Abstract
Cell responses are actuated by tightly controlled signal transduction pathways. Although the concept of an integrated signaling network replete with interpathway cross-talk and feedback regulation is broadly appreciated, kinetic data of the type needed to characterize such interactions in conjunction with mathematical models are lacking. In mammalian cells, the Ras/ERK pathway controls cell proliferation and other responses stimulated by growth factors, and several cross-talk and feedback mechanisms affecting its activation have been identified. In this work, we take a systematic approach to parse the magnitudes of multiple regulatory mechanisms that attenuate ERK activation through canonical (Ras-dependent) and non-canonical (PI3K-dependent) pathways. In addition to regulation of receptor and ligand levels, we consider three layers of ERK-dependent feedback: desensitization of Ras activation, negative regulation of MEK kinase (e.g. Raf) activities, and up-regulation of dual-specificity ERK phosphatases. Our results establish the second of these as the dominant mode of ERK self-regulation in mouse fibroblasts. We further demonstrate that kinetic models of signaling networks, trained on a sufficient diversity of quantitative data, can be reasonably comprehensive, accurate, and predictive in the dynamical sense.
Keywords: Computer Modeling, Dual Specificity Phosphoprotein Phosphatase, Growth Factors, Kinetics, MAP Kinases (MAPKs), Phosphatidylinositol 3-Kinase, Raf, Ras, Receptor Tyrosine Kinase
Introduction
Mammalian cells recognize and respond to chemical stimuli through ligation of specific receptors at the cell surface, which in turn activate highly conserved intracellular signal transduction pathways. These pathways elicit growth and proliferation, polarization and migration, differentiation, and other responses by actuating cell gene-regulatory and cytoskeletal systems. Obviously, signal transduction must be tightly regulated, as spurious intracellular signaling is associated with autonomous cell proliferation, invasive cell migration, and other molecular signatures of cancer progression (1–4).
The concept of a signaling pathway provides a useful framework for understanding the flow of information as an ordered series of activation processes, exemplified by the Ras → Raf → mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) kinase (MEK) → ERK pathway and other MAPK cascades, which control diverse responses in cells stimulated by various growth factors and cytokines (5–7). Our current understanding of signal transduction, however, encompasses the concept of signaling networks, in which the canonical pathways interact with and thus affect one another (cross-talk); the sequential pathway concept is further challenged by the regulation of signaling through negative feedback and, in some cases, reinforcement of signaling through positive feedback (8–11). These complexities of signaling networks have proven difficult to characterize, and most of the data that has accumulated about such mechanisms are qualitative in nature and scattered across different experimental contexts. Although kinetic models of signal transduction processes have steadily appeared over the past decade, and recently published models of the epidermal growth factor receptor system in particular have been more tightly integrated with biochemical data to establish quantitative features of signaling networks (12–14), a more comprehensive data acquisition effort is needed to better constrain models at the network scale of complexity.
We previously conducted a quantitative analysis of cross-talk in the platelet-derived growth factor (PDGF) receptor network (15). The major signaling modes mediated by PDGF receptors are the phosphoinositide 3-kinase (PI3K) pathway and the aforementioned Ras/ERK pathway, which are most closely associated with chemotaxis and cell proliferation, respectively (6, 16). Through measurements of PDGF-stimulated signaling in mouse fibroblasts, systematically covering a diverse array of stimulation and molecular perturbation conditions and building upon other quantitative studies (17–20), we showed that PDGF-stimulated ERK activation requires signaling through either of two pathways: the canonical, Ras-dependent pathway or PI3K-dependent cross-talk. PI3K-dependent signaling positively modulates the ERK pathway, while the PI3K pathway is not significantly affected by endogenous Ras signaling. Through quantitative analysis of a coarse-grained kinetic model, we estimated that the magnitudes of the Ras- and PI3K-dependent contributions to MEK/ERK activation are comparable; the PI3K-dependent pathway was found to be only moderately more potent (1.6:1 ratio), once negative feedback desensitization of Ras-GTP loading was taken into account (15).
We have since refined our kinetic model and acquired additional data to quantify negative feedback regulation of ERK signaling through multiple feedback loops (Fig. 1). Although there is a sound theoretical understanding of (9, 21, 22), and quantitative information about (23–25), the magnitude and kinetics of ERK pathway adaptation in mammalian cells, it has not heretofore been demonstrated that one can parse the contributions of multiple cross-talk and feedback interactions as a function of time and dose of stimulus. To do so will be critically important if we are to understand and predict naturally occurring and interventional modifications of signaling networks (26, 27). Indeed, paradoxical effects of Raf expression and pharmacological inhibition on ERK signaling have been reported recently (28, 29) and highlight the need to characterize the balance between activation and desensitization of the cascade more quantitatively.
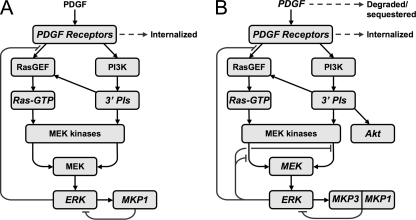
FIGURE 1.
Refined model of the ERK signaling network. A, our previous model focused on quantifying cross-talk from PI3K-dependent signaling impinging upon the canonical Ras/ERK pathway and included two ERK-dependent negative feedback loops: desensitization of Ras-GEF recruitment and up-regulation of MKP/DUSP activity. The model also quantitatively accounted for endocytosis of activated PDGF receptor dimers as an ERK-independent attenuation mechanism. B, present study additionally accounts for ERK-dependent feedback impinging on Ras- and PI3K-dependent MEK kinase activities and includes depletion of PDGF from the extracellular medium. The experimental readouts used to constrain each model are indicated in italics (A and B).
Three layers of ERK-dependent feedback are included in the current model: 1) desensitization of Ras-guanine nucleotide exchange factor (GEF)2 recruitment through hyperphosphorylation of Sos (30–32), 2) desensitization of MEK kinases, especially isoforms of Raf (Raf-1, B-Raf, and A-Raf) through phosphorylation on known regulatory sites (33–36), and 3) transcriptional up-regulation of MAPK phosphatases (MKPs)/dual specificity phosphatases (DUSPs) that dephosphorylate ERK (37). Our analysis shows that the second of these, directly affecting MEK phosphorylation, is in fact the dominant layer of ERK self-regulation in our cells, accounting for >90% of the signal attenuation. We additionally found significant depletion of growth factor from the extracellular medium, which affects signaling at subsaturating growth factor concentrations. Support for the refined mathematical model, trained by alignment to the superset of old and new data (>300 distinct experimental measurements), is demonstrated through its ability to quantitatively predict the enhancement of PDGF-stimulated MEK phosphorylation in cells with both ERK1 and ERK2 expression knocked down. A more surprising model prediction, also confirmed experimentally, is a lack of effect of MKP3/DUSP6 knockdown on ERK phosphorylation.
EXPERIMENTAL PROCEDURES
Reagents
All tissue culture reagents were from Invitrogen (Carlsbad, CA). Human recombinant PDGF-BB was from Peprotech (Rocky Hill, NJ). Antibodies against total ERK1/2 and MKP3 and phosphospecific antibodies against Akt pSer473, ERK pThr202/pTyr204, MEK pSer217/pSer221, and Raf-1 pSer289/pSer296/pSer301 were from Cell Signaling Technology (Beverly, MA); antibodies against total Akt1/2 were from Santa Cruz Biotechnology (Santa Cruz, CA). Human PDGF-BB ELISA kit, with PDGF β-receptor/Fc chimera as the capture reagent, was from R&D Systems (Minneapolis, MN). Pharmacological inhibitors were from CalBiochem or, in the case of MG-132, Sigma-Aldrich; where applicable, cells were pre-incubated with the inhibitor for 30–60 min prior to PDGF stimulation. The siGENOME siRNA reagents and siGENOME SMARTpool siRNAs against mouse MKP3 (GeneID: 67603), ERK1 (GeneID: 26417), and ERK2 (GeneID: 26413) and siGENOME Non-Targeting siRNA Pool #2 were purchased from Dharmacon (Lafayette, CO). Unless otherwise noted, all other reagents were from Sigma-Aldrich.
Cell Culture and siRNA Transfection
NIH 3T3 fibroblasts (American Type Culture Collection, Manassas, VA) were cultured at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and the antibiotics penicillin and streptomycin. Where applicable, NIH 3T3 cells were serially infected with retrovirus bearing empty vector or S17N H-Ras and selected using puromycin prior to each experiment, as described previously (15, 20). NIH 3T3 cells were transfected with siRNAs according to the manufacturer's protocol and incubated for 3 days prior to the experiment.
Lysate Preparation and Quantitative Immunoblotting
Cells were serum-starved for 4 h prior to stimulation. Detergent lysates were prepared for quantitative immunoblotting, and immunoblots were performed using enhanced chemiluminescence, as described previously (17). Blots comparing lysates prepared on the same day, representing either different inhibitor treatments or different cell variants and respective control conditions, were performed in parallel and exposed at the same time. The Bio-Rad Fluor S-Max system, which gives a linear response with respect to light output, was used, and band intensity was quantified using local background subtraction. Immunoblot data were first normalized by an appropriate loading control and then further normalized to evaluate the consistency of relative trends across independent experiments, based on the 1 nm time course for the control condition, as described in detail previously (15).
Kinetic Model and Computational Analysis
The refined mathematical model of the PDGF receptor network is illustrated conceptually in Fig. 1B and described in detail in supplemental Text S1. PDGF receptor binding, dimerization, and endocytosis, and the production of 3′ phosphoinositides by receptor-recruited PI3K, are modeled essentially as described previously (17, 19). This portion of the model was supplemented with a differential equation accounting for depletion of PDGF-BB from the extracellular medium. Our previous coarse-grained model of Ras- and PI3K-dependent MEK kinase/MEK/ERK activation and regulation (15) was supplemented with ERK-dependent negative feedbacks affecting the MEK kinase activities and ERK-dependent regulation of MKP3 expression. It also allows that MKP1, MKP3, and/or a constitutive level of dual-specificity phosphatase activity contribute(s) to the dephosphorylation of ERK. Other phosphatases, such as those acting on phosphorylated MEK and MEK kinases, appear in the model with constant activities assumed.
The parameter estimation approach used is related to the algorithm described previously (15), with certain modifications as described in detail in supplemental Text S1; both are Monte Carlo-based and generate a large (n = 10,000) ensemble of “good” parameter sets (supplemental Table S1) rather than one “best” fit, but here a modified simulated annealing protocol was designed. After acquiring the ensemble, the model output is recalculated for each parameter set, and at each time point, an ensemble mean and standard deviation are calculated.
RESULTS
MEK Phosphorylation Kinetics Reveal a Potent, Intermediate Layer of Negative Feedback Regulation
We first show that MEK phosphorylation is regulated in a manner that cannot be explained by feedback loops impinging upstream of Ras or at the level of ERK phosphatases. Whereas our previous model was constrained by quantitative measurements of Ras-GTP loading and ERK phosphorylation, measurements of MEK phosphorylation kinetics in the same cell backgrounds provide critical data and mechanistic insights about the regulation of the pathway (Fig. 2). Referring to the diagrams in Fig. 1, it is clear why: the activation of both MEK and ERK reflect the integration of Ras- and PI3K-dependent inputs to the pathway, but unlike ERK, MEK is not directly affected by modulation of MKP/DUSP levels.
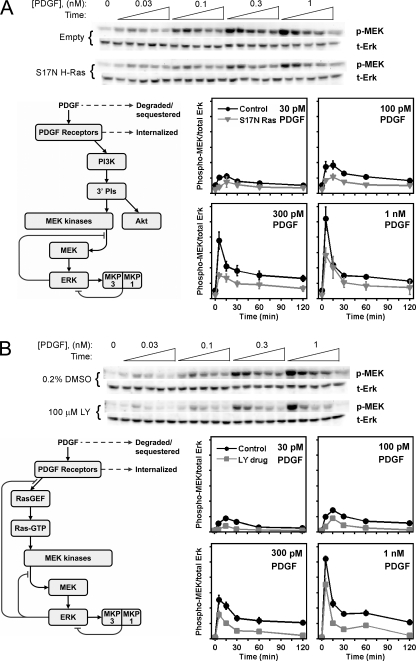
FIGURE 2.
PDGF-stimulated MEK phosphorylation is strongly regulated by negative feedback and is sensitive to ablation of Ras or PI3K signaling. PDGF-stimulated MEK1/2 phosphorylation (p-MEK) kinetics in NIH 3T3 cells were assessed by quantitative immunoblotting and normalized by total ERK1 (t-Erk). The blots shown are representative of three independent experiments; samples were drawn from lysates used previously to probe phosphorylation of ERK and Akt (15). Values are normalized as previously described and are reported as mean ± S.E. in arbitrary units (n = 3). A, comparison of cells expressing dominant-negative (S17N) H-Ras to the empty vector control. B, comparison of PI3K inhibition (100 μm LY294002) to the 0.2% DMSO vehicle control.
Samples were obtained from among the same NIH 3T3 cell lysates used previously to quantify ERK and Akt phosphorylation. Systematic quantification of Ras- and PI3K-dependent contributions to MEK activation was achieved through inhibition of PI3K and Ras, by incubation with LY294002 compound and expression of dominant-negative (S17N) H-Ras, respectively. The results show that PDGF-stimulated MEK phosphorylation is generally transient and sensitive to ablation of either Ras (emphasizing the PI3K-dependent pathway; Fig. 2A) or PI3K (emphasizing the Ras-dependent pathway; Fig. 2B) signaling. Conceptually, the transience of MEK phosphorylation might seem to be consistent with the previously reported ERK phosphorylation kinetics (15); however, our previous model, using parameter values fit without the benefit of MEK data, predicts sustained MEK phosphorylation with only a small overshoot (supplemental Fig. S1). Indeed, the previous scheme cannot possibly explain how MEK phosphorylation is transient in Ras-inhibited cells stimulated with a high dose of PDGF; in the previous model, partial adaptation of ERK phosphorylation under those conditions had been solely attributed to up-regulation of MKP activity, downstream of MEK. The new results identify desensitization of MEK phosphorylation, downstream of Ras and PI3K, as an important regulatory mechanism in the ERK signaling network.
Dual Specificity Phosphatases MKP3 and MKP1 Are Modulated with Distinct Kinetics in PDGF-stimulated Cells, but Their Expression Levels Do Not Affect ERK Dephosphorylation
We next present evidence that feedback at the level of modulating two DUSP isoforms, MKP1/DUSP1 and MKP3/DUSP6, does not significantly impact ERK phosphorylation kinetics. We showed previously that high doses of PDGF elicit 3–5-fold up-regulation of MKP1 in our cells (15). In the context of the previous model, this negative feedback loop was important for explaining partial adaptation of the ERK phosphorylation response, especially as activated by the PI3K-dependent pathway as explained above; although the potential importance of ERK-MKP feedback has also been emphasized in a number of mathematical models of ERK signaling, it must be acknowledged that expression of MKP1 in particular might not be a quantitative indicator of ERK dephosphorylation.
Indeed, other DUSP isoforms, especially MKP3, are thought to be more important in that regard (37), and we found that MKP3 and MKP1 expression levels are modulated quite differently in our cells. Whereas MKP1 expression increases sharply (after a time lag) and plateaus in response to high PDGF doses (15), MKP3 expression rapidly decays and then recovers, as quantified in Fig. 3, A and B. These kinetics are consistent with ERK-dependent modulation of both synthesis and degradation of MKP3 (38).
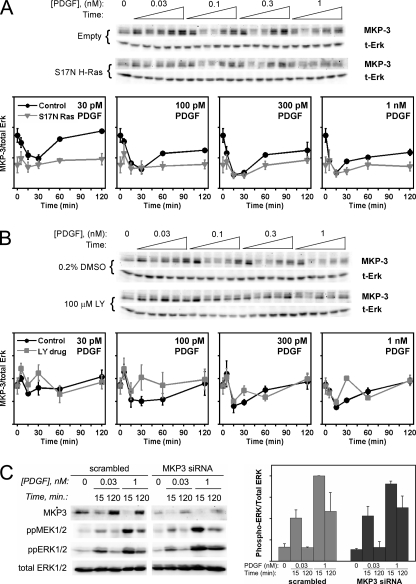
FIGURE 3.
MKP3 expression is regulated in response to PDGF stimulation but does not affect ERK phosphorylation. MKP3/DUSP6 expression in PDGF-stimulated NIH 3T3 cells was monitored by quantitative immunoblotting. The blots shown are representative of three independent experiments; samples were drawn from lysates used previously to probe phosphorylation of ERK and Akt (15). Values are normalized as previously described and are reported as mean ± S.E. (n = 3) in arbitrary units. A, comparison of cells expressing dominant-negative (S17N) H-Ras to the empty vector control. B, comparison of PI3K inhibition (100 μm LY294002) to the 0.2% DMSO vehicle control. C, siRNA knockdown of MKP3 does not affect MEK or ERK phosphorylation stimulated by PDGF in our cells. For each immunoblot, control and MKP3 siRNA bands were cropped from the same blot, which is representative of two independent experiments. The graph shows quantification of the ERK phosphorylation results (normalized by the maximum value and reported as mean ± S.E.).
Surprisingly, despite the complex regulation of these two DUSPs, we found that ERK phosphorylation is not sensitive to changes in either of their endogenous expression levels. Reduction of MKP3 expression by RNA interference (≈60–70% knockdown) had no significant effect on the kinetics or dose responsiveness of PDGF-stimulated ERK phosphorylation, as compared with cells treated with a scrambled oligonucleotide control (Fig. 3C); these results stand in contrast with published data using porcine aortic endothelial cells with heterologous expression of PDGF receptors (38). Similarly, the expectation that ERK phosphorylation might be negatively correlated with changes in MKP1 expression does not hold in cells treated with MG-132, a proteasome inhibitor that amplifies MKP1 up-regulation, or SP600125, an inhibitor of c-Jun N-terminal kinase activity that has the opposite effect (supplemental Fig. S2). Although these results do not rule out the possibility that ERK phosphorylation is shaped by feedback regulation of other DUSP isoforms, they do further suggest that the primary determinant of ERK adaptation in this system is the transience of MEK activation. Indeed, using our refined mathematical model, we will show that this sufficiently and quantitatively explains the kinetics of the ERK network under all conditions tested.
Adaptation of Subsaturated PDGF Receptor-mediated Signaling Is Also Affected by PDGF Depletion from the Extracellular Medium
To round out the data needed to accurately quantify the mechanisms that contribute to adaptation of signaling, we sought to ensure that dynamics affecting PDGF receptor activation were sufficiently characterized. Because our previous framework already accounted for endocytosis as a mechanism for PDGF receptor down-regulation (17, 20), we speculated that depletion of PDGF from the external medium might significantly temper prolonged responses to low PDGF doses (Fig. 4).
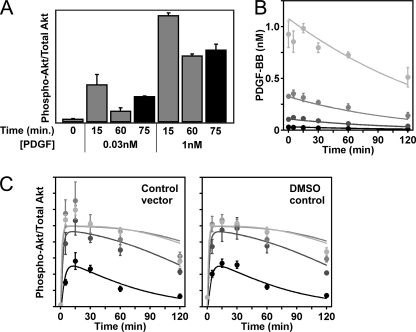
FIGURE 4.
Accounting for PDGF depletion from the extracellular medium as a secondary mode of signal adaptation. A, PDGF add-back experiments. NIH 3T3 cells were stimulated with a low (30 pm) or high (1 nm) dose of PDGF-BB for the indicated times, and phosphorylated and total Akt levels were quantified by immunoblotting (cropped from the same blot). For the 75 min. time point (black bars), the PDGF solution was replaced at 60 min. Values are mean ± S.E. (two independent experiments) in arbitrary units. B and C, PDGF depletion kinetics (B) were measured by PDGF-BB ELISA (mean ± S.E., three independent experiments) and reconciled against Akt phosphorylation kinetics (C) for different control conditions (empty vector and DMSO vehicle controls) taken from (15). PDGF concentrations are: 30% gray, 1 nm; 50% gray, 300 pm; 70% gray, 100 pm; black, 30 pm. Solid curves are kinetic model calculations (supplemental Text S1) representing the best global fit to both data types.
To evaluate the significance of PDGF depletion, we measured time courses of PI3K-dependent Akt phosphorylation for low (30 pm) and high (1 nm) doses of PDGF-BB, and prior to the final time point of 75 min, the medium was aspirated (at 60 min) and replaced with the same initial PDGF concentration. The results confirm that the Akt phosphorylation level recovers substantially (by roughly 2-fold) once the low PDGF dose is replenished (Fig. 4A). As expected, this is not true of the high PDGF dose, because the PI3K/Akt pathway is maximally activated (saturated) as long as the external PDGF concentration exceeds roughly 0.5 nm (17).
The kinetics of PDGF-BB depletion were directly quantified by enzyme-linked immunosorbent assay (ELISA) (Fig. 4B). The results show that, for initial doses ranging from 0.03–1 nm, the extent of depletion over 2 h is 40–50%. To demonstrate the consistency of these data with those of PDGF-stimulated Akt phosphorylation reported previously (15), we achieved a satisfactory global fit of both data types to a minimal submodel of the pathway kinetics (Fig. 4, B and C). The two types of experiments were performed using roughly the same cell densities and the same volume of medium, and the fit to the data were highly constrained, as only 5 of the submodel parameters were adjusted (see supplemental Text S1 for details).
The Refined Mathematical Model of the PDGF Receptor Network Reconciles All Existing Measurements and Allows a Better Fit to Previously Acquired Data
The current model is illustrated conceptually in Fig. 1B and described in detail in supplemental Text S1. It has a total of 22 state variables and 57 adjustable rate parameters; of the parameters, 14 are fixed at constant values, based on previous work and the constrained fit to the ligand depletion and Akt phosphorylation data described in the previous section. The remaining 43 parameters were estimated by direct and global alignment with the rest of the data, which included the kinetics of Ras-GTP loading, ERK phosphorylation, and MKP1 levels reported previously (15) and the newly acquired MEK phosphorylation and MKP3 expression data. Thus, whereas our previous analysis required fewer fit parameters, the present analysis further constrains the model fit with a disproportionately higher number of readouts and nearly double the number of data points for comparison (Table 1). As in our previous work, the approach is not designed to identify a single set of “best” parameter values but rather a large ensemble (n = 10,000) of parameter sets that perform almost equally well in fitting the data. Analysis of those parameter sets (supplemental Table S1) indicates which parameters are constrained well by the data and which are less so.
TABLE 1.
Comparison of the current and previous PDGF receptor signaling network models
The fit refers to the Monte Carlo parameter fitting of phosphorylated ERK, Ras-GTP, MKP1 expression, and (in the case of this work) MEK phosphorylation and MKP3 expression readouts, as shown in Fig. 5 and supplemental Fig. S3. Data points fit refers to the number of distinct experimental measurements, i.e. not considering experimental replicates. It also does not include the PDGF depletion and Akt phosphorylation data shown in Fig. 4, which were fit separately. The sum of squared deviations (SSD) for each readout is reported as the mean ± S.D. for the 10,000 parameter sets in each ensemble.
| This work | Wang et al. (15) | |
|---|---|---|
| Variables | 23 | 18 |
| Parameters (fit) | 57 (43) | 44 (34) |
| Data points fit | 337 | 169 |
| SSDERK, n = 104 | 3.05 ± 0.18 | 4.30 ± 0.25 |
| SSDRas, n = 21 | 1.09 ± 0.14 | 1.13 ± 0.17 |
| SSDMKP1, n = 44 | 1.44 ± 0.12 | 1.62 ± 0.18 |
| SSDMEK, n = 84 | 1.96 ± 0.14 | |
| SSDMKP3, n = 84 | 2.38 ± 0.09 |
The results show the quality of the fit and the full array of quantitative data used for alignment (Fig. 5A and supplemental Fig. S3). Unlike the previous version, the current model accurately captures the newly quantified MEK phosphorylation kinetics, and it outperforms the previous version in fitting ERK phosphorylation kinetics (Table 1). Notably, the current model properly “spreads” the peak ERK phosphorylation levels for the four PDGF doses, and it captures the ERK phosphorylation kinetics of the S17N Ras, 30 pm PDGF time course that was missed by the previous model (15).
FIGURE 5.
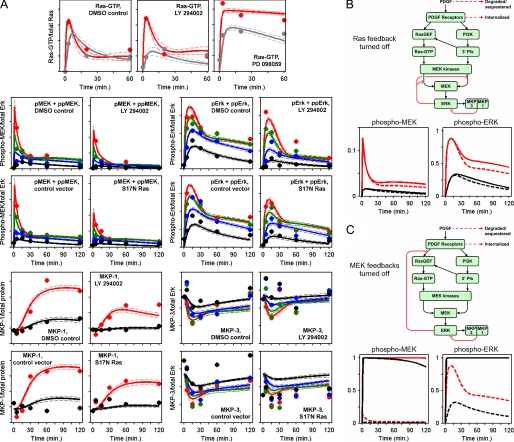
The refined network model reconciles all experimental data acquired to date. A, simulated annealing algorithm was used to align the kinetic model to the data as indicated, thus collecting an ensemble of parameter sets that fit our data set well (supplemental Text S1). Solid curves are ensemble means, and the dashed curves are mean ± S.D. (n = 10,000). The mean data values used to constrain the model (symbols) are also shown. PDGF concentrations are: red, 1 nm; green, 300 pm; blue, 100 pm; gray, 50 pm; black, 30 pm. B and C, feedback desensitization of MEK kinase activities is the dominant mode of ERK self-regulation in the model. Model predictions (ensemble means) of MEK and ERK phosphorylation are shown. PDGF concentrations are: red, 1 nm; black, 30 pm. Solid curves are hypothetical scenarios in which each of the following ERK-dependent feedback loops is selectively turned off: Ras-GEF desensitization (B) or MEK kinase desensitization (C). Dashed curves are with all ERK-dependent feedback loops intact, as shown in A.
Among the insights that we can glean directly from the parameter statistics, relevant to feedback regulation of the network, is the tendency of the fit to marginalize the contributions of both MKP1 and MKP3 (parameters β1 and β3), in relation to a third, time-invariant ERK phosphatase activity. This is a bona fide model prediction, because the experimental data showing the same were not used to constrain the model. Quantitatively, the model accurately predicts the results of the MKP3 knockdown experiment shown in Fig. 3C (supplemental Fig. S4).
The lack of sensitivity of ERK phosphorylation to MKP1 and MKP3 dynamics, which are characterized by 11 of the 43 global fit parameters in the current model, together with considerations of fast reactions and enzymatic reactions or complexes operating far from saturation (supplemental Table S1), indicate that the model can be simplified without significantly affecting predictions about ERK signaling. It is conservatively estimated that less than half of the parameters are needed to achieve essentially the same fit of the Ras, MEK, and ERK data.
Further analysis of the computational model reveals the relative magnitudes of the negative feedbacks impinging upstream and downstream of Ras (Fig. 5, B and C), which reconcile the constraints imposed by the experimentally determined Ras-GTP loading and MEK phosphorylation kinetics. We simulated a scenario in which Ras-GEF desensitization is selectively and completely turned off in each of the 10,000 parameter sets, with all other feedbacks intact (Fig. 5B). This enhances the rates of MEK and ERK phosphorylation through the Ras-dependent pathway; however, the predicted increases in MEK/ERK phosphorylation levels are rather modest. By comparison, selectively turning off MEK kinase desensitization in the model results in nearly stoichiometric activation of both MEK and ERK (Fig. 5C). This analysis suggests that negative feedback at the point of MEK phosphorylation is the dominant mode of ERK pathway self-regulation.
The Refined Network Model Successfully Predicts the Collective Strength of ERK-dependent Negative Feedback
To test this hypothesis and thus establish with greater confidence the magnitudes of ERK-dependent negative feedback loops, we abrogated ERK activity by RNA interference and measured the attendant effect on MEK phosphorylation (Fig. 6). This experimental test of the model probes the desensitization of MEK phosphorylation almost directly, because both the previous and current models quantitatively account for ERK-dependent desensitization of Ras-GTP loading, based on experiments using a MEK inhibitor (15). Such inhibitors function by binding MEK1/2 and preventing their activation, and thus are likely to obscure effects on MEK phosphorylation, motivating the siRNA approach used here.
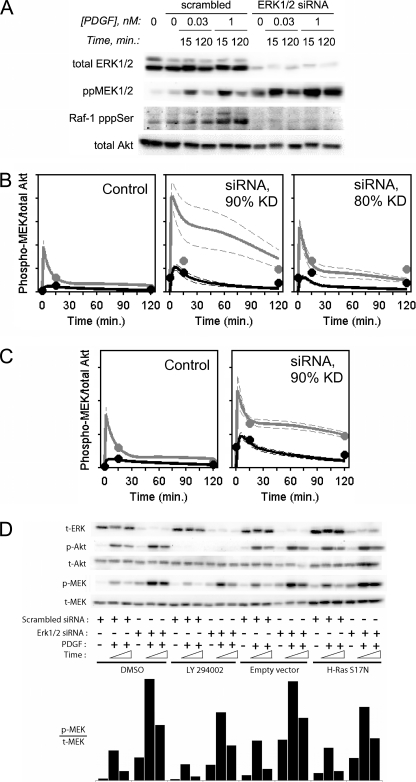
FIGURE 6.
siRNA knockdown of ERK1 and ERK2 enhances Ras- and PI3K-dependent MEK phosphorylation, as quantitatively predicted by the current model. A, NIH 3T3 cells were transfected with siRNAs directed against ERK1 and ERK2; their pan-ERK expression and PDGF-stimulated MEK1/2 phosphorylation were measured by quantitative immunoblotting in parallel with a scrambled siRNA control. Raf-1 phosphorylation on negative regulatory sites controlled by ERK (Ser289/Ser296/Ser301) and total Akt (as a loading control) were also assessed. The results are representative of two independent experiments. B, the quantified results from A are overlaid with a priori kinetic model predictions of MEK phosphorylation kinetics, assuming 90 or 80% knockdown of ERK in the model; solid curves represent ensemble means, and dashed curves are mean ± S.D. (n = 10,000). PDGF concentrations are: gray, 1 nm; black, 30 pm. C, same as B, except that the model was refit with the additional data included in the alignment, assuming 90% ERK knockdown by siRNA treatment (Ensemble 2; see supplemental Fig. S5). D, NIH 3T3 cells were transfected with siRNAs directed against ERK1 and ERK2 or with a scrambled siRNA control; PI3K activity and Ras-GTP accumulation were blocked using 100 μm LY294002 and expression of dominant-negative (S17N) H-Ras, alongside their appropriate controls, to isolate Ras- and PI3K-dependent pathways, respectively. The cells were either unstimulated or stimulated with 1 nm PDGF-BB for 15 or 120 min as indicated. Lysates were probed for total ERK expression and phosphorylation of Akt and MEK1/2 by quantitative immunoblotting, with total Akt and MEK levels as loading controls. Quantification of phospho-MEK/total MEK ratio is shown. The results are representative of two independent experiments.
Using siRNAs targeting both ERK1 and ERK2, we achieved 80–90% knockdown of both isoforms and confirmed that ERK1/2 depletion yields a dramatic increase in MEK phosphorylation, consistent with relief of ERK-dependent negative feedback. Accordingly, PDGF-stimulated phosphorylation of Raf-1 on known negative regulatory sites (34) was abrogated in the ERK1/2-depleted cells (Fig. 6A). Corresponding a priori predictions of MEK phosphorylation kinetics were generated using our quantitative model, assuming reductions of ERK expression by 80 and 90%, and those predictions show nice agreement with the experimental data (Fig. 6B). We note that the calculated enhancement of MEK phosphorylation is sensitive to the extent of ERK knockdown, as the residual ERK retains a certain potency of feedback regulation in the model, and this contributes to the uncertainty of the prediction. Nevertheless, the model could be re-fit, incorporating the ERK siRNA data (assuming 90% knockdown) in the alignment without compromising the overall quality of fit for the rest of the data (Fig. 6C and supplemental Fig. S5). MEK phosphorylation was also enhanced in cells with ERK1/2 depleted and either PI3K or Ras signaling blocked (Fig. 6D), in semi-quantitative agreement with corresponding model predictions (supplemental Fig. S6). The effect of ERK knockdown in dominant-negative Ras-expressing cells is especially telling, as it rules out the possibility that ERK-dependent negative feedback acts predominantly upstream of Ras. We conclude that the current model is refined to the extent that it accurately accounts for the dynamics of ERK-dependent negative feedback at multiple levels of the pathway.
DISCUSSION
Adaptation of intracellular signaling has long been recognized as a cornerstone of cell regulation. The concept is well known in the field of chemotaxis, for example, where exact or nearly complete adaptation of the sensory output is thought to enable cells to respond to chemoattractant gradients spanning a broad range of concentrations (39–43). Coupled with ultrasensitivity or positive feedback, it is well understood that negative feedback can produce spiking/oscillatory responses, as in calcium signaling and regulation of the cell cycle (44, 45). At least in principle, the ERK pathway is also capable of oscillations (21, 46); however, in the context of growth factor receptor-mediated ERK signaling, the more plausible role of negative feedback regulation is that of partial adaptation, modulating what is ultimately a biologically meaningful (quasi-) steady state (47, 48). Indeed, more than 30 years ago, it was shown that PDGF stimulation renders cells competent for (but not necessarily committed to) DNA synthesis, and that this process requires exposure to PDGF for varying lengths of time, on the scale of hours, depending on the dose of growth factor (49).
The topology of a negative regulatory mechanism imposes certain limitations on its kinetic properties. The direct inhibition of upstream signaling components by a MAPK does not readily foster strong adaptation of its output response, because MAPK activation and onset of the feedback are essentially the same process (9). Thus, in our experiments as well as in our mathematical model, ERK phosphorylation exhibits a much less dramatic peak and decline compared with MEK phosphorylation. Some degree of adaptation is attributed to the desensitization of Ras-GTP loading, the magnitude of which was established from our previous experiments (15); however, our new results revealed that the predominant level of feedback regulation lies downstream of Ras, for example through ERK-dependent phosphorylation of Raf isoforms (33–36). This seems to be especially important in the context of the PDGF receptor network, because signaling through Ras is not the only pathway to ERK, nor is it necessarily the dominant one. By the same token, regulation of the MEK kinase layer allows for differential regulation of parallel signaling pathways that branch off from Ras (50).
The issue of quantifying cross-talk (PI3K-dependent) and canonical (Ras-dependent) pathways to ERK, which was the primary focus of our previous model, raises a generally important question about model refinement. As additional data come to light, and regulatory mechanisms are added to models, will we find the conclusions drawn from previous analyses to be invalid or obsolete? The ratio of PI3K-dependent/Ras-dependent signaling inputs, the MEK activation comparator (MAC), had been estimated to be roughly 1.6 once the negative feedback affecting Ras-GEF activity had been taken into account (15); for the present model, we calculated an analogous, time-varying quantity, the “dynamic MAC” (supplemental Fig. S7). Under maximal stimulation conditions and with all feedbacks intact, the ensemble-averaged dynamic MAC varies within the approximate range between 1 and 2, consistent with the previously estimated, “static” MAC value cited above; in terms of the overall inputs to MEK, incorporating feedback regulation of upstream components, the models are in semi-quantitative agreement. Where the models differ is in the nature of the desensitization. With Ras-GEF desensitization selectively turned off in the present model, the value of the dynamic MAC hovers between 1–1.3 for maximal stimulation, whereas the corresponding static MAC estimate derived from the previous model was much lower (median value ≈0.2); the discrepancy reflects the new finding that the feedback regulation of MEK has two distinct layers, of which Ras-GEF desensitization plays the subordinate role.
Faced with the many mechanisms by which signaling pathways might be attenuated, it is easy to neglect the most basic of regulatory processes, namely those that affect availability of ligand and receptor molecules. In previous work we carefully quantified PDGF receptor phosphorylation kinetics for stimulation times up to 20 min, characterizing the rates of PDGF binding, receptor dimerization, and receptor endocytosis (17). Here we found that, for somewhat longer times (∼1 h or more), depletion of PDGF from the extracellular medium also needs to be taken into account, as it clearly affects receptor-mediated signaling at low doses of PDGF (for the number of cells/volume of medium used). At higher PDGF concentrations, Ras and PI3K signaling are both saturated; thus, it requires more time for the effects of ligand depletion to be felt, an example of dose-to duration encoding (51). To the extent that cells effectively integrate growth factor-stimulated signals over a fairly long period of time, ligand depletion ought to affect cell proliferation and other functional responses. Indeed, under certain conditions it has been observed that the total amount of PDGF added, rather than its initial concentration, dictates the overall extent of cell proliferation in culture (52). Limitations on cell growth imposed by depletion of EGF-family ligands have also been documented (53, 54).
Mathematical modeling of biological processes has a rich history and has emerged in recent years as a valuable tool for characterizing intracellular signal transduction. In general, the utility of modeling spans a spectrum bracketed by the definition of what is possible and the interpretation of what is (26, 55). The present analysis is squarely at the latter extreme, as it is driven by an expanding set of quantitative data, which affords some measure of confidence in model predictions of network dynamics. Looking forward, further data-driven refinements of mathematical models will need to be directed toward a more detailed understanding of the molecular mechanisms that govern network dynamics, especially feedback regulation of Raf isoforms and other MEK kinases (56).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM088987 and through a graduate fellowship (to M. C.) from the NCSU Biomanufacturing Training & Education Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Text, and Figs. S1–S7.
- GEF
- guanine nucleotide exchange factor
- MKP
- mitogen-activated protein kinase phosphatase
- DUSP
- dual specificity phosphatase.
REFERENCES
- 1.Condeelis J., Singer R. H., Segall J. E. (2005) Annu. Rev. Cell Dev. Biol. 21, 695–718 [DOI] [PubMed] [Google Scholar]
- 2.Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 3.Dhillon A. S., Hagan S., Rath O., Kolch W. (2007) Oncogene 26, 3279–3290 [DOI] [PubMed] [Google Scholar]
- 4.Engelman J. A. (2009) Nat. Rev. Cancer 9, 550–562 [DOI] [PubMed] [Google Scholar]
- 5.Cuevas B. D., Abell A. N., Johnson G. L. (2007) Oncogene 26, 3159–3171 [DOI] [PubMed] [Google Scholar]
- 6.Meloche S., Pouysségur J. (2007) Oncogene 26, 3227–3239 [DOI] [PubMed] [Google Scholar]
- 7.Whitmarsh A. J. (2007) Biochim. Biophys. Acta 1773, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 8.McKay M. M., Morrison D. K. (2007) Oncogene 26, 3113–3121 [DOI] [PubMed] [Google Scholar]
- 9.Behar M., Hao N., Dohlman H. G., Elston T. C. (2007) Biophys. J. 93, 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandman O., Meyer T. (2008) Science 322, 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legewie S., Herzel H., Westerhoff H. V., Blüthgen N. (2008) Mol. Syst. Biol. 4, article no. 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyatkin A., Aksamitiene E., Markevich N. I., Borisov N. M., Hoek J. B., Kholodenko B. N. (2006) J. Biol. Chem. 281, 19925–19938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birtwistle M. R., Hatakeyama M., Yumoto N., Ogunnaike B. A., Hoek J. B., Kholodenko B. N. (2007) Mol. Syst. Biol. 3, article no. 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W. W., Schoeberl B., Jasper P. J., Niepel M., Nielsen U. B., Lauffenburger D. A., Sorger P. K. (2009) Mol. Syst. Biol. 5, article no. 239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C. C., Cirit M., Haugh J. M. (2009) Mol. Syst. Biol. 5, article no. 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider I. C., Haugh J. M. (2006) Cell Cycle 5, 1130–1134 [DOI] [PubMed] [Google Scholar]
- 17.Park C. S., Schneider I. C., Haugh J. M. (2003) J. Biol. Chem. 278, 37064–37072 [DOI] [PubMed] [Google Scholar]
- 18.Schneider I. C., Haugh J. M. (2004) Biophys. J. 86, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider I. C., Haugh J. M. (2005) J. Cell Biol. 171, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur H., Park C. S., Lewis J. M., Haugh J. M. (2006) Biochem. J. 393, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kholodenko B. N. (2000) Eur. J. Biochem. 267, 1583–1588 [DOI] [PubMed] [Google Scholar]
- 22.Shin S. Y., Rath O., Choo S. M., Fee F., McFerran B., Kolch W., Cho K. H. (2009) J. Cell Sci. 122, 425–435 [DOI] [PubMed] [Google Scholar]
- 23.Bhalla U. S., Ram P. T., Iyengar R. (2002) Science 297, 1018–1023 [DOI] [PubMed] [Google Scholar]
- 24.Santos S. D., Verveer P. J., Bastiaens P. I. (2007) Nat. Cell Biol. 9, 324–330 [DOI] [PubMed] [Google Scholar]
- 25.Shankaran H., Ippolito D. L., Chrisler W. B., Resat H., Bollinger N., Opresko L. K., Wiley H. S. (2009) Mol. Syst. Biol. 5, article # 332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyengar R. (2009) Sci. Signal. 2, eg3. [DOI] [PubMed] [Google Scholar]
- 27.Kreeger P. K., Lauffenburger D. A. (2010) Carcinogenesis 31, 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karreth F. A., DeNicola G. M., Winter S. P., Tuveson D. A. (2009) Mol. Cell 36, 477–486 [DOI] [PubMed] [Google Scholar]
- 29.Poulikakos P. I., Zhang C., Bollag G., Shokat K. M., Rosen N. (2010) Nature 464, 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters S. B., Holt K. H., Ross S. E., Syu L. J., Guan K. L., Saltiel A. R., Koretzky G. A., Pessin J. E. (1995) J. Biol. Chem. 270, 20883–20886 [DOI] [PubMed] [Google Scholar]
- 31.Langlois W. J., Sasaoka T., Saltiel A. R., Olefsky J. M. (1995) J. Biol. Chem. 270, 25320–25323 [DOI] [PubMed] [Google Scholar]
- 32.Klarlund J. K., Cherniack A. D., McMahon M., Czech M. P. (1996) J. Biol. Chem. 271, 16674–16677 [DOI] [PubMed] [Google Scholar]
- 33.Wartmann M., Hofer P., Turowski P., Saltiel A. R., Hynes N. E. (1997) J. Biol. Chem. 272, 3915–3923 [DOI] [PubMed] [Google Scholar]
- 34.Dougherty M. K., Müller J., Ritt D. A., Zhou M., Zhou X. Z., Copeland T. D., Conrads T. P., Veenstra T. D., Lu K. P., Morrison D. K. (2005) Mol. Cell 17, 215–224 [DOI] [PubMed] [Google Scholar]
- 35.Rushworth L. K., Hindley A. D., O'Neill E., Kolch W. (2006) Mol. Cell. Biol. 26, 2262–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritt D. A., Monson D. M., Specht S. I., Morrison D. K. (2010) Mol. Cell. Biol. 30, 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 38.Jurek A., Amagasaki K., Gembarska A., Heldin C. H., Lennartsson J. (2009) J. Biol. Chem. 284, 4626–4634 [DOI] [PubMed] [Google Scholar]
- 39.Segel L. A., Goldbeter A., Devreotes P. N., Knox B. E. (1986) J. Theor. Biol. 120, 151–179 [DOI] [PubMed] [Google Scholar]
- 40.Alon U., Surette M. G., Barkai N., Leibler S. (1999) Nature 397, 168–171 [DOI] [PubMed] [Google Scholar]
- 41.Parent C. A., Devreotes P. N. (1999) Science 284, 765–770 [DOI] [PubMed] [Google Scholar]
- 42.Yi T. M., Huang Y., Simon M. I., Doyle J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4649–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levchenko A., Iglesias P. A. (2002) Biophys. J. 82, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai T. Y., Choi Y. S., Ma W., Pomerening J. R., Tang C., Ferrell J. E., Jr. (2008) Science 321, 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novák B., Tyson J. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao L., Nachbar R. B., Kevrekidis I. G., Shvartsman S. Y. (2007) PLoS Comput. Biol. 3, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knauer D. J., Wiley H. S., Cunningham D. D. (1984) J. Biol. Chem. 259, 5623–5631 [PubMed] [Google Scholar]
- 48.Marshall C. J. (1995) Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]
- 49.Pledger W. J., Stiles C. D., Antoniades H. N., Scher C. D. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 4481–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitin N., Rossman K. L., Der C. J. (2005) Curr. Biol. 15, R563–R574 [DOI] [PubMed] [Google Scholar]
- 51.Behar M., Hao N., Dohlman H. G., Elston T. C. (2008) PLoS Comput. Biol. 4, article no. e1000197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel A., Ross R., Raines E. (1980) J. Cell Biol. 85, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy C. C., Wells A., Lauffenburger D. A. (1994) Biotechnol. Prog. 10, 377–384 [DOI] [PubMed] [Google Scholar]
- 54.Reddy C. C., Wells A., Lauffenburger D. A. (1996) J. Cell. Physiol. 166, 512–522 [DOI] [PubMed] [Google Scholar]
- 55.Mogilner A., Wollman R., Marshall W. F. (2006) Dev. Cell 11, 279–287 [DOI] [PubMed] [Google Scholar]
- 56.Kolch W., Calder M., Gilbert D. (2005) FEBS Lett. 579, 1891–1895 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.