Abstract
Background
The Nme gene family is involved in multiple physiological and pathological processes such as cellular differentiation, development, metastatic dissemination, and cilia functions. Despite the known importance of Nme genes and their use as clinical markers of tumor aggressiveness, the associated cellular mechanisms remain poorly understood. Over the last 20 years, several non-vertebrate model species have been used to investigate Nme functions. However, the evolutionary history of the family remains poorly understood outside the vertebrate lineage. The aim of the study was thus to elucidate the evolutionary history of the Nme gene family in Metazoans.
Methodology/Principal Findings
Using a total of 21 eukaryote species including 14 metazoans, the evolutionary history of Nme genes was reconstructed in the metazoan lineage. We demonstrated that the complexity of the Nme gene family, initially thought to be restricted to chordates, was also shared by the metazoan ancestor. We also provide evidence suggesting that the complexity of the family is mainly a eukaryotic innovation, with the exception of Nme8 that is likely to be a choanoflagellate/metazoan innovation. Highly conserved gene structure, genomic linkage, and protein domains were identified among metazoans, some features being also conserved in eukaryotes. When considering the entire Nme family, the starlet sea anemone is the studied metazoan species exhibiting the most conserved gene and protein sequence features with humans. In addition, we were able to show that most of the proteins known to interact with human NME proteins were also found in starlet sea anemone.
Conclusion/Significance
Together, our observations further support the association of Nme genes with key cellular functions that have been conserved throughout metazoan evolution. Future investigations of evolutionarily conserved Nme gene functions using the starlet sea anemone could shed new light on a wide variety of key developmental and cellular processes.
Introduction
The Nme family, initially called NDPK or Nm23, was named after the identification of a novel gene associated with low metastatic potential [1]. In humans, NME genes are involved in a wide variety of physiological or pathological cellular processes including development, metastatic potential, ciliary functions, and cell differentiation and proliferation at various tissular and subcellular localization (see [2] for recent review). Despite their critical role in key developmental and pathological processes, the molecular functions of Nme genes remain poorly documented [3]. In vertebrates, Nme genes can be separated in 2 groups – group I and group II – based on their evolutionary history and protein domains [4]. Nme genes of the group I (Nme1-4) originate from a unique gene of the chordate ancestor while Nme genes of the group II (Nme 5-8) are present throughout chordate evolution [4]. Nme-related genes have been sporadically reported in Archaea [5], Eubacteria [6], [7], and in several eukaryotic lineages including fungi [8], plants [9], and bilaterians [10], [11]. However, the evolutionary history of the family that has led to a repertoire of 5 Nme genes in the chordate ancestor remains poorly understood. Indeed, the complexity of the Nme gene repertoire outside the chordate lineage was previously uncharacterized and existing literature suggested that the complexity of the Nme gene family was much more limited in non-chordate species. In Dictyostelium discoideum, two Nme-related proteins, named NdkC-2 and NdkM, and expressed in the cytosol and in the mitochondria, respectively, were used for biochemical and structural studies [12]–[14]. In Drosophila melanogaster, only one Nme-related gene, named awd, had been reported and intensively studied for its role in aberrant development [10]. In Caenorhabditis elegans, one Nme-related gene had been shown to be associated with severe developmental defects [15]. As recently stressed, the use of model species to decipher Nme gene functions is extremely beneficial and needs to be further supported [16]. However, a better understanding of the evolutionary links between the Nme genes that are found in non-vertebrate model species and their mammalian counterparts is required to allow this comparative biology approach. In order to gain insight into putatively conserved key functions of Nme genes that would have been retained throughout evolution, the aim of the present study was thus to characterize gene family complexity and protein features among metazoans. We were able to show that the complexity of the Nme family predates the metazoan radiation. We also provided evidence supporting the association of Nme genes with key cellular functions that have been conserved throughout metazoan evolution.
Results/Discussion
Nme gene family complexity predates Metazoan radiation
Using available sequenced genomes of 21 eukaryote species ranging from amoebozoans to humans, we were able to reconstruct, in opisthokonts – the metazoan/choanoflagellate/fungi phylum – the evolutionary history of the Nme genes that had previously been identified in the chordate ancestor [4]. We were able to show that the metazoan and chordate ancestors share a similar Nme gene repertoire. A similar repertoire was also found in Monosiga brevicollis, a choanoflagellate, but not in fungi (Figures 1– 5).
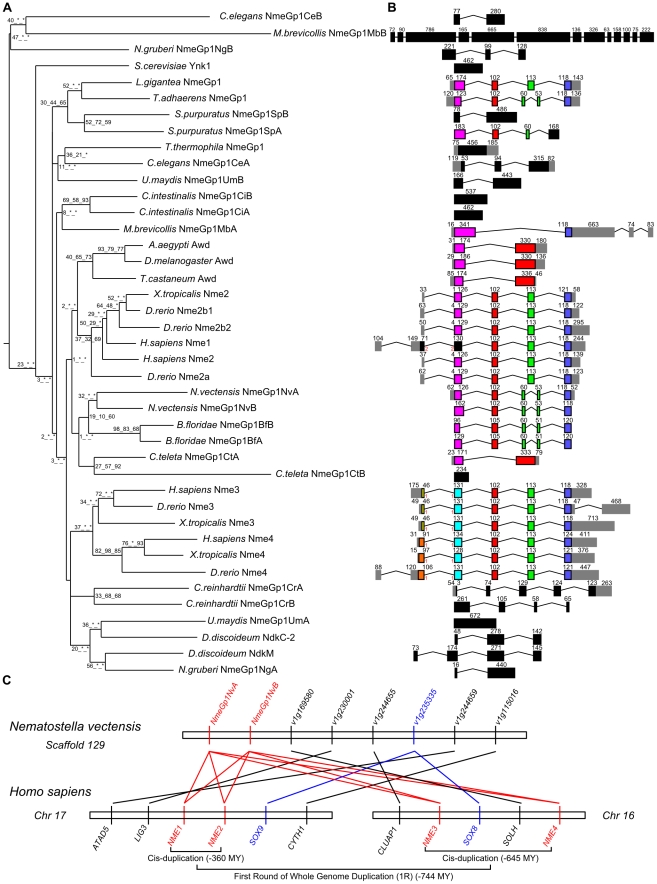
Figure 1. Phylogenetic tree and exon-intron structure of Group I Nme in Eukaryotes and synteny conservation between human and starlet sea anemone Group I Nme.
A. The phylogenetic tree was constructed from a single multiple sequence alignment. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. Asterisks (*) indicate that the node was not recovered by the corresponding phylogenetic method. The consensus midpoint-rooted tree was calculated using the FIGENIX automated phylogenomic annotation pipeline [41]. For each sequence, species and corresponding name are shown. B. Corresponding exon/intron structures of genes coding for the proteins used for the phylogenetic tree estimation. Exon/intron structure was obtained through Ensembl, NCBI or JGI databases. When exon boundaries correspond to identical amino acid positions, the exons are displayed in color. Otherwise, exons are displayed in black. Non-coding exons are shown in grey. Exon size (in nucleotides) is indicated above each box. C. Synteny analysis between Nematostella vectensis NmeGp1 genes and human group I NME genes. Nematostella vectensis gene names are NCBI Entrez gene symbols. For clarity reasons “NEMVEDRAFT_” was removed from all gene symbols. Duplication time estimations are from TimeTree [46].
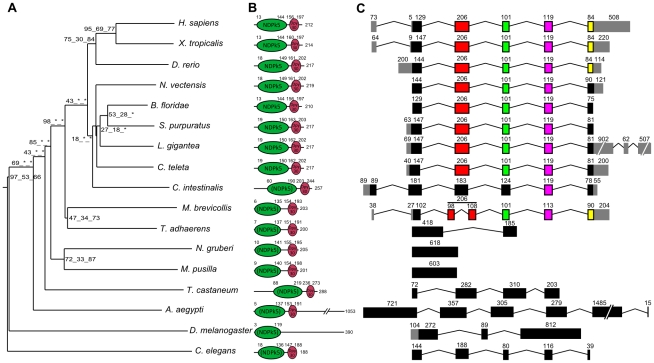
Figure 2. Phylogenetic tree, protein domains, and exon-intron structure of Nme5 in Eukaryotes.
A. The phylogenetic midpoint-rooted tree was constructed as described in Figure 1A. B. Corresponding protein domain structure of Nme proteins. Protein domain information was obtained using Genbank Conserved Domain Database [40]. Parentheses indicate that the domain type is the best hit given by NCBI CDD but is not a specific hit according to CDD default parameters. C. Exon/intron gene structure was obtained as described in Figure 1B.
Figure 3. Phylogenetic tree, protein domains, and exon-intron structure of Nme6 in Eukaryotes.
A. The phylogenetic midpoint-rooted tree was constructed as described in Figure 1A. B. Corresponding protein domain structure was obtained as described in Figure 2A. C. Exon/intron gene structure was obtained as described in Figure 1B.
Figure 4. Phylogenetic tree and protein domains of Nme7 in Eukaryotes.
A. The phylogenetic midpoint-rooted tree was constructed as described in Figure 1A. B. Corresponding protein domain structure was obtained as described in Figure 2A.
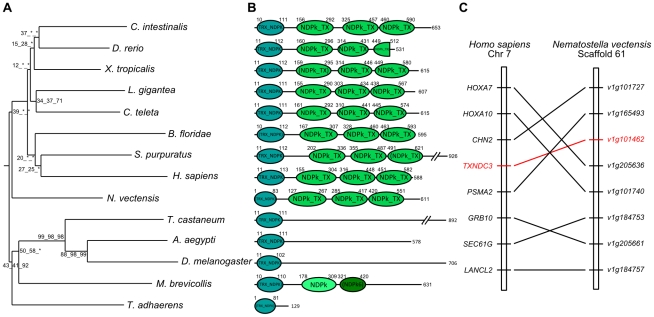
Figure 5. Phylogenetic tree and protein domains of Nme8 in Metazoans.
A. The phylogenetic midpoint-rooted tree was constructed as described in Figure 1A. B. Corresponding protein domain structure was obtained as described in Figure 2A. C. Synteny analysis between Nematostella vectensis Nme8 genes and human TXNDC3 (NME8) genes. Nematostella vectensis gene names correspond to the NCBI Entrez Gene identification. For clarity reasons the unchanged part “NEMVEDRAFT_” was removed.
All non-vertebrate metazoans species displayed 2 Nme genes of the group I with the exception of Trichoplax adhaerens, Drosophila melanogaster, Aedes aegypti, Tribolium castaneum, and Lottia gigantea, in which a single gene could be identified (Figure 1A–B). In all studied non-metazoan eukaryotes, two Nme genes of the group I could also be identified with the exception of Saccharomyces cerevisiae and Tetrahymena thermophila. In starlet sea anemone Nematostella vectensis, Caenorhabditis elegans and Branchiostoma floridae, the two genes are located tandemly and thus originate from a cis-duplication of an ancestral gene (Table 1). In addition, a surprisingly well conserved gene synteny of group I Nme sequences was found between sea anemone and humans (Figure 1C). The location of the Sox8/9 ancestor gene in the vicinity of Nme genes in the sea anemone and the location of SOX8 and SOX9 on human chromosomes 17 and 16, respectively, are consistent with the first round of whole genome duplication (1R) that gave rise to Nme2 and Nme3/4 in vertebrates [4]. In Ciona intestinalis, we have obtained evidence suggesting a duplication of a large portion of genomic DNA resulting in duplicated genes on two different chromosomes (Table 1, Figure S1). Two duplicated Nme genes of the group I originating from a common group I ancestral gene were found on chromosomes 2q and 8q (Figure S1). In all studied non-metazoan eukaryotes, Capitela teleta, and Strongylocentrotus purpuratus, two Nme genes of the group I can be found on different scaffolds. Because of the limited size of these scaffolds and the level of assembly of corresponding draft genomes, it is not currently possible to speculate on the nature of the duplication event that has led to 2 genes. In Dictyostelium discoideum, 2 Nme genes of the group I could be identified on different chromosomes. The topology of the phylogenetic tree displaying – for Chlamydomonas reinhardtii, Nematostella vectensis, Capitela teleta, Strongylocentrotus purpuratus, Branchiostoma floridae, and Ciona intestinalis – the group I Nme sequences more closely related within a species than between species strongly suggests that corresponding duplication events are independent and lineage-specific. It is noteworthy that for the above species, duplicated sequences remained closely related, whereas in other species, Naegleria gruberi, Ustilago maydis, M. brevicollis, and C. elegans one sequence is highly divergent in comparison to the other one. Altogether, this strongly suggests independent and lineage specific gene duplications. It was previously shown that a single Nme gene of the group I was present in the vertebrate ancestor [4]. This ancestor gene subsequently duplicated differently in the different vertebrates lineages and resulted in 2 to 5 genes, depending on the species [4]. Together, our data indicate that a single Nme gene of the group I was present in the opisthokont ancestor. Our observations also suggest that a single Nme gene of the group I was present in the eukaryote ancestor. Understanding the selective factors that have led to multiple independent duplications events in the Nme family and the role of these proteins in non-vertebrate species would provide major insights into the evolution and functions of the family.
Table 1. GroupI Nme proteins: names and symbols by species, accession numbers and corresponding chromosomal location.
| Species | Name | GenBank Acc. no. | Ensembl or JGI Acc. no. | Localisation | Position | |
| N. gruberi | NmeGp1NgA | XP_002672581 | 72250 | Scaffold_52 | 70,213–70,711 | |
| N. gruberi | NmeGp1NgB | XP_002678151 | 60551 | Scaffold_19 | 18,232–18,965 | |
| T. thermophila | NmeGp1 | XP_001018488 | Scaffold_8254597 | 472,318–473,033 | ||
| D. discoideum | NdkC-2 | XP_644519 | DDB0238334 | Chr 2 | 3,427,961–3,428,694 | |
| D. discoideum | NdkM | XP_641417 | DDB0214817 | Chr 3 | 2,740,248–2,741,470 | |
| C. reinhardtii | NmeGp1CrA | XP_001698246 | 58944 | Chr 16 | 365,385–367,897 | |
| C. reinhardtii | NmeGp1CrB | XP_001702884 | 292075 | Scaffold_26 | 140,833–142,142 | |
| U. maydis | NmeGp1UmA | XP_759114 | 2967 | Contig_1_101 | 162,394–163,065 | |
| U. maydis | NmeGp1UmB | XP_758923 | 2776 | Contig_1_94 | 34,001–34,701 | |
| S. cerevisiae | Ynk1 | NP_012856 | YKL067W | Chromosome XI | 314,456–314,917 | |
| M. brevicollis | NmeGp1MbA | XP_001742597 | 19073 | Scaffold_2 | 878,808–880,474 | |
| M. brevicollis | NmeGp1MbB | XP_001749207 | 28652 | Scaffold_28 | 66,018–71,920 | |
| T. adhaerens | NmeGp1 | XP_002115688 | 49508 | Scaffold_11 | 2,334,568–2,336,179 | |
| N. vectensis | NmeGp1NvA | XP_001630018 | 169568 | Scaffold_129 | 58,094–60,224 | |
| N. vectensis | NmeGp1NvB | XP_001630017 | 115087 | Scaffold_129 | 51,034–52,881 | |
| C. elegans | NmeGp1CeA | NP_492761 | F25H2.5.1 | Chr I | 10,553,902–10,553,101 | |
| C. elegans | NmeGp1CeB | NP_492819 | F55A3.6 | Chr I | 10,784,780–10,783,989 | |
| T. castaneum | Awd | XP_967503 | Linkage Group 3 | 32,141,388–32,142,070 | ||
| Group1 | D. melanogaster | Awd | NP_476761 | FBpp0085223 | Chr 3R | 27,570,894–27,571,706 |
| Nme | A. aegypti | Awd | XP_001662512 | AAEL012359-PA | SuperContig1.684 | 87,333–88,186 |
| L. gigantea | NmeGp1 | 205662 | Scaffold_3 | 3,942,249–3,946,053 | ||
| C. teleta | NmeGp1CtA | 21725 | Scaffold_758 | 80,222–84,888 | ||
| C. teleta | NmeGp1CtB | 116635 | Scaffold_527 | 188,815–189,048 | ||
| S. purpuratus | NmeGp1SpA | XP_799145 | Scaffold71291 | 17,770–21,754 | ||
| S. purpuratus | NmeGp1SpB | XP_785384 | Scaffold42444 | 298,840–301,211 | ||
| C. intestinalis | NmeGp1CiA | XP_002123476 | ENSCINP00000011619 | Chr 8q | 5,908,347–5,908,808 | |
| C. intestinalis | NmeGp1CiB | XP_002121438 | ENSCINP00000002194 | Chr 2q | 7,888,026–7,888,562 | |
| B. floridae | NmeGp1BfA | XP_002598285 | 57540 | Scaffold_24 | 2,074,549–2,076,844 | |
| B. floridae | NmeGp1BfB | XP_002598283 | 69640 | Scaffold_24 | 2,064,578–2,066,435 | |
| H. sapiens | NME1 | NP_937818 | ENSP00000337060 | Chr 17 | 49,230,937–49,239,422 | |
| H. sapiens | NME2 | NP_001018149 | ENSP00000376888 | Chr 17 | 49,243,639–49,249,108 | |
| X. tropicalis | Nme2 | NP_001005140 | ENSXETP00000024764 | Scaffold_673 | 77,640–81,451 | |
| Nme2 | D. rerio | Nme2a | NP_956264 | ENSDARP00000064338 | Scaffold Zv8_scaffold3117 | 528,950–537,135 |
| D. rerio | Nme2b1 | NP_571001 | ENSDARP00000099319 | Chr 19 | 48,501,402–48,504,320 | |
| D. rerio | Nme2b2 | NP_571002 | ENSDARP00000098065 | Chr 19 | 48,507,488–48,510,647 | |
| H. sapiens | NME3 | NP_002504 | ENSP00000219302 | Chr 16 | 1,820,321–1,821,710 | |
| Nme3 | X. tropicalis | Nme3 | NP_001005115 | ENSXETP00000022770 | Scaffold_27 | 933,363–937,018 |
| D. rerio | Nme3 | NP_571003 | ENSDARP00000075112 | Chr 3 | 11,995,596–12,045,134 | |
| H. sapiens | NME4 | NP_005000 | ENSP00000219479 | Chr 16 | 447,209–450,759 | |
| Nme4 | X. tropicalis | Nme4 | NP_001039239 | ENSXETP00000022726 | Scaffold_27 | 1,305,654–1,312,714 |
| D. rerio | Nme4 | NP_957489 | ENSDARP00000103207 | Chr 3 | 10,909,410–10,922,804 |
Protein names were retrieved from Ensembl and NCBI or proposed according to the evolutionary history of the genes. Chromosomal/genomic location was obtained using Ensembl genome browser, JGI databases, or NCBI Entrez Gene when not available on Ensembl or JGI.
All studied metazoan species displayed a full set of group II Nme genes with the exception of the 4 ecdysozoan species and T. adhaerens (Figures 2– 5). The 2 other group II Nme genes, Nme9 and Nme10, that have been shown to be eutherian and vertebrate innovations, respectively [4], will not be discussed here. No Nme7 homolog could be identified in the C. elegans genome thus indicating a possible gene loss after the nematode radiation. In M. brevicollis the Nme7 protein is structurally highly divergent as shown but the topology of the phylogenetic tree (Figure 4A) and displays a unique and incomplete domain (Figure 4B). In insects, Nme7 proteins also displayed specific domain structure (Figure 4B) resulting in a divergent position of the corresponding group in the phylogenetic tree (Figure 4A). As for all phylogenetic analyses reported here, the tree topology remained unchanged whether we used the full length protein sequence or only domains for the phylogenetic reconstruction. Similarly, very divergent Nme8-related sequences were identified in insects and T. adhaerens (Figure 5). In these 4 species, the Nme8-related proteins have lost the 3 NDPK_TX domains that are found in all other metazoan species, including the starlet sea anemone (Figure 5). In M. brevicollis, the Nme8 protein does not display typical Nme8 NDPK_TX domains but 2 different domains of the NDPk superfamily. It is also noteworthy that the exon/intron structure corresponding to the Thioredoxin TRX_NDPK domain is very well conserved among all studied choanoflagellate and metazoan species. In contrast, the exon/intron structure corresponding to the NDPK domains is highly divergent in insects, placozoans and choanoflagellates (Figure S2). Together, our data demonstrate that Nme5, Nme6, Nme7, and Nme8 genes were already present in the genome of the common ancestor of choanoflagellates and metazoans. In non-choanoflagellate/metazoan species, all Nme proteins of the group II were found in low branching eukaryotic lineages such as heteroloboseans, green plants, amoebozoans, alveolates, and fungi (Figures 2– 4, Table 2) with the exception of Nme8. In contrast, we failed to identify Nme genes of the group II outside the eukaryotic lineage. Together, our results strongly suggest that Nme5, Nme6, and Nme7 emerged around Eukaryote radiation. Interestingly, none of the studied non-choanoflagellate/metazoan species display all 3 proteins suggesting lineage specific loss of Nme 5, Nme6, and Nme7 genes. The Nme8 protein typically displays 1 Thioredoxin (TRX) domain followed by 2 or 3 complete NDPk domains. In non-choanoflagellates/metazoan species investigated, very few sequences displaying one thioredoxin domain could be identified but were never associated with an NDPk domain. We thus hypothesize that Thioredoxin domains already existed in the opisthokont ancestor and that Nme8 emerged in the choanoflagellate/metazoan ancestor by domain shuffling of 2 or 3 NDPk domains. This would, however, require further analysis.
Table 2. GroupII Nme proteins: names and symbols by species, accession numbers and corresponding chromosomal location.
| Species | Name | GenBank Acc. no. | Ensembl or JGI Acc. no. | Localisation | Position | |
| Nme5 | N. gruberi | Nme5 | XP_002682323 | 29950 | Scaffold_3 | 679,761–680,378 |
| M. pusilla | Nme5 | XP_003056664 | 15249 | Scaffold_3 | 169,341–169,943 | |
| M. brevicollis | Nme5 | XP_001749876 | 38924 | Scaffold_34 | 201,106–202,764 | |
| T. adhaerens | Nme5 | XP_002112439 | 50304 | Scaffold_5 | 3,451,699–3,452,497 | |
| N. vectensis | Nme5 | XP_001631264 | 244029 | Scaffold_105 | 447,049–449,367 | |
| C. elegans | Nme5 | NP_501212 | R05G6.5 | Chr IV | 7,508,567–7,509,459 | |
| T. castaneum | Nme5 | XP_001811234 | Linkage Group 2 | 13,741,320–13,742,593 | ||
| D. melanogaster | Nme5 | NP_651833 | FBpp0085077 | Chr 3R | 26,707,514–26,709,225 | |
| A. aegypti | Nme5 | XP_001662993 | AAEL003030-PA | SuperContig1.75 | 2,400,537–2,515,468 | |
| L. gigantea | Nme5 | 226200 | Scaffold_168 | 228,601–233,346 | ||
| C. teleta | Nme5 | 177417 | Scaffold_116 | 97,523–98,941 | ||
| S. purpuratus | Nme5 | XP_790390 | Scaffold25557 | 320,279–323,900 | ||
| C. intestinalis | Nme5 | NP_001154961 | ENSCINP00000008954 | Chr 7q | 1,031,987–1,036,756 | |
| B. floridae | Nme5 | XP_002596479 | 61845 | Scaffold_430 | 334,501–336,787 | |
| H. sapiens | NME5 | NP_003542 | ENSP00000265191 | Chr 5 | 137,450,866–137,475,104 | |
| X. tropicalis | Nme5 | NP_001072619 | ENSXETP00000008322 | Scaffold_65 | 2,613–8,494 | |
| D. rerio | Nme5 | NP_001002516 | ENSDARP00000060997 | Chr 14 | 6,455,589–6,462,855 | |
| Nme6 | D. discoideum | Nme6 | XP_629447 | DDB0191701 | Chr 6 | 2,282,811–2,283,293 |
| C. reinhardtii | Nme6 | XP_001698136 | 139197 | Chr 16 | 825,573–828,346 | |
| U. maydis | Nme6 | XP_760135 | 3988 | Contig_1_139 | 15,223–16,271 | |
| M. brevicollis | Nme6 | XP_001744091 | 15435 | Scaffold_5 | 413,751–414,530 | |
| T. adhaerens | Nme6 | XP_002114036 | 58087 | Scaffold_7 | 2,675,108–2,676,755 | |
| N. vectensis | Nme6 | XP_001622626 | 140474 | Scaffold_525 | 60,519–64,497 | |
| C. elegans | Nme6 | NP_001021779 | Y48G8AL.15 | Chr I | 1,254,771–1,258,597 | |
| D. melanogaster | Nme6 | NP_572965 | FBpp0073750 | Chr X | 14,477,710–14,478,649 | |
| T. castaneum | Nme6 | XP_972639 | Linkage Group 8 | 6,083,034–6,083,586 | ||
| A. aegypti | Nme6 | XP_001648448 | AAEL004107-PA | SuperContig1.107 | 2,016,668–2,033,334 | |
| L. gigantea | Nme6 | 112601 | Scaffold_15 | 3,040,725–3,042,846 | ||
| C. teleta | Nme6 | 91319 | Scaffold_32 | 665,868–668,578 | ||
| S. purpuratus | Nme6 | XP_001200902 | Scaffold35436 | 11,834–18,064 | ||
| C. intestinalis | Nme6 | XP_002129729 | ENSCINP00000027945 | Scaffold_1779 | 5,509–6,021 | |
| B. floridae | Nme6 | XP_002589414 | 279926 | Scaffold_243 | 2,320,842–2,322,329 | |
| H. sapiens | NME6 | NP_005784 | ENSP00000416658 | Chr 3 | 48,334,754–48,342,848 | |
| X. tropicalis | Nme6 | NP_001123709 | ENSXETP00000034257 | Scaffold_857 | 287,305–293,502 | |
| D. rerio | Nme6 | NP_571672 | ENSDARP00000094574 | Chr 20 | 18,803,906–18,812,975 | |
| Nme7 | N. gruberi | Nme7 | XP_002677897 | 33146 | Scaffold_20 | 272,036–273,256 |
| C. reinhardtii | Nme7 | XP_001702841 | 180221 | Chr 12 | 8,939,035–8,941,564 | |
| T. thermophila | Nme7 | XP_001015884 | Scaffold_8254649 | 122,957–124,735 | ||
| M. brevicollis | Nme7 | XP_001749106 | 11267 | Scaffold_27 | 519,462–520,004 | |
| T. adhaerens | Nme7 | XP_002108466 | 51403 | Scaffold_1 | 1,598,731–1,601,855 | |
| N. vectensis | Nme7 | XP_001626602 | 125694 | Scaffold_222 | 17,379–24,092 | |
| T. castaneum | Nme7 | XP_974333 | Linkage Group 7 | 18,619,008–18,620,171 | ||
| D. melanogaster | Nme7 | NP_649926 | FBpp0081561 | Chr 3R | 5,505,663–5,507,224 | |
| A. aegypti | Nme7 | XP_001661412 | AAEL011098-PA | SuperContig1.541 | 206,988–220,408 | |
| L. gigantea | Nme7 | 187020 | Scaffold_18 | 1,179,880–1,185,739 | ||
| C. teleta | Nme7 | 160391 | Scaffold_422 | 28,192–33,650 | ||
| S. purpuratus | Nme7 | XP_795051 | Scaffold9766 | 108,673–117,014 | ||
| C. intestinalis | Nme7 | NP_001155162 | ENSCINP00000025129 | Chr 1p | 3,423,461–3,424,231 | |
| B. floridae | Nme7 | XP_002588622 | 287848 | Scaffold_254 | 342,459–356,708 | |
| H. sapiens | NME7 | NP_037462 | ENSP00000356785 | Chr 1 | 169,101,769–169,337,205 | |
| X. tropicalis | Nme7 | NP_988903 | ENSXETP00000005150 | Scaffold_169 | 1,646,165–1,680,298 | |
| D. rerio | Nme7 | NP_571004 | ENSDARP00000073091 | Chr 6 | 31,135,867–31,196,533 | |
| Nme8 | M. brevicollis | Nme8 | XP_001746342 | 32671 | Scaffold_12 | 606,463–609,773 |
| T. adhaerens | Nme8 | XP_002110931 | 22954 | Scaffold_3 | 3,392,460–3,393,060 | |
| N. vectensis | Nme8 | XP_001634297 | 101462 | Scaffold_61 | 602,301–615,863 | |
| T. castaneum | TRX-Nme8 | XP_972627 | Linkage Group 5 | 12,145,420–12,148,468 | ||
| D. melanogaster | TRX-Nme8 | NP_572772 | FBpp0073425 | Chr X | 11,884,151–11,886,801 | |
| A. aegypti | TRX-Nme8 | XP_001652618 | AAEL007253-PA | SuperContig1.245 | 180,440–198,050 | |
| L. gigantea | Nme8 | 107502 | Scaffold_7 | 645,416–655,143 | ||
| C. teleta | Nme8 | 96991 | Scaffold_940 | 8,079–11,527 | ||
| S. purpuratus | Nme8 | XP_001181827 | Scaffold66657 | 21,472–41,768 | ||
| C. intestinalis | Nme8 | NP_001027618 | ENSCINP00000013583 | Chr 9q | 3,694,058–3,703,935 | |
| B. floridae | Nme8 | XP_002597926 | 280761 | Scaffold_152 | 66,164–77,826 | |
| H. sapiens | NME8 | NP_057700 | ENSP00000199447 | Chr 7 | 37,888,199–37,940,003 | |
| X. tropicalis | Nme8 | NP_001121456 | ENSXETP00000002355 | Scaffold_664 | 444,670–467,036 | |
| D. rerio | Nme8 | NP_001082944 | ENSDARP00000103107 | Chr9 | 51,493,472–51,569,898 |
Protein names were retrieved from Ensembl and NCBI or proposed according to the evolutionary history of the genes. Chromosomal/genomic location was obtained using Ensembl genome browser, JGI databases, or NCBI Entrez Gene when not available on Ensembl or JGI.
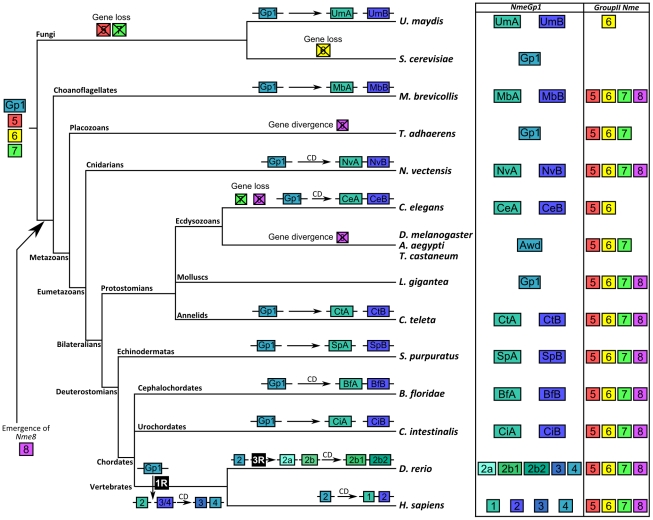
Here, we have shown that the Nme gene repertoire of the metazoan ancestor was similar to that of the vertebrate ancestor (Figure 6). In agreement with prior studies reporting major gene losses and genomic rearrangement experienced by ecdysozoans [17]–[20] we show that Nme genes have highly diverged in this phylogenetic group, resulting in the loss of either functional NDPK domains or entire genes. Furthermore, we demonstrate that the complexity of the family predates metazoan radiation. We also provide evidence suggesting that the complexity of the family is mainly a eukaryotic innovation, with the exception of Nme8 that is likely to be a choanoflagellate/metazoan innovation. This unexpectedly ancient complexity of the eukaryotic Nme gene family is in striking contrast with the existing hypothesis associating the emergence of Nme family complexity with the differentiation of Bilateralian [21] lineage. It should be stressed that this burst of complexity in the Nme family is much more ancient than what has been reported for many genes in which the expansion of the family is thought to have occurred around the metazoan radiation [18], [19], [22], [23]. This would be in favor of the participation of Nme genes in ancestral functions and would also be consistent with their known involvement in key biological processes such as cell proliferation and development.
Figure 6. Schematic depiction of Nme genes evolution in Opisthokonts.
The gene repertoire is shown at the root of the tree for the opisthokont ancestor. For all lineages, duplication events are shown and cis-duplications (CD) indicated. For the vertebrate lineage, 1R and 3R whole genome duplications events are shown and duplication events of group I Nme genes redrawn from [4]. Nme 9 and Nme10 that have been shown to be vertebrate innovations [4] are not displayed here for clarity reasons. The gene repertoire is given for all studied species. For clarity reasons, the complete name of the gene is not given and only the gene number is given and the color kept consistent with the corresponding gene in the opisthokont ancestor.
Highly conserved Nme sequence features exist among Metazoans and Eukaryotes
The topology of the phylogenetic tree (Figure 1A) obtained using group I Nme proteins does not match the topology of the tree of life. Indeed, N. vectensis sequences appear, along with B. floridae and C. teleta sequences, as a sister group of the vertebrate and insect sequences, in contrast to T. adhaerens, S. purpuratus, and C. intestinalis sequences that appear much more divergent. The topology of the tree is, however, consistent with the genomic structure of group I Nme genes reported on Figure 1B. A highly conserved exon/intron structure is observed between T. adhaerens, N. vectensis, L. gigantea, and vertebrates, while ecdysozoans, C. teleta, C. intestinalis, and S. purpuratus exhibit a totally different genomic organization that reflects the high gene divergence observed in these species. Interestingly, the D. discoideum mitochondrial NdkM shows gene and protein features that are similar (Figure 1B) to its cytosolic paralog NdkC-2, but displays a longer N-terminus sequence containing a mitochondrial assignation signal. In vertebrates, the Nme4 protein also displays a mitochondrial assignation signal in N-terminus sequence and was shown to be a gnathostome innovation [4]. In contrast, no mitochondrial assignation signal is found in any other Nme protein. This feature is thus likely to be a functional convergence between amoebozoan NdkM and vertebrate Nme4.
As indicated above, orthologs of Human Nme5, Nme6, Nme7, and Nme8 are found in metazoans (Figure S3). The topology of the phylogenetic tree (Figure 2A) obtained using Nme5 proteins showed that ecdysozoan sequences are highly divergent. Ecdysozoan Nme5 proteins appear to be even more divergent than the early diverging eukaryotes N. gruberi and M. pusilla. Similarly to group I, gene exon/intron structure and protein domains of N. vectensis, B. floridae, S. purpuratus, C. teleta, and L. gigantea Nme 5 proteins (Figures 2B and 2C) were highly similar to their human counterpart, while ecdysozoan sequences exhibited different protein length and/or domains. In agreement with these observations was the remarkably conserved exon/intron structure observed between humans and sea anemone Nme5 genomic sequences (Figure 2C). Similar observations on tree topology, protein domains, and genomic structure were made for Nme6 (Figure 3). For Nme7, no ortholog could be identified in C. elegans, while very divergent Nme7 sequences were identified in non-metazoan eukaryote species, M. brevicollis, and insects as shown by the topology of the phylogenetic tree (Figure 4A). In contrast to all other studied species, no NDPk7A domain was found (Figure 4B) in M. brevicollis and insects sequences whereas they could be identified in several non-opisthokont eukaryote species, thus suggesting a high divergence of ecdysozoan genes within the metazoan lineage. In addition, an incomplete NDPk7B domain was found in A. aegypti (Figure 4B). It should be stressed that, in contrast to insects, the Nme7 sequences of T. adhaerens, N. vectensis, and B. floridae were remarkably similar to their human counterpart in terms of genomic exon/intron structure, protein size, and functional domains (Figures 4B and S4). No synteny analysis could be performed between starlet sea anemone and humans for Nme5, Nme6, and Nme7 genes due to the limited number of genes present on corresponding sea anemone scaffolds (Table 2). Similarly to Nme7, no Nme8 ortholog was identified in C. elegans. In insects and T. adhaerens, the Nme8-related sequences that could be identified were extremely divergent (Figure 5A, Figure S2) and lacked the 3 NDPK_TX domains found in all eumetazoan Nme8 proteins, including the starlet sea anemone (Figure 5B). A conserved synteny was also identified between human and sea anemone (Figure 5C). Together our data suggest that, in contrast to Nme5 and Nme6 that have been identified in all investigated metazoan species, Nme7 and Nme8 have been lost in C. elegans. In insects, very divergent Nme7 and Nme8 genes remain. For Nme8, the high divergence associated with the loss of the 3 NDPK_TX domains in insects and T. adhaerens suggests a fast evolution of the gene possibly associated with a loss of ancestral metazoan Nme8 function.
We have shown that, in addition to the complexity of the Nme family, several highly conserved gene structures and protein domains are also conserved throughout metazoan evolution, some features being also conserved throughout the eukaryotic lineage. When considering the entire Nme family, the starlet sea anemone is the metazoan species exhibiting the most conserved gene and protein sequence features with humans.
Starlet sea anemone as a model to investigate evolutionarily conserved Nme functions
Several non vertebrate model species, mainly C. elegans and D. melanogaster, have been used to investigate Nme functions. This fruitful approach has shed light on evolutionary conserved mechanisms involved in Alzheimer's disease [15], ciliary function [24], or epithelial integrity [25]. Nevertheless, the need for studies of Nme functions in non-vertebrate model species was recently stressed by the scientific community [16]. Here, we show that the starlet sea anemone exhibits a full set of metazoan Nme genes and shares remarkably conserved gene and protein sequence features with humans that were lost in flies and C. elegans. The starlet sea anemone is an emerging model [26] offering several biological features such as separate sexes, inducible spawning, flagellated sperm, and external fertilization. This model species thus offers significant opportunities to investigate Nme gene functions and thus shed new light on Nme functions that remain poorly understood [16].
Nme proteins of the group I are involved in a wide variety of cellular and physiological processes including tumor metastatic potential. Using a reciprocal BLASTP strategy, the starlet sea anemone genome was searched for homologs of human proteins known to interact with group I NME proteins. Among the 48 human proteins known to interact with NME1, NME2, NME3, or NME4, 44 had a homolog in sea anemone (Table S1). For instance, homologs of proteins involved in cancer and cell cycle control, such as MIF [27] and Rac1 [28], were clearly identified in sea anemone (Table S1). In addition, NME1 and NME2 have been demonstrated to regulate the expression of specific genes such as c-MYC [29], [30] and p53 [31]. Interestingly, c-MYC and p53 homologs could also be identified in the starlet sea anemone genome thus suggesting that at least some down-stream targets of Nme proteins are also present in this species (Table S1). Interestingly, the importance of p53 in sea anemone development was recently stressed and found to be similar to its known function in vertebrate development [32].
As previously documented, most Nme proteins of the group II, with the exception of Nme6, have been associated with ciliary functions. They have been shown to play critical roles in spermatogenesis [33]–[35], sperm motility [2], development [36], and human conditions associated with primary ciliary dyskinesia [37]. The existence of orthologs in non-metazoan species was however unsuspected with the exception of the report of Nme7 in C. reinhardtii and T. thermophila [24]. In the present study, we have demonstrated that Nme5, Nme6, Nme7, and Nme8 genes were present in the metazoan ancestor, while Nme5, Nme6, and Nme7 were most likely present in the eukaryote ancestor. To date, only 7 proteins are known to interact with NME proteins of the group II. We were however able to identify homologs for 6 of these interacting partners in starlet sea anemone (Table S1).
As indicated above, functional evidence exist demonstrating the importance of Nme gene for key biological processes, including development, cell proliferation, ciliary function, and cancer. We have shown here that the complexity of the Nme family predates metazoan radiation and that all Nme proteins display functional domains that have been conserved throughout evolution. Using the starlet sea anemone we were able to show that most proteins known to interact with human NME were also found in the eumetazoan ancestor. Together, these observations suggest a participation of Nme genes in key cellular functions that have been conserved throughout evolution. In this context, the starlet sea anemone that exhibits a full set of highly conserved Metazoan group II Nme genes and appropriate biological features - such as separate sex, flagellated sperm, and asymmetrical expression patterns during development -offers major opportunities to investigate Nme functions.
Conclusion
In summary, we demonstrated that the complexity of the Nme gene family initially thought to be restricted to chordates was also shared by the Metazoan ancestor. We also provide evidence suggesting that the complexity of the family is mainly a eukaryotic innovation, with the exception of Nme8 that is likely to be a choanoflagellate/metazoan innovation. Remarkably conserved gene structures, genomic linkage, and protein domains were identified among metazoans, some features being also conserved in eukaryotes. When considering the entire Nme family, the starlet sea anemone is the studied metazoan species exhibiting the most conserved gene and protein sequence features with humans. In addition, we were able to show that most of the proteins known to interact with human NME proteins were also found in the starlet sea anemone. Together, our observations further support the association of Nme genes with key cellular functions that have been conserved throughout metazoan evolution.
Materials and Methods
Sequence analysis
All Nme sequences were identified using the following genome assemblies: human (Homo sapiens, Assembly GRCh37), xenopus (Xenopus tropicalis, Assembly V.4.1), zebrafish (Danio rerio, Assembly ZV8), tunicate (Ciona intestinalis, Assembly V.2.0), florida lancelet (Branchiostoma floridae, Assembly V.2.0), purple sea urchin (Strongylocentrotus purpuratus, Assembly NCBI V.2.1), fruit fly (Drosophila melanogaster, Assembly BDGP5), yellow fever mosquito (Aedes aegypti, Assembly AaegL1), red flour beetle (Tribolium castaneum, Assembly Tcas 3.0), nematode (Caenorhabditis elegans, Assembly WS214), polychaete worm (Capitella teleta, Assembly V1.0), owl limpet (Lottia gigantea, Assembly V.1.0), starlet sea anemone (Nematostella vectensis, Assembly V.1.0), placozoan (Trichoplax adhaerens, Assembly Grell-BS-1999 V.1.0), marine choanoflagellate (Monosiga brevicollis, Assembly V1.0), fungi (Saccharomyces cerevisiae, Assembly EF 2; and Ustilago maydis, Assembly 1), amoebozoa (Dictyostelium discoideum, Assembly V.2.1), alveolate (Tetrahymena thermophila, Assembly 1.1), green plants (Micromonas pusilla, Assembly V.2.0; and Chlamydomonas reinhardtii, Assembly V.4.0) and heterolobosean (Naegleria gruberi, Assembly V.1.0). A large number of sequences were obtained from the NCBI NR database using human or zebrafish protein sequences as a query. When more than one sequence was obtained, the RefSeq one was preferentially selected. When no RefSeq sequence was available, the longest sequence was used. When no sequences were available in the NR database, BLASTP was used on the Ensembl [38] and DoE Joint Genome Institute databases. The chromosomal localization of Nme genes was established using the Ensembl genome browser or JGI gene information, or when not available, using the UCSC Genome Bioinformatics BLAT [39] and the NCBI Sequence Viewer. Exon/intron structure was obtained from the Ensembl, NCBI, or JGI databases. The protein domain structure of Nme proteins was obtained from the GenBank Conserved Domain Database [40].
Phylogenetic analysis of Nme proteins
Phylogenetic reconstructions were performed using the automated genomic annotation platform FIGENIX [41]. For each phylogenetic tree reconstruction, all selected protein sequences were added to a single multiple sequence alignment. Sequence alignment was performed automatically by the FIGENIX pipeline using MUSCLE v3.6 [42], [43]. The pipeline used is based on three different methods of phylogenetic tree reconstruction (i.e. neighbour-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML)). The substitution model was calculated from data for ML while BLOSUM was used for NJ. Bootstrapping was carried out to assess node support with 1000 pseudoreplicates [44]. Support values were mapped onto a midpoint-rooted 50% majority rule consensus tree for each optimality criterion. Bootstrap values are reported for the nodes that are present in all three phylogenetic reconstruction methods. Asterisks denote the absence of a node for a given phylogenetic method.
Synteny analysis
The synteny relationships of starlet sea anemone and human Nme genes were analyzed by reciprocal BLASTP on the NCBI NR database using surrounding genes of Nme genes in Nematostella vectensis. Homologous genes were considered in the analysis only when reciprocal BLASTP returned the couple as best hit. For the Ciona intestinalis paralogy analysis, synteny relationships were obtained using the Synteny Database [45] and putative paralogs were validated by reciprocal BLASTP on the NCBI NR database.
Identification of Nme partners homologs
Validated human NME partners were obtained though NCBI Entrez Gene Interactions (http://www.ncbi.nlm.nih.gov/gene) information. Homologous genes in Nematostella vectensis were identified by reciprocal BLASTP on the NCBI NR database. Only BLASTP hits with an E-value lower than 10−10 were considered significant.
Supporting Information
Ciona intestinalis genomic region paralogy relationships between chromosomes 2q and 8q. For Ciona intestinalis paralogy analysis, synteny relationships were inquired using the Synteny Database [45] and putative paralogs were validated by reciprocal BLASTP on NCBI NR databases. (TIF)
Exon/intron structure of Nme 8 genes. Exon/intron structure was obtained through Ensembl, NCBI, or JGI databases. When exon boundaries correspond to similar amino acid positions, the exons are displayed in color. Otherwise, exons are displayed in black. Non-coding exons are shown in grey. Numbers indicate exon size in nucleotides. (TIF)
Phylogenetic reconstruction of the Nme protein family in eumetazoans. Phylogenetic tree was constructed from a single multiple alignment. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated using the FIGENIX [41] automated phylogenomic annotation pipeline. Only Homo sapiens, Danio rerio, Ciona intestinalis and Nematostella vectensis sequences were used in this phylogenetic tree reconstruction because of the highly divergent ecdysozoans sequences greatly modifying the tree topology. (TIF)
Exon/intron structure of Nme 7 genes. Exon/intron structure was obtained through Ensembl, NCBI, or JGI databases. When exon boundaries correspond to similar amino acid positions, the exons are displayed in color. Otherwise, exons are displayed in black. Non-coding exons are shown in grey. Numbers indicate exon size in nucleotides. (TIF)
BLASTP hits of human NME partners against starlet sea anemone sequences. Partners of human NME proteins were listed from NCBI Entrez Gene interaction information. BLASTP hits were considered significant for e-values lower than 10− 10. Accession numbers and BLASTP e-values are also given for c-Myc and p53, two downstream targets of human NME1 and NME2 proteins. (XLS)
Acknowledgments
Authors thank Alexis Fostier for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by an INRA - IFREMER PhD fellowship to TD. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 222719 - LIFECYCLE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 2.Boissan M, Dabernat S, Peuchant E, Schlattner U, Lascu I, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. 2009;329:51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- 3.Postel EH. Multiple biochemical activities of NM23/NDP kinase in gene regulation. J Bioenerg Biomembr. 2003;35:31–40. doi: 10.1023/a:1023485505621. [DOI] [PubMed] [Google Scholar]
- 4.Desvignes T, Pontarotti P, Fauvel C, Bobe J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256. doi: 10.1186/1471-2148-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polosina YY, Jarrell KF, Fedorov OV, Kostyukova AS. Nucleoside diphosphate kinase from haloalkaliphilic archaeon Natronobacterium magadii: purification and characterization. Extremophiles. 1998;2:333–338. doi: 10.1007/s007920050076. [DOI] [PubMed] [Google Scholar]
- 6.Hama H, Almaula N, Lerner CG, Inouye S, Inouye M. Nucleoside diphosphate kinase from Escherichia coli; its overproduction and sequence comparison with eukaryotic enzymes. Gene. 1991;105:31–36. doi: 10.1016/0378-1119(91)90510-i. [DOI] [PubMed] [Google Scholar]
- 7.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 8.Yang M, Jarrett SG, Craven R, Kaetzel DM. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–78. doi: 10.1016/j.mrfmmm.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammargren J, Sundstrom J, Johansson M, Bergman P, Knorpp C. On the phylogeny, expression and targeting of plant nucleoside diphosphate kinases. Physiologia Plantarum. 2007;129:79–89. [Google Scholar]
- 10.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, et al. Is outer arm dynein intermediate chain 1 multifunctional? Mol Biol Cell. 1996;7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troll H, Winckler T, Lascu I, Muller N, Saurin W, et al. Separate nuclear genes encode cytosolic and mitochondrial nucleoside diphosphate kinase in Dictyostelium discoideum. J Biol Chem. 1993;268:25469–25475. [PubMed] [Google Scholar]
- 13.Cherfils J, Morera S, Lascu I, Veron M, Janin J. X-ray structure of nucleoside diphosphate kinase complexed with thymidine diphosphate and Mg2+ at 2-A resolution. Biochemistry (Mosc) 1994;33:9062–9069. doi: 10.1021/bi00197a006. [DOI] [PubMed] [Google Scholar]
- 14.Lascu L, Giartosio A, Ransac S, Erent M. Quaternary structure of nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:227–236. doi: 10.1023/a:1005580828141. [DOI] [PubMed] [Google Scholar]
- 15.Napolitano F, D'Angelo F, Bimonte M, Perrina V, D'Ambrosio C, et al. A differential proteomic approach reveals an evolutionary conserved regulation of Nme proteins by Fe65 in C. elegans and mouse. Neurochem Res. 2008;33:2547–2555. doi: 10.1007/s11064-008-9683-z. [DOI] [PubMed] [Google Scholar]
- 16.Mehta A, Orchard S. Nucleoside diphosphate kinase (NDPK, NM23, AWD): recent regulatory advances in endocytosis, metastasis, psoriasis, insulin release, fetal erythroid lineage and heart failure; translational medicine exemplified. Mol Cell Biochem. 2009;329:3–15. doi: 10.1007/s11010-009-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, McDougall C, Troscianko J, Chen WC, Jayaraman-Nagarajan A, et al. An EST screen from the annelid Pomatoceros lamarckii reveals patterns of gene loss and gain in animals. BMC Evol Biol. 2009;9:240. doi: 10.1186/1471-2148-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolaidis N, Chalkia D, Watkins DN, Barrow RK, Snyder SH, et al. Ancient origin of the new developmental superfamily DANGER. PLoS ONE. 2007;2:e204. doi: 10.1371/journal.pone.0000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, et al. Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE. 2007;2:e153. doi: 10.1371/journal.pone.0000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada A, Pang K, Martindale MQ, Tochinai S. Surprisingly complex T-box gene complement in diploblastic metazoans. Evol Dev. 2007;9:220–230. doi: 10.1111/j.1525-142X.2007.00154.x. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa N, Shimada N, Takagi Y, Ishijima Y, Fukuda M, et al. Molecular evolution of nucleoside diphosphate kinase genes: conserved core structures and multiple-layered regulatory regions. J Bioenerg Biomembr. 2003;35:7–18. doi: 10.1023/a:1023433504713. [DOI] [PubMed] [Google Scholar]
- 22.Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda T. NDP kinase 7 is a conserved microtubule-binding protein preferentially expressed in ciliated cells. Cell Struct Funct. 2010;35:23–30. doi: 10.1247/csf.09016. [DOI] [PubMed] [Google Scholar]
- 25.Woolworth JA, Nallamothu G, Hsu T. The Drosophila metastasis suppressor gene Nm23 homolog, awd, regulates epithelial integrity during oogenesis. Mol Cell Biol. 2009;29:4679–4690. doi: 10.1128/MCB.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 27.Jung H, Seong HA, Ha H. Direct interaction between NM23-H1 and macrophage migration inhibitory factor (MIF) is critical for alleviation of MIF-mediated suppression of p53 activity. J Biol Chem. 2008;283:32669–32679. doi: 10.1074/jbc.M806225200. [DOI] [PubMed] [Google Scholar]
- 28.Otsuki Y, Tanaka M, Yoshii S, Kawazoe N, Nakaya K, et al. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci U S A. 2001;98:4385–4390. doi: 10.1073/pnas.071411598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 30.Postel EH, Berberich SJ, Rooney JW, Kaetzel DM. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J Bioenerg Biomembr. 2000;32:277–284. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 31.Jung H, Seong HA, Ha H. NM23-H1 tumor suppressor and its interacting partner STRAP activate p53 function. J Biol Chem. 2007;282:35293–35307. doi: 10.1074/jbc.M705181200. [DOI] [PubMed] [Google Scholar]
- 32.Pankow S, Bamberger C. The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. . PLoS ONE. 2007;2:e782. doi: 10.1371/journal.pone.0000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munier A, Feral C, Milon L, Pinon VP, Gyapay G, et al. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 1998;434:289–294. doi: 10.1016/s0014-5793(98)00996-x. [DOI] [PubMed] [Google Scholar]
- 34.Hwang KC, Ok DW, Hong JC, Kim MO, Kim JH. Cloning, sequencing, and characterization of the murine nm23-M5 gene during mouse spermatogenesis and spermiogenesis. Biochem Biophys Res Commun. 2003;306:198–207. doi: 10.1016/s0006-291x(03)00916-1. [DOI] [PubMed] [Google Scholar]
- 35.Choi YJ, Cho SK, Hwang KC, Park C, Kim JH, et al. Nm23-M5 mediates round and elongated spermatid survival by regulating GPX-5 levels. FEBS Lett. 2009;583:1292–1298. doi: 10.1016/j.febslet.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Vogel P, Read R, Hansen GM, Freay LC, Zambrowicz BP, et al. Situs inversus in Dpcd/Poll-/-, Nme7-/-, and Pkd1l1-/- mice. Vet Pathol. 2010;47:120–131. doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- 37.Duriez B, Duquesnoy P, Escudier E, Bridoux AM, Escalier D, et al. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc Natl Acad Sci U S A. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, et al. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence-limits on phylogenies - An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ciona intestinalis genomic region paralogy relationships between chromosomes 2q and 8q. For Ciona intestinalis paralogy analysis, synteny relationships were inquired using the Synteny Database [45] and putative paralogs were validated by reciprocal BLASTP on NCBI NR databases. (TIF)
Exon/intron structure of Nme 8 genes. Exon/intron structure was obtained through Ensembl, NCBI, or JGI databases. When exon boundaries correspond to similar amino acid positions, the exons are displayed in color. Otherwise, exons are displayed in black. Non-coding exons are shown in grey. Numbers indicate exon size in nucleotides. (TIF)
Phylogenetic reconstruction of the Nme protein family in eumetazoans. Phylogenetic tree was constructed from a single multiple alignment. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated using the FIGENIX [41] automated phylogenomic annotation pipeline. Only Homo sapiens, Danio rerio, Ciona intestinalis and Nematostella vectensis sequences were used in this phylogenetic tree reconstruction because of the highly divergent ecdysozoans sequences greatly modifying the tree topology. (TIF)
Exon/intron structure of Nme 7 genes. Exon/intron structure was obtained through Ensembl, NCBI, or JGI databases. When exon boundaries correspond to similar amino acid positions, the exons are displayed in color. Otherwise, exons are displayed in black. Non-coding exons are shown in grey. Numbers indicate exon size in nucleotides. (TIF)
BLASTP hits of human NME partners against starlet sea anemone sequences. Partners of human NME proteins were listed from NCBI Entrez Gene interaction information. BLASTP hits were considered significant for e-values lower than 10− 10. Accession numbers and BLASTP e-values are also given for c-Myc and p53, two downstream targets of human NME1 and NME2 proteins. (XLS)