Abstract
Background
Genetic variants on chromosome 4q25 are associated with atrial fibrillation (AF). We sought to determine whether there is more than one susceptibility signal at this locus.
Methods and Results
34 haplotype-tagging single nucleotide polymorphisms (SNPs) at the 4q25 locus were genotyped in 790 case and 1,177 control subjects from Massachusetts General Hospital and tested for association with AF. We replicated SNPs associated with AF after adjustment for the most significantly associated SNP in 5,066 case and 30,661 referent subjects from the German Competence Network for Atrial Fibrillation, Atherosclerosis Risk in Communities Study, Cleveland Clinic Lone AF study, Cardiovascular Health Study, and Rotterdam Study. All subjects were of European ancestry. A multimarker risk score comprised of SNPs tagging distinct AF susceptibility signals was constructed and tested for association with AF, and all results were meta-analyzed. The previously reported SNP, rs2200733, was most significantly associated with AF (minor allele odds ratio 1.80, 95%CI 1.50-2.15, P=1.2×10−20) in the discovery sample. Adjusting for rs2200733 genotype revealed 2 additional susceptibility signals marked by rs17570669 and rs3853445. A graded risk of AF was observed with an increasing number of AF risk alleles at SNPs tagging these 3 susceptibility signals.
Conclusions
We identified 2 novel AF susceptibility signals on chromosome 4q25. Consideration of multiple susceptibility signals at chromosome 4q25 identifies individuals with an increased risk of AF and may localize regulatory elements at the locus with particular biological relevance in the pathogenesis of AF.
Keywords: atrial fibrillation, electrophysiology, genetics, epidemiology, risk factors
Atrial fibrillation (AF) is the most common arrhythmia encountered in clinical practice, and is associated with substantial morbidity1 and societal healthcare costs.2 Whereas many risk factors for AF have been identified, the recognition of a common heritable component underlying AF3,4 indicates that genetic variation may play a role in its pathogenesis.
We recently participated in a genome-wide association study that identified a disease susceptibility locus for AF on chromosome 4q25 in individuals of European and Asian descent.5 We replicated the association between the most significantly associated single nucleotide polymorphism (SNP), rs2200733, and AF in a subsequent study of 3,508 subjects with AF and 12,173 controls from 4 additional cohorts of European ancestry.6 A meta-analysis of the results from both studies revealed an odds ratio (OR) of 1.9 for the rs2200733 risk allele (95% confidence interval [CI] 1.60-2.26, P=3.3×10−13).6 We and others have again replicated the chromosome 4q25 AF susceptibility locus in subsequent genome-wide association studies for AF.7-9
In the present study, we sought to identify whether there are multiple AF susceptibility signals at the 4q25 locus in individuals of European ancestry by performing fine mapping of common SNPs in the region and replicating associations in independent study samples. We further sought to determine whether the consideration of multiple markers associated with AF at this locus could further refine the association signal.
METHODS
Study samples
Detailed descriptions of the study cohorts are provided in the supplement. Individuals in the discovery stage of the analysis were drawn from 2 different samples at the Massachusetts General Hospital (MGH) and pooled for analysis. These samples included patients with lone AF referred to the Cardiac Arrhythmia Service starting in June 2001 in whom AF was documented by electrocardiogram before 66 years of age, and patients with AF by electrocardiogram or history who were admitted to the MGH Stroke service between January 1998 and July 2006 with an acute ischemic or hemorrhagic stroke. Referent subjects from MGH were from a large, primary care practice of greater than 18,000 patients serving the hospital catchment area. Absence of AF was documented through interview and from review of medical records including all available electrocardiograms.
Genetic variants associated with AF in the discovery sample, after adjustment for the top SNP (see statistical analysis below), were genotyped in an independent replication study sample comprised of subjects from the German Competence Network for Atrial Fibrillation (AFNET), a national registry of AF patients.10 AF was confirmed by electrocardiogram, and DNA samples were collected from patients with AF in whom onset occurred before 60 years of age. Referent subjects were derived from a community-based epidemiologic survey study conducted between 1999 and 2001 of persons living in or near the city of Augsburg, Southern Germany (KORA S4), and were excluded if they reported a history of AF, had signs or symptoms of AF on physical examination, or absence of sinus rhythm on a required electrocardiogram.11
We performed in silico replication of associations between AF and SNPs representing distinct susceptibility signals in MGH and AFNET in 4 additional study samples with previously genotyped subjects. The Atherosclerosis Risk in Communities (ARIC) study is a prospective population-based study of cardiovascular disease in the United States consisting of participants aged 45 to 64 years at enrollment. Subjects included in this analysis included those recruited from 3 United States communities (suburbs of Minneapolis, Minnesota; Washington County, Maryland; and Forsyth County, North Carolina) between 1987-1989.12 The Cleveland Clinic Lone AF Study (CCAF) is comprised of case subjects with AF in the absence of significant structural heart disease. Referent subjects were population controls from Studies 64, 65, 66 and 67 in the Illumina iControl database, a publicly accessible database of genotype and phenotype data from control genome-wide association study populations. The Cardiovascular Health Study (CHS) is a prospective population-based study of cardiovascular disease in individuals 65 years or older recruited from 4 Field Centers in the United States (Forsyth County, NC; Sacramento County, CA; Washington County, MD; Pittsburgh, PA).13 The Rotterdam Study (RS) is a community-based longitudinal study of elderly individuals from a suburb of Rotterdam founded in 1990 with a focus on identifying determinants of health and cardiovascular, neurogeriatric, bone, and eye diseases.14
Prevalent AF was defined as events that occurred in individuals prior to an individual’s DNA collection in cohort studies and on the basis of AF ascertainment in case-control studies. Incident AF was defined as events that occurred after DNA collection among participants free of AF at DNA collection in cohort studies. Subjects were restricted to those of self-reported European descent.
SNP selection and genotyping
A 200 kilobase (kb) region extending from the PITX2 gene to approximately 50 kb beyond the previously reported SNP rs2200733 was considered for fine mapping of the chromosome 4q25 locus. All SNPs on chromosome 4 between positions 111,780,000 and 111,985,000 with a minor allele frequency of 5% or greater were identified from the HapMap CEU dataset release 22 (NCBI build 36, dbSNP build 126). We identified 35 haplotype-tagging SNPs (r2 ≥ 0.8) in this region using the Tagger program within Haploview version 4.0.15 Additionally, 6 SNPs that were moderately correlated with rs2200733 (r2 between 0.2 and 0.8) also were selected.
We extracted DNA from whole blood of each subject using standard techniques. In the MGH and AFNET samples, genotyping was performed using PCR, iPlex single base primer extension, and matrix-assisted laser desorption/ionization-time of flight mass spectrometry in a 384-well-format (Sequenom, San Diego, CA) according to the manufacturer’s instructions. Data were analyzed with SpectroTYPER 3.4 software and cluster plots were visually inspected and manually curated to confirm genotyping calls. The genotyping platforms for the remaining cohorts were Affymetrix 6.0 (ARIC), Illumina Hap550 v3 and Illumina Hap610 v1 (CCAF cases), Illumina Hap550 v1 or v3 (CCAF referents), Illumina 370 CNV (CHS), and Illumina Infinium HumanHap550 v3 (RS). Only directly genotyped SNPs were included in the analysis, with the exception of rs17570669 during the replication stage in the CCAF sample. Imputation in CCAF was performed using MACH v1.0.1616 with the HapMap CEU reference panel (NCBI built 36) (Rsq 0.6144 for the cases and 0.7302 for the iControlDB controls). In CHS, genotypes for rs17570669 were imputed using BIMBAM v0.99,17 but were not included in the analyses owing to poor imputation quality (ratio of observed to expected genotype variance of 0.11).
The Institutional Review Board or Medical Ethics Committee, as appropriate for participating institutions, approved study procedures. Written informed consent was obtained from all study subjects or their proxies, including consent to use DNA for genetic analyses of cardiovascular disease.
Statistical Analysis
We tested each SNP included in the discovery stage for deviation from Hardy-Weinberg Equilibrium using an exact test18 and excluded the SNP if the P Value was ≤1×10−4 in referent subjects. This corresponds to an experiment-wide Hardy-Weinberg significance threshold of (0.005/41≈1×10−4). We tested the remaining SNPs for association with AF in the MGH sample using logistic regression assuming an additive genetic model, and subsequently adjusted for the genotype of the top SNP in order to identify independent associations with AF. Significance thresholds were adjusted for multiple testing using the Bonferroni method with an experiment-wide error rate of 0.05. Thirty-four SNPs passed quality control measures, and therefore the adjusted significance threshold was P<0.001 (0.05/34). We then tested SNPs significantly associated with AF after adjusting for the top SNP in the MGH discovery sample for association with AF in the AFNET sample.
In order to identify SNPs representing distinct AF susceptibility signals, we calculated pairwise linkage disequilibrium measures r2 and D’ in the MGH and AFNET samples and constructed linkage disequilibrium blocks using Haploview15 with previously described definitions.20 We inferred haplotypes comprised of SNPs located on the same block from unphased data using an expectation-maximization algorithm, and tested haplotypes for association with AF using a weighted logistic regression model adjusting for genotypes of the remaining SNPs marking separate AF susceptibility signals.21 Haplotypes were weighted according to the posterior probability of possible haplotype pairs for each individual subject.21
The SNPs associated with AF in both the MGH and AFNET samples that were markers for independent signals were then assessed for association with AF in the additional replication cohorts using logistic regression in samples with prevalent AF (CCAF, CHS, RS) and Cox proportional hazards regression in samples with incident AF (ARIC, CHS, RS). Individuals were censored at death, loss to follow-up, or date of last contact. Person-time for the incident analyses began at the time of DNA collection. Associations were adjusted for significant principal components of race for those studies in which population structure was associated with AF. Both prevalent and incident associations were meta-analyzed using an inverse variance weighted fixed effects method.22
As indirectly measured haplotype phasing is accompanied by uncertainty, we used the combination of genotypes at SNPs marking each distinct AF susceptibility signal to define a multimarker variable for each individual. We assessed the association between each multimarker combination of genotypes and AF relative to the most common combination of genotypes for these SNPs, allowing for separate effects for each genotype combination. The effects were meta-analyzed as described above. In samples with incident AF, the time-dependent area under the receiver operating characteristic curve was estimated for models with and without the multimarker variables included.23
For the discovery stage of the analysis in the MGH sample of 790 cases and 1,177 referent subjects, we estimated that we would have 41% power to detect ORs of 1.5 for risk alleles with frequencies of 5%, and 80% power to detect ORs of 1.5 for risk alleles with frequencies of at least 10%, assuming a two-sided alpha level of 0.001 and population disease prevalence of 1%.24
Statistical analyses were performed using PLINK version 1.06,25 SAS version 9.1.3 (SAS Institute, Cary, NC), and R version 2.11.26 Regional association plots were prepared using SNAP.19
RESULTS
A total of 790 subjects with AF and 1,177 referent subjects from MGH were included in the discovery stage of the analysis. Among the 790 case subjects, 488 were from the MGH lone AF cohort, and 302 from the MGH stroke cohort (Table 1). The overall call rate for the 34 SNPs tested for association with AF was 98.9%.
Table 1. Characteristics of study subjects.
| Stage Analysis Cohort |
Discovery Prevalent MGH |
Replication | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prevalent | Incident | |||||||||||||||
| AFNET | CCAF | CHS | RS | ARIC | CHS | RS | ||||||||||
| Affection status |
AF | No AF | AF | No AF | AF | No AF | AF | No AF | AF | No AF | AF | No AF | AF | No AF | AF | No AF |
| Number | 790 | 1,177 | 2,145 | 4,073 | 496 | 2,971 | 66 | 3,205 | 309 | 5,665 | 743 | 7,184 | 765 | 2,440 | 542 | 5,123 |
| Age (yrs) | 63±15 | 67±13 | 49±14 | 61±12 | 58±11 | 28±22 | 76±6 | 72±5 | 76±8 | 69±9 | 57±5 | 54±6 | 73±6 | 72±5 | 72±8 | 69±9 |
| Female | 31 | 47 | 27 | 51 | 24 | 62 | 52 | 39 | 53 | 60 | 40 | 54 | 45 | 37 | 54 | 60 |
| Hypertension | 49 | 57 | 56 | 18 | 54 | Unknown | 52 | 52 | 42 | 33 | 44 | 25 | 59 | 50 | 45 | 32 |
Data presented as mean ± standard deviation or %.
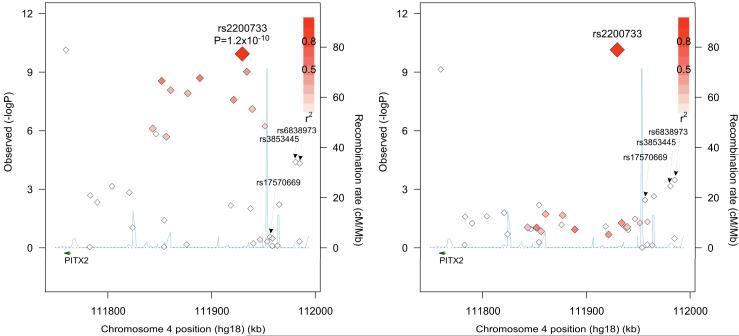
There was a strong association between AF and SNPs on chromosome 4q25 (Table 2, Figure 1A, and Supplemental Table 1). Among the 34 SNPs examined, 15 exceeded the significance threshold of P<0.001 after adjusting for age, sex, and hypertension. The most significant association with AF observed at this locus was with the previously reported SNP, rs2200733, with an OR for the minor T allele of 1.80, 95% CI 1.50-2.15, P=1.2 ×10−10. A second SNP previously reported to confer an independent risk of AF, rs10033464,5 was not significantly associated with AF in our sample (OR for minor T allele 1.07, 95% CI 0.84-1.35, P=0.59).
Table 2. Fine mapping of the locus for AF on chromosome 4q25 in the discovery sample from MGH.
| Single nucleotide polymorphism |
Position | Minor/ major allele |
Minor allele frequency (%) |
Adjusted OR (95% CI)* |
P Value | |
|---|---|---|---|---|---|---|
| AF | No AF | |||||
| rs17554590 | 111,782,351 | G/C | 1.7 | 2.0 | 0.97 (0.59-1.61) | 0.91 |
| rs2595098 | 111,782,931 | A/T | 4.8 | 7.1 | 0.63 (0.46-0.84) | 2.9×10−3 |
| rs1448818 | 111,789,672 | C/A | 30.1 | 25.7 | 1.24 (1.07-1.44) | 4.7×10−3 |
| rs12498374 | 111,803,868 | T/C | 25.0 | 19.9 | 1.32 (1.12-1.55) | 7.0×10−4 |
| rs1448822 | 111,820,547 | A/G | 36.0 | 30.6 | 1.26 (1.09-1.45) | 1.5×10−3 |
| rs13120244 | 111,823,793 | A/G | 10.0 | 12.4 | 0.83 (0.67-1.03) | 0.09 |
| rs1900827 | 111,843,188 | T/C | 40.0 | 31.2 | 1.41 (1.23-1.62) | 7.8×10−7 |
| rs4371683 | 111,846,216 | A/C | 40.1 | 31.6 | 1.40 (1.22-1.61) | 1.5×10−6 |
| rs17042026 | 111,851,823 | A/G | 26.1 | 16.8 | 1.64 (1.39-1.92) | 2.8×10−9 |
| rs12646859 | 111,854,082 | G/T | 14.1 | 14.7 | 0.99 (0.82-1.19) | 0.89 |
| rs10222783 | 111,854,275 | T/C | 3.6 | 2.3 | 1.56 (1.02-2.37) | 0.04 |
| rs2595085 | 111,856,222 | G/C | 40.2 | 31.8 | 1.40 (1.22-1.60) | 2.0×10−6 |
| rs1448817 | 111,860,502 | G/A | 38.0 | 27.6 | 1.51 (1.31-1.74) | 8.4×10−9 |
| rs11098090 | 111,875,857 | C/T | 14.8 | 14.2 | 1.04 (0.86-1.26) | 0.68 |
| rs4307025 | 111,876,952 | A/T | 37.8 | 27.4 | 1.50 (1.31-1.73) | 1.2×10−8 |
| rs2634071 | 111,888,669 | T/C | 28.9 | 19.2 | 1.60 (1.37-1.87) | 2.0×10−9 |
| rs2723333 | 111,918,540 | A/G | 8.9 | 12.0 | 0.74 (0.59-0.92) | 6.7×10−3 |
| rs1906615 | 111,921,247 | T/G | 30.1 | 20.6 | 1.54 (1.32-1.80) | 2.6×10−8 |
| rs2200733 | 111,929,618 | T/C | 21.5 | 11.7 | 1.80 (1.50-2.15) | 1.2×10−10 |
| rs13143308 | 111,933,868 | T/G | 31.7 | 21.1 | 1.60 (1.38-1.86) | 9.5×10−10 |
| rs13105878 | 111,937,596 | A/C | 7.8 | 10.8 | 0.73 (0.58-0.93) | 9.7×10−3 |
| rs11931959 | 111,939,134 | G/A | 38.2 | 28.5 | 1.47 (1.28-1.69) | 7.8×10−8 |
| rs10033464 | 111,940,210 | T/G | 9.2 | 8.5 | 1.07 (0.84-1.35) | 0.59 |
| rs3855819 | 111,946,612 | G/C | 14.3 | 13.2 | 1.09 (0.90-1.32) | 0.38 |
| rs6533531 | 111,951,414 | G/T | 47.3 | 37.0 | 1.43 (1.24-1.64) | 5.9×10−7 |
| rs3853444 | 111,953,585 | C/T | 29.7 | 30.8 | 0.95 (0.82-1.10) | 0.49 |
| rs17570669 | 111,956,331 | T/A | 7.2 | 8.5 | 0.87 (0.67-1.12) | 0.28 |
| rs13130446 | 111,958,605 | T/C | 49.8 | 49.8 | 1.02 (0.89-1.16) | 0.80 |
| rs10516564 | 111,958,741 | G/A | 29.1 | 30.0 | 0.93 (0.80-1.08) | 0.33 |
| rs3866834 | 111,963,462 | A/G | 33.9 | 34.0 | 1.02 (0.89-1.18) | 0.79 |
| rs4124163 | 111,965,048 | G/A | 3.0 | 5.0 | 0.61 (0.42-0.87) | 6.1×10−3 |
| rs3853445 | 111,980,936 | C/T | 20.3 | 26.2 | 0.71 (0.61-0.84) | 4.1×10−5 |
| rs6838901 | 111,984,764 | C/G | 13.6 | 14.6 | 0.93 (0.77-1.13) | 0.47 |
| rs6838973 | 111,984,944 | T/C | 34.6 | 41.5 | 0.75 (0.66-0.86) | 4.8×10−5 |
Genomic position from NCBI build 36. OR is the odds ratio corresponding to the minor allele.
Adjusted for age, sex, and hypertension.
Figure 1. Regional plot of SNPs associated with prevalent AF in the MGH discovery sample.
Panel A displays the associations between SNPs included in the analysis and AF in the MGH sample, adjusted for age, sex and hypertension. Panel B displays the associations after additional adjustment for rs2200733 genotype, with the rs2200733 position corresponding to the unadjusted association significance level. SNPs are plotted according to their genomic position (NCBI Build 36) and –log10 P value for the association. The intensity of shading for each SNP corresponds to the strength of linkage disequilibrium (r2) relative to rs2200733. Estimated recombination rates are shown by the blue line. PITX2 is indicated by the dark green arrow. LD and recombination rates are based on the CEU HapMap release 22. SNPs that were associated with AF after adjusting for rs2200733 genotype and meta-analyzing results from both the MGH and AFNET samples are labeled. Figures were prepared using SNAP.19
We then performed analyses adjusting for rs2200733 genotype. The 4 SNPs most significantly associated with AF were all located within 30 kb of one another and telomeric to rs2200733 (Figure 1, and Supplemental Table 2). Associations between these 4 SNPs and AF after adjusting for age, sex, hypertension and rs2200733 genotype (rs17570669 OR for minor T allele 0.60, 95% CI 0.46-0.78, P=2.0×10−4; rs4124163 OR for minor G allele 0.56, 95% CI 0.39-0.81, P=2.3×10−3; rs6838973 OR for minor T allele 0.77, 95% CI 0.67-0.89, P=3.4×10−4; and rs3853445 OR for minor C allele 0.75, 95% CI 0.64-0.89, P=6.9×10−4) were similar to unadjusted associations (Supplemental Table 2). The minor alleles for each of these 4 SNPs were associated with a lower risk of AF. While rs3853445 and rs6838973 were both associated with AF prior to adjusting for rs2200733, the association between rs17570669 and AF was not evident until after adjusting for rs2200733. There was a suggestion of a separate signal associated with AF centromeric to rs2200733; however, no SNPs in this region were significantly associated with AF after adjusting for multiple comparisons (Figure 1 and Supplemental Table 2).
Characteristics of the 2,145 case and 4,073 referent subjects in the AFNET sample are displayed in Table 1. In addition to rs2200733, 3 of the 4 SNPs associated with AF in the MGH discovery cohort analysis passed quality control measures with an overall call rate of 94.4% and were tested for association with AF. As in the MGH sample, when adjusting for age, sex, and hypertension, rs2200733 (P=3.8×10−52), rs3853445 (P=2.14×10−7), and rs6838973 (P=1.45×10−8) were associated with AF but rs17570669 (P=0.28) was not. After additional adjustment for rs2200733 genotype, significant associations with AF were observed for the remaining 3 SNPs (rs17570669 minor T allele OR 0.64, 95% CI 0.54-0.77, P=5.3×10−7; rs3853445 minor C allele OR 0.82, 95% CI 0.74-0.91, P=1.1×10−4; rs6838973 minor T allele OR 0.81, 95% CI 0.74-0.89, P=5.4×10−6).
Pairwise linkage disequilibrium measures revealed that SNPs rs3853445 and rs6838973 were moderately correlated (r2 0.43 and 0.41 in MGH and AFNET, respectively) and located on the same haplotype block in both samples, suggesting that associations between each of these 2 SNPs and AF represented the same signal (Supplemental Table 3). As rs6838973 was not directly genotyped in the remaining replication samples, rs3853445 was used for subsequent analyses as a marker for this susceptibility signal. In contrast, there was a very low level of correlation between the remaining SNPs (r2 < 0.11 for all pairwise comparisons), as expected based on the haplotype-tagging SNP selection method.
We therefore tested the three independent susceptibility signals for association with AF by modeling the two independent SNPs along with inferred rs3853445 and rs6838973 haplotypes (Table 3). The haplotype consisting of the minor alleles for both rs3853445 and rs6838973 (CT) occurred with a frequency of 23% in the MGH sample and 26% in the AFNET sample, and conferred a reduced odds of AF relative to the major allele haplotype (TC) after adjusting for rs2200733 and rs17570669 genotype (combined OR 0.78, 95%CI 0.71-0.87, P=1.75×10−6). The remaining two susceptibility signals marked by rs2200733 and rs17570669 remained significantly associated with AF in the combined analysis, though the association for rs17570669 was attenuated in the MGH sample.
Table 3. Associations between AF and rs2200733, rs17570669, and common rs3853445 | rs6838973 haplotypes in the MGH and AFNET samples.
| MGH | AFNET | Combined† | ||||||
|---|---|---|---|---|---|---|---|---|
| Variant | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | ||
| rs2200733 | 1.87 (1.54-2.27) | 2.4×10−10 | 2.64 (2.32-3.00) | 3.4×10−49 | 2.37 (2.13-2.64) | 3.1×10−56 | ||
| rs17570669 | 0.74 (0.55-1.01) | 0.06 | 0.70 (0.58-0.85) | 2.8×10−4 | 0.71 (0.61-0.84) | 4.3×10−5 | ||
|
| ||||||||
| rs3853445 | rs6838973 Haplotype |
Haplotype
Frequency |
Haplotype
Frequency |
||||||
| TC | 0.60 | Reference | - | 0.56 | Reference | - | Reference | |
| TT | 0.16 | 0.92 (0.75-1.13) | 0.45 | 0.17 | 0.99 (0.86-1.13) | 0.89 | 0.97 (0.87-1.09) | 0.59 |
| CC | 0.01 | 1.24 (0.58-2.66) | 0.57 | 0.01 | 1.87 (1.21-2.91) | 5.0×10−3 | 1.69 (1.16-2.47) | 7.0×10−3 |
| CT* | 0.23 | 0.75 (0.63-0.90) | 1.6×10−3 | 0.26 | 0.80 (0.71-0.90) | 2.7×10−4 | 0.78 (0.71-0.87) | 1.75×10−6 |
Adjusted for age, sex, and hypertension. OR is the odds ratio corresponding to the minor allele or specified haplotype.
Comprised of the minor alleles for each respective SNP.
Meta-analysis performed using a fixed effects method.
Associations between SNPs representing distinct AF susceptibility signals in the MGH and AFNET samples were then tested for replication in the ARIC, CCAF, CHS, and RS study samples. Characteristics of these samples are displayed in Table 1. In general, the associations replicated in the prevalent AF samples, but not in the incident AF samples, though effect estimates tended to be in the same direction as those observed in the discovery sample (Figure 2).
Figure 2. SNPs associated with AF after adjusting for rs2200733 genotype.
SNPs associated with AF after adjusting for rs2200733 genotype in the MGH discovery sample were tested for association in the replication samples. The meta-analyzed effects are plotted according to prevalent (odds ratio), incident (hazard ratio), or pooled (relative risk) analysis status. Associations are adjusted for age, sex, and hypertension (MGH, AFNET, ARIC, CHS, RS) or sex only (CCAF). Samples with prevalent AF included MGH, AFNET, CHS, CCAF, and RS. Samples with incident AF included ARIC, CHS, and RS.
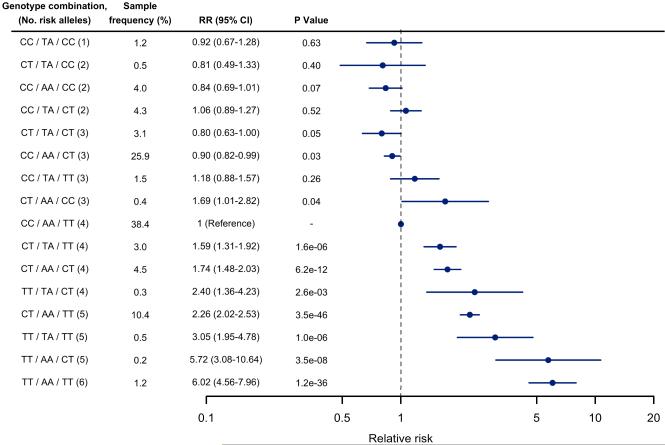
We then calculated the relative risk of AF for each multimarker combination of genotypes for SNPs tagging each of the 3 AF susceptibility signals relative to the most common combination of genotypes in each of the study samples (Figure 3). The multimarker analysis indicated a graded risk of AF generally corresponding to the number of AF risk alleles, though the risks varied within strata defined by numbers of risk alleles, and confidence intervals overlap for many of the multimarker groups owing to small sample sizes. The relative risks (RR) of AF for the 3 most frequent rs2200733 / rs17570669 / rs3853445 genotype combinations, relative to the most common genotype combination CC/AA/TT comprised of 4 AF risk alleles (38% of subjects), were 0.90 (95% CI 0.82-0.99, CC/AA/CT [3 risk alleles, 26% of subjects]), 1.74 (95% CI 1.48-2.03, CT/AA/CT [4 risk alleles, 5% of subjects]), and 2.26 (95% CI 2.02-2.53, CT/AA/TT [5 risk alleles, [10% of subjects]). The greatest RR was observed with the combination comprised of both AF risk alleles at each of the 3 SNPs, TT/AA/CC, which occurred in approximately 1% of subjects (RR 6.02, 95% CI 4.56-7.96). The associations stratified by prevalent or incident status are displayed in (Supplemental Figure 2).
Figure 3. Multimarker risk score for AF based on combined rs2200733, rs17570669, and rs3853445 genotypes.
The meta-analyzed relative risk of AF for each multimarker combination of rs2200733, rs17570669, and rs3853445 genotypes relative to the most common multimarker combination are shown. Only multimarker combinations with an average sample frequency of ≥ 0.2% are displayed, though the effects are adjusted for all potential combinations, as well as age, sex, and hypertension, or sex only (CCAF). Individuals with incomplete genotypes were not included. Risk alleles for AF were the minor T allele for rs2200733, the major A allele for rs17570669, and the major T allele for rs3853445.
In samples with incident AF, the time-dependent area under the curve for a model with age, sex and hypertension was 0.70 (95% CI 0.69-0.72) and 0.68 (95% CI 0.66-0.70) over the follow-up periods in ARIC and RS, respectively. The AUC increased to 0.72 (95% CI 0.70-0.73) and 0.70 (95% CI 0.68-0.72), respectively, after addition of the multimarker allele combinations.
DISCUSSION
In our sample of subjects from MGH, the previously reported SNP, rs2200733, remained the variant most significantly associated with AF even after consideration of other SNPs at the chromosome 4q25 locus. In addition to this signal, we identified 2 novel AF susceptibility signals after adjustment for rs2200733 genotype in a meta-analysis of 5,856 subjects with AF and 31,838 without AF, all of whom were of European ancestry. A multimarker risk-score comprised of SNPs tagging each of these 3 AF susceptibility signals on chromosome 4q25 identified individuals at varying risks of developing AF which approximately corresponded to the number of AF risk alleles present.
Our results reinforce the association between chromosome 4q25 and AF and extend knowledge by defining the genetic architecture of this locus and its relation to AF.5-8 Specifically, SNPs rs17570669, rs3853445, and rs6838973 were confined to a 30 kb region out of the 200 kb region assayed, and located within 50 kb telomeric of rs2200733. The locus studied in our analysis is marked by regions that appear to be phylogenetically conserved (Supplemental Figure 3).27 Indeed, there is emerging evidence that highly conserved non-coding regions may act as regulatory elements, and underlie phenotypic diversity.28,29 However, the mechanism by which genetic variation at the chromosome 4q25 locus leads to AF remains unknown.
The lack of significant association in the incident AF samples between AF and SNPs rs17570669 and rs3853445, after adjustment for rs2200733 genotype, may reflect reduced power in the incident stratum, an absence of true association when modeled in this fashion, interactions between variants and unmeasured clinical factors that differ between prevalent and incident AF, or other phenotypic heterogeneity between prevalent and incident AF. A multimarker risk-score comprised of AF risk alleles at SNPs tagging these 3 susceptibility signals identified individuals predisposed to the development of AF, raising the possibility that consideration of multiple genetic variants at the chromosome 4q25 locus may help improve risk stratification of individuals at risk for AF.30 Whether the consideration of the 3 signals identified in our analysis will contribute to AF prediction in the context of additional variants associated with AF7-9,31,32 is presently unclear but merits examination in larger prospective datasets.
Limitations
Our analysis was restricted to individuals of European descent, and therefore the findings may not be generalizable to individuals of other races and ethnicities. Although we could not assess for population stratification in the MGH and AFNET samples, the relative homogeneity of our cohorts limits the likelihood of such confounding. Furthermore, we adjusted analyses for population structure in replication cohorts when there was evidence of association with AF. We did not restrict the age of subjects included in the analysis. Although the chromosome 4q25 region has been associated with AF in subjects with a diverse spectrum of presumed etiologies,7,9,33,34 including those with lone9 as well as typical forms of AF,7 sources of heterogeneity in the discovery cohort that were not accounted for may have affected the SNPs selected for replication. The haplotype and multi-marker risk score analyses represent multiple testing, and several subgroups were of small sample size as reflected by the wide confidence intervals accompanying some effect estimates; positive associations may represent winners’ curse.35 Although we did not adjust for multiple testing at this stage, the observed associations are supported by the fact that SNPs tested in these analyses were selected after conservative Bonferroni-adjusted association thresholds in the discovery analysis. Moreover, patterns of association for these subgroups were consistent across independent study samples. We are unable to rule out longer distance associations at the locus beyond the boundaries of our SNP selection region. Lastly, while we cannot exclude the possibility that all of the identified SNPs merely tag a single, common element associated with AF, the low correlation between the SNPs identified in our analysis and the haplotype association analysis suggest that these SNPs represent three independent signals at the chromosome 4q25 locus that affect AF risk.
Conclusion
We confirmed the strong association between AF and rs2200733 at the chromosome 4q25 locus and identified 2 novel disease susceptibility signals telomeric to this SNP that are associated with AF. Simultaneous consideration of these signals identifies individuals with an increased risk for AF.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of the AFNET/KORA, ARIC, CHS, CCAF, MGH, and RS studies for their important contributions.
Funding Sources
MGH: This study was funded by grants to Dr. Ellinor from the Deane Institute for Integrative Research in Atrial Fibrillation and Stroke, and grants from the NIH to Dr. Rosand (R01NS059727), Dr. Furie (P50NS051343), Dr. Lubitz (T32HL007575), and Drs. Ellinor, Benjamin, and Lunetta (RO1HL092577, RC1HL101056, DA027021).
AFNET: The study was also supported by the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN), the German National Competence network on atrial fibrillation (AFNET), the Leducq Foundation (07-CVD 03), the D.W. Reynolds Foundation Clinical Cardiovascular Research Center at Johns Hopkins University, and the Bioinformatics for the Functional Analysis of Mammalian Genomes program (BFAM) by grants to Stefan Kääb (01GS0499, 01GI0204, 01GS0838). The Cooperative Health Research in the Region of Augsburg (KORA) platform is funded by the Helmholtz Zentrum München, the BMBF partly in the context of the NGFN, the State of Bavaria and as part of LMUinnovativ.
ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367, R01HL086694, RC1 HL101056; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. This analysis was also supported by American Heart Association grant 09SDG2280087 and National Heart Lung and Blood Institute/NIH grant 1RC1HL099452.
CCAF: The Cleveland Clinic Lone AF Study is supported by an R01 HL090620 from the National Heart, Lung, and Blood Institute (Drs. Chung, Barnard, and Smith); NIH/National Center for Research Resources, Clinical Translational Science Award 1UL-RR024989 (Dr. Chung); Heart and Vascular Institute, Department of Cardiovascular Medicine, Cleveland Clinic (Dr. Chung). P50 HL077107 from the National Heart, Lung, and Blood Institute (Dr. Barnard)
CHS: The Cardiovascular Health Study research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
RS: The GWA database of the Rotterdam Study was funded through the Netherlands Organization of Scientific Research NWO (nr. 175.010.2005.011). The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Scientific Research (NWO), the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
Footnotes
Conflict of Interest Disclosures
None to report.
Journal Subject Codes: [5] Arrhythmias, clinical electrophysiology, drugs; [109] Clinical genetics; [8] Epidemiology; [89] Genetics of cardiovascular disease
Clinical Trial Registration Information: NA
References
- 1.Fuster V, Ryden LE, Cannom DS, Crijns HJ, Curtis AB, Ellenbogen KA, Halperin JL, Le Heuzey JY, Kay GN, Lowe JE, Olsson SB, Prystowsky EN, Tamargo JL, Wann S, Smith SC, Jr., Jacobs AK, Adams CD, Anderson JL, Antman EM, Halperin JL, Hunt SA, Nishimura R, Ornato JP, Page RL, Riegel B, Priori SG, Blanc JJ, Budaj A, Camm AJ, Dean V, Deckers JW, Despres C, Dickstein K, Lekakis J, McGregor K, Metra M, Morais J, Osterspey A, Tamargo JL, Zamorano JL. ACC/AHA/ESC 2006 Guidelines for the Management of Patients with Atrial Fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Circulation. 2006;114:e257–354. doi: 10.1161/CIRCULATIONAHA.106.177292. [DOI] [PubMed] [Google Scholar]
- 2.Lee WC, Lamas GA, Balu S, Spalding J, Wang Q, Pashos CL. Direct treatment cost of atrial fibrillation in the elderly American population: a Medicare perspective. J Med Econ. 2008;11:281–298. doi: 10.3111/13696990802063425. [DOI] [PubMed] [Google Scholar]
- 3.Fox CS, Parise H, D’Agostino RB, Sr., Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 4.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 6.Kaab S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, Schnabel R, Makino S, Sinner MF, Kannankeril PJ, Beckmann BM, Choudry S, Donahue BS, Heeringa J, Perz S, Lunetta KL, Larson MG, Levy D, MacRae CA, Ruskin JN, Wacker A, Schomig A, Wichmann HE, Steinbeck G, Meitinger T, Uitterlinden AG, Witteman JC, Roden DM, Benjamin EJ, Ellinor PT. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D’Agostino RB, Sr., Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiriksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Kottgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kaab S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinor PT, Lunetta KL, G NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel R, Soliman EZ, Rice K, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen S, Steinbeck G, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WH, Vasan RS, Nothen MM, MacRae CA, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Psaty BM, Roden D, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR. Common Variants in KCNN3 are Associated with Lone Atrial Fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. T M. Ka□a□b S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabauer M, Gerth A, Limbourg T, Schneider S, Oeff M, Kirchhof P, Goette A, Lewalter T, Ravens U, Meinertz T, Breithardt G, Steinbeck G. The Registry of the German Competence NETwork on Atrial Fibrillation: patient characteristics and initial management. Europace. 2009;11:423–434. doi: 10.1093/europace/eun369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wichmann HE, Gieger C, Illig T. KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 12.The ARIC investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 13.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O’Leary DH, Psaty BM, Rautaharju P, Tracy RP, Weiler PG. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 14.Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, Tiemeier H, Uitterlinden AG, Vingerling JR, Witteman JC. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.MACH 1.0. 2008 Accessed at http://www.sph.umich.edu/csg/abecasis/MaCH/index.html.
- 17.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Sinnwell JP, Schaid DJ, Yu Z. Haplo Stats 1.4.4. 2009 Accessed at http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm.
- 22.Egger M, Smith GD, Altman DG. Systematic reviews in health care: meta-analysis in context. 2 ed. BMJ; London: 2001. [Google Scholar]
- 23.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Development Core Team R: A language and environment for statistical computing. 2010 http://www.R-project.org.
- 27.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 29.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 30.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB, Sr., Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 32.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WH, Kottgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BH, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Muller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kaab S, Witteman JC, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, Perry TE, Muehlschlegel JD, Aranki S, Donahue BS, Pretorius M, Estrada JC, Ellinor PT, Newton-Cheh C, Seidman CE, Seidman JG, Herman DS, Lichtner P, Meitinger T, Pfeufer A, Kaab S, Brown NJ, Roden DM, Darbar D. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viviani Anselmi C, Novelli V, Roncarati R, Malovini A, Bellazzi R, Bronzini R, Marchese G, Condorelli G, Montenero AS, Puca AA. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart. 2008;94:1394–1396. doi: 10.1136/hrt.2008.148544. [DOI] [PubMed] [Google Scholar]
- 35.Nakaoka H, Inoue I. Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner’s curse. J Hum Genet. 2009;54:615–623. doi: 10.1038/jhg.2009.95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.