Abstract
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family of neurotrophic factors, has important functions in the peripheral and central nervous system of vertebrates. We have generated bacterial artificial chromosome (BAC) transgenic mice harboring 207 kb of the rat BDNF (rBDNF) locus containing the gene, 13 kb of genomic sequences upstream of BDNF exon I, and 144 kb downstream of protein encoding exon IX, in which protein coding region was replaced with the lacZ reporter gene. This BDNF-BAC drove transgene expression in the brain, heart, and lung, recapitulating endogenous BDNF expression to a larger extent than shorter rat BDNF transgenes employed previously. Moreover, kainic acid induced the expression of the transgenic BDNF mRNA in the cerebral cortex and hippocampus through preferential activation of promoters I and IV, thus recapitulating neuronal activity-dependent transcription of the endogenous BDNF gene. genesis 48:214–219, 2010. © 2010 Wiley-Liss, Inc.
Keywords: neurotrophin, transcription, promoter, BAC, transgenic mouse, kainic acid
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family of proteins, supports the survival and differentiation of certain neuronal populations during development (Bibel and Barde, 2000; Binder and Scharfman, 2004). In the adult, BDNF regulates long-term potentiation of synapses, thus playing a key role in long-term memory formation (Lu et al., 2008). BDNF was originally isolated from the brain, but it is also expressed in the peripheral nervous system and non-neural tissues (Binder and Scharfman, 2004). Changes in BDNF gene expression accompany and contribute to the development of various disorders of the nervous system (Bibel and Barde, 2000).
The BDNF gene contains multiple promoters that initiate the transcription of a number of distinct mRNAs, each of which contains an alternative 5′ untranslated exon spliced to a common 3′ protein coding exon. In addition, the protein coding exon employs two different polyadenylation sites that give rise to mRNA species with 3′ untranslated regions (UTRs) of different lengths. Alternative promoter usage, differential splicing, and the use of two different polyadenylation sites within each of the transcription units generate at least 22 different BDNF mRNAs in rodents and 34 BDNF mRNAs in human that encode the same mature BDNF protein (Aid et al., 2007; Pruunsild et al., 2007). It has been shown that the subcellular localization of BDNF mRNAs and its regulation by neuronal activity depends on the 5′ exon and 3′ UTRs used in the transcript (An et al., 2008; Chiaruttini et al., 2008). In addition, it has been shown that BDNF mRNAs containing the short 3′ UTRs are more enriched in polysomal fraction isolated from total brain than BDNF mRNAs with the long 3′ UTRs suggesting that they are more efficiently translated (Timmusk et al., 1994). Numerous regulatory elements involved in the regulation of BDNF expression in vitro and in vivo have been identified and characterized in different BDNF promoters. Transcription factors such as REST (Timmusk et al., 1999; Zuccato et al., 2003), CREB (Shieh et al., 1998; Tao et al., 1998), NFkB (Lipskyet al., 2001), MEF2 (Flavell et al., 2008), NPAS4 (Lin et al., 2008), bHLHB2 (Jiang et al., 2008), and MeCP2 (Chen et al., 2003; Marti-nowich et al., 2003) have been shown to regulate BDNF expression in a promoter-specific manner. However, the genomic regions including all necessary cis-acting elements responsible for the tissue-specific and activity-dependent BDNF gene regulation in vivo remain poorly characterized. A few studies have addressed these issues using transgenic mouse models (Funakoshi et al., 1998; Guillemot et al., 2007; Koppel et al., 2009; Timmusk etal., 1995, 1999).
In the present study, we have generated a transgenic mouse line using a bacterial artificial chromosome (BAC) clone containing 207 kb of rat BDNF (rBDNF) locus, encompassing the genomic region from 13 kb upstream of rBDNF exon I to 144 kb downstream of rBDNF coding exon. Neighboring genes of the rBDNF gene lie 151 kb upstream (Ifna4) and 190 kb downstream (Sqrdl) from it and therefore no additional genes/promoters were included in the BAC construct. To facilitate detection of transgene expression, we replaced the protein coding region of exon IX in the rBDNF-BAC with lacZ reporter gene (Fig. 1a). This should lead to the expression of functional β-galactosidase protein but not a BDNF-lacZ fusion protein. Functional β-galactosidase protein encoded by the lacZ reporter gene in rBDNF-lacZ-BAC was detected by transient expression in COS-7 cells (data not shown).
FIG. 1.
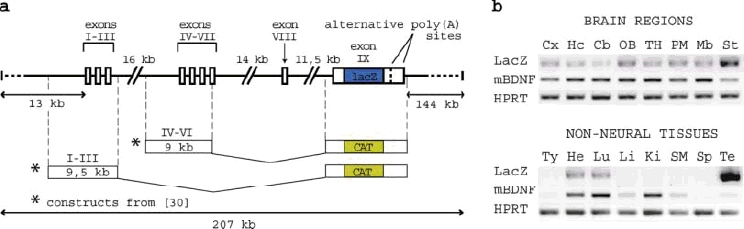
(a) Schematic diagram of the BAC construct used for generating rBDNF-lacZ-BAC transgenic mice (thick lines). White boxes represent untranslated sequences and the blue filled box represents lacZ reporter gene that replaces the BDNF coding sequence. rBDNF-CAT constructs (I–III and IV–VI) used by Timmusk et al. (1995) to generate rBDNF transgenic mice are shown with asterisks. (b) RT-PCR analysis of rBDNF-lacZ mRNA expression driven by rBDNF promoters in transgenic mouse tissues. Abbreviations: mBDNF, mouse BDNF; HPRT, hypoxanthine phosphoribosyltransferase 1; Cx, cortex; Hc, hippocampus; Cb, cerebellum; OB, olfactory bulb; TH, thalamus and hypothala-mus; PM, pons/medulla; Mb, midbrain; St, striatum; Ty, thymus; He, heart; Lu, lung; Li, liver; Ki, kidney; SM, skeletal muscle; Sp, spleen; Te, testis. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
In the rBDNF-lacZ-BAC transgenic line, the expression of rBDNF-lacZ mRNA was detected by RT-PCR in several brain regions and peripheral organs expressing endogenous mouse BDNF (mBDNF) mRNA (Fig. 1b). Specifically the expression of rBDNF-lacZ mRNA was detected in the brain regions of cortex, hippocampus, cerebellum, olfactory bulb, thalamus/hypothalamus, pons/medulla, midbrain, striatum, and also in the heart and lung. rBDNF-lacZ mRNA expression levels were not detected by RT-PCR in the thymus, liver, kidney, spleen, and skeletal muscle. Particularly high expression of the transgene was observed in the testis.
In the adult brain of the rBDNF-lacZ-BAC transgenic mice, in situ hybridization analysis revealed intense labeling of both rBDNF-lacZ and endogenous mBDNF mRNAs in the cerebral cortex (Figs. 2a–f and 3g,h), olfactory nucleus (Fig. 2ab,), hippocampus (Figs. 2e,f and 3a–f), amygdala (Fig. 2e–f), nucleus of the lateral olfactory tract (Fig. 2i,j), and hypothalamic nuclei (Fig. 2e,f, and 2k–n) including mamillary nuclei (Fig. 2k,l). In the granular cell layer of the olfactory bulb (Fig. 2a,b), caudate putamen, and nucleus accumbens (Fig. 2c,d), high levels of rBDNF-lacZ mRNA were detected, whereas labeling of the endogenous mBDNF mRNA was indistinguishable from background signal. In the claustrum (Fig. 2c,d) and hypothalamus (Fig. 2e,f), rBDNF-lacZ mRNA expression levels were relatively lower than mBDNF mRNA levels. In the hippocampus, intensive rBDNF-lacZ labeling over scattered neurons in the CA1 and CA3 subfields (Fig. 3a,c) mirrored the expression of the endogenous mBDNF (Fig. 3b,d). However, in the granule cells of dentate gyrus that showed high expression of mBDNF mRNA (Figs. 2f and 3f) no expression of rBDNF-lacZ was detected (Figs. 2e and 3e). In the cortex, rBDNF-lacZ expression was observed in cingulate and somatosensory areas in layers II–III and V–VI (Figs. 2c,e and 3g), whereas endogenous mBDNF was expressed throughout layers II–VI (Figs. 2d,f and 3h). Expression of rBDNF-lacZ (Fig. 2g,o) and mBDNF (Fig. 2h,p) mRNA was detected also in cardiac blood vessels but not in ventricular myocardium (Fig. 2g,h). In lung tissue, the levels of both rBDNF-lacZ and mBDNF mRNA were below detection limits of our in situ hybridization analysis (data not shown).
FIG. 2.
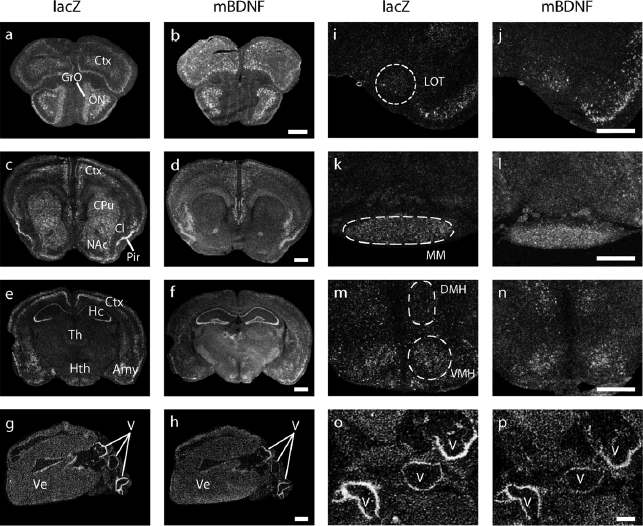
In situ hybridization analysis of rBDNF-lacZ mRNA expression in adult rBDNF-lacZ-BAC transgenic mouse brain and heart. Photomicrographs of 16 lm coronal brain (a–f; i–n) and transverse heart sections (g,h,o,p) hybridized with 35S-labeled lacZ or mouse endogenous BDNF (mBDNF) cRNA. The brain sections shown are at the levels of olfactory bulb (a,b), striatum (c,d), and hippocampus (e,f). (i–n) Magnifications of selected brain regions: LOT, nucleus of the lateral olfactory tract; MM, medial mammillary nucleus; DMH, dorsomedial hypothala-mic nucleus; VMH, ventromedial hypothalamic nucleus. (o,p) Magnifications of cardiac blood vessels. Scale bars: 1 mm (a-h) and 0.5 mm (i–p). Abbreviations: Ctx, cortex; GrO, olfactory bulb, granular cell layer; ON, olfactory nuclei; CPu, caudate putamen; Cl, claustrum; NAc, nucleus accumbens; Pir, piriform cortex; Hc, hippocampus; Th, thalamus; Hth, hypothalamus; Amy, amygdala; Ve, ventricle; V, cardiac blood vessel.
FIG. 3.
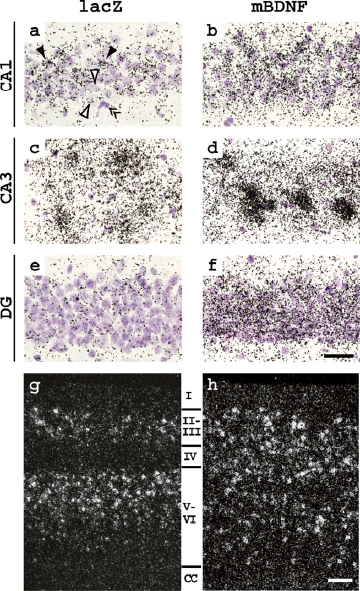
Cellular expression of rBDNF-lacZ mRNA in adult transgenic mouse brain: in situ hybridization analysis. (a–f) Bright-field photomicrographs of hippocampal subfields CA1, CA3, and dentate gyrus (DG). Hybridization probes are indicated above the columns; closed arrowheads indicate neurons with strong labeling; open arrowheads indicate neurons with weak or absent labeling; double arrowheads indicate a glial cell showing no labeling. (g,h) Distribution of lacZ and mouse BDNF labeling in cortical layers I–VI. Abbreviation: CC, corpus callosum. Scale bars: 20 μm (a–f) and 100 μm (g,h).
We also analyzed the expression and enzymatic activity of β-galactosidase protein in rBDNF-lacZ-BAC mouse tissues. Reporter activity was not detected in the brain or testis of the analyzed rBDNF-lacZ-BAC mouse line using X-gal staining assay. In addition, no expression of β-galactosidase protein was detected in the hippocampus, cortex, and testis of the transgenic animals using Western blot analysis (data not shown). These results suggest that β-galactosidase protein was either not translated from BAC-driven rBDNF-lacZ mRNAs or the levels of expression of the reporter protein remained below detection limits of the methods used in this study.
Kainic acid has been shown to induce BDNF mRNA expression in the adult rodent hippocampus and cerebral cortex (Zafra et al., 1990) in a promoter-specific manner (Aid et al., 2007; Timmusk et al., 1993). Three hours after systemic injection of kainic acid, the levels of transgenic rBDNF-lacZ mRNA were increased in rBDNF-lacZ-BAC mice similarly to endogenous mBDNF mRNA (see Fig. 4). The elevated levels of rBDNF-lacZ and mBDNF mRNA expression were observed in cortical layers II–III and V–VI, hippocampal subfields CA1 and CA3, and in the amygdala. However, in contrast to endogenous mBDNF, induction of rBDNF-lacZ mRNA expression in the granule cells of the dentate gyrus was not observed (Fig. 4e,f). Quantitative real-time PCR analysis showed that induction pattern of different rBDNF-lacZ transcripts by kainic acid largely followed that of the endogenous BDNF: both transgenic and endogenous exon I and exon IV mRNAs transcribed from promoters I and IV, respectively, showed higher levels of induction than exon VI mRNAs transcribed from promoter VI (Fig. 4g,h). Similarly to untreated mice, β-galactosidase activity and protein expression was not detected in the cortex, hippocampus, and testis of kainate-treated rBDNF-lacZ-BAC mice (data not shown).
FIG. 4.
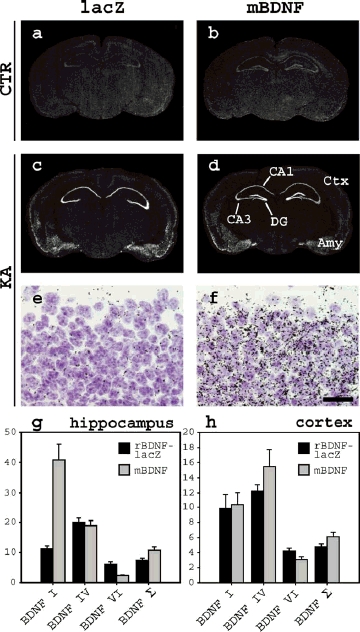
Induction of rBDNF-lacZ mRNA in transgenic mouse brain by kainic acid treatment. (a–f) In situ hybridization analysis with probes for transgenic rBDNF-lacZ and mouse endogenous (mBDNF) mRNA. Autoradiographs of sections from vehicle-treated (a,b) and kainate-treated animals (c–f) are shown. Dark-field autoradiographs of coronal sections (a–d); high magnification bright-field photomicrographs of the dentate gyrus (e,f). Scale bar: 20 μm (e,f). (g,h) Quantitative real-time PCR analysis of rBDNF-lacZ and endogenous mBDNF mRNA expression in the hippocampus (g) and cerebral cortex (h) of transgenic mice, expressed as fold difference relative to mRNA levels in vehicle-treated mice. Shown are transcripts containing exons I, IV, VI, and total BDNF mRNA (BDNF Σ). Error bars represent standard deviation of three RT-PCR experiments. Abbreviations: CTR, vehicle-treated control mice; KA, kainate-treated mice; CA1, CA3, hippocampal subfields; DG, dentate gyrus; Ctx, cortex; Amy, amygdala.
Transgenic mice expressing reporter genes under the control of various regulatory regions of the rBDNF gene have been described previously. rBDNF-CAT transgenic mice carrying 9 kb of genomic sequence comprising one or more BDNF 5′ untranslated exons were reported in (Timmusk et al., 1995). These transgenic mice (Fig. 1a) recapitulated BDNF expression in most brain regions and in the thymus. However, BDNF IV–VI construct failed to recapitulate BDNF expression in the cerebellum, heart, and other peripheral tissues (Timmusk et al., 1995) where BDNF transcripts IV and VI are endogenously expressed (Aid et al., 2007; Pruunsild et al., 2007; Timmusk et al., 1993). Here we demonstrate that rBDNF-lacZ-BAC including 50 kb of the rBDNF gene, 13 kb of upstream and 144 kb of downstream sequences are not sufficient to drive EGFP (enhanced green fluorescent protein) reporter gene expression in the heart (Koppel et al., 2009). Expression of rBDNF-lacZ mRNA in the heart of rBDNF-lacZ-BAC transgenic mice reported here (with 144 kb region 3′ of the rBDNF contains regulatory elements necessary for recapitulation of endogenous BDNF expression in the brain, heart, and lung, indicating that regulatory elements governing BDNF mRNA expression in these tissues are located within the 207 kb rat genomic sequence of the transgene. In addition, neuronal activity induced expression of rBDNF-lacZ mRNA in a promotor-specific manner in the rBDNF-lacZ-BAC mice, mimicking induction of the respective 5′ exon-specific transcripts of endogenous BDNF.
Recently, we have shown that human BDNF-EGFP-BAC covering 67 kb of the human BDNF (hBDNF) gene, 84 kb of upstream and 17 kb of downstream sequences gene) suggests that a heart-specific regulatory element is located within 18-144 kb 3′ of BDNF gene. However, this prediction should be treated with caution as regulatory regions of BDNF genes of different species are compared. On the other hand, neither hBDNF-EGFP-BAC (Koppel et al., 2009) nor rBDNF-lacZ-BAC could direct transgene expression to hippocampal dentate granule cells suggesting that the respective regulatory regions are located in genomic regions further than 84 kb upstream of BDNF exon I and 144 kb downstream of BDNF coding exon. Existence of remote cis-acting elements controlling BDNF transcription has been demonstrated by recent studies describing a regulatory region 850 kb upstream of human and mouse BDNF genes, disruption of which causes obesity, cognitive impairment, and hyperactivity (Gray et al., 2006; Sha et al., 2007).
In conclusion, we have generated transgenic mice containing rBDNF-lacZ-BAC transgene that recapitulated the expression of endogenous BDNF mRNA in the brain and peripheral tissues and neuronal activity-dependent regulation of BDNF mRNA in the adult cerebral cortex and hippocampus. This mouse model represents a useful tool for further mapping of proximal and distal regulatory elements in rodent BDNF gene in vivo.
METHODS
rBDNF-lacZ-BAC transgenic mice were generated using BAC clone CH230-106M15 (Chori BACPAC Resources, Oakland, CA) modified to replace rBDNF coding sequence with the lacZ reporter gene (Red®/ET® homologous recombination technology, Gene Bridges, Heidelberg, Germany) (Muyrers et al., 1999). The BAC clone contains 207 kb of the rBDNF genomic locus (GenBank: AC108236) including 50 kb of rBDNF gene, 13 kb of 5′ and 144 kb of 3′ flanking sequences (Fig. 1a). Purified rBDNF-lacZ-BAC was transfected into COS-7 cells by DEAE-dextran and tested for reporter activity using β-galactosidase assay. Transgenic mice were generated at the Karolinska Center for Transgene Technologies (Stockholm, Sweden) by injection of NotI-linearized rBDNF-lacZ-BAC into CBA x C57Bl/6 mouse pronuclei. One transgenic founder mouse was obtained and bred to establish a transgenic mouse line. Integration of two copies of rBDNF-lacZ-BAC transgene was estimated by slot-blot hybridization of genomic DNA with [α-32P]dCTP-labeled lacZ-specific probe.
RNA isolation and analysis of rBDNF-lacZ mRNA expression in transgenic mouse tissues with RT-PCR was performed as described (Pruunsild et al., 2007). Quantitative real-time PCR was performed on LightCycler 2.0 (Roche Diagnostics, Mannheim, Germany) using qPCR Core Kit for SYBR(r) Green I No ROX (Eurogentec, Liège, Belgium). qPCR reactions were processed in triplicate and all expression data were normalized to hypoxan-thine phosphoribosyltransferase 1 (HPRT1) mRNA levels. For primer sequences see Table 1. In situ hybridization analysis with [α-35S]UTP-labeled cRNA probes for rBDNF-lacZ and endogenous mouse BDNF mRNA was performed as described in Timmusk et al. (1993). Kainic acid (KA; 30 mg/kg) or phosphate-buffered saline was administered intraperitoneally to adult rBDNF-lacZ-BAC mice weighing 20–25 g. Two kainic acid-treated and two vehicle-treated animals were used for qRT-PCR analysis. Four kainic acid-treated animals and one vehicle-treated animal were used for in situ hybridization analysis. Only animals with induced tonic-clonic seizures were selected for analysis and results are shown for individuals showing highest induction of transgenic and endogenous BDNF mRNA. All animal procedures were carried out in compliance with the local ethics committee.
Table 1.
PCR Primers Used in This Study
| BAC modification | |
| mrBDNF_rpsLneo_F | TGTCTGTCTCTGCTTCCTTCCCACAGTTCCACCAGGTGAGAAGAGTGGGCCTGGTGATGATGGCGGGATCG |
| rBDNF_rpsLneo_R | ATACAAATAGATAATTTTTGTCTCAATATAATCTATACAACATAAATCCATCAGAAGAACTCGTCAAGAAGG |
| BDNF_lacZ_300_F | GCCGTCACTTGCTTAGAAACCGTT |
| BDNF_lacZ_300_R | GAGTACTAACAAGAACGAAGATACT |
| Genotyping/RT-PCR | |
| rBDNF_LacZ_F | CCCTGCAGCTGGAGTGGATCAGTAAG |
| rBDNF_LacZ_R | GAAGATCGCACTCCAGCCAGCTTTCC |
| mBDNF_F | GTATGTTCGGGCCCTTACTATGGATAGC |
| mBDNF_R | AAGTTGTGCGCAAATGACTGTTTC |
| HPRT1_F | CTTTGCTGACCTGCTGGATTAC |
| HPRT1_R | GTCCTTTTCACCAGCAAGCTTG |
| Quantitative real-time RT-PCR | |
| Mouse endogenous mRNAs | |
| mBDNFq_I_F | TTGAAGCTTTGCGGATATTGCG |
| mBDNFq_IV_F | GAAATATATAGTAAGAGTCTAGAACCTTG |
| mBDNFq_VI_F | GCTTTGTGTGGACCCTGAGTTC |
| mBDNFq_RT_IXcod_R | AAGTTGCCTTGTCCGTGGAC |
| mBDNFq_cod_F | GGCCCAACGAAGAAAACCAT |
| mBDNFq_cod_R | AGCATCACCCGGGAAGTGT |
| HPRT1q_F | CAGTCCCAGCGTCGTGATTA |
| HPRT1q_R | AGCAAGTCTTTCAGTCCTGTC |
| Rat BDNF-lacZ mRNAs | |
| rBDNFq_I_F | AGTCTCCAGGACAGCAAAGC |
| rBDNFq_IV_F | GAAATATATAGTAAGAGTCTAGAACCTTG |
| rBDNFq_VI_F | GCTTTGTGTGGACCCTGAGTTC |
| LacZq_F | CGAAGTGACCAGCGAATACCTGT |
| LacZq_R1 | CAACTGTTTACCTTGTGGAGCGACA |
| LacZq_R2 (with I_F) | CAAGGCGATTAAGTTGGGTAAC |
| LacZq_R3 (with IV,VI_F) | GTTTTCCCAGTCACGACGTT |
Acknowledgments
We thank Epp Väli for technical assistance and Priit Pruunsild for critical comments on the manuscript.
LITERATURE CITED
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi H, Risling M, Carlstedt T, Lendahl U, Timmusk T, Metsis M, Yamamoto Y, Ibanez CF. Targeted expression of a multifunctional chimeric neurotrophin in the lesioned sciatic nerve accelerates regeneration of sensory and motor axons. Proc Natl Acad Sci USA. 1998;95:5269–5274. doi: 10.1073/pnas.95.9.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Yeo GS, Cox JJ, Morton J, Adlam AL, Keogh JM, Yanovski JA, El Gharbawy A, Han JC, Tung YC, Hodges JR, Raymond FL, O'Rahilly S, Farooqi IS. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–3371. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Cerutti I, Auffray C, Devignes MD. A transgenic mouse model engineered to investigate human brain-derived neurotrophic factor in vivo. Transgenic Res. 2007;16:223–237. doi: 10.1007/s11248-006-9060-0. [DOI] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu XZ, Xu K, Kenney H, Egan SE, Turley H, Harris AL, Marini AM, Lipsky RH. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci. 2008;28:1118–1130. doi: 10.1523/JNEUROSCI.2262-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel I, Aid-Pavlidis T, Jaanson K, Sepp M, Pruunsild P, Palm K, Timmusk T. Tissue-specific and neural activity-regulated expression of human BDNF gene in BAC transgenic mice. BMC Neurosci. 2009;10:68. doi: 10.1186/1471-2202-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Xu K, Zhu D, Kelly C, Terhakopian A, Novelli A, Marini AM. Nuclear factor kappaB is a critical determinant in N-methyl-d-aspartate receptor-mediated neuroprotection. J Neurochem. 2001;78:254–264. doi: 10.1046/j.1471-4159.2001.00386.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Christian K, Lu B. BDNF: A key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learn Mem. 2008;89:312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, Xu J, Tang J, Ding J, Gong J, Ge X, Kong D, Gao X. Disruption of a novel regulatory locus results in decreased Bdnf expression, obesity, and type 2 diabetes in mice. Physiol Genomics. 2007;31:252–263. doi: 10.1152/physiolgenomics.00093.2007. [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Lendahl U, Funakoshi H, Arenas E, Persson H, Metsis M. Identification of brain-derived neurotrophic factor promoter regions mediating tissue-specific, axotomy-, and neuronal activity-induced expression in transgenic mice. J Cell Biol. 1995;128:185–199. doi: 10.1083/jcb.128.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Lendahl U, Metsis M. Brain-derived neurotrophic factor expression in vivo is under the control of neuron-restrictive silencer element. J Biol Chem. 1999;274:1078–1084. [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Persson H, Metsis M. Analysis of transcriptional initiation and translatability of brain-derived neurotrophic factor mRNAs in the rat brain. Neurosci Lett. 1994;177:27–31. doi: 10.1016/0304-3940(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Zafra F, Hengerer B, Leibrock J, Thoenen H, Lindholm D. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. EMBO J. 1990;9:3545–3550. doi: 10.1002/j.1460-2075.1990.tb07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]


