Abstract
Estimation of genewise variance arises from two important applications in microarray data analysis: selecting significantly differentially expressed genes and validation tests for normalization of microarray data. We approach the problem by introducing a two-way nonparametric model, which is an extension of the famous Neyman-Scott model and is applicable beyond microarray data. The problem itself poses interesting challenges because the number of nuisance parameters is proportional to the sample size and it is not obvious how the variance function can be estimated when measurements are correlated. In such a high-dimensional nonparametric problem, we proposed two novel nonparametric estimators for genewise variance function and semiparametric estimators for measurement correlation, via solving a system of nonlinear equations. Their asymptotic normality is established. The finite sample property is demonstrated by simulation studies. The estimators also improve the power of the tests for detecting statistically differentially expressed genes. The methodology is illustrated by the data from MicroArray Quality Control (MAQC) project.
Keywords and phrases: Genewise variance estimation, gene selection, local linear regression, nonparametric model, correlation correction, validation test
1. Introduction
Microarray experiments are one of widely used technologies nowadays, allowing scientists to monitor thousands of gene expressions simultaneously. One of the important scientific endeavors of microarray data analysis is to detect statistically differentially expressed genes for downstream analysis (Cui, Hwang, and Qiu, 2005; Fan, Tam, Vande Woude and Ren, 2004; Fan and Ren, 2006; Storey and Tibshirani, 2003; Tusher, Tibshirani, and Chu, 2001). Standard t-test and F-test are frequently employed. However, due to the cost of the experiment, it is common to see a large number of genes with a small number of replications. Even in customized arrays where only several hundreds of genes expressions are measured, the number of replications is usually limited. As a result, we are facing a high dimensional statistical problem with a large number of parameters and a small sample size.
Genewise variance estimation arises at the heart of microarray data analysis. To select differentially expressed genes among thousands of genes, the t-test is frequently employed with a stringent control of type I errors. The degree of freedom is usually small due to limited replications. The power of the test can be significantly improved if the genewise variance can be estimated accurately. In such a case, the t-test becomes basically a z-test. A simple genewise variance estimator is the sample variance of replicated data, which is not reliable due to a relatively small number of replicated genes. They have direct impact on the sensitivity and specificity of t-test (Cui et al., 2005). Therefore, novel methods for estimating the genewise variances are needed for improving the power of the standard t-test.
Another important application of genewise variance estimation arises from testing whether systematic biases have been properly removed after applying some normalization method, or selecting the most appropriate normalization technique for a given array. Fan and Niu (2007) developed such validation tests (see Section 4), which require the estimation of genewise variance. The methods of variance estimation, like pooled variance estimator, and REML estimator (Smyth, Michaud, and Scott, 2005), are not accurate enough due to the small number of replications.
Due to the importance of genewise variance in microarray data analysis, conscientious efforts have been made to accurately estimate it. Various methods have been proposed under different models and assumptions. It has been widely observed that genewise variance is to a great extent related to the intensity level. Kamb and Ramaswami (2001) proposed a crude regression estimation of variance from microarray control data. Tong and Wang (2007) discussed a family of shrinkage estimators to improve the accuracy.
Let Rgi and Ggi respectively be the intensities of red (Cy3) and green (Cy5) channels for the ith replication of the gth gene on a two-color microarray data. The log-ratios and log-intensities are computed respectively as
where I is the number of replications for each gene and N is the number of genes with replications. For the purpose of estimating genewise variance, we assume that there is no systematic biases or the systematic biases have been removed by a certain normalization method. This assumption is always made for selecting significantly differentially expressed genes or validation test under the null hypothesis. Thus we have
with αg denoting the log-ratio of gene expressions in the treatment and control samples. Here, (εg1, ···, εgI)T follows a multivariate normal distribution with εgi ~ N (0, 1) and Corr(εgi, εgj) = ρ when i ≠ j. It is also assumed that observations from different genes are independent. Such a model was used in Smyth et al. (2005).
In the papers by Wang, Ma, and Carroll (2008) and Carroll and Wang (2008), nonparametric measurement-error models have been introduced to aggregate the information of estimating the genewise variance:
| (1) |
The model is intended for the analysis of the Affymetrix array (one-color array) data in which αg represents the expected intensity level, and Ygi is the ith replicate of observed expression level of gene g. When it is applied to the two-color microarray data as in our setting, in which αg is the relative expression profiles between the treatment and control, several drawbacks emerge: (a) The model is difficult to interpret as the genewide variance is a function of the log-ratio of expression profiles. (b) Errors-in-variable methods have a very slow rate of convergence for the nonparametric problem and the observed intensity information Xgi is not used. (c) They are usually hard to be implemented robustly and depend sensitively on the distribution of σ(αg)εgi and the i.i.d assumption on the noise. (d) In many microarray applications, αg = 0 for most g and hence σ(αg) are the same for most genes, which is unrealistic. Therefore, our model (2) below is complementary to that of Wang et al. (2008) and Carroll and Wang (2008), with focus on the applications to two-color microarray data.
To overcome these drawbacks in the applications to microarray data and to utilize the observed intensity information, we assume that σgi = σ(Xgi) for a smooth function σ(·). This leads to the following two-way nonparametric model
| (2) |
for estimating genewise variance. This model is clearly an extension of the Neyman-Scott problem (Neyman and Scott, 1948), in which the genewise variance is a constant. The Neyman-Scott problem has many applications in astronomy. Note that the number of nuisance parameters {αg} is proportional to the sample size. This imposes an important challenge to the nonparametric problem. It is not even clear whether the function σ(·) can be consistently estimated.
To estimate the genewise variance in their microarray data analysis, Fan et al. (2004) assumed a model similar to (2). But in the absence of other available techniques, they had to impose that the treatment effect {αg} is also a smooth function of the intensity level so that they can apply nonparametric methods to estimate genewise variance (Ruppert et al., 1997). However, this assumption is not valid in most microarray applications, and the estimator of genewise variance incurs big biases unless {αg} is sparse, a situation that Fan et al. (2004) hoped. Fan and Niu (2007) approached this problem in another simple way. When the noise in the replications is small, i.e., Xgi ≈ X̄g, where X̄g is the sample mean for the g-th gene. Therefore, they simply smoothed the pair {(X̄g, r̄g)}, where . This also leads to a biased estimator, which is denoted as ξ̂2(x). One asks naturally whether the function σ(·) is estimable and how it can be estimated in the general two-way nonparametric model.
We propose a novel nonparametric approach to estimate the genewise variance. We first study a benchmark case when there is no correlation between replications, i.e., ρ = 0. This corresponds to the case with independent replications across arrays (Fan et al., 2005; Huang et al., 2005). It is also applicable to those dealt by the Neyman-Scott problem. By noticing E{(Ygi − Ȳg)2|Xgi} is a linear combination of σ2(Xgi), we obtain a system of linear equations. Hence, σ2(·) can be estimated via nonparametric regression of a proper linear combination of {(Ygi − Ȳg)2, i = 1, ···, I} on {Xgi}. The asymptotic normality of the estimator is established. In the case that the replication correlation does not vanish, the system of equations becomes nonlinear and can not be analytically solved. However, we are able to derive the correlation corrected estimator, based on the estimator without genewise correlation. The genewise variance function and the correlation coefficient of repeated measurements are simultaneously estimated by iteratively solving a nonlinear equation. The asymptotic normality of such estimators is established.
Model (2) can be applied to the microarrays in which within-array replications are not available. In that case, we can aggregate all the microarrays together and view them as a super array with replications (Fan et al., 2005; Huang et al., 2005). In other words, i in (2) indexes arrays and ρ can be taken as 0, namely (2) is the across-array replication with ρ = 0.
The structure of this paper is as follows. In Section 2 we discuss the estimation schemes of the genewise variance and establish the asymptotic properties of the estimators. Simulation studies are given in Section 3 to verify the performance of our methods in the finite sample. Applications to the data from MicroArray Quality Control (MAQC) project are showed in Section 4 to illustrate the proposed methodology. In Section 5, we give a short summary. Technical proofs are relegated to the Appendix.
2. Nonparametric Estimators of Genewise Variance
2.1. Estimation without correlation
We first consider the specific case where there is no correlation among the replications Yg1, ···, YgI of the same gene g under model (2). This is usually applicable to the across-array replication and stimulates our procedure for the more general case with the replication correlation. In the former case, we have
We will discuss in §2.4 the case that I = 2. For I > 2, we have I different equations with I unknowns σ2(Xg1), σ2(Xg2), ···, σ2(XgI) for a given gene g. Solving these I equations, we can express the unknowns in terms of , estimable quantities. Let
Then, it can easily be shown that , where B is the coefficient matrix:
with I being the I × I identity matrix and E the I × I matrix with all elements 1. Define
Then, we have
| (3) |
Note that the left hand side of (3) depends only on Xgi, not other variables. By the the double expectation formula, it follows that the variance function σ2(·) can be expressed as the univariate regression:
| (4) |
Using the synthetic data {(Xgi, Zgi), g = 1, ···, N} for each given i, we can apply the local linear regression technique (Fan and Gijbels, 1996) to obtain a nonparametric estimator of σ2(·). Explicitly, for a given kernel K and bandwidth h,
| (5) |
with
where Kh(u) = h−1 K(u/h) and , whose dependence on i is suppressed. Thus we have I estimators for the same genewise variance function σ(·). Each of these I estimators is a consistent estimator of σ2(x). To optimally aggregate those I estimators, we need the asymptotic properties of .
Denote
Assume that Xgi are i.i.d. with marginal density fX(·) and εgi are i.i.d. random variables from the standard normal distribution. In the following result, we assume that I is fixed, but N diverges.
Theorem 1
Under the regularity conditions in the Appendix, for a fixed point x, we have
provided that h → 0 and N h → ∞, where e = (1, 1, ···, 1)T and
with ,
Note that V2 is one order of magnitude smaller than V1. Hence, the estimators are asymptotically independently distributed as N(σ2(x) + b(x), V1). Their dependence is only in the second order. The best linear combination of I estimators is
| (6) |
with the asymptotic distribution
| (7) |
See also the aggregated estimator (16) with ρ = 0, which has the same asymptotic property as the estimator (8). See Remark 1 below for additional discussion.
Theorem 1 gives the asymptotic normality of the proposed nonparametric estimators under the presence of a large number of nuisance parameters . With the newly proposed technique, we do not have to impose any assumptions on αg such as sparsity or smoothness. This kind of local linear estimator can be applied to most two-color microarray data, for instance, customized arrays and Agilent arrays.
2.2. Variance estimation with correlated replications
2.2.1. Aggregated Estimator
We now consider the case with correlated with-array replications. There is a lot of evidence that correlation among within-array replicated genes exists (Smyth et al., 2005; Fan and Niu, 2007). Suppose that within-array replications have a common correlation corr(Ygi, Ygj|X) = ρ when i ≠ j. Observations across different genes or arrays are independent. Then, the conditional variance of (Ygi − Ȳg) can be expressed as
| (8) |
This is a complex system of nonlinear equations and the analytic form can not be found. Innovative ideas are needed.
Using the same notation as that in the previous section, it can be calculated that
Taking the expectation with respect to Xgj for all j ≠ i, we obtain
| (9) |
where σ1 = E[σ(X)].
Here, we can directly apply the local linear approach to all aggregated data , due to the same regression function (9). Let be the local linear estimator of η2(·), based on the aggregated data. Then,
| (10) |
with
where . There are two solutions to (9)
| (11) |
Notice that given the sample X and Y, σ̂A(x, ρ)(1),(2) are continuous in both x and ρ. For ρ < 0, σ̂A(x, ρ)(1) should be used since the standard deviation should be nonnegative. Since σ̂A(x, ρ)(1) > σ̂A(x, ρ)(2) for every x and ρ, by the continuity of the solution in ρ, we can only use the same solution when ρ changes continuously. Then, σ̂A(x, ρ)(1) should always be used regardless of ρ. From now on, we drop the superscript and denote:
| (12) |
This is called the aggregated estimator. Note that in (12), ρ, σ1 and σ(·) are all unknown.
2.2.2. Estimation of Correlation
To estimate ρ, we assume that there are J independent arrays (J ≥ 2). In other words, we observed data from (2) independently J times. In this case, the residual maximum likelihood (REML) estimator introduced by Smyth et al. (2005) is as follows:
| (13) |
where with and is the between-arrays variance and is the within-array variance:
As discussed in Smyth et al. (2005), the estimator ρ̂0 of ρ is consistent when var(Ygij|X) = σg is the same for all i = 1, ···, I and j = 1, ···, J. However, this assumption is not valid under the model (2) and a correction is needed. We propose the following estimator:
| (14) |
The consistency of ρ̂ is given by the following theorem.
Theorem 2
Under the regularity condition in the Appendix, the estimator ρ̂ of ρ is -consistent:
With a consistent estimator of ρ, σ1, σ2 and σA(·) can be solved by the following iterative algorithm:
Step 1. Se as an initial estimate of
- Step 2. With σ̂A(·), compute
(15) Step 3. With σ̂1, σ̂2, and ρ̂, compute σ̂A(·) using (12).
Step 4. Repeat Steps 2 and 3 until convergence.
This provides simultaneously the estimators σ̂1, σ̂2, ρ̂, and σ̂A(·). From our numerical experience, this algorithm converges quickly after a few iterations. When the algorithm converges, the estimator is given by
| (16) |
Note that the presence of multiple arrays is only used to estimate the correlation ρ for the replications. It is not needed for estimating the genewise variance function. In the case of the presence of J arrays, we can take the average of the J estimates from each array.
2.2.3. Asymptotic properties
Following a similar idea as the case without correlation, we can derive the asymptotic property of .
Theorem 3
Under the regularity conditions in the Appendix, for a fixed point x, we have:
provided that h → 0 and Nh → ∞, with and
where
with coefficients C2, ···, C4, D0, ···, D4 defined in the Appendix.
The asymptotic normality of can be derived from that of . More specifically, with . The derivative of ϕ(·) with respect to z is . Then, by the delta method, we have
Remark 1
An alternative approach when correlation exists is to apply the same correlation correction idea to for every replication i, resulting in the estimator . In this case, it can be proved that the best linear combination of the estimator is
| (17) |
This estimator has the same asymptotic performance as the aggregated estimator. However, we prefer the aggregated estimator due to the following reasons: The equation (16) only needs to be solved once by using the algorithm in §2.2.2, all data are treated symmetrically, and can be estimated more stably.
2.2.4. Two replications
The aforementioned methods apply to the case when there are more than two replications. For the case I = 2, the equations for var[(Ygi − Ȳg)|X] collapse into one. In this case, it can be shown using the same arguments before that
| (18) |
where σ2 = E[σ2(Xgi)]. In this case, the left hand side is always equal to var[(Yg1 − Yg2)/2|Xgi = x].
Let η̂2(x) be the local linear estimator of the function on the right hand side by smoothing on and . Then, the genewise variance is a solution to the following equation
| (19) |
. The algorithm in §2.2.2 can be applied directly.
3. Simulations and comparisons
In this section, we conduct simulations to evaluate the finite sample performance of different variance estimators ξ̂2(x), η̂2(x) and . First, the bias problem of the naive non-parametric variance estimator ξ̂2(x) is demonstrated. It is shown that this bias issue can be eliminated by our newly proposed methods. Then, we consider the estimators η̂2(x) and under different configurations of the within-array replication correlation.
3.1. Simulation design
In all the simulation examples, we set the number of genes N = 2000, each gene having I = 3 within-array replications, and J = 4 independent arrays. For the purpose of investigating the genewise variance estimation, the data are generated from model (2). The details of simulation scheme are summarized as follows:
αg: The expression levels of the first 250 genes are generated from the standard double exponential distribution. The rest are 0’s. These expression levels are the same over 4 arrays in each simulation, but may vary over simulations.
X: The intensity is generated from a mixture distribution: with probability 0.7 from the distribution .0004(x − 6)3I(6 < x < 16) and 0.3 from the uniform distribution over [6, 16].
ε: εgi is generated from the standard normal distribution.
σ2(·): The genewise variance function is taken as
The parameters are taken from Fan et al. (2005). The kernel function is selected as . In addition, we fix the bandwidth h = 1 for all the numerical analysis.
For every setting, we repeat the whole simulation process for T times and evaluate the estimates of σ2(·) over K = 101 grid points on the interval [6, 16]. For the k-th grid point, we define
and . Let f(·) be the density function of intensity X. Let
and
be the integrated squared bias (Bias2), the integrated variance (VAR), and the integrated mean squared error (MISE) of the estimate σ̂2(·), respectively. For the t-th simulation experiment, we define
be the integrated squared error for the t-th simulation.
3.2. The bias of naive nonparametric estimator
A naive approach is to regard αg in (2) as a smooth function of Xgi, namely, αg = α(Xgi). The function α(·) can be estimated by a local linear regression estimator, resulting in an estimated function α̂(·). The squared residuals is then further smoothed on to obtain an estimate ξ̂2(x) of the variance function σ2(·), where rgi = Ŷgi − α̂(Xgi) (Ruppert et al., 1997).
To provide a comprehensive view of the performances of the naive and the new estimators, we first compare the performances of ξ̂2(x) and η̂2(x) under the smoothness assumption of the gene effect αg. Data from the naive nonparametric regression model is also generated with
This allows us to understand the loss of efficiency when αg is continuous in Xgi. This usually does not occur for microarray data, but can appear in other applications. Note that α(·) is zero in most of the region and thus is reasonably sparse. Here, the number of simulations is taken to be T = 100. The data is generated with the assumption that ρ = 0, in which case the variance estimators η̂2(x) and have the same performance (See also Table 2 below). Thus, we only report the performance of η̂2(x).
Table 2.
Mean integrated squared bias (Bias2), mean integrated variance (VAR), mean integrated squared error (MISE) over 1000 simulations for different variance estimators η̂2(x) and . Seven different correlation schemes are simulated: ρ = −0.4, ρ = −0.2, ρ = 0, ρ = 0.2, ρ = 0.4, ρ = 0.6, and ρ = 0.8. All quantities are multiplied by 1000.
| ρ = | −0.4 | −0.2 | 0 | 0.2 | 0.4 | 0.6 | 0.8 | |
|---|---|---|---|---|---|---|---|---|
| Bias2 | η̂2(x) | 5.93 | 1.48 | 0.00 | 1.48 | 5.91 | 13.31 | 23.67 |
|
|
0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
|
|
0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | |
| VAR | η̂2(x) | 0.44 | 0.33 | 0.24 | 0.16 | 0.10 | 0.05 | 0.02 |
|
|
0.27 | 0.25 | 0.24 | 0.22 | 0.20 | 0.19 | 0.20 | |
|
|
0.27 | 0.25 | 0.24 | 0.22 | 0.20 | 0.18 | 0.23 | |
| MISE | η̂2(x) | 6.37 | 1.81 | 0.24 | 1.64 | 6.01 | 13.37 | 23.69 |
|
|
0.27 | 0.25 | 0.24 | 0.22 | 0.21 | 0.19 | 0.20 | |
|
|
0.27 | 0.25 | 0.24 | 0.22 | 0.20 | 0.18 | 0.24 | |
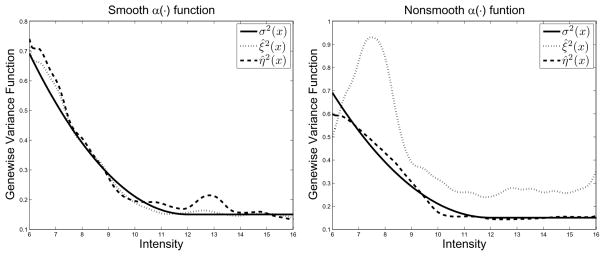
In Table 1, we report the mean integrated squared bias (Bias2), the mean integrated variance (VAR), and the mean integrated squared error (MISE) of ξ̂2(x) and η̂ 2(x) with and without the smoothness assumption on the gene effect αg. From the left panel of Table 1, we can see that when the smoothness assumption is valid, the estimator ξ̂2(x) outperforms η̂2(x). The reason is that the mean function α(Xgi) depends on the replication and is not a constant. Therefore, model (2) fails and η̂2(x) is biased. One should compare the results with those on the second row of the right panel where the model is right for η̂2(x). In this case, η̂2(x) performs much better. Its variance is about 3/2 as large as the variance in the case that mean is generated from a smooth function α(Xgi). This is expected. In the latter case, to eliminate αg, the degree of freedom reduces from I = 3 to 2, whereas in the former case, α(Xgi) can be estimated without losing the degree of freedom, namely the number of replications is still 3. The ratio 3/2 is reflected in Table 1. However, when the smoothness assumption does not hold, there is serious bias in the estimator ξ̂2(x), even though that αg is still reasonably sparse. The bias is an order of magnitude larger than those in the other situations.
Table 1.
Mean integrated squared bias (Bias2), mean integrated variance (VAR), mean integrated squared error (MISE) over 100 simulations for variance estimators ξ̂2(x) and η̂2(x). Two different gene effect functions α(·) are implemented. All quantities are multiplied by 1000.
| Smooth Gene Effect |
Non-Smooth Gene Effect |
|||||
|---|---|---|---|---|---|---|
| Bias2 | VAR | MISE | Bias2 | VAR | MISE | |
| ξ̂2(x) | 0.01 | 0.14 | 0.15 | 16.00 | 1.47 | 17.47 |
| η̂2(x) | 0.57 | 0.24 | 0.80 | 0.00 | 0.22 | 0.23 |
To see how variance estimators behave, we plot typical estimators ξ̂2(x) and η̂2(x) with median ISE value among 100 simulations in Figure 1. The solid line is the true variance function while the dotted and dashed lines represent ξ̂2(x) and η̂2(x) respectively. On the left panel of Figure 1, we can see that estimator ξ̂2(x) outperforms the estimator η̂2(x) when the smoothness assumption is valid. The region where the biases occur has already been explained above. However, ξ̂2(x) will generate substantial bias when the nonparametric regression model does not hold, and at the same time, our nonparametric estimator η̂2(x) corrects the bias very well.
Fig 1.
Variance estimators ξ̂2(x) and η̂2(x) with median performance when different gene effect function α(·) are implemented. Left Panel: Smooth α(·) function. Right panel: Non-smooth α(·) function.
3.3. Performance of new estimators
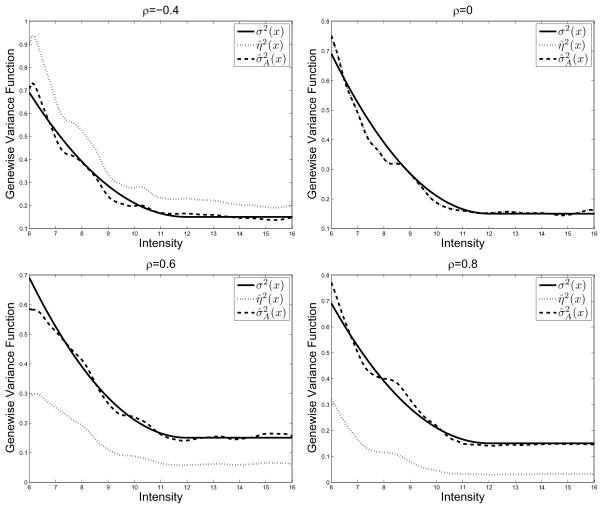
In this example, we consider the setting in §3.1 that the smoothness assumption of the gene effect αg is not valid. For comparison purpose only, we add an oracle estimator in which we assume that σ1, σ2 and ρ are all known. We now evaluate the performance of the estimators η̂2(x), , and when the correlation between within-array replications varies. To be more specific, seven different correlation settings are considered: ρ = −0.4, −0.2, 0, 0.2, 0.4, 0.6, 0.8, with ρ = 0 representing across-array replications. In this case, we increase the number of simulations to T = 1000. Again, we report Bias2, VAR and MISE of the three estimators for each correlation setting in Table 2. When ρ = 0, all the three estimators give the same bias and variance. This is consistent with our theory. We can see clearly from the table that, when ρ ≠ 0, the estimator produces much smaller biases than η̂2(x). In fact, when |ρ| as small as 0.2, the bias of η̂2(x) already dominates the variance.
It is worth noticing that the performance of and are almost always the same, which indicates that our algorithm for estimating ρ, σ1 and σ2 is very accurate. To see this more clearly, the squared bias, variance, and MSE of the estimator ρ, σ1 and σ2 in under the seven correlation settings are reported in Table 3. Here the true value of σ1 and σ2 is 0.4217 and 0.1857. For example, when ρ = 0.8, the bias of ρ̂ is less than 0.002 for , which is acceptable because the convergence threshold in the algorithm is set to be 0.001.
Table 3.
Squared bias, variance and MSE of ρ̂, σ̂1 and σ̂2 in the estimate . All quantities are multiplied by 106.
| ρ = | −0.4 | −0.2 | 0 | 0.2 | 0.4 | 0.6 | 0.8 | ||
|---|---|---|---|---|---|---|---|---|---|
| ρ̂ | Bias2 | 0.07 | 0.04 | 0.01 | 0.00 | 0.00 | 0.00 | 3.90 | |
| VAR | 7.90 | 16.91 | 28.65 | 36.17 | 35.68 | 27.21 | 20.44 | ||
| MSE | 7.97 | 16.95 | 28.66 | 36.17 | 35.68 | 27.21 | 24.35 | ||
| σ̂1 | Bias2 | 0.24 | 0.23 | 0.19 | 0.14 | 0.11 | 0.05 | 2.47 | |
| VAR | 11.65 | 11.52 | 11.79 | 12.46 | 13.64 | 15.55 | 18.66 | ||
| MSE | 11.89 | 11.75 | 11.99 | 12.60 | 13.75 | 15.59 | 21.12 | ||
| σ̂2 | Bias2 | 0.14 | 0.14 | 0.12 | 0.09 | 0.08 | 0.05 | 0.67 | |
| VAR | 10.34 | 10.17 | 10.45 | 11.12 | 12.24 | 13.96 | 16.16 | ||
| MSE | 10.47 | 10.31 | 10.57 | 11.20 | 12.32 | 14.00 | 16.83 | ||
In Figure 2, we render the estimates η̂2(x) and with the median ISE under four different correlation settings: ρ = −0.4, ρ = 0, ρ = 0.6 and ρ = 0.8. We omit the other correlation schemes since they all have similar performance. The solid lines represent the true variance function. The dotted lines and dashed lines are for η̂2(x) and respectively. For the case ρ = 0, the two estimators are indistinguishable. When ρ < 0, η̂2(x) overestimates the genewise variance function, whereas when ρ > 0, it underestimates the genewise variance function.
Fig 2.
Median performance of variance estimators η̂2(x), σ̂2(x), and when ρ =−0.4, 0, 0.6, and 0.8.
4. Application to human total RNA samples using Agilent arrays
Our real data example comes from Microarray Quality Control (MAQC) project (Patterson et al., 2006). The main purpose of the original paper is on comparison of reproducibility, sensitivity and specificity of microarray measurements across different platforms (i.e., one-color and two-color) and testing sites. The MAQC project use two RNA samples, Stratagene Universal Human Reference total RNA and Ambion Human Brain Reference total RNA. The two RNA samples have been assayed on three kinds of arrays: Agilent, CapitalBio and TeleChem. The data were collected at five sites. Our study focuses only on the Agilent arrays. At each site, 10 two-color Agilent microarrays are assayed with 5 of them dye swapped, totalling 30 microarrays.
4.1. Validation Test
In the first application, we revisit the validation test as considered in Fan and Niu (2007). For the purpose of the validation tests, we use gProcessedSignal and rProcessedSignal values from Agilent Feature Extraction software as input. We follow the preprocessing scheme described in Patterson et al. (2006) and get 22144 genes from a total of 41675 non-control genes. Among those, 19 genes with each having 10 replications are used for validation tests. Under the null hypothesis of no experimental biases, a reasonable model is
| (20) |
We use the notation G to denote the number of genes that have I replications. For our data, G = 19 and I = 10. Note that G can be different from N, the total number of different genes. The validation test statistics in Fan and Niu (2007) include weighted statistics
and unweighted test statistics
where and . Under the null hypothesis, the test statistic T1 is χ2 distributed with degree of freedom (I − 1)G and T2, T3 and T4 are all asymptotically normally distributed. As a result, the corresponding p-values can be easily computed.
Here, we apply the same statistics T1, T2, T3 and T4 but we replace the pooled sample variance estimator by the aggregated local linear estimator
where f̂ is the estimated density function of Xgi. The difference between the new variance estimator and the simple pooled variance estimator is that we consider the genewise variance as a nonparametric function of the intensity level. The latter estimator may drag small variances of certain arrays to much higher levels by averaging, resulting in a larger estimated genewise variance and smaller test statistics or bigger p-values.
In the analysis here, we first consider all thirty arrays. The estimated correlation among replicated genes is ρ̂ = 0.69. The p-values based on the newly estimated genewise variance are depicted in Table 4. As explained in Fan and Niu (2007), T4 is the most stable test among the four. It turns out that none of the arrays needs further normalization, which is the same as Fan and Niu (2007). Furthermore, we separate the analysis into two groups: the first group using 15 arrays without dye-swap, which has the estimated correlation ρ̂ = 0.66, and the second group using 15 arrays with dye-swap, resulting in an estimated correlation ρ̂ = 0.34. The p-values are summarized in Table 5. Results show that array AGL-2-D3 and array AGL-2-D5 need further normalization if 5% significance level applies. The difference is due to decreased estimated ρ for the dye swap arrays and p-values are sensitive to the genewise variance. We also did analysis by separating data into 6 groups: with and without dye swap, and three sites of experiments. Due to the small sample size, the six estimates of ρ range from 0.08 to 0.74, and we also find that array AGL-2-D3 needs further normalization.
Table 4.
Comparison of p-values for T1, ···, T4 for MAQC project data considering all 30 arrays together
| p-values | ||||
|---|---|---|---|---|
| slide name | T1 | T2 | T3 | T4 |
| AGL-1-C1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-D1 | 1.0000 | 0.9999 | 0.9996 | 0.9999 |
| AGL-2-D2 | 0.8387 | 0.9011 | 0.8953 | 0.9182 |
| AGL-2-D3 | 0.3525 | 0.1824 | 0.3902 | 0.1905 |
| AGL-2-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-D5 | 0.8820 | 0.8070 | 0.8848 | 0.7952 |
| AGL-3-C1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-C2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-C3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-C4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-C5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
Table 5.
Comparison of p-values for T1, ···, T4 for MAQC project data considering the arrays with and without dye-swap separately
| p-values | ||||
|---|---|---|---|---|
| slide name | T1 | T2 | T3 | T4 |
| AGL-1-C1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C3 | 1.0000 | 1.0000 | 0.9999 | 1.0000 |
| AGL-1-C4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-C5 | 1.0000 | 1.0000 | 0.9999 | 1.0000 |
| AGL-1-D1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-1-D5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C1 | 1.0000 | 1.0000 | 0.9943 | 1.0000 |
| AGL-2-C2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-C4 | 0.0152 | 0.9493 | 0.3931 | 0.9136 |
| AGL-2-C5 | 1.0000 | 1.0000 | 0.8060 | 1.0000 |
| AGL-2-D1 | 0.7806 | 0.8074 | 0.6622 | 0.6584 |
| AGL-2-D2 | 0.2170 | 0.2984 | 0.1651 | 0.2217 |
| AGL-2-D3 | 0.0002 | 0.0000 | 0.0001 | 0.0000 |
| AGL-2-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-2-D5 | 0.1236 | 0.0662 | 0.0669 | 0.0300 |
| AGL-3-C1 | 1.0000 | 1.0000 | 0.9996 | 1.0000 |
| AGL-3-C2 | 1.0000 | 1.0000 | 0.9988 | 1.0000 |
| AGL-3-C3 | 1.0000 | 1.0000 | 0.9977 | 1.0000 |
| AGL-3-C4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-C5 | 1.0000 | 1.0000 | 0.9999 | 1.0000 |
| AGL-3-D1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| AGL-3-D5 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
4.2. Gene selection
To detect the differentially expressed genes, we follow the filter instruction and get 19,802 genes out of 41,000 unique non-control genes as in Patterson et al. (2006), i.e., I = 1. The dye swap result was averaged before doing the one-sample t-test. Thus at each site, we have five microarrays.
For each site, significant genes are selected based on these 5 dye-swaped average arrays. For all N = 19, 802 genes, there are no within-array replications. However, model (2) is still reasonable, in which i indexes the array. Hence, the “within-array correlation” becomes “between-array correlation” and is reasonably assumed as ρ = 0.
In our nonparametric estimation for the variance function, all the 19,802 genes are used to estimate the variance function, which gives us enough reason to believe that the estimator is close to the inherent true variance function σ2(x).
We applied both the t-test and z-test to each gene to see if the logarithm of the expression ratio is zero, using the five arrays collected at each location. The number of differentially expressed genes detected by using the two different tests under three Fold Changes (FC) and four significant levels are given in Table 6. Large numbers of genes are identified as differentially expressed, which is expected when comparing a brain sample and a tissue pool sample (Patterson et al., 2006). We can see clearly that the z-test associated with our new variance estimator leads to more differentially expressed genes. For example, at site 1, using α = 0.001, among the fold changes at least 2, t-test picks 8231 genes whereas z-test selects 8875 genes. This gives an empirical power increase of (8875–8231)/19802 ≈ 3.25% in the group with observed fold change at least 2.
Table 6.
Comparison of the number of significantly differentially expressed genes
| p < 0.05 | p < 0.01 | p < 0.005 | p < 0.001 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| t-test | z-test | t-test | z-test | t-test | z-test | t-test | z-test | ||
| Agilent 1 | FC>1.5 | 12692 | 12802 | 12464 | 12752 | 12313 | 12722 | 11744 | 12646 |
| FC>2 | 8802 | 8879 | 8654 | 8872 | 8556 | 8869 | 8231 | 8858 | |
| FC>4 | 3493 | 3493 | 3431 | 3493 | 3376 | 3493 | 3231 | 3493 | |
| Agilent 2 | FC>1.5 | 12282 | 12678 | 11217 | 12587 | 10502 | 12536 | 8270 | 12421 |
| FC>2 | 8644 | 8877 | 7908 | 8875 | 7452 | 8861 | 6125 | 8828 | |
| FC>4 | 3600 | 3649 | 3188 | 3649 | 2964 | 3649 | 2422 | 3649 | |
| Agilent 3 | FC>1.5 | 12502 | 12692 | 11994 | 12576 | 11694 | 12519 | 10788 | 12374 |
| FC>2 | 8689 | 8832 | 8344 | 8810 | 8150 | 8800 | 7591 | 8762 | |
| FC>4 | 3585 | 3603 | 3378 | 3602 | 3278 | 3602 | 2985 | 3600 | |
To verify the accuracy of our variance estimation in the z-test, we compare the empirical power increase with the expected theoretical power increase. The expected theoretical power increase is computed as
| (21) |
taking the average of power increases across all μg ≠ 0. However, in the absence of the availability, we replace μg by its estimate, which is the sample average of n = 5 observed log-expression ratios. Table 7 depicts the results at three different sites, in which the columns “Theo” refer to the expected theoretical power increase defined by (21), with μg replaced by Ȳg and σg replaced by its estimate from the genewise variance function, and the columns “Emp” refer to the empirical power increase.
Table 7.
Comparison of Expected Theoretical and Empirical Power Difference (In percentage)
| α = 0.05 | α = 0.01 | α = 0.005 | α = 0.001 | |||||
|---|---|---|---|---|---|---|---|---|
| Theo | Emp | Theo | Emp | Theo | Emp | Theo | Emp | |
| Agilent 1 | 2.52 | 0.61 | 6.08 | 3.75 | 8.06 | 5.59 | 13.66 | 11.74 |
| Agilent 2 | 4.03 | 7.56 | 10.11 | 17.61 | 13.61 | 22.86 | 23.75 | 37.63 |
| Agilent 3 | 3.02 | 2.56 | 7.14 | 7.39 | 9.42 | 10.19 | 15.94 | 18.18 |
| Average | 3.19 | 3.58 | 7.78 | 9.58 | 10.36 | 12.88 | 17.79 | 22.51 |
There are two things worth noticing here. First, for expected theoretical power increase, we use the sample mean Ȳg = μg + ε̄g instead of the real gene effect μg, which is not observable, so it inevitably involves the error term ε̄g. Second, the power functions Pz(μ) and Pt(μ) depend sensitively on μ and the tails of the assumed distribution. Despite of these, the expected theoretical and empirical power increases are in the same bulk and the averages are very close. This provides good evidence that our genewise variance estimation has an acceptable accuracy.
We also apply SIMEX and permutation SIMEX methods in Wang and Carroll (2008) to the MAQC data, to illustrate its utility. As mentioned in the introduction, their model is not really intended for the analysis of two-color microarray data. Should we only use the information on log-ratios (Y), the model is very hard to interpret. In addition, one might question why the information on X (observed intensity levels) is not used at all. Nevertheless, we apply the SIMEX methods of Wang and Carroll (2008) to only the log-ratios Y in the two-color data and produce similar tables to the Table 6 and 7.
From the results, we have the following understandings. First, all the numbers for z-test in Tables 8 and 9 at all significance levels are approximately the same. In fact, the p-values are very small so that numbers at different significance levels are about the same. That indicates that both SIMEX and permutation SIMEX method are tending to estimate genewise variance very small, making the test statistics large for all the time. On the other hand, our method estimates the genewise variance moderately so that the numbers are not exactly the same for different significance levels. Second, in the implementation, we found that the SIMEX and permutation SIMEX is computationally expensive (more than one hour) while our method only takes a few minutes. Third, from Tables 10 and 11 we can see that the expected theoretical power increase and the empirical ones are reasonably close, which are in lines with our method.
Table 8.
Comparison of the number of significantly differentially expressed genes using SIMEX method
| p < 0.05 | p < 0.01 | p < 0.005 | p < 0.001 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| t-test | z-test | t-test | z-test | t-test | z-test | t-test | z-test | ||
| Agilent 1 | FC>1.5 | 12692 | 12820 | 12464 | 12820 | 12313 | 12820 | 11744 | 12820 |
| FC>2 | 8802 | 8879 | 8654 | 8879 | 8556 | 8879 | 8231 | 8879 | |
| FC>4 | 3493 | 3493 | 3431 | 3493 | 3376 | 3493 | 3231 | 3493 | |
| Agilent 2 | FC>1.5 | 12282 | 12721 | 11217 | 12721 | 10502 | 12721 | 8270 | 12721 |
| FC>2 | 8644 | 8878 | 7908 | 8878 | 7452 | 8878 | 6125 | 8878 | |
| FC>4 | 3600 | 3649 | 3188 | 3649 | 2964 | 3649 | 2422 | 3649 | |
| Agilent 3 | FC>1.5 | 12502 | 12760 | 11994 | 12760 | 11694 | 12760 | 10788 | 12760 |
| FC>2 | 8689 | 8836 | 8344 | 8836 | 8150 | 8836 | 7591 | 8836 | |
| FC>4 | 3585 | 3603 | 3378 | 3603 | 3278 | 3603 | 2985 | 3603 | |
Table 9.
Comparison of the number of significantly differentially expressed genes using permutation SIMEX
| p < 0.05 | p < 0.01 | p < 0.005 | p < 0.001 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| t-test | z-test | t-test | z-test | t-test | z-test | t-test | z-test | ||
| Agilent 1 | FC>1.5 | 12692 | 12820 | 12464 | 12820 | 12313 | 12820 | 11744 | 12820 |
| FC>2 | 8802 | 8879 | 8654 | 8879 | 8556 | 8879 | 8231 | 8879 | |
| FC>4 | 3493 | 3493 | 3431 | 3493 | 3376 | 3493 | 3231 | 3493 | |
| Agilent 2 | FC>1.5 | 12282 | 12721 | 11217 | 12721 | 10502 | 12721 | 8270 | 12721 |
| FC>2 | 8644 | 8878 | 7908 | 8878 | 7452 | 8878 | 6125 | 8878 | |
| FC>4 | 3600 | 3649 | 3188 | 3649 | 2964 | 3649 | 2422 | 3649 | |
| Agilent 3 | FC>1.5 | 12502 | 12760 | 11994 | 12760 | 11694 | 12760 | 10788 | 12760 |
| FC>2 | 8689 | 8836 | 8344 | 8836 | 8150 | 8836 | 7591 | 8836 | |
| FC>4 | 3585 | 3603 | 3378 | 3603 | 3278 | 3603 | 2985 | 3603 | |
Table 10.
Comparison of Expected Theoretical and Empirical Power Difference using SIMEX method (In percentage)
| α = 0.05 | α = 0.01 | α = 0.005 | α = 0.001 | |||||
|---|---|---|---|---|---|---|---|---|
| Theo | Emp | Theo | Emp | Theo | Emp | Theo | Emp | |
| Agilent 1 | 2.43 | 2.06 | 7.17 | 5.42 | 10.30 | 7.34 | 20.71 | 13.44 |
| Agilent 2 | 7.16 | 3.41 | 19.20 | 12.06 | 26.17 | 16.90 | 43.46 | 30.42 |
| Agilent 3 | 4.18 | 2.88 | 11.71 | 7.38 | 16.45 | 9.89 | 31.38 | 17.57 |
| Average | 4.59 | 2.78 | 12.69 | 8.29 | 17.64 | 11.38 | 31.85 | 20.48 |
Table 11.
Comparison of Expected Theoretical and Empirical Power Difference using permutation SIMEX method (In percentage)
| α = 0.05 | α = 0.01 | α = 0.005 | α = 0.001 | |||||
|---|---|---|---|---|---|---|---|---|
| Theo | Emp | Theo | Emp | Theo | Emp | Theo | Emp | |
| Agilent 1 | 1.89 | 2.86 | 5.66 | 6.43 | 8.19 | 8.59 | 16.75 | 15.07 |
| Agilent 2 | 4.84 | 7.37 | 13.44 | 17.22 | 18.97 | 22.50 | 36.90 | 37.26 |
| Agilent 3 | 2.89 | 4.91 | 8.34 | 10.13 | 11.87 | 13.11 | 23.44 | 21.31 |
| Average | 3.20 | 5.05 | 9.15 | 11.26 | 13.01 | 14.74 | 25.70 | 24.55 |
5. Summary
The estimation of genewise variance function is motivated by the downstream analysis of microarray data such as validation test and selecting statistically differentially expressed genes. The methodology proposed here is novel by using across-array and within-array replications. It does not require any specific assumptions on αg such as sparsity or smoothness, and hence reduces the bias of the conventional nonparametric estimators. Although the number of nuisance parameters is proportional to the sample size, we can estimate the main interest (variance function) consistently. By increasing the degree of freedom largely, both the validation tests and z-test using our variance estimators are more powerful in identifying arrays that need to be normalized further and more capable of selecting differentially expressed genes.
Our proposed methodology has a wide range of applications. In addition to the microarray data analysis with within-array replications, it can be also applied to the case without within-array replications, as long as the model (2) is reasonable. Our two-way nonparametric model is a natural extension of the Neyman-Scott problem. Therefore, it is applicable to all the problems where the Neyman-Scott problem is applicable.
There are possible extensions. For example, the SIMEX idea can be applied on our model in order to take into account the measurement error. We can also make adaptations to our methods when we have a prior correlation structure among replications other than the identical correlation assumption.
Acknowledgments
The authors thank the Editor, the Associate Editor and two referees, whose comments have greatly improved the scope and presentation of the paper.
APPENDIX A: APPENDIX
The following regularity conditions are imposed for the technical proofs:
The regression function σ2(x) has a bounded and continuous second derivative.
The kernel function K is a bounded symmetric density function with a bounded support.
h → 0, Nh → ∞.
E[σ8(X)] exists and the marginal density fX(·) is continuous.
We need the following conditional variance-covariance matrix of the random vector Zg in our asymptotic study.
Lemma 1
Let Ω be the variance-covariance matrix of Zg conditioning on all data X. Then respectively the diagonal and off-diagonal elements are:
| (22) |
| (23) |
Proof of Lemma 1
Let A be the variance-covariance matrix of rg conditioning on all data X. By direct computation, the diagonal elements are given by
| (24) |
and the off-diagonal elements are given by
| (25) |
Using Ω = BABT, we can obtain the result by direct computation.
The proofs for Theorems 1 and 3 follow a similar idea. Since Theorem 1 doesn’t involve a lot of coefficients, we will show the proof of Theorem 1 and explain the difference in the proof of Theorem 3.
Proof of Theorem 1
First of all, the bias of comes from the local linear approximation. Since is an i.i.d. sequence, by (4) and the result in Fan and Gijbels (1996), it follows that
Similarly, the asymptotic variance of also follows from Fan and Gijbels (1996).
We now prove the off-diagonal elements in matrix var[η|X]
| (26) |
Recalling that , we have
The equality follows by the fact that cov[(Zgi, Zg′j)|X] = 0 when g ≠ g′. Recall Ωij = cov[(Zgi, Zgj)|X] and define RN,g = N · WN,j((Xgj − x)/h)Ωij. Thus
| (27) |
The right hand side of (27) can be seen as local linear smoother of the synthetic data . Although RN,g involves N at the first glance, its conditional expectation E[RN,g|Xgi = x] and conditional variance var[RN,g|Xgi = x] do not grow with N. Since is an i.i.d. sequence, by the results in Fan and Gijbels (1996), we obtain
To calculate E[RN,g|Xgi = x], we apply the approximation WN,i(u) = K(u)(1 + oP (1))/(NhfX(x)) in the example of Fan and Gijbels (1996, p. 64) and have the following arguments
where s represents all the integrating variables corresponding to Xg1, ···, XgI except Xgi and Xgj. That justifies (26).
To prove the multivariate asymptotic normality
| (28) |
we employ Cramér-Wold device: for any unit vector a = (a1, ···, aI)T in ℝI,
Denote by Qg,i = WN,i((Xgi − x)/h)(Zgi− σ2(Xgi)) and . Note that the sequence is i.i.d. distributed. To show the asymptotic normality of F*, it is sufficient to check Lyapunov’s condition:
To facilitate the presentation, we first note that sequences are i.i.d. and satisfy Lyapunov’s condition for each fixed i. Denote . And recall that . Let c* be a generic constant which may vary from one line to another. We have the following approximation
Therefore . By the marginal Lyapunov conditions, we have the following inequality
For the denominator, we have the following arguments
Note that the second to last equality holds by the asymptotic conditional variance-covariance matrix Σ. Therefore Lyapunov’s condition is justified. That completes the proof.
Proof of Theorem 2
First of all, for each given g,
Note that by (8), we have
Thus, for all g, we have
Since { } and { } are i.i.d sequences across the N genes, by the central limit theorem, we have
Therefore,
Proof of Theorem 3
Note that
Following similar steps in the proof of Theorem 1, one can verify , where the coefficients C2, ···, C4, D0, ···, D4 are as follows:
Footnotes
The financial support from NSF grant DMS-0714554 and NIH grant R01-GM072611 are greatly acknowledged.
Contributor Information
Jianqing Fan, Email: jqfan@princeton.edu, Department of Operations Research and Financial Engineering, Princeton University, Princeton, NJ 08544.
Yang Feng, Email: yangfeng@princeton.edu, Department of Operations Research and Financial Engineering, Princeton University, Princeton, NJ 08544.
Yue S. Niu, Email: yueniu@math.arizona.edu, Department of Mathematics, University of Arizona, 617 N. Santa Rita Ave., P.O. Box 210089, Tucson, AZ 85721-0089.
References
- Carroll RJ, Wang Y. Nonparametric variance estimation in the analysis of microarray data: a measurement error approach. Biometrika. 2008 doi: 10.1093/biomet/asn017. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modelling and Its Applications. Chapman & Hall; 1996. [Google Scholar]
- Fan J, Niu Y. Selection and validation of normalization methods for c-DNA microarrays using within-array replications. Bioinformatics. 2007;23:2391–2398. doi: 10.1093/bioinformatics/btm361. [DOI] [PubMed] [Google Scholar]
- Fan J, Peng H, Huang T. Semilinear high-dimensional model for normalization of microarray data: a theoretical analysis and partial consistency (with discussion) Journal of the American Statistical Association. 2005;100:781–813. [Google Scholar]
- Fan J, Ren Y. Statistical analysis of DNA microarray data in cancer research. Clinical Cancer Research. 2007;12:4469–4473. doi: 10.1158/1078-0432.CCR-06-1033. [DOI] [PubMed] [Google Scholar]
- Fan J, Tam P, Vande Woude G, Ren Y. Normalization and analysis of cDNA micro-arrays using within-array replications applied to neuroblastoma cell response to a cytokine. Proceedings of the National Academy of Sciences, USA. 2004;101(5):1135–1140. doi: 10.1073/pnas.0307557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang D, Zhang C. A two-way semi-linear model for normalization and significant analysis of cDNA microarray data. Journal of the American Statistical Association. 2005;100:814–829. [Google Scholar]
- Kamb A, Ramaswami A. A simple method for statistical analysis of intensity differences in microarray-deried gene expression data. BMC Biotechnology. 2001:1–8. doi: 10.1186/1472-6750-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyman J, Scott E. Consistent estimates based on partially consistent observations. Econometrica. 1948;16:1–32. [Google Scholar]
- Patterson T, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nature Biotechnology. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- Ruppert D, Wand MP, Holst U, Hössjer O. Local polynomial variance function estimation. Technometrics. 1997;39:262–273. [Google Scholar]
- Smyth G, Michaud J, Scott H. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proceedings of the National Academy of Sciences, USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Wang Y. Optimal Shrinkage Estimation of Variances with Applications to Microarray Data Analysis. Journal of the American Statistical Association. 2007;102:113–122. [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ma Y, Carroll RJ. Variance Estimation in the Analysis of Microarray Data. Journal of the Royal Statistical Society, SerB. 2008 doi: 10.1111/j.1467-9868.2008.00690.x. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]