Abstract
Arabidopsis microRNA (miRNA) genes (MIR) give rise to 20- to 22-nt miRNAs that are generated predominantly by the type III endoribonuclease Dicer-like 1 (DCL1) but do not require any RNA-dependent RNA Polymerases (RDRs) or RNA Polymerase IV (Pol IV). Here, we identify a novel class of non-conserved MIR genes that give rise to two small RNA species, a 20- to 22-nt species and a 23- to 27-nt species, at the same site. Genetic analysis using small RNA pathway mutants reveals that the 20- to 22-nt small RNAs are typical miRNAs generated by DCL1 and are associated with Argonaute 1 (AGO1). In contrast, the accumulation of the 23- to 27-nt small RNAs from the miRNA-generating sites is dependent on DCL3, RDR2 and Pol IV, components of the typical heterochromatic small interfering RNA (hc-siRNA) pathway. We further demonstrate that these MIR-derived siRNAs associate with AGO4 and direct DNA methylation at some of their target loci in trans. In addition, we find that at the miRNA-generating sites, some conserved canonical MIR genes also produce siRNAs, which also induce DNA methylation at some of their target sites. Our systematic examination of published small RNA deep sequencing datasets of rice and moss suggests that this type of dual functional MIRs exist broadly in plants.
INTRODUCTION
Small non-coding RNAs (sRNAs) serve as sequence-specific negative regulators that control expression of a wide variety of genes in almost all cellular processes of eukaryotes (1,2). In plants, sRNAs are classified into microRNAs (miRNAs) and small interfering RNAs (siRNAs) based on their precursor structures and biogenesis pathways. miRNAs are originated from hairpin-folded single-stranded RNAs transcribed from miRNA genes (MIR) (3–5), while siRNAs are produced usually from long double-stranded RNAs (dsRNAs) (6–8).
miRNAs are 20- to 22-nt in length and are processed predominantly by the type III endoribonuclease Dicer-like 1 (DCL1) in Arabidopsis (3–5). A recent work also identified 23- to 27-nt long miRNAs (lmiRNAs) generated by DCL3 (9). Canonical miRNAs mediate gene silencing mainly at the post-transcriptional level by mRNA cleavage or translational repression (3–5), while the function of the long miRNAs has not yet been unraveled. The role of miRNA in mediating DNA methylation was observed only in a few cases. The only example reported in Arabidopsis is miR165/166, which induces DNA methylation downstream of its target sites (10). In moss Physcomitrella patens, miRNAs induce DNA methylation under the hormone abscisic acid treatment or in the DCL1b mutant that abolished the cleavage activity of miRNAs. The authors proposed that this epigenetic gene silencing is triggered by miRNA to target RNA ratios (11).
Several classes of plant endogenous siRNAs have been documented (1). Among them, the heterochromatic siRNAs (hc-siRNAs) are predominantly 24-nt in length and are mainly derived from repeats or transposons (8,12,13). They safeguard genome integrity by promoting heterochromatin formation via DNA methylation and/or histone modifications. The biogenesis of hc-siRNAs is dependent on DCL3, RDR2 and Pol IV (12,13).
Here, we present the discovery of a novel class of Arabidopsis MIR genes that give rise to both 20- to 22-nt and 23- to 27-nt sRNAs at the same site. Biogenesis analysis shows that the 20- to 22-nt species are miRNAs that are DCL1 dependent and RDR- or Pol IV independent, whereas the 23- to 27-nt species are siRNAs that depend on DCL3, RDR2 and Pol IV, components of a typical hc-siRNA pathway. We further demonstrate that these 23- to 27-nt siRNAs generated from the miRNA sites can associate with AGO4 and guide DNA methylation at some of their target loci in trans. In agreement with this finding, some canonical MIR genes could also generate both sRNA classes and the 23- to 27-nt siRNAs could also mediate de novo DNA methylation in trans at their target site. Our systematic analysis of published sRNA deep sequencing datasets shows that 43% of rice (Oryza sativa) miRNA sites and 36% of moss (P. patens) miRNA sites produce 23- to 26-nt sRNAs, suggesting that this type of siRNAs from miRNA loci exist broadly in plants.
MATERIALS AND METHODS
Plant materials, small RNA library construction and deep sequencing
A total number of 13 small RNA libraries were prepared from 4- to 5-week-old short-day grown Arabidopsis Col-0 inoculated with mock (10 mM MgCl2) and a series of bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 strains, a type III secretion system mutated strain Pst DC3000 hrcC, a virulent strain Pst DC3000 carrying an empty vector (EV) and an avirulent strain Pst DC3000 carrying an effector gene (avrRpt2). Bacteria infiltration was carried out on the leaves as described previously (14) at a concentration of 2 × 107 cfu/ml. Plants were grown in the green house at 22°C with 12 h light and infiltrated leaves were harvested at 6- and 14-h post-inoculation (hpi).
From the harvested leaves, total RNA was isolated using Trizol reagent (Invitrogen) and it was fractionated on 15% denaturing polyacrylamide (PAGE) gel. The sRNA library for deep sequencing was constructed using RNA molecules ranging from 18- to 26-nt and ligated to 5′- and 3′-RNA adaptors by 5′-phosphate-dependent method as described in detail (15). The sRNA libraries were sequenced by Illumina Inc. and UCR core facility.
For the sRNA biogenesis, the following Arabidopsis thaliana mutants dcl2-1, dcl3-1, dcl4-2, hen1-1, hyl1-2, dcl1-7/fwf2, rdr1-1, rdr2-2, rdr6-15, nrpd1-3, ago1-27, ago4-1 and their corresponding wild-type ecotypes, Columbia-0 and Landsberg erecta (Ler) were used in this study.
Processing of deep sequencing data
The 13 libraries of sequencing reads have been deposited into NCBI/GEO databases (GSE19694). Raw sequence reads were parsed to remove the 3′-adaptors. The sequencing reads from each of the small RNA libraries, with adaptors trimmed, were mapped to the Arabidopsis nuclear, chloroplast and mitochondrial genome sequences and cDNA sequences, which were all retrieved from TAIR (version 9, http://www.arabidopsis.org). The reads that match to these sequences with 0 mismatch (the raw labeled ‘mapped’ in Supplementary Table S1) were retained for further analysis. Sequencing reads were aligned to the precursors of the annotated Arabidopsis miRNAs in miRBase (release 13.0, http://microrna.sanger.ac.uk), with Novoalign version 2.04 (http://www.novocraft.com). Those sequencing reads that can be mapped to a miRNA precursor with 0 mismatch were retained for further analysis.
MIR2831-derived long siRNA cloning
MIR2831 gene-derived long siRNA cloning was achieved by fractionating 26- to 40-nt RNAs from total RNAs and ligated to 5′- and 3′-RNA adaptors using the same sRNA cloning protocol as the construction of sRNA library, and reverse transcribed using 5′-miR2831 specific primers and 3′-reverse complementary sequence of adapters. Primer pairs for amplifying 5′-end of the long siRNA were: 5′-adaptor-F: CAGAGTTCTACAGTCCGACGA and miR2831 specific reverse complementary primer miRNA2831R: AGAAGTGGATGGGCCAAGAAAA. Primer pairs for amplifying 3′-end of the long siRNA were: miRNA2831F: TTTTCTTGGCCCATCCACTTCT and 3′-adaptor-R: CAAGCAGAAGACGGCATACGA. The sRNA PCR fragments were cloned and sequenced.
Northern blot analysis
RNA was separated on 15% denaturing PAGE gel and blotted onto Hybond-NX membrane (GE Health Care). RNA was cross-linked to membrane using EDC as described (16). Pre-hybridization and hybridization were carried out in PerfectHyb Plus Hybridization Buffer (Sigma) supplemented with sheared salmon sperm DNA (100 µg/ml). LNA or DNA oligos complementary to miRNAs were labeled at their 5′-end with gamma P-32 ATP (6000 Ci/mmol; Perkin Elmer) using T4 polynucleotide kinase (NEB). After overnight hybridization, post-hybridization washes were performed in 2 × SSC and 0.1% SDS, two washes each 20 min. Blots were exposed to phosphorscreens, scanned using PhosphorImager (Molecular dynamics).
The following LNA oligos were used to detect new miRNAs.
CandNew_2883: c+tt c+gt t+gt c+at c+ac a+aa g+tt
CandNew_2328: c+cg a+gt c+gt c+at t+tt g+ct t+ct
CandNew_2831-5P: gaa+gtg+gat+ggg+cca+aga+aaa
The following DNA oligos were used to detect known miRNAs.
miR156-AS: GTGCTCACTCTCTTCTGTC
miR390-AS: GGCGCTATCCCTCCTGAGCTT
miR164-AS: TGCACGTGCCCTGCTTCTCCA
miR402-AS: CAGAGGTTTAATAGGCCTCGAA
Target gene expression analysis by quantitative RT–PCR
Total RNA was extracted with phenol/chloroform and was treated with DNase I (Invitrogen) to remove DNA contamination. About 5 µg of RNA was used for reverse transcription with oligo dT primer using Superscript II (Invitrogen). Quantitative PCR was performed using SensiMix SYBR (Quantace) using specific pair of primers listed below. Minimum cycles were carried out based on their expression levels as we standardized to see the detection. The comparative threshold cycle (Ct) method was used for determining relative transcript levels (Bulletin 5279, Real-time PCR applications Guide, Bio-Rad). Actin was used as an internal control.
At5g08490F: CACAGATGCCAGTTGAACCT
At5g08490R: CACCACAGGCTCTTTCATCT
At4g16580F: TGTATCTGGTCCGGCCTTTC
At4g16580R: CTCTGAGTCTCATACTGATT
ActinF: AGTGGTCGTACAACCGGTATT
ActinR: GATGGCATGAGGAAGAGAGAA
Bisulfite sequence analysis
Total genomic DNA was isolated from Arabidopsis leaves using CTAB method from Col-0 and nrpd1-3 mutant. The total DNA was subjected to bisulfite treatment as described in (17). Primers are designed to amplify specific regions flanking the miRNA-binding sites. The following primers were used to amplify target regions for newly identified miRNAs.
BisAt4g16580Fp-TTGTTGATTAACGAAAGGAGGAAT
BisAt4g16580Rp-GCTGATTCAAATAAATCCATCTCTTA
BisAt5g08490-F: GGAAACCACGTTCTAATATCGAATATGTAT G
BisAt5g08490-R: AGGCTCTTTCATCTGAAGATATAAAGCATTTA
BisAt5g43270-SPL2F: AAGTTCGAAGCACGTACCATCAAGAGTG
BisAt5g43270-SPL2R: GTAGTTTTCATTAAACTGGCCTACCCCATT
To analyze the methylation status of the target genes, we selected three locations—at the binding site, 50–100-bp up- and downstream of the miRNA target sites—for investigation.
Identification of putative miRNA precursor loci
We extended the method that we developed previously (18) to identify novel miRNA genes based on the reads in the 13 small RNA sequencing libraries, which are available at NCBI/GEO under accession number GSE19694. Briefly, in our method, all sRNA reads were first mapped to the Arabidopsis genome. The genome loci that have reads mapped to were clustered if they were overlapped or adjacent to each other. For each cluster with no longer than 50 nt, we extracted two sequences to further analyze their folding structures; one extended from 160-nt upstream to 30-nt downstream of the cluster, and the other started from 30-nt upstream and ended at 160-nt downstream of the cluster. These parameters were chosen based on the lengths of the known Arabidopsis miRNA precursors, which range from 63- to 689-nt, with a mean of 171-nt. More than 80% of these precursors are shorter than 210-nt. Hence we extract ∼210-nt sequences surrounding the (clustered) reads for further analysis of their secondary structures using the RNA-fold program (19). The rest steps of our method followed the same procedure in (18), except the features used to build the support vector machines (SVM) classification model. All the features that we used in our miRank method (20) were adopted in the current study. In addition, we introduced several extra features based on the patterns of sRNA reads mapped to the known miRNA precursors. The first feature was the ratio between the number of the most frequent reads mapped to the two arms of a hairpin structure and the total number of reads on the precursor. The second feature was the ratio between the numbers of reads mapped to both arms (the larger number over the smaller one). The third and the fourth features were the width of the (clustered) reads on the arm with majority reads and the width of the (clustered) reads on the opposite arm.
Identification of MIRs generating two sRNA species from deep sequencing data
We extensively searched for mature miRNA loci generating two types of sRNAs in publicly available small RNA deep-sequencing datasets on O. sativa (rice, GEO Access number: GSE14462, 2 datasets) and P. patens [moss, (21) six datasets). Raw sequence reads were parsed to remove low quality reads and the 3′-adaptors. The mature miRNAs of the annotated miRNA precursors of the corresponding species in miRBase release 14.0 (http://microrna.sanger.ac.uk) were extended 50-nt upstream and 50-nt downstream. The sRNA reads from sRNA sequencing libraries on rice and moss, with adaptors trimmed, were mapped (with no mismatch) separately to these ∼120-nt regions of the corresponding genomes. Two criteria were used to select mature miRNAs that have potential siRNA and miRNA generated at the same loci. First, there are 23- to 26-nt sequencing reads that overlap with mature miRNAs by >18-nt. Second, reads mapped to mature miRNAs do not form a laddering pattern.
Prediction of putative sRNA targets
We implemented a target prediction algorithm for plant miRNA and siRNAs, which extended and improved upon the methods proposed by Zhang (22) and Jones-Rhoades and Bartel (23), which is in principle similar to the TargetFinder method (24). Briefly, a score function was first used to minimize the number of mismatches, G-U wobbles and bulges along the alignment of miRNAs and their putative target sites, where the penalty for a G:U wobble pairing is 0.5, the penalty for a insertions/deletions is 2.0, and that for a mismatch is 1.0. The algorithm then considered all possible arrangements of mismatches and bulges. Random permutations of each miRNA or siRNA with first-order hidden Markov model were applied to test the signal–noise ratio of its putative target sites. We used the cutoff of maximal score of 4.5 for target prediction.
RESULTS
Identification of candidate new MIR genes
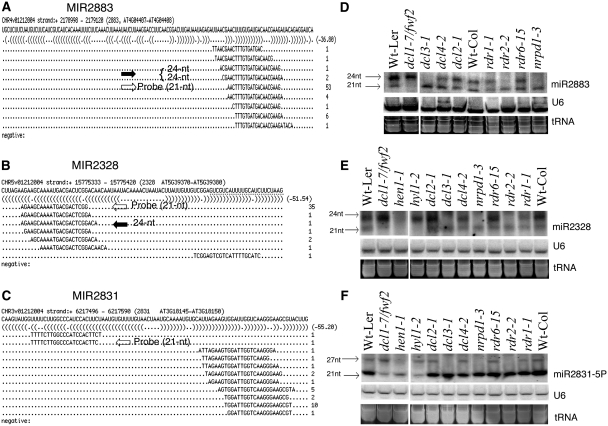
Most MIR genes are transcribed by RNA polymerase II and the resulting miRNA precursors can fold back to form hairpin structures that are recognized and processed predominantly by DCL1 to generate miRNAs. A few young miRNAs derived from precursors with long stem-loop structures are processed by DCL4 (25), whereas the 23- to 27-nt lmiRNAs are processed by DCL3 (9). To identify new MIR genes in Arabidopsis, we first searched for putative miRNA precursor loci that could fold into stem-loop structures from the Arabidopsis intergenic, intronic and UTR regions (see ‘Materials and Methods’ section). We then mapped the small RNA sequencing reads that we obtained from bacterial pathogen-challenged Arabidopsis leaves onto the newly predicted hairpin regions. We chose the candidate MIR genes that have perfectly matched small RNA reads that predominantly mapped at the stem regions of the hairpin structures with no or very low reads matching the negative strand (Supplementary Figure S1). We followed in general the proposed criteria for miRNA annotation (26), although a few of the candidate miRNAs we identified did not have corresponding miRNA* reads. We found a total of 10 candidate MIR genes, eight of which were from intergenic regions, one from intron and one from 5′ UTR, respectively (Supplementary Figure S1). Notably, eight out of the 10 candidate MIR genes have 23- to 27-nt reads at the same site of the 20- to 22-nt reads (Supplementary Figure S1 and Table S1; Table 1).
Table 1.
Summary of total number of MIR genes that give rise to 23- to 26-nt sRNAs in our Arabidopsis dataset, and in published rice and moss datasets as indicated in the respective columns
|
Arabidopsis |
Rice |
Moss |
|||||
|---|---|---|---|---|---|---|---|
| Known MIR | Known MIRs with 23- to 26-nt reads (%) | New MIR | New MIRs with 23- to 26-nt reads (%) | MIR | MIR with 23- to 26-nt reads (%) | MIR | MIR with 23- to 26-nt reads (%) |
| 191 | 81 (42%) | 10 | 8 (80%) | 414 | 176 (43%) | 230 | 83 (36%) |
Novel MIR genes that give rise to both miRNAs and siRNAs at the same site
To confirm these candidate MIR genes, we examined the expression of these candidate miRNAs in various mutants of DCLs, RDRs and the Pol IV large subunit NRPD1. The biogenesis of miRNAs is predominantly dependent on DCL1 and does not require RDRs and Pol IV. Interestingly, for some of the candidates, we detected two sRNA species, a 20- to 22-nt species and a 23- to 27-nt species, using a single probe. Figure 1 shows three such examples, MIR2328, MIR2883 and MIR2831. The accumulation of the 21-nt bands of these three candidates was unaffected in rdr1-1, rdr2-2, rdr6-15, nrpd1-3. However, they were drastically reduced in the dcl1-7/fwf2 double mutant (Figure 1D, E and F), hen1-1 and hyl1-2 as compared with the wild-type (Figure 1E and F). The dcl1-7/fwf2 double mutant rescued the pleiotropic phenotype of dcl1-7 (27), which ruled out the possibility that the dependence on DCL1 was due to a secondary effect of the strong morphological phenotype of dcl1. HYL1 encodes a dsRNA-binding protein that functions with DCL1 for miRNA processing and HEN1 encodes a methyltransferase that methylates plant sRNAs, including miRNAs (28–30). Thus, the 21-nt species were bona fide new miRNAs that require DCL1 but not any RDRs or Pol IV. However, to our surprise, the 24-nt sRNA species of miR2883 and miR2328 (Figure 1D and E) were absent not only in dcl3-1, but also in rdr2-2 and nrpd1-3. These results clearly indicate that the 24-nt sRNA species are not miRNAs, but rather siRNAs generated by DCL3/RDR2/Pol IV that represent the typical hc-siRNA pathway. For miR2831, we could only detect miR2831-5P but not miR2831-3P by northern blot analysis (Figure 1F; Supplementary Figure S2). We observed a larger sRNA band of ∼26–30 nt in length at miR2831-5P site in addition to the 21-nt miRNA band (Figure 1F). Biogenesis analysis showed that this ∼26- to 30-nt sRNA was also dependent on DCL3, RDR2 and Pol IV (Figure 1F). Note that ∼26- to 30-nt reads were not obtained from the deep sequencing data because the sRNA libraries were prepared from the size-fractionated sRNAs of 18–26 nt in length. To determine the sequence of the long siRNA, we performed sRNA cloning by RNA adapter ligation-based RT–PCR and sequencing (see ‘Materials and Methods’ section) and identified a 27-nt siRNA at the miR2831-5P site (Supplementary Figure S3). Thus, our results suggest that there is a novel class of MIR genes that give rise to two sRNA species, a 20- to 22-nt miRNA species and a 23- to 27-nt siRNA species.
Figure 1.
New MIR genes give rise to both miRNAs and siRNAs. (A–C) Candidate MIR genes MIR2883, MIR2328 and MIR2831, and the predicted precursor sequence with perfectly matched sRNA sequences. Solid arrows indicate 24-nt siRNAs and open arrows indicate the most abundant 21-nt miRNA used for probing. No sRNAs matched negative strand of these regions. (D–F) Accumulation of miR2883, miR2328 and miR2831-5P in various sRNA pathway mutants. U6 RNA and ethidium bromide-stained tRNA were used as the loading control.
MIR-derived siRNAs associate with AGO4
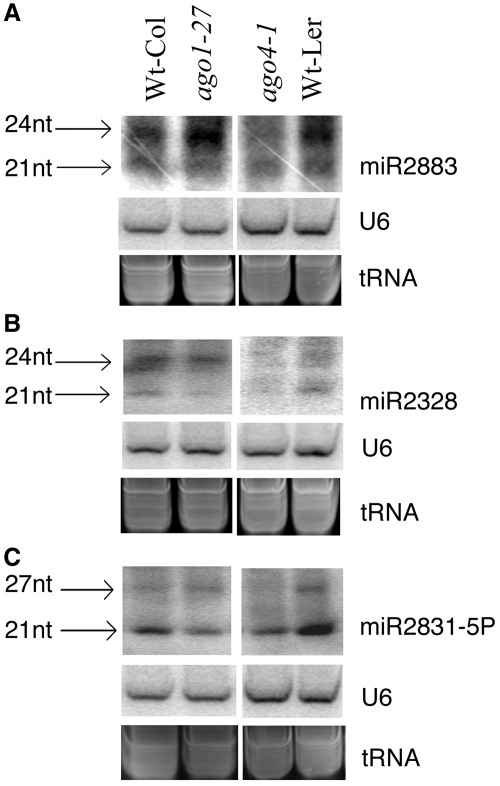
hc-siRNAs are preferentially associated with AGO4, whereas miRNAs are mainly associated with AGO1 (13,31,32). To determine whether these siRNAs generated from miRNA sites are also loaded into AGO4, we examined their accumulation in AGO1 and AGO4 mutants. While the 20- to 22-nt bands were mainly dependent on AGO1 (Figure 2), the level of the 23- to 27-nt siRNAs was clearly reduced in ago4-1 mutant, suggesting that these siRNAs were mainly associated with AGO4 (Figure 2). We also confirmed this result by analyzing the published Arabidopsis datasets of AGO-associated small RNAs (33–35). Out of the 10 newly identified MIR candidates, 7 have 23- to 26-nt reads found in these datasets and six of them were associated exclusively with AGO4, including both miR2012 and miR2012* (Table 2; Supplementary Table S2). While the number of reads for these siRNAs in the AGO4-coimmunoprecipitation were generally small such that we cannot rule out the possibility that they may also associate with other AGOs. For example, miR2812 was also associated with AGO7 (Table 2; Supplementary Table S2). These observations taken together suggest strongly that these siRNAs can associate with AGO4. Thus, these MIR-derived siRNAs are bona fide hc-siRNAs that are dependent on DCL3/RDR2/Pol IV/AGO4 for their biogenesis and function.
Figure 2.
AGO4 is required for the accumulation of siRNAs from the newly identified MIR genes MIR2883 (A), MIR2328 (B) and MIR2831-5P (C). AGO1 and AGO4 are essential for the accumulation of 21-nt and 23- to 27-nt sRNA species, respectively. U6 and ethidium bromide-stained tRNA served as the loading control.
Table 2.
MIR gene-derived siRNAs are mainly associated with AGO4 in Arabidopsis
| Total MIRs with sRNAs from our dataset |
MIR gene-derived 23- to 26-nt siRNAs that associated with AGOs |
|||||
|---|---|---|---|---|---|---|
| Total | AGO1 | AGO2 | AGO4 | AGO5 | AGO7 | |
| 191 known MIR | 51 | 28/51 (55%) | 1/51 (2%) | 40/51 (78%) | 9/51 (18%) | 22/51 (43%) |
| 10 new MIR | 7 | 0/7 (0%) | 0/7 (0%) | 7/7 (100%) | 0/7 (0%) | 1/7 (14%) |
The Arabidopsis datasets of AGO-associated small RNAs from (33–35) were used in this analysis.
MIR-derived siRNAs can direct DNA methylation at some of their target loci
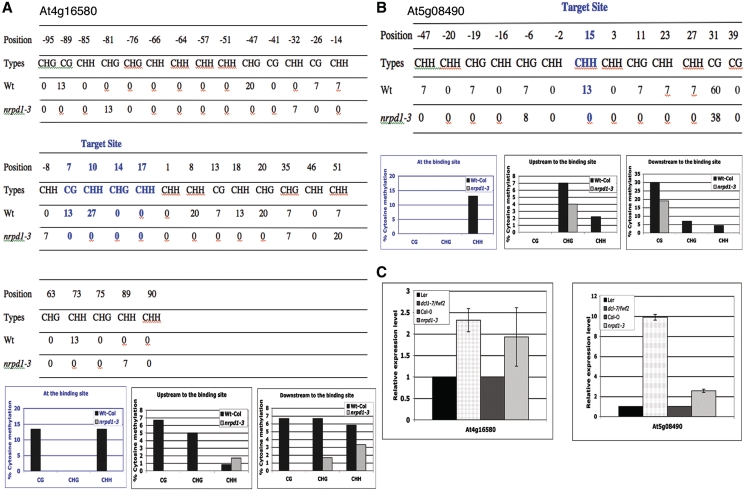
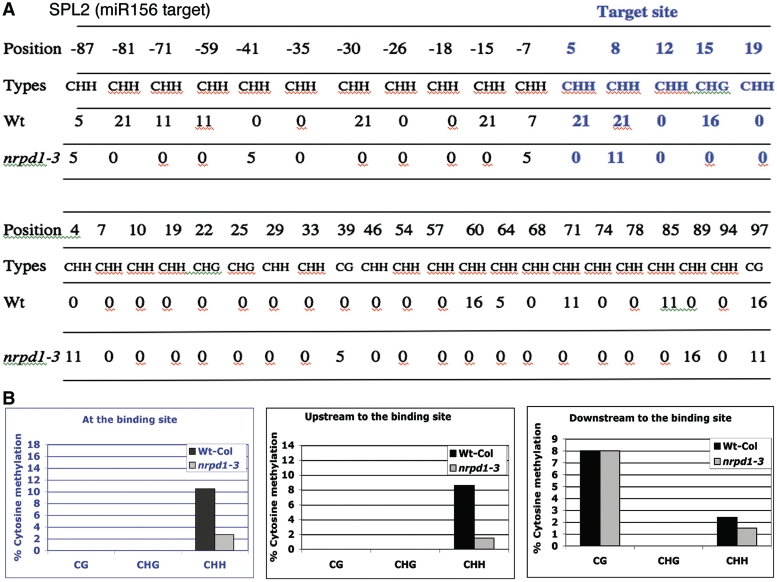
It is known that hc-siRNAs are generated by the Pol IV/DCL3/RDR2 pathway and subsequently associate with AGO4 to direct DNA methylation. To study the potential function of these Pol IV/DCL3/RDR2-dependent siRNAs generated from the miRNA sites, we predicted their putative targets (see ‘Materials and Methods’ section). The dependence of these MIR-derived siRNAs on Pol IV/DCL3/RDR2 and the association of these siRNAs with AGO4 suggested that they might also guide DNA methylation at their sites of origin in cis or their target sites in trans. We analyzed the DNA methylation level of the three new MIR gene loci and their predicted target sites in both wild-type and nrpd1-3 by bisulfite sequencing (17). If these MIR-derived siRNAs indeed direct DNA methylation, we would expect reduced DNA methylation in the nrpd1-3 mutant, where these MIR gene-derived siRNAs were absent. We did not observe an obvious reduction in DNA methylation at the siRNA-generating sites of the new MIR genes, MIR2883 and MIR2831, in nrpd1-3 (Supplementary Figure S4), suggesting these siRNAs had little effect on the methylation level of their generating sites. MIR2328 gene was hardly methylated in both wild-type and nrpd1-3 (Supplementary Figure S4). On the contrary, DNA methylation at some of their target sites was clearly affected. DNA methylation level, especially the asymmetric CHH methylation level of At4g16580 (a target of miR2328) and At5g08490 (a target of miR2831-5P) was clearly reduced or eliminated at the siRNA target sites in nrpd1-3 (Figure 3A and B). In At4g16580, the upstream region of the target site was hardly methylated, and the methylation level of the downstream region was modestly reduced in nrpd1-3 (Figure 3A). In At5g08490, reduction in CHH and CHG methylation was observed in both the upstream and downstream regions of the target sites in nrpd1-3, although to a lesser degree as compared with the reduction at the target site (Figure 3B). These results suggest that the MIR-derived siRNAs direct the DNA methylation at their target sites.
Figure 3.
MIR-derived siRNAs direct DNA methylation at their target sites. (A and B) Cytosine methylation was examined by bisulfite sequence analysis at the siRNA-targeting sites of At4g16580 (miR2328 target) and At5g08490 (miR2831-5P target). About 50–100 bp up- and downstream regions of the target sites in wild-type and nrpd1-3 mutant were also analyzed. Percentage of methylation level of each cytosine is indicated in the table and sRNA target sites are in blue. For At4g16580 (miR2328 target), 15 clones for each wild-type and nrpd1-3 were analyzed. In the case of At5g08490 (miR2831-5P target), 15 and 13 clones were used for wild-type and nrpd1-3, respectively. Overall average percentage of cytosine methylation status in each context at the binding sites, and up- and downstream regions is present in the diagrams below. (C) Relative expression levels of At4g16580 (miR2328 target) and At5g08490 (miR2831-5P target) in dcl1-7/fwf2 and nrpd1-3 mutants as compared to their respective controls. The comparative threshold cycle (Ct) method was used for calculating relative transcript levels (Bulletin 5279, real-time PCR applications Guide, Bio-Rad). Actin was used as an internal control.
We then asked whether DNA methylation mediated by these MIR-derived siRNAs has an effect on the expression of the target genes. We examined the expression of At4g16580 and At5g08490 in nrpd1-3 mutant using quantitative real-time RT–PCR. Expression level of these targets in dcl1, where the 21-nt miRNAs were absent, was examined and used as a positive control. Increased level of the two targets was observed in nrpd1-3 mutant, as compared with the corresponding wild-type (Figure 3C), similar results were observed in dcl1 mutant (Figure 3C). This result suggests that these MIR genes could regulate target expression in dual modes, possibly via siRNA-mediated DNA methylation and miRNA-mediated RNA degradation.
Some canonical MIR genes can also give rise to siRNAs at the miRNA sites
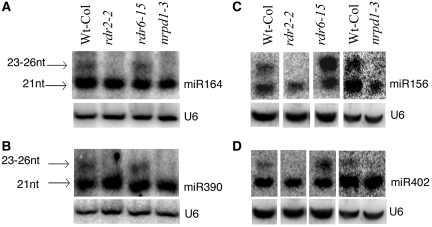
The involvement of DCL3 in generating 23- to 26-nt so-called long miRNAs from 41 known miRNA families was reported in Arabidopsis (9). However, the dependency of these 23- to 26-nt sRNAs on RDRs and Pol IV was not examined, and these sRNAs were presumed to be long miRNAs according to their biogenesis exclusively from the positive strand of MIR genes. From our sRNA dataset, we detected the expression of 191 of the 207 Arabidopsis miRNAs listed in miRBase release 11.0 (data not shown), and 81 of them (42%) have 23- to 26-nt sRNA reads (Table 1; Supplementary Table S3). We examined the accumulation of the 23- to 26-nt sRNA species of miR156, miR164, miR390 and miR402 in the mutants of RDR2 and NRPD1 using the corresponding antisense probes. We found that the 23- to 26-nt sRNA bands were absent in rdr2-2 and nrpd1-3 (Figure 4), which indicates that these sRNA species from the canonical miRNA loci are also siRNAs rather than long miRNAs. Furthermore, to determine which AGO proteins these canonical MIR-derived siRNAs are mainly associated with, we analyzed the published Arabidopsis datasets of AGO-associated small RNAs (33–35). The result showed that these 23- to 26-nt siRNAs were preferentially associated with AGO4 (Table 2; Supplementary Table S2). Out of 81 known MIR genes with 23- to 26-nt reads, 51 MIRs have 23- to 26-nt reads in the AGO pull down datasets. Out of 51 (78%), 40 MIR genes have AGO4-associated sRNA reads, and 13 of them (26%) were only present in AGO4. In addition, 28, 1, 9 and 22 MIR genes had 23- to 26-nt reads co-immunoprecipitated with AGO1, AGO2, AGO5 and AGO7, respectively (Table 2; Supplementary Table S2). For those MIR genes with 23- to 26-nt sRNAs associated with more than one AGO, the majority of the reads were associated with AGO4. These results suggest that these canonical MIR-derived siRNAs likely function through AGO4.
Figure 4.
miR164 (A), miR390 (B), miR156 (C) and miR402 (D) give rise to both 21-nt miRNAs and 23- to 26-nt siRNAs. Accumulation of the siRNAs at these miRNA loci was abolished in rdr2-2 and nrpd1-3 mutants. U6 RNA served as the loading control.
We asked whether these 23- to 26-nt siRNAs from canonical MIR genes could also direct DNA methylation at their generating sites in cis or their target sites in trans. We chose miR156 and a miR156 target, SPL2 as a case study. We found that the level of DNA methylation, especially the asymmetric CHH methylation, at the target site of SPL2 was clearly reduced in nrpd1-3 where the generation of 23- to 26-nt siRNA was impaired (Figure 5A and B). Reduced DNA methylation was detected also within the 81-bp upstream and 100-bp downstream regions of the miR156 target site of SPL2 in nrpd1-3 mutant (Figure 5A and B). This result suggests that the 23- to 26-nt siRNAs from canonical miRNA sites could also direct DNA methylation at their target sites. However, the DNA methylation level at most MIR loci are generally very low (based on Anno-J database http://neomorph.salk.edu/epigenome/epigenome.html), we chose MIR156a that has the highest DNA methylation level among all the MIR156 genes for bisulfite sequence analysis. We found that DNA methylation level at MIR156a was not reduced in nrpd1-3 (Supplementary Figure S4D), suggesting that these MIR156-derived siRNAs had little effect on the DNA-methylation level at their own generating site.
Figure 5.
The siRNA from miR156 site directs DNA methylation of its target SPL2. (A) Cytosine methylation was examined for SPL2 at the siRNA-target site, as well as the neighboring regions of 100-bp upstream and 100-bp downstream from the target site in wild-type and nrpd1-3. miRNA target site is indicated in blue. Table shows percentage methylated cytosines at each position. Nineteen clones for wild-type and 18 clones for nrpd1-3 were analyzed. (B) Overall average percentage of cytosine methylation in each context at the binding sites, and up- and downstream regions.
Thus, in addition to our newly identified candidate MIR genes, some conserved canonical MIR genes also have dual function of generating both miRNAs and siRNAs, which mediate gene silencing using dual modes of action—mRNA cleavage/degradation and DNA methylation.
MIR genes generate siRNAs predominantly at the miRNA-generating loci
In general, hc-siRNAs are generated by DCL3 from both DNA strands and can spread along the heterochromatic regions. To determine whether the siRNAs we detected in MIR genes are just part of the hc-siRNAs generated from these genomic regions that overlap with the MIR genes, we examined sRNA sequence reads mapped to the regions of 60-nt up- and downstream of the miRNA generating sites (Supplementary Figures S5 and S6). We found that the reads of 23–26-nt were only from the positive strand of miRNA precursors and predominantly originated from the miRNA-generating sites instead of spreading along the surrounding regions. To confirm this result, we performed northern blot analysis to detect any sRNAs generated from 100-bp up- and downstream of miR2831 site, as well as the loop region within MIR2831 precursor. Both sense and antisense probes were used and no signal was detected in these regions (Supplementary Figure S2). These results suggest that these dual-function MIR genes give rise to siRNAs only at the same sites from which the canonical miRNAs are produced.
Dual function MIR genes in rice and moss
To determine whether such MIR genes that give rise to both 20- to 22-nt and 23- to 27-nt sRNA species exist in other plant species, we analyzed publicly available small RNA deep-sequencing datasets from two additional plants: O. sativa (rice, GSM361264) and P. patens (moss, GSM313212) (21). We found that 176 of the 414 rice miRNA loci (43%) and 83 of the 230 moss miRNA loci (36%) can generate 23- to 26-nt sRNA reads (Table 1; Supplementary Tables S4 and S5). Among them, 54 rice miRNA loci (49.1%) have more 23- to 26-nt reads than 21-nt reads, whereas Arabidopsis and moss have fewer miRNA loci with more 23- to 26-nt reads than 21-nt reads. This result suggests that these 23- to 26-nt sRNAs may play more important roles in rice than in Arabidopsis and moss. An alignment analysis further confirmed that, just like in Arabidopsis, these 23- to 26-nt sRNAs in rice and moss also predominantly localized to the miRNA-generating sites and were derived from the positive strand of the miRNA precursors (Supplementary Figures S7 and S8), which suggests that these 23- to 26-nt sRNAs are likely generated from MIR genes. Thus, it is likely that it is a widespread phenomenon in plants that many MIR genes have the dual function of generating two sRNA species, each of which may require its own biogenesis pathway and have its own mode of function in gene regulation.
DISCUSSION
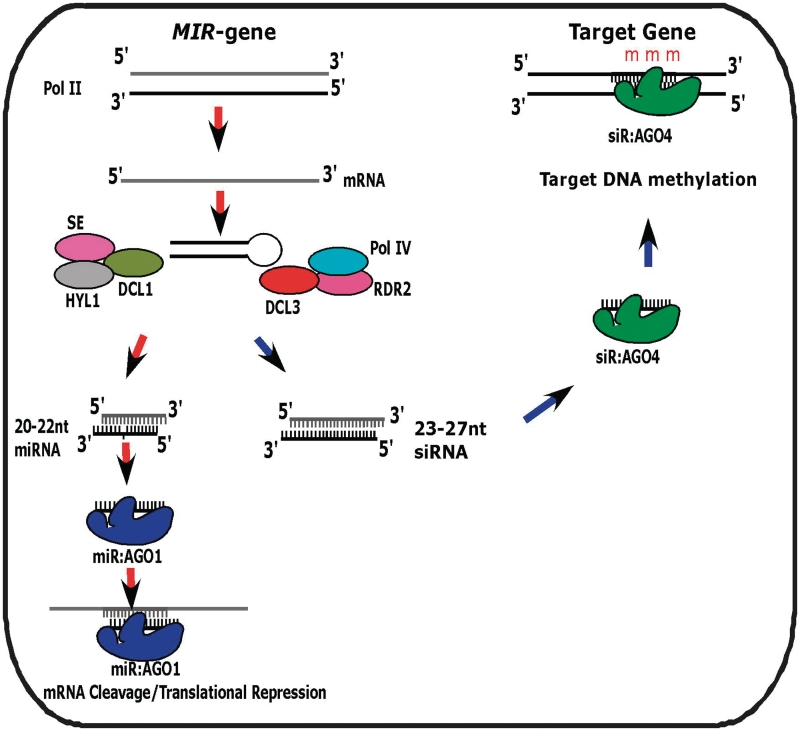
In this study, we have identified new MIR genes in Arabidopsis that play a dual role of generating both 20- to 22-nt miRNAs and 23- to 27-nt siRNAs at the same sites. The biogenesis of these 23- to 27-nt siRNAs is dependent on the DCL3/RDR2/Pol IV pathway. As presented in the model, we proposed based on our results (Figure 6), the miRNA precursors are recognized and processed by two major pathways generating two species of sRNAs: miRNAs (20–22 nt) are processed by the DCL1/HYL1/SERRATE pathway and siRNAs (23–27 nt) are produced by the Pol IV/RDR2/DCL3 pathway. From our RNA-gel blot analyses, it is clear that mutations in any of the genes in each pathway would affect the accumulation of the corresponding sRNAs species. Furthermore, the DCL1-dependent 20- to 22-nt miRNAs are associated with AGO1 and DCL3-generated 23- to 27-nt siRNAs associate with AGO4, which are indicative of their difference in modes of action on the same targets. AGO1-associated 20- to 22-nt miRNAs repress gene expression at the post-transcriptional level by mRNA cleavage or translation inhibition, whereas the 23- to 27-nt siRNAs associated with AGO4 regulate gene expression at the transcriptional (36) level by directing de novo DNA methylation (Figure 6). After the submission of this article, three classes of AGO4-associated 24-nt miRNAs were identified in rice (37). They can guide DNA methylation at some of their generation sites and their target sites. Two classes of these 24-nt miRNAs arise from the 21-nt miRNA sites, one class requires both DCL1 and DCL3 for its biogenesis, while the other requires only DCL3. The level of rice lmiRNAs was not reduced in RDR2 RNAi lines, which suggests that their biogenesis does not require RDR2. It is not clear whether these lmiRNAs also depend on Pol IV. In Arabidopsis, MIR-derived 23- to 27-nt siRNAs that we identified required RDR2 and PolIV. These studies suggest that Arabidopsis and rice have distinct biogenesis pathways for generating MIR-derived small RNAs for directing DNA methylation. Although we cannot absolutely rule out the possibility that some of these lmiRNAs from rice may still be siRNAs because it is not clear if other Rice RDRs function redundantly with RDR2 for generating hc-siRNAs for guiding DNA methylation.
Figure 6.
A model for the MIR genes of dual function. The miRNA precursors generate two species by following two pathways: (i) 20- to 22-nt miRNAs are processed predominantly by DCL1/HYL1/SE pathway from pri-miRNA transcripts, and (ii) siRNAs of 23- to 27-nt species are generated by the activities of Pol IV/RDR2/DCL3 pathway. Impairment of any protein in the pathway would affect the biogenesis and function of the particular species of sRNAs. The miRNAs are associated with AGO1 and mediated target mRNA cleavage or translational repression, whereas the siRNAs are associated with AGO4 and direct de novo DNA methylation at target loci.
Our initial hypothesis was that the dual-function MIR genes may be transcribed by both Pol II and Pol IV. The Pol II transcripts form fold-back structures and are processed by DCL1 to produce miRNAs, whereas the Pol IV transcripts serve as templates for RDR2 to generate dsRNAs, which are subsequently processed by DCL3 to produce siRNAs. However, our systemic analysis in Supplementary Figures S5–S8 showed that these MIR gene-derived siRNAs were generated predominantly at the miRNA generation sites from the positive strand. This result cannot be well explained by our initial hypothesis because the majority of DCL3/RDR2/Pol IV pathway products are known to originate from both DNA strands and spread along the precursor region rather than exhibiting such site-specific patterns as those shown in (Supplementary Figures S5 and S6). It is possible that these MIR-derived siRNAs could be generated from the whole MIR regions, but only the siRNAs at the miRNA sites are protected and stable. The mature miRNA may help determine the position of the stable siRNAs. Alternatively, it is likely that the miRNA precursors or the mature miRNAs help determine the site-specific generation of these siRNAs (Figure 6). We suggest that generation of these MIR-derived siRNAs is initiated from Pol II transcripts, which serve as templates for Pol IV and RDR2. The secondary structures of miRNA precursors generated by Pol II may limit the siRNA generation by Pol IV/RDR2/DCL3 to the site of miRNAs; or the miRNAs may interact with the Pol IV/RDR2-generated dsRNAs and this interaction may somehow limit DCL3-mediated generation of siRNAs to the site of miRNA interaction. Recent report revealed the influence of precursor structures in plant primary-miRNA processing (36,38–41). Pol II has been shown to recruit Pol IV to type II heterochromatic loci with low copy-number repeats to generate siRNAs (42). It is plausible that Pol II could also recruit Pol IV to these MIR gene loci to generate siRNAs. To determine whether Pol II-generated pri-miRNA transcripts could be exploited by the hc-siRNA biogenesis pathway to form these siRNAs, we checked the accumulation levels of these MIR-derived siRNAs in the recently identified weak allele of the second largest subunit of Pol II (nrpb2-3) (42). However, nrpb2-3 is too weak, and we did not detect any expression change of either the MIR-derived siRNAs or the 21-nt miRNAs from the same loci at the dual function new MIRs or canonical MIRs we tested (data not shown). This question remains open until a stronger Pol II mutant becomes available.
Many MIR genes were shown to generate DCL3-dependent 23- to 27-nt long miRNAs in Arabidopsis (9), although their dependence on RDRs and Pol IV was not tested. Subsequently, 11 families of atypical MIR genes were found to generate 21- to 22-nt miRNAs and 23- to 26-nt sRNAs from the opposite strand of the hairpin in Medicago (43). However, the biogenesis feature of these 23- to 26-nt sRNAs was not examined either. We speculate that some of these 23- to 26-nt sRNAs might be siRNAs. miR165/166 were shown to direct DNA methylation downstream of their target sites on PHABULOSA (PHB) and PHAVOLUTA (PHV) coding regions, and the methylation level was not affected in dcl1 and ago1 mutants (10). Both MIR165 and MIR166 can give rise to 23- to 26-nt sRNAs, and the majority of which are associated with AGO4 and AGO7 (Supplementary Table S2). Furthermore, the existence of both sRNA species at miR165 site has been revealed by northern blot analysis (9). It is likely that the real players that are responsible for DNA methylation in PHB and PHV genes are the 23- to 26-nt siRNAs generated from the miR165/miR166 sites, which explained why the DNA methylation level was not altered in dcl1 and ago1 mutants. The miRNAs and siRNAs derived from these dual role MIR genes likely have different modes of action. The 21-nt miRNAs are mainly associated with AGO1 and may mediate post-transcriptional gene silencing by mRNA cleavage or translational inhibition. On the other hand, the MIR gene-derived siRNAs are mainly associated with AGO4 and likely mediate DNA methylation at some of their target loci.
We previously reported that AtlsiRNA-1 is generated by DCL1 and requires RDR6 and Pol IV for its biogenesis (27). AtlsiRNA-1 suppresses gene expression by triggering mRNA decapping and 5′-3′-degradation. Here, we found another 27-nt lsiRNA, which was generated from the MIR2831-5P locus and was DCL3 dependent. This result suggests that different DCL proteins could be potentially involved in generating lsiRNAs. These lsiRNAs are unlikely to be generated by the imprecise dicing activity of DCLs because if that were the case, we would expect to have precise dicing products in the same region. However, no precise dicing products were observed: no 21-nt band products were observed in the case of the DCL1-dependent AtlsiRNA-1 (27), and no 24-nt band products were observed in the case of the DCL3-dependent lsiRNA from the MIR2831-5P locus (Figure 1C). Thus, these lsiRNAs appear to be true products of DCL proteins. DCL proteins may be involved in processing one end of the lsiRNAs, and the other end of lsiRNAs may involve other unidentified ribonucleases. piRNAs are good examples of products from two different ribonucleases (44–46).
Here, we show that a significant number of MIR genes in Arabidopsis have the dual function of generating both miRNAs and siRNAs from the same site. Our systematic analysis using rice and moss sRNA deep sequencing datasets suggests that these dual-function MIR genes are broadly present in plant species. Note that moss is among the earliest land plants on earth, and Arabidopsis and rice are evolutionarily distant from each other—the former is a dicotyledonous plant while the latter belongs to monocotyledons. The existence of dual-function MIR genes in these three plants suggests that the underlying mechanism is conserved. The fact that such genes exist in moss alludes to their possible evolutionary origin in this ancient land plant. These dual-function MIRs should be evolutionally beneficial, because they regulate target gene expression at both transcriptional and post-transcriptional levels by using dual modes of action—siRNA-mediated DNA methylation and miRNA-mediated mRNA degradation and translational inhibition.
After this paper was published on line new names were assigned by miRBASE for the following microRNAs:
ath-MIR2883a has changed to ath-MIR3932a
ath-MIR2883b has changed to ath-MIR3932b
ath-MIR2328 has changed to ath-MIR3933
ath-MIR2831 has changed to ath-MIR3440b
ACCESSION NUMBER
NCBI/GEO (GSE19694).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSF Career Award MCB-0642843; National Institute of Health R01GM093008-01; University of California Discovery (grant Bio06-10566); AES-CE Research Allocation Award PPA-7517H (to H.J.); NSF (grant DBI-0743797); National Institute of Health (grants RC1AR05868101 and U54AI05716006S1); Monsanto Company (to W.Z.); Swiss National Science Foundation Ambizione (grant PZ00P3_126329/1 to F.V.). Funding for open access charge: NSF Career Award (MCB-0642843).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Baulcombe, Jim Carrington, Herve Vaucheret, Jian-Kang Zhu, Steve Jacobsen, John Clarke, Xuemei Chen, Adam Vivian-Smith and Zhixin Xie for providing seeds of various mutants. We thank Daisuke Miki for his advice on bisulfite sequencing.
REFERENCES
- 1.Vazquez F, Legrand S, Windels D. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 2010;15:337–345. doi: 10.1016/j.tplants.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S37–S37. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 4.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez F, Blevins T, Ailhas J, Boller T, Meins FJ. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:629–630. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu JK, Staskawicz BJ, Jin HL. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chellappan P, Jin H. Discovery of plant microRNAs and short-interfering RNAs by deep parallel sequencing. Methods Mol. Biol. 2009;495:121–132. doi: 10.1007/978-1-59745-477-3_11. [DOI] [PubMed] [Google Scholar]
- 16.Pall GS, Servat-C C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:E60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu J-K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinformatics. 2008;24:i50–i58. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SH, Addo-Quaye C, Coruh C, Arif MA, Ma Z, Frank W, Axtell MJ. Physcomitrella patens DCL3 is required for 22-24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 2008;4:e1000314. doi: 10.1371/journal.pgen.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones-Rhoades MW, Bartel DP. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 24.Fahlgren N, Carrington JC. miRNA Target Prediction in Plants. Methods Mol. Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 25.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gen. Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Gen. Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl Acad. Sci. USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu B, Yang ZY, Li JJ, Minakhina S, Yang MC, Padgett RW, Steward R, Chen XM. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 33.Qi YJ, He XY, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 34.Mi SJ, Cai T, Hu YG, Chen Y, Hodges E, Ni FR, Wu L, Li S, Zhou H, Long CZ, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery TA, Howell MD, Cuperus JT, Li DW, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 36.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol. Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl Acad. Sci. USA. 2010;107:466–471. doi: 10.1073/pnas.0913203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 40.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of MicroRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010;20:49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 41.Meyers BC, Simon SA, Zhai J. MicroRNA processing: battle of the bulge. Curr. Biol. 2010;20:R68–R70. doi: 10.1016/j.cub.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Zheng B, Wang Z, Li S, Yu B, Liu J, Chen X. Intergenic transcription by RNA Polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, Gouzy J, Crespi M. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 45.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.