adjacent to the nucleoid. To quantify these phenotypes, we binned the cell division events into four classes (Figure 4C). The first class corresponded to septa between well-segregated nucleoids. The second class was septa that had DNA closely abutting both sides of the division plane. This arrangement most likely arose from a septation event that bisected the DNA mass. However, we could not rule out the possibility that at least in some cases the nucleoids had segregated sufficiently to allow division between them. The third class was septa that unambiguously bisected the nucleoid. Finally, the fourth class was septa that generated an anucleate compartment. These divisions could have arisen from a septum forming adjacent to the nucleoid or from septation on top of the DNA followed by translocation of the bisected DNA into the adjacent daughter cells and/or degradation of the DNA. Consistent with their proposed mechanism of action, YneA mutants exhibited increased septation adjacent to the DNA mass (Class 4), while Noc mutants displayed increased division on top of the nucleoid (Classes 2 and 3) (Figure 4C). Interestingly, cells lacking both YneA and Noc displayed a phenotype similar to the Noc single mutant. This suggests that, under our assay conditions, Noc-mediated nucleoid occlusion plays a more significant role in preventing cell division than YneA. Altogether these results confirm that both Noc and YneA function in cell division inhibition when all replication elongation is blocked and indicate that if they acted to inhibit division during the replication roadblock, our assays would have been sensitive enough to observe it. Thus, these experiments strongly support the idea that during arrest of a single replication fork another mechanism that prevents cell division is at play.
Figure 4.
YneA and Noc inhibit cell division when all replication is blocked by the inhibitor HPUra. Wild-type (BDR11), ΔyneA (BRB12), Δnoc (BRB73) and the double mutant (BRB89) were subjected to HPUra treatment and visualized by fluorescence microscopy. (A) Representative fields of cells 80 min after addition of HPUra. Membranes were stained with FM4-64 (red) and DNA was stained with DAPI (false-colored green). Septation events adjacent to or on top of the nucleoid are highlighted (yellow carets). White bar is 1 μm. (B) Average cell length measurements following HPUra treatment. Values are the average from 3 independent experiment (±SD), >1500 cells were measured for each strain and time point. (C) Histogram quantifying different septation events 80 min after addition of HPUra. Septations were binned into 4 classes described in the text. A representative picture of each class is shown below the x-axis. The fraction of each class was calculated relative to the total numbers of septation events monitored (n=500). Values and standard deviations are based on data from 3 independent experiments.
FtsZ-ring formation is inhibited on top of the DNA mass during the replication roadblock
In a complementary approach aimed at identifying the mechanism of cell division inhibition during the replication roadblock, we investigated the step at which the division process was affected. Since most regulation of cell division is mediated at the level of FtsZ ring formation, we wondered whether Z-rings were present during the replication roadblock. To investigate this, we used a functional ZapA-YFP fusion as a surrogate marker for FtsZ. ZapA interacts directly with FtsZ and is recruited by it to the septal ring (Gueiros-Filho & Losick, 2002). The ZapA-YFP reporter was introduced into a strain expressing TetR-CFP and harboring the (tetO)120 array at +7° and the cells were analyzed by fluorescence microscopy before and after the induction of the replication roadblock. Prior to fork arrest, cells were of normal length, averaging 3.2 ± 1 μm, and contained compact nucleoids (Figure 5). As expected, the ZapA-YFP fusion formed regularly spaced ring-like structures at future division sites. Strikingly, 75 minutes after induction of the roadblock, when the DNA mass had begun to occupy the cell filaments, ZapA-YFP rings were still present in greater than 80% of the cells. Importantly, these rings were positioned near the edges of the DNA masses but almost never on top of the DNA (Figure 5). The same pattern was observed at later times after the induction of the roadblock in even longer filaments (data not shown). Moreover, similar results were obtained when FtsZ was monitored directly using an IPTG-inducible FtsZ-GFP fusion (data not shown). The Z-rings that form during the roadblock are likely to be either immature or not fully functional because two essential division proteins (FtsL and FtsW) that are recruited to the septal ring after FtsZ assembly (Daniel et al., 2006, Gamba et al., 2009) had a diffuse localization in most cells during the roadblock (data not shown). These results and those in the previous sections are consistent with the idea that a nucleoid occlusion mechanism independent of Noc is involved in preventing cell division during the replication roadblock.
Figure 5.
Septal rings do not form on top of the DNA during the replication roadblock. (A) Localisation of the FtsZ-associated protein ZapA (Zap-YFP) before and 90 minutes after induction of the replication roadblock. Images show strain BRB291. Images show membranes stained with FM4-64 (red), DAPI-stained DNA (blue) and ZapA-YFP (false-colored green). ZapA-YFP rings that are present at the edges of the DNA mass are indicated (yellow carets). White bar is 1μM.
Altering chromosome organization or compaction during fork arrest allows cell division on top of the DNA
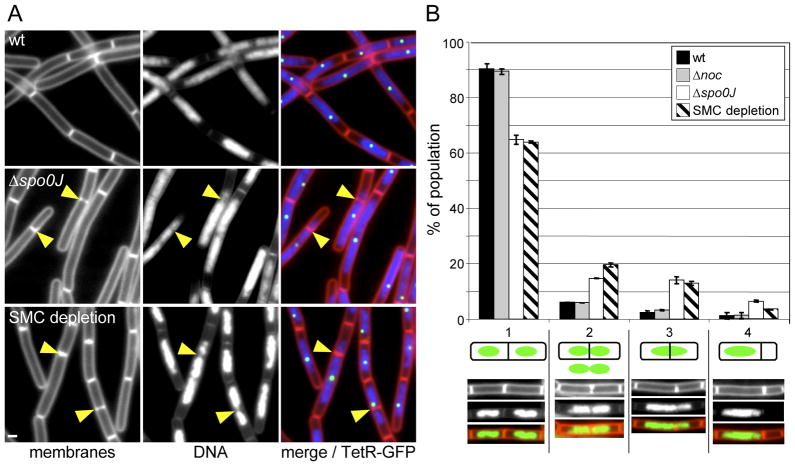
We reasoned that if nucleoid occlusion was preventing cell division during the roadblock, then alterations in the organization or compaction of the DNA would allow septation on top of it. To investigate this, we sought a condition in which chromosome organization and/or compaction was disrupted. It has recently been shown that the B. subtilis partitioning protein ParB (called Spo0J) recruits the SMC chromosome condensation complex to the origin of replication (Sullivan et al., 2009, Gruber & Errington, 2009). Origin-localized SMC appears to help organize the origin region and promote efficient chromosome segregation. Cells lacking Spo0J have a relatively mild defect in both chromosome organization and segregation (Ireton et al., 1994). Accordingly, the Spo0J mutant was an ideal genetic background to investigate whether nucleoid occlusion was preventing cell division during the replication roadblock. We induced the replication roadblock in wild-type cells and cells lacking Spo0J and monitored the increase in cell length and the frequency of divisions on top of and adjacent to the nucleoid. Although the average increase in cell length in the Spo0J mutant was only slightly less than wild type (7.8 ± 4 μm compared to 9 ± 2 μm, 90 minute after induction of the roadblock), the distribution of cell sizes was significantly larger in the absence of Spo0J (Figure S8). For example, at 90 minutes after induction of the roadblock there were 9-fold more cells that were less than 5 μm in length in the Spo0J mutant compared to wild-type. The differences in cell length distribution in the Spo0J mutant reflected an increase in septation on top of or adjacent to the DNA mass (Figure 6A). To quantify the effect, we binned the cell division events into four classes as described above. For these experiments we analyzed cell division 45 minutes after the induction of the roadblock in wild-type cells and cells lacking Spo0J or Noc. In wild-type cells and cells lacking Noc ~90% of the divisions were between well-segregated nucleoids (Class 1) (Figure 6B). By contrast, in the Spo0J mutant only 62% were in this class of divisions. The second class of septa corresponded to those that had DNA closely abutting both sides of the division plane. In wild-type and Noc mutants ~5% of the septa fell into this class compared to 14% for the Spo0J mutant. The third class of septa represented those in which the division plane was unambiguously bisecting DNA. Wild-type and Noc mutant had 2–3% septa of this variety compared to 13% in the Spo0J mutant. The fourth class were septa that generated an anucleate compartment. Again, there were more of these septa in the Spo0J mutant than in wild-type or cells lacking Noc. Finally, in accordance with our analysis of the Noc mutant (Figure 3) and consistent with the idea that Noc plays little role in inhibiting cell division during the replication roadblock, cells lacking both Spo0J and Noc divided on top of or adjacent to the DNA mass at frequencies that were similar to the Spo0J mutant (data not shown).
Figure 6.
Altering chromosome compaction and/or organization relieves cell division inhibition on top of the DNA mass during fork arrest. (A) Representative images obtained 45 min after induction of the replication roadblock in wild-type (strain BRB150), Δspo0J (strain BRB225), and during SMC depletion (strain BRB359). Membranes were stained with FM4-64 (red), DNA was stained with DAPI (blue) and TetR-GFP foci are shown in green. Septa that form on top of the DNA are highlighted (yellow carets). White bar is 1μM. (B) Histogram quantifying the different septation events in the indicated strains 45 min after induction of the replication roadblock. Septations were binned into 4 classes described in the text. A representative picture of each class is shown below the x-axis. The fraction of each class was calculated relative to the total numbers of septation events monitored (n=600). Average values and standard deviations are from 3 independent experiments.
Our results are consistent with the idea that cells lacking Spo0J fail to properly position SMC leading to alterations in chromosome organization and/or compaction and thus promote division on top of the nucleoid. In support of this idea, the localization of a subunit (ScpB-YFP) of the SMC condensin complex (Mascarenhas et al., 2002) was impaired in the absence of Spo0J during the replication roadblock (Figure S9). However, to more directly test this model, we used a strain containing an IPTG-inducible promoter fusion to SMC in which we could deplete the chromosome compaction protein. Depletion of SMC in rich medium leads to highly unstructured nucleoids, the production of anucleate cells, and ultimately cell lysis (Britton et al., 1998)(K.A.M and DZR, unpublished). Cells grown for ~3.5 hours in the absence of IPTG begin to display aberrant nucleoid morphologies (K.AM. and D.Z.R. unpublished). Accordingly, for our experiments, we grew cells in the absence of IPTG for only one hour prior to induction of the roadblock. At this time point the nucleoid morphology was indistinguishable from wild-type (data not shown). We then examined the cells 60 minutes later. Under these conditions the average cell length was similar to wild-type cells undergoing the roadblock. However, we could easily detect septation events adjacent to and on top of the nucleoid (Figure 6A). The increase in these classes of cells was similar to the Spo0J mutant (Figure 6B). Thus, mild perturbations to DNA organization and/or compaction result in relief of cell division inhibition during the roadblock. Altogether, these results indicate that, during fork arrest, a Noc-independent nucleoid occlusion mechanism prevents cell division on top of the DNA.
Discussion
In this study we have employed arrays of tet operators and fluorescent fusions to the tet repressor protein to examine the consequences of arresting a single replication fork in Bacillus subtilis. Although it is unlikely that the replisome would encounter a tandem array of repressor proteins in vivo, transient roadblocks leading to replication stalling are likely to be quite common (Mirkin & Mirkin, 2007, Azvolinsky et al., 2009, Deshpande & Newlon, 1996). Importantly, the arrest of a single replication fork (rather than inhibition of all replication) due to DNA lesions or roadblocks is probably the most common type of replication inhibition experienced by the cell. Accordingly, these unnatural roadblocks have something to teach us about how bacteria respond to and/or manage fork arrest. The two principal findings from our characterization of these roadblocks are i) B. subtilis manages fork arrest rather than inducing a stress response and ii) cell division inhibition during fork arrest is not mediated by any of the previously characterized pathways that prevent division during replication stress. Specifically, cell division inhibition during the roadblock was not mediated by up-regulation of YneA or down regulation of ftsL, nor was it due to the nucleoid occlusion protein Noc. Our data showing that relatively mild perturbations to chromosome organization or compaction lead to division on top of the DNA (and chromosome bisection) during fork arrest, strongly supports the idea that nucleoid occlusion is preventing division.
The division inhibition during replication roadblocks that we report here is perhaps the most striking example of nucleoid occlusion that is independent of Noc, however, it is not the first. A Noc-independent pathway for nucleoid occlusion was originally hypothesized by Wu and Errington based on their observations that FtsZ-ring assembly remained biased toward internucleoid regions in cells mutant for both Noc and the Min system (Wu & Errington, 2004). A similar proposal was put forth by Bernhardt and de Boer based on their analysis of E. coli cells lacking SlmA and the Min system (Bernhardt & de Boer, 2005). Moreover, recent evidence from Harry and co-workers suggests that in B. subtilis mechanisms in addition to Noc control Z-ring positioning during the early stages of DNA replication (Moriya et al., 2010).
How the nucleoid occludes the cell division machinery was the subject of speculation for many years. The discovery of Noc in B. subtilis and SlmA in E. coli provided a compelling solution and largely put to rest much of this speculation. However, our data force us to return to the question of how the nucleoid can prevent cell division. Two types of models have been put forth previously. The first, originally proposed by Mulder and Woldringh (Mulder & Woldringh, 1989) involves steric crowding at the membrane due to coupled transcription-translation and membrane protein insertion (called, transertion). In this model, the chromosome (through the distribution of actively transcribed genes encoding co-translationally inserted membrane proteins) imparts an organization to ribosomes coupled to the sec translocons. These localized and well-distributed “areas of transertion” would serve to disrupt the division machinery (Woldringh, 2002, Mulder & Woldringh, 1989). Although definitive proof for the existence of transertion from chromosomal loci remains lacking, it seems plausible that active translocons interacting with ribosomes could be organized by the underlying DNA. Moreover, indirect evidence suggests that plasmids actively transcribing genes encoding membrane proteins are tethered (or constrained) at the membrane (Lynch & Wang, 1993). In the case of the replication roadblock, it is also possible that the increase in total DNA due to ongoing replication of the unimpeded arm, could result in an increase in the volume of the nucleoid and in a more direct fashion cause crowding at the membrane. The second type of model involves additional DNA binding proteins analogous to Noc and SlmA that inhibit cell division on top of the DNA. To date, only Noc and SlmA have been identified as protein mediators of nucleoid occlusion. However, it is possible that functionally redundant DNA binding protein have inhibitory activity. Accordingly, individual mutants would not have significant phenotypic consequences and therefore would have eluded discovery. Moreover, if the activity of these inhibitors requires site-specific DNA binding as is the case for Noc (Wu & Errington, 2004), this could buffer strong cell division inhibitory phenotypes upon overexpression. Thus, these genes would not have been identified through overexpression screens. Either model: crowding due to transertion or DNA binding proteins that inhibit division could explain the relief of cell division inhibition upon loss of compaction or organization of the chromosome. Under these conditions, transertion areas and/or nucleoid occlusion proteins might loose their inhibitory distribution leading to bisection of the DNA mass. Distinguishing between these two models is the challenge for the future. However, whatever the molecular mechanism, our data indicate that Noc-independent nucleoid occlusion plays a vital role in preventing chromosome bisection during fork arrest.
Finally, we have shown that the replication roadblock generates robust RecA-GFP foci and/or filaments in virtually all cells. These structures are similar to those generated after DNA double-strand breaks and in cells blocked for replication elongation by HPUra (Kidane & Graumann, 2005, Simmons et al., 2007). Single-stranded DNA bound by RecA at the stalled fork could arise from any number of reasons including fork regression, gaps in the lagging strand, or uncoupling of the replicative helicase from the replisome (Courcelle & Hanawalt, 2003, Freidberg et al., 1995). However and interestingly, despite the generation of these filaments, SOS was not significantly induced. These results are in accordance with the observations of Simmons and colleagues who compared RecA-GFP foci formation and SOS induction in cells in which double strand breaks were generated by a restriction endonuclease or by ionizing radiation (Simmons et al., 2009). In both cases, double-strand breaks efficiently triggered the formation of RecA-GFP foci and filaments yet only a sub-population of the B. subtilis cells induced SOS. By contrast, in E. coli most cells that generated RecA-GFP foci/filaments induced SOS (Simmons et al., 2009). Collectively, these results indicate that B. subtilis has set a higher threshold than E. coli for inducing this response. Replication roadblocks in E. coli similarly generate blocks to cell division (Possoz et al., 2006), it will be interesting to see whether or not SOS is induced. Our data and those of Simmons suggest that B. subtilis has evolved mechanisms to repair damage and manage fork arrest without assuming the mutagenic cost of inducing this response. We hypothesize that avoidance of mutagenic repair processes associated with these responses may be a more common strategy among microbes than previously appreciated.
Experimental procedures
General Methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al., 1983). E. coli strains were TG1, DH5α or AB1157. To visualize the localization of fluorescent fusions during vegetative growth, strains were grown in CH (casein hydrolysate) medium (Harwood & Cutting, 1990) at 37°C. Unless otherwise indicated, B. subtilis strains harbouring tetO arrays and constitutively expressing TetR-GFP were grown in the presence of aTC (1 ng/ml). Cells grown in the presence of aTC exhibited weak TetR-GFP foci; no foci were observed in strains lacking the tetO arrays. Importantly, there was no discernable effect of aTC on wild-type cells. Mitomycin C (Sigma) was used at 1 μg/ml. HPUra [6-(p-HydroxyPhenylazo)-Uracil] a generous gift from William Burkholder was used at 40 μg/ml. Spontaneous suppressors of B. subtilis strains harbouring (tetO)120 arrays and constitutively expressing TetR-GFP were analyzed by PCR to assess the size of the array. In all cases, the PCR products were less than 950 bp indicating that the array had shrunken to 24 or fewer copies of tetO. A description of strains (Table S2), plasmids (Table S3), and oligonucleotide sequences (Table S4) can be found in supplemental material.
Induction of the replication roadblock
To induce the replication roadblock, cells expressing fluorescent fusions to TetR and harboring a (tetO)120 array were pre-cultured in CH medium containing 1 ng/ml aTC at 22°C. Cells in mid-logarythmic growth were washed 4 times in fresh CH medium lacking aTC and inoculated in the same medium at an OD600=0.05. The culture was then incubated at 37°C. This was defined as time zero.
Genomic microarrays
Genomic microarray analysis was carried as described previously (Wang et al., 2007). Briefly, genomic DNA was isolated from the strains of interest and cleaved to completion with the restriction endonuclease HaeIII. The reference DNA was from a temperature sensitive replication initiation mutant (BNS1733, dnaB134) grown at the non-permissive temperature (42°C) for 1 hr to ensure that all rounds of replication were complete. The HaeIII-cleaved reference DNA was used as a template to generate Cy3-labelled probes and the experimental DNAs were used as template to generate Cy5-labelled probes (GE Healthcare). The fluorescently labelled probes were hybridized to an oligonucleotide microarray (Sigma Genosys) containing a 70mer for each gene in the B. subtilis genome. The hybridized array was scanned using a Genepix 4000B scanner (Axon, Software v. 5.1), and data was analyzed in Microsoft Excel. Intensities from the 532 nm channel were normalized to the reference (635 nm channel) prior to subsequent analysis. Each data point on the smooth line in Figures 1C and S2 represents an average of the normalized ratios of the 25 genes on either side of each locus. The oriC/ter ratios were calculated by dividing the average normalized intensities of the 25 genes on either side of dnaA (oriC) by the normalized intensities of 25 genes on either side of rtp (ter).
Transcriptional profiling
Samples of cells taken for RNA isolation were pelleted, flash frozen in N2(l) and stored at −76°C. RNA was extracted by a hot acid-phenol isolation method as previously described (Fawcett et al., 2000). Labelled cDNA was generated by incorporation of Cy3- or Cy5-labelled dUTP. Briefly, random hexamers (1 μg) (Invitrogen) were annealed to purified RNA (25 μg) at 70°C for 10 min and cooled on ice for 2 min. cDNA was prepared using Superscript III Reverse Transcriptase (Invitrogen), 1mM dATP, 1mM dGTP, 1mM dCTP and 0.4mM dTTP and 2mM aminoallyl-dUTP (Ambion). The reaction was incubated at 42°C for 1 hr followed by the addition of NaOH to hydrolyze the RNA. The reaction was then inactivated at 70°C for 15 min. After neutralization with HCl, the cDNAs were purified using a Qiagen MinElute column. Approximately 2 μg of cDNA was obtained per sample. The cDNA was labelled with Alexa Fluor 555 (Cy3) or Alexa Fluor 647 (Cy5) (Invitrogen) for 2 hr in the dark, as described by the manufacturer. Differentially labelled cDNAs were mixed and unincorporated nucleotides were removed using a Qiagen MinElute column. The final elution volume was 12 μl and contained 200–400 ng of labelled Cy-dUTP*-cDNAs. The entire sample was used to probe a B. subtilis oligonucleotide microarray as described previously (Wang et al., 2006).
Microarrays were scanned using a Genepix 4000B scanner (Axon, Software v. 5.1) and the data were analyzed using the Rosetta Resolver software (Agilent). After combining data from 4 independent experiments, only the genes that matched the following criteria were considered significantly affected: the fold change was found above a cut-off of 1.6 in at least two of the four experiments and the P-value was below 0.01.
Fluorescence microscopy
Fluorescence microscopy was performed as previously described (Doan et al., 2005). Fluorescent signals were visualized with a phase contrast objective UplanFLN 100X and captured with a monochrome CoolSnapHQ digital camera (Photometrics) using Metamorph software version 6.1 (Universal Imaging). Exposure times were typically 500–1000 ms for GFP, CFP and YFP protein fusions. Membranes were stained with FM4-64 (Molecular Probes) at a final concentration of 3 μg/ml and imaged with exposure times of 300 ms. DNA was stained with DAPI (Molecular Probes) at a final concentration of 2 μg/ml and imaged with a typical exposure time of 200 ms. Fluorescence images were analysed, adjusted and cropped using Metamorph v 6.1 software (Molecular Devices).
Constructions of B. subtilis strains expressing fluorescent TetR derivatives
All constructs containing constitutively expressed TetR-GFP (or CFP or YFP) were generated by direct transformation of ligation products into B. subtilis.
BKM830 [amyE::Pspac(C)-tetR-gfp (spec)] was generated by direct transformation into B. subtilis (PY79) of the following ligation product: a HindIII-BamHI fragment containing tetR-gfp from pDR151 and pMF35 cut with HindIII and BamHI. pMF35 [amyE::Pspac(c)-gfp] (Fujita & Losick, 2002) contains the Pspac promoter but lacks the lacI gene rendering the promoter constitutive. pDR151 [amyE::PspoIID-tetR-gfp (spec)] (Marquis et al., 2008) contains tetR-gfp and an optimized ribosome-binding site (Vellanoweth & Rabinowitz, 1992).
BRB132 [amyE::Pspac(C)-tetR-cfp (spec)] was generated by direct transformation into B. subtilis (PY79) of the following ligation product: an EcoRI-HindIII fragment containing Pspac(C) from pMF35 and pNS112 cut with EcoRI and HindIII. pNS112 [amyE::PspoIIE-tetR-cfp (spec)] was a generous gifts from N. Sullivan.
BRB139 [amyE::Pspac(C)-tetR-yfp (spec)] was generated by direct transformation into B. subtilis (PY79) of the following ligation product: an EcoRI-HindIII fragment containing Pspac(C) from pMF35 and pNS114 cut with EcoRI and HindIII. pNS114 [amyE::PspoIIE-tetR-yfp (spec)] was a generous gifts from N. Sullivan.
BRB321 [yhdG::Ppen-tetR-gfp (spec)] was generated by direct transformation into B. subtilis of the following ligation product: a HindIII-BamHI fragment containing tetR-gfp from pDR151 and pRB045 cut with HindIII and BamHI. pRB045 [yhdG::Ppen (spec)] was generated in a two-way ligation with an EcoRI-HindIII PCR product containing the Ppen promoter (oligonucleotide primers oDR699 and oDR700, and pKL190 [thrC::Ppen-lacI(Δ11)-cfp (JF)] as a template (Lemon & Grossman, 2000), and pBB278 cut with EcoRI and HindIII. pBB278 [yhdG::spec] is an ectopic integration vector for double cross-over insertions into the nonessential yhdG locus (B. Burton and D.Z.R., unpublished).
BRB355 [yhdG::Pspac(c)-tetR-cfp (erm)] was generated by direct transformation into B. subtilis of the following 3-way ligation product: an EcoRI-HindIII fragment containing Pspac(C) from pMF35, a HindIII-BamHI fragment containing tetR-cfp from pNS112 and pBB279 cut with EcoRI and BamHI. pBB279 [yhdG::erm] is an ectopic integration vector for double cross-over insertions into the nonessential yhdG locus (Burton and D.Z.R., unpublished).
Plasmid constructions
pRB012 [yneA-yneB-ynzC::phleo] was generated in a two-way ligation using a SalI-BamHI fragment containing a phleomycin resistance cassette (from pKM080, K.A.M. and D.Z.R. unpublished) and pRB002 cut with SalI and BamHI. pRB002 [yneA-yneB-ynzC::cat] was generated in a two-way ligation using a SalI-EagI PCR product containing the genomic region downstream of the yneA-yneB-ynzC operon (oligonucleotide primers oRB3 and oRB4 and wild-type genomic DNA as a template) and pRB001 cut with SalI and EagI. pRB001 was generated in a two-way ligation using a EcoRI-BamHI PCR product containing the genomic region upstream of the yneA-yneB-ynzC operon (oligonucleotide primers oRB1 and oRB2 and wild-type genomic DNA as a template) and pKM074 cut with EcoRI and BamHI. pKM074 is a plasmid containing restriction sites flanking the cat gene (K.A.M. and D.Z.R., unpublished). Oligonucleotides oRB5 and oRB6 were used to verify the correct disruption of the yneA-yneB-ynzC operon once pRB12 was inserted into the Bacillus subtilis chromosome.
pRB013 [yneA-yneB-ynzC::erm] was generated in a two-way ligation using a SalI-BamHI fragment containing an erythromycin cassette (from pKM082, K.A.M. and D.Z.R. unpublished) and pRB002 [yneA-yneB-ynzC::cat] cut with SalI and BamHI.
pRB015 [sacA::PyneA-cfp (erm)] was generated in a three-way ligation with an EcoRI-HindIII PCR product containing the yneA promoter (oligonucleotide primers oRB14 and oRB15 and wild-type genomic DNA as a template), a HindIII-BamHI fragment containing cfp with codons optimized for expression in B. subtilis (Doan et al., 2005) and pKM062 cut with EcoRI and BamHI. pKM062 is an ectopic integration vector for double cross-over insertions into the nonessential sacA locus based on pRM158 (Middleton & Hofmeister, 2004) (K.A.M. and D.Z.R., unpublished).
pRB020 [+7° Ω (tetO)120 (cat)] was generated in a two-way ligation with an XhoI-HindIII fragment containing (tetO)120 (from pKM195 [yycR::(tetO)120 (erm)]) and pRB017 cut with XhoI and HindIII. pRB17 [+7° Ω (cat)] was generated in a two-way ligation with an EcoRI-BamHI PCR product containing the ftsH-yacB intergenic region (oligonucleotide primers oRB18 and oRB19 and wild-type genomic DNA as a template) and pER19 cut with EcoRI and BamHI. pER19 is a pUC19 derivative containing a cat cassette (Ricca, Losick 1992). pKM195 [yycR::(tetO)120 (erm)] was generated in a two-way ligation with a HindIII-XhoI PCR product containing the (tetO)120 sequence (oligonucleotides oDR458 and oDR459 and pLAU44 as template (Lau et al., 2003)) and pNS043 cut with HindIII and XhoI. pNS043 [yycR::erm] is an ectopic integration vector for double cross-over insertions into the nonessential yycR locus (N. Sullivan. and D.Z.R., unpublished).
pRB032 [+7° Ω (tetO)120 (phleo)] was generated in a two-way ligation with an XhoI-HindIII fragment containing (tetO)120 from pKM195 and pRB030 cut with XhoI and HindIII. pRB030 [+7° Ω (phleo)] was generated in a two way ligation with an EcoRI-HindIII fragment containing the ftsH-yacB intergenic region from pRB017 and pUPh19 cut with EcoRI and HindIII. pUPh19 was a generous gift from Thierry Doan and is a pUC19 derivative containing a phleomycin resistance cassette.
pRB033 [yndN Ω kan] was generated in a two-way ligation with an EcoRI-BamHI PCR product containing the 3′ end of the yndN gene (oligonucleotide primers oRB035 and oRB036 and wild-type genomic DNA as a template) and pUK19 cut with EcoRI and BamHI. pUK19 is a pUC19 derivative containing a kanamycin resistance cassette (Roels & Losick, 1995). The yndN gene is adjacent to the lexA gene and provides a linked antibiotic marker to lexA and lexA(G92D) (Fabret et al., 2002).
pRB046 [yhdG::PxylA-zapA-yfp (erm)] was generated in a three-way ligation with a HindIII-XhoI PCR product containing zapA and an optimized RBS (oligonucleotide primers oRB039 and oRB040, and wild-type genomic DNA as a template), an XhoI-BamHI PCR product containing yfp (oligonucleotide primers oDR78 and oDR79, and pKL183 as a template, (Lemon & Grossman, 2000), and pRB038 cut with HindIII and BamHI. pRB038 [yhdG::PxylA-xylR (erm)] was generated in a two-way ligation with an EcoRI-BamHI fragment containing PxylA promoter and xylR from pDR150 [amyE::PxylA- xylR (spec)] (D.Z.R., unpublished) and pBB279 cut with EcoRI and BamHI. pBB279 [yhdG::erm] is an ectopic integration vector for double cross-over insertions into the nonessential yhdG locus (Burton and Rudner, unpublished).
Supplementary Material
Acknowledgments
We thank members of the Rudner lab past and present and Sandra Castang for valuable discussions and support. We thank Lyle Simmons, Tom Bernhardt, Johannes Walter for valuable discussions, Nora Sullivan for plasmids and help with the genomic microarrays, Reddy Gali for help with data analysis, Eric Rubin for the use of his fluorescent scanner, William Burkholder for HPUra, Alan Grossman and Jeff Errington for strains. Support for this work comes in part from the National Institute of Health Grants GM086466 and GM073831and funds from a John and Virginia Kaneb Award.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, O’Brien TC, Shah A, Tierney JT, Tomm LL, O’Gara TM, Goranov AI, Grossman AD, Lovett CM. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak I, Wilkinson AJ. Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol Rev. 2007;31:311–326. doi: 10.1111/j.1574-6976.2007.00067.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Burkholder WF. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol Microbiol. 2009;74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NC. 6-(p-hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci U S A. 1970;67:1454–61. doi: 10.1073/pnas.67.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- Cozzarelli NR, Low RL. Mutational alteration of Bacillus subtilis DNA polymerase 3 to hydroxyphenylazopyrimidine resistance: polymerase 3 is necessary for DNA replication. Biochem Biophys Res Commun. 1973;51:151–157. doi: 10.1016/0006-291x(73)90521-4. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol Microbiol. 2000;36:278–289. doi: 10.1046/j.1365-2958.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Noirot-Gros MF, Noirot P, Errington J. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J Bacteriol. 2006;188:7396–7404. doi: 10.1128/JB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Timing of FtsZ assembly in Escherichia coli. J Bacteriol. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C, Ehrlich SD, Noirot P. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol Microbiol. 2002;46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Freidberg EC, Walker GC, Siede W. DNA Repair. ASM Press; Washington, DC: 1995. [Google Scholar]
- Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass KB, Low RL, Cozzarelli NR. Inhibition of a DNA polymerase from Bacillus subtilis by hydroxyphenylazopyrimidines. Proc Natl Acad Sci U S A. 1973;70:103–107. doi: 10.1073/pnas.70.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci U S A. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Kuester-Schoeck E, Wang JD, Grossman AD. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J Bacteriol. 2006;188:5595–5605. doi: 10.1128/JB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E, Monahan L, Thompson L. Bacterial cell division: the mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Wiley; New York: 1990. [Google Scholar]
- Huisman O, D’Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Ireton K, Gunther NWt, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Moriya S, Ogasawara N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol Microbiol. 2003;47:1113–1122. doi: 10.1046/j.1365-2958.2003.03360.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Ogasawara N. Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division. Microbiology. 2006;152:1129–1141. doi: 10.1099/mic.0.28497-0. [DOI] [PubMed] [Google Scholar]
- Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J Cell Biol. 2005;170:357–366. doi: 10.1083/jcb.200412090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Lynch AS, Wang JC. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J Bacteriol. 1993;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis KA, Burton BM, Nollmann M, Ptacin JL, Bustamante C, Ben-Yehuda S, Rudner DZ. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev. 2008;22:1786–1795. doi: 10.1101/gad.1684008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R, Hofmeister A. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid. 2004;51:238–245. doi: 10.1016/j.plasmid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Miller MC, Resnick JB, Smith BT, Lovett CM., Jr The bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein. J Biol Chem. 1996;271:33502–33508. [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo AH, Burkholder WF. YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. J Bacteriol. 2010;192:3159–3173. doi: 10.1128/JB.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S, Rashid RA, Rodrigues CD, Harry EJ. Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Mol Microbiol. 2010;76:634–647. doi: 10.1111/j.1365-2958.2010.07102.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E, Woldringh CL. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Filipe SR, Grainge I, Sherratt DJ. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- Robson SA, Michie KA, Mackay JP, Harry E, King GF. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol Microbiol. 2002;44:663–674. doi: 10.1046/j.1365-2958.2002.02920.x. [DOI] [PubMed] [Google Scholar]
- Roels S, Losick R. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J Bacteriol. 1995;177:6263–6275. doi: 10.1128/jb.177.21.6263-6275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Goranov AI, Kobayashi H, Davies BW, Yuan DS, Grossman AD, Walker GC. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J Bacteriol. 2009;191:1152–1161. doi: 10.1128/JB.01292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Grossman AD, Walker GC. Replication is required for the RecA localization response to DNA damage in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:1360–1365. doi: 10.1073/pnas.0607123104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Wake RG. Definition and polarity of action of DNA replication terminators in Bacillus subtilis. J Mol Biol. 1992;227:648–657. doi: 10.1016/0022-2836(92)90214-5. [DOI] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Berkmen MB, Grossman AD. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:5608–5613. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Cheo D, Bayles KW. The SOB system of Bacillus subtilis: a global regulon involved in DNA repair and differentiation. Res Microbiol. 1991;142:885–892. doi: 10.1016/0923-2508(91)90069-m. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, O’Sullivan A, Sueoka N. Sequential Replication of the Bacillus Subtilis Chromosome. 3. Regulation of Initiation. Proc Natl Acad Sci U S A. 1964;52:973–980. doi: 10.1073/pnas.52.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.