Abstract
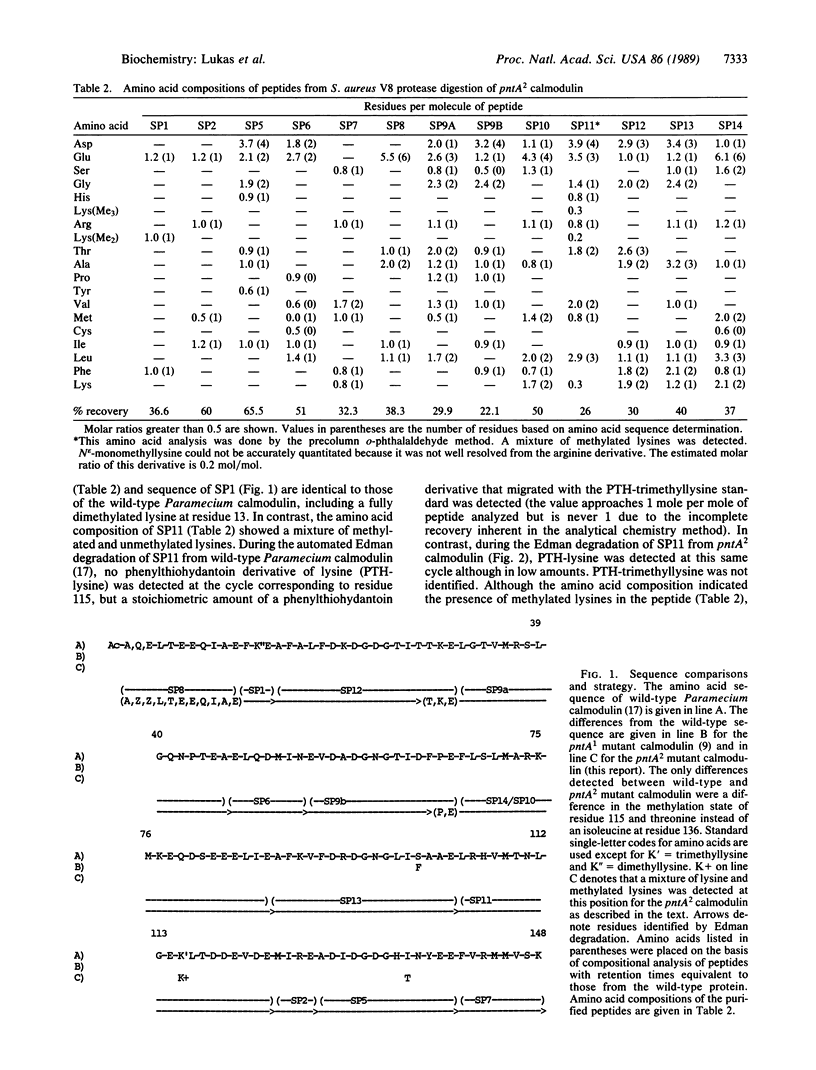
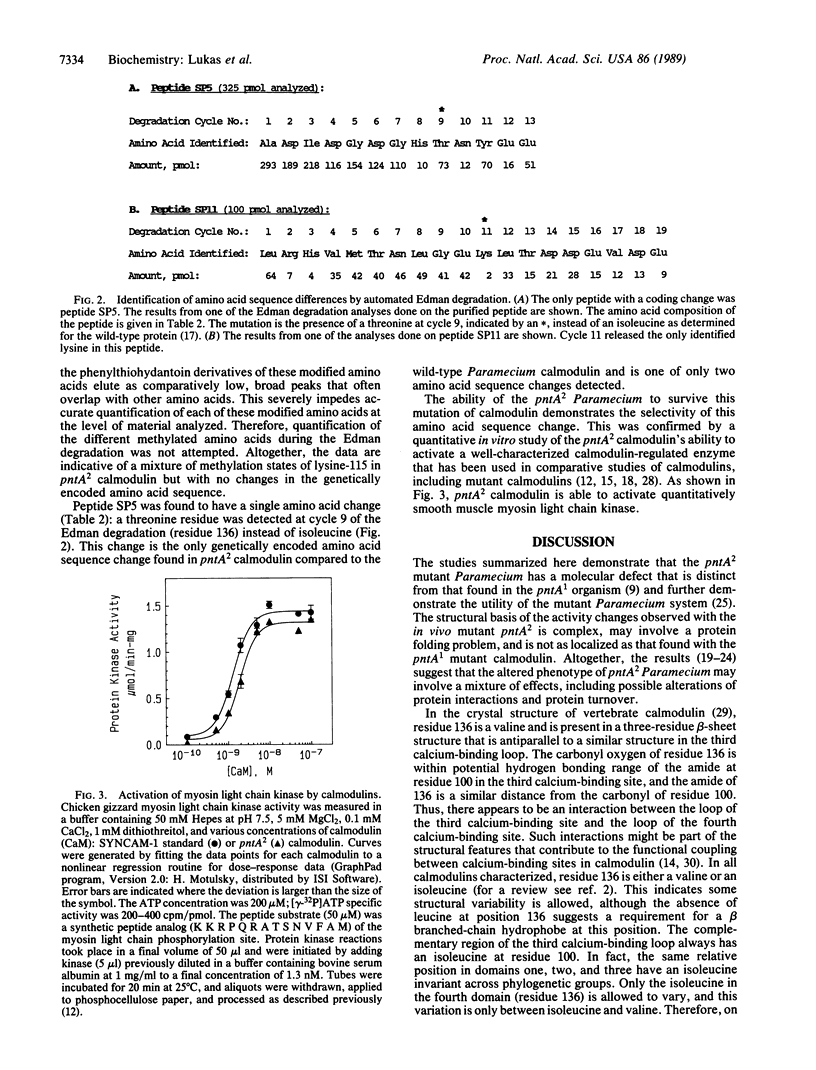
The Paramecium tetraurelia mutants termed pantophobiacs have altered behavior due to perturbed calcium activation of ion channel activity. The calmodulin from pantophobiac A1 (pntA1) was shown in previous studies to have a single amino acid change at residue 101 that is selective in its effects on activity. This change has no effect on posttranslational modifications. However, the calmodulin from the phenotypically related mutant pantophobiac A2 (pntA2) has a threonine residue at position 136, in the fourth calcium-binding domain, instead of an isoleucine or valine like all other calmodulins. This region of the calmodulin structure is within 4 A of a complementary hydrophobic structure in the third calcium-binding domain, raising the possibility of a perturbation of interdomain interactions in the pntA2 mutant. This possibility is supported by the heterogenous methylation state of lysine-115 in the pntA2 calmodulin. This lysine residue, located in the peptide connecting calcium-binding domains three and four, is fully trimethylated in the wild-type and pntA1 calmodulins. The functional selectivity of these structural changes is demonstrated by the conservation of calmodulin activator activity with a calmodulin-regulated protein kinase that has been used as a standard of comparison. Overall, these results indicate the degree to which the calmodulin can be mutated in vivo without being lethal to the organism, and they provide genetic evidence suggesting that the post-translational methylation state of residue 115 requires the appropriate conformation in addition to the local amino acid sequence.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babu Y. S., Bugg C. E., Cook W. J. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988 Nov 5;204(1):191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Burgess-Cassler A., Hinrichsen R. D., Maley M. E., Kung C. Biochemical characterization of a genetically altered calmodulin in Paramecium. Biochim Biophys Acta. 1987 Jul 7;913(3):321–328. doi: 10.1016/0167-4838(87)90142-7. [DOI] [PubMed] [Google Scholar]
- Craig T. A., Watterson D. M., Prendergast F. G., Haiech J., Roberts D. M. Site-specific mutagenesis of the alpha-helices of calmodulin. Effects of altering a charge cluster in the helix that links the two halves of calmodulin. J Biol Chem. 1987 Mar 5;262(7):3278–3284. [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Hinrichsen R. D., Burgess-Cassler A., Soltvedt B. C., Hennessey T., Kung C. Restoration by calmodulin of a Ca2+-dependent K+ current missing in a mutant of Paramecium. Science. 1986 Apr 25;232(4749):503–506. doi: 10.1126/science.2421410. [DOI] [PubMed] [Google Scholar]
- Kilhoffer M. C., Roberts D. M., Adibi A. O., Watterson D. M., Haiech J. Investigation of the mechanism of calcium binding to calmodulin. Use of an isofunctional mutant with a tryptophan introduced by site-directed mutagenesis. J Biol Chem. 1988 Nov 15;263(32):17023–17029. [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Lukas T. J., Wiggins M. E., Watterson D. M. Amino Acid sequence of a novel calmodulin from the unicellular alga chlamydomonas. Plant Physiol. 1985 Jul;78(3):477–483. doi: 10.1104/pp.78.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak D. R., Clarke M., Roberts D. M., Watterson D. M. Structural and functional properties of calmodulin from the eukaryotic microorganism Dictyostelium discoideum. Biochemistry. 1984 Jun 19;23(13):2891–2899. doi: 10.1021/bi00308a007. [DOI] [PubMed] [Google Scholar]
- Putkey J. A., Ono T., VanBerkum M. F., Means A. R. Functional significance of the central helix in calmodulin. J Biol Chem. 1988 Aug 15;263(23):11242–11249. [PubMed] [Google Scholar]
- Roberts D. M., Burgess W. H., Watterson D. M. Comparison of the NAD Kinase and Myosin Light Chain Kinase Activator Properties of Vertebrate, Higher Plant, and Algal Calmodulins. Plant Physiol. 1984 Jul;75(3):796–798. doi: 10.1104/pp.75.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. M., Crea R., Malecha M., Alvarado-Urbina G., Chiarello R. H., Watterson D. M. Chemical synthesis and expression of a calmodulin gene designed for site-specific mutagenesis. Biochemistry. 1985 Sep 10;24(19):5090–5098. doi: 10.1021/bi00340a020. [DOI] [PubMed] [Google Scholar]
- Roberts D. M., Rowe P. M., Siegel F. L., Lukas T. J., Watterson D. M. Trimethyllysine and protein function. Effect of methylation and mutagenesis of lysine 115 of calmodulin on NAD kinase activation. J Biol Chem. 1986 Feb 5;261(4):1491–1494. [PubMed] [Google Scholar]
- Saimi Y., Hinrichsen R. D., Forte M., Kung C. Mutant analysis shows that the Ca2+-induced K+ current shuts off one type of excitation in Paramecium. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5112–5116. doi: 10.1073/pnas.80.16.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saimi Y., Kung C. Behavioral genetics of Paramecium. Annu Rev Genet. 1987;21:47–65. doi: 10.1146/annurev.ge.21.120187.000403. [DOI] [PubMed] [Google Scholar]
- Schaefer W. H., Hinrichsen R. D., Burgess-Cassler A., Kung C., Blair I. A., Watterson D. M. A mutant Paramecium with a defective calcium-dependent potassium conductance has an altered calmodulin: a nonlethal selective alteration in calmodulin regulation. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3931–3935. doi: 10.1073/pnas.84.11.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer W. H., Lukas T. J., Blair I. A., Schultz J. E., Watterson D. M. Amino acid sequence of a novel calmodulin from Paramecium tetraurelia that contains dimethyllysine in the first domain. J Biol Chem. 1987 Jan 25;262(3):1025–1029. [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eldik L. J., Zendegui J. G., Marshak D. R., Watterson D. M. Calcium-binding proteins and the molecular basis of calcium action. Int Rev Cytol. 1982;77:1–61. doi: 10.1016/s0074-7696(08)62463-8. [DOI] [PubMed] [Google Scholar]


