Abstract
Thermodynamic analysis of metabolic networks has recently generated increasing interest for its ability to add constraints on metabolic network operation, and to combine metabolic fluxes and metabolite measurements in a mechanistic manner. Concepts for the calculation of the change in Gibbs energy of biochemical reactions have long been established. However, a concept for incorporation of cross-membrane transport in these calculations is still missing, although the theory for calculating thermodynamic properties of transport processes is long known. Here, we have developed two equivalent equations to calculate the change in Gibbs energy of combined transport and reaction processes based on two different ways of treating biochemical thermodynamics. We illustrate the need for these equations by showing that in some cases there is a significant difference between the proposed correct calculation and using an approximative method. With the developed equations, thermodynamic analysis of metabolic networks spanning over multiple physical compartments can now be correctly described.
Introduction
Theoretical and computational studies of metabolic network function are increasingly applying thermodynamic principles. Recent applications include analysis of the thermodynamic feasibility of reaction fluxes in pathways (1–6), cellular-scale networks (7–18), and the thermodynamic feasibility of metabolite concentrations in large-scale networks (19,20). Theoretical analyses of the relationships between metabolic flux and Gibbs energy have been developed (21–23); and thermodynamics has also been used as constraints in kinetic models (24–29).
Thermodynamic analyses of biochemical systems are complicated by a number of factors. One major complicating factor is that biochemical reactions occur in aqueous solutions, and thus biochemical reactants (e.g., ATP) are typically made up of a number of rapidly interconverting proton-bound or metal cation-bound species (e.g., ATP4−, HATP3−, and H2ATP2−). A concentration of a biochemical reactant is therefore typically a lumped sum of the concentrations of individual species. Concentrations of the individual species are usually neither measured nor reported (i.e., even with state-of-the art metabolomics platforms as described in Büscher et al. (30)), requiring a concept that can predict the distribution of a biochemical reactant into multiple species, given a certain pH, metal ion concentrations, temperature, and ionic strength. Alberty's (31) Legendre transform formalism provides such a concept and allows us to make thermodynamics calculations based on reactant concentrations. Another approach uses binding polynomials to calculate species concentrations from reactant concentrations (32,33).
Another factor that complicates thermodynamic analysis of metabolic networks is that these systems are often distributed over multiple compartments with different physiochemical properties, such as pH or ionic strength. To calculate the change in Gibbs energy of cross-membrane transport processes, the electrochemical potential of that process needs to be considered (34). To make such a calculation, one has to consider the specific species that are transported. Thus, the equations of biochemical thermodynamics that use reactant concentrations must be extended to allow description of thermodynamics of transport processes. Unfortunately, to our knowledge, this does not yet exist, but it is required for correct thermodynamic analysis of genome-scale metabolic networks.
In this article, we develop a framework for the thermodynamic description of biochemical transport and reaction processes based on reactant concentrations. We illustrate how this framework may be practically applied for the calculation of thermodynamics of reactions, transport processes, and of coupled reaction and transport processes. Furthermore, we show the effect that the correct consideration of species has on the calculated change in Gibbs energy for transport/reaction processes. We envision that the proposed framework provides the necessary tool for thermodynamic analysis of metabolic networks with compartments.
Results
To derive a formulation for the calculation of the change in Gibbs energy (ΔG) of any transport/reaction process based on the concentrations of the involved reactants, we start from the basic equation to calculate the ΔG of a chemical reaction,
| (1) |
where ΔfG0i values are the Gibbs energies of formation for each involved species i, si values are the stoichiometric coefficients for the reacting species, and ci values are the concentrations of each species. To calculate the ΔG of a biochemical process using Eq. 1, we would need to know the species concentration ci. Because the concentrations of the species are usually not measured, we require a formulation to calculate the ΔG of a reaction in terms of reactant concentrations.
Alberty (31) developed such a concept, in which the ΔG of a reaction is calculated with the equation,
| (2) |
where the values are the so-called transformed Gibbs energies of formation for a reactant j and Cj values are the reactant concentrations. Sj values are the stoichiometric coefficients of reactants and are the sum of the stoichiometric coefficients of the constituting species that participate in the reaction,
For example, when 1 mol of a reactant participates in a reaction as 0.5 mol of species 1 and 0.5 mol of species 2, the stoichiometric coefficients s1 and s2 are 0.5 and 0.5, respectively. The transformed Gibbs energy of formation is a formation energy that incorporates the pH of the environment, in which the reaction takes place. In the derivation of Eq. 2, the pH is introduced by a Legendre transformation (see Eq. A2i in the Supporting Material). In biochemical reactions, a proton is not considered as a reactant. In Eq. 2, the protons are considered in the transformed Gibbs energies of formation. See Section 1 of the Supporting Material for a complete derivation of Eq. 2 and the equation to calculate the transformed Gibbs energies of formation for reactants.
An equivalent way to calculate the ΔG of a reaction in terms of reactant concentrations is by using the concept of binding polynomials (32,33). By using binding polynomials, we can calculate a species concentration from the reactant concentration and can then use this concentration with Eq. 1. The binding polynomials can be incorporated in Eq. 1 as
| (3) |
where values are the Gibbs energies of formation for the reference species, Cj values are the reactant concentrations, Pj([H+]) values are the binding polynomials, and ΔfG0H+ values the Gibbs energy of formation for a hydrogen ion. Because in biochemical reactions protons are not considered as a reactant, the last two terms of Eq. 3 are added to incorporate the effect of the proton concentration, or pH. See Section 1 in the Supporting Material for a complete derivation of Eq. 3. With Eqs. 2 and 3, the ΔG of any biochemical reaction can be calculated based on reactant concentrations.
We would now like to incorporate the effect of transport of a specific species across membranes into the calculation of the ΔG, because metabolites are often transported by proteins with a structure that allows only a specific species to be transported (35). We can start again from the species level with Eq. 1. To calculate the contribution of transport on the ΔG, Eq. 1 can be extended to incorporate the effect of the electrochemical potential (34),
| (4) |
where F is the Faraday constant, zi is the charge of species i, and Δϕm is the membrane potential difference defined as the difference of the inside minus the outside electric potential for transport to the inside. The sum over the stoichiometric coefficients of the transported species, with charge zi, in the inside compartment () is introduced to calculate the charge that is transferred over the membrane.
To translate Eq. 4 into an equation that only uses reactant concentrations, we must make a distinction between the reacting biochemical reactants, as in Eqs. 2 and 3, and the transported chemical species. To make this distinction, we define the species in Eq. 4 as a group of three types of species,
| (5) |
where i ε I (j) are the constituting species of reactants j, i ε protons are the free protons participating in a reaction, and i ε transported are the species that are transported. With Eq. 5, we separate the different species that can participate in a transport/reaction process and classify them into the three different groups. In Fig. 1A, the process of ATP synthesis is illustrated on a species level. In Fig. 1 B, it is then shown which species belong to the different groups of stoichiometric coefficients from Eq. 5.
Figure 1.
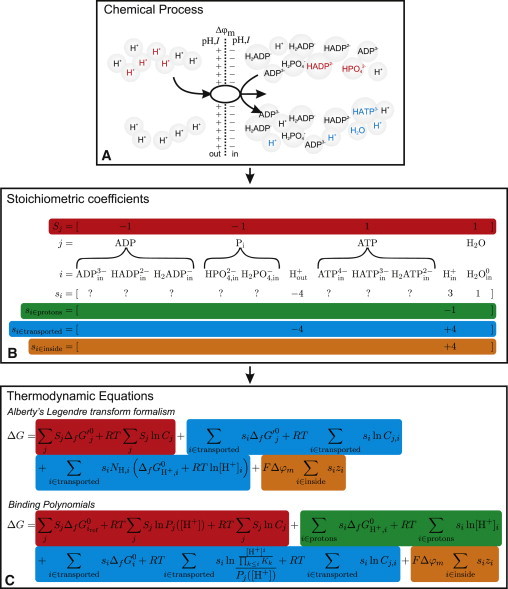
(A) The biochemical transport/reaction process ATP synthase, where ATP is generated from the transport of four protons from the cytosol (out) to the mitochondrium (in), is represented on a species level, with species that have a negative stoichiometric coefficient in red and species that have a positive stoichiometric coefficient in blue. Several species, with each having their own concentration (ci), are shown for each reactant. (B) Because we do not know for each reactant which species reacts, we define a vector of stoichiometries for reactants (Sj). For the protons that participate in the reaction, a separate vector (siεprotons) is defined. For the transported species, the vector (siεtransported) is defined, and the vector (siεinside) is the equivalent of the transported species vector but only contains the stoichiometric coefficients of the inside species. (C) Then we can apply the two equations that can be used to calculated the ΔG of the process. Where in the reaction equation the information for the ΔG equations is obtained is indicated by similar colors. See Section 2 in the Supporting Material for a detailed derivation of the equations.
Now, we can define equations to calculate the ΔG of transport/reaction processes by using the two different ways of dealing with biochemical thermodynamics. The derivation of Eqs. 6 and 7 is shown in detail in Section 2 in the Supporting Material. Using Alberty's Legendre transform formalism, we obtain the following equation for a transport/reaction process,
| (6) |
and by using the concept of binding polynomials we obtain the following equation,
| (7) |
where [H+]i and values are the concentration and the transformed Gibbs energy of formation of a hydrogen ion in the compartment of species i. The value Cj,i is the concentration of the reactant of which the transported species i is part. Equations 6 and 7 are now written only in terms of concentrations of reactants but do account for the specific species that are being transported.
Examples
To show how these equations can be applied, we consider the process of succinate transport over a membrane into a cell. Here we assume that all biochemical conditions (ionic strength, temperature, and pressure) other than pH are equal on both sides of the membrane. Furthermore, we assume that succinate may associate with hydrogen ions but not with other dissolved cations such as potassium or magnesium. The biochemical process can be described as
| (8) |
where Succ0 indicates the uncharged form of succinate (i.e., the acid with two protons bound), which is transported from the outside to the inside compartment. From Eq. 8, we can define the stoichiometric coefficients as defined in Eq. 5,
| (9) |
where the indices represent and , respectively. Now using the information from Table 1, we have all the information to calculate the ΔG of the transport process. In Fig. 2 a (solid line), the value of the ΔG is shown as a function of inside pH with outside pH held constant. When the pH of the inside compartment changes, the relative abundances of the different species of succinate (e.g., Succ−2, Succ−1, and Succ0) will change. The difference in relative abundances of the species in each compartment has an effect on the ΔG of the transport process because the gradients of the species over the membrane are different than the gradient of the reactants.
Table 1.
Data used in the example calculations for the processes shown in Eqs. 8 and 10
| ATP | ADP | Pi | H2O | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔfG0 (kJ mol−1) | |||||||||
| Species charge | −4 | −2768.1 | |||||||
| −3 | −2811.48 | −1906.13 | |||||||
| −2 | −2838.18 | −1947.1 | −1069.1 | −690.44 | −690.44 | ||||
| −1 | −1971.98 | −1137.3 | −722.62 | −722.62 | |||||
| 0 | −237.19 | −746.64 | −746.64 | ||||||
| P | 1.28 | 1.42 | 1.54 | 1.00 | 2.54 | 1.14 | |||
| Cj (mM) | 1 | 0.5 | 10 | 1000 | 0.05 | 0.5 | |||
| pK1 | 6.44 | 6.62 | 6.73 | 1 | 5.15 | 5.15 | |||
| pK2 | 3.87 | 3.94 | 3.96 | 3.96 | |||||
Figure 2.
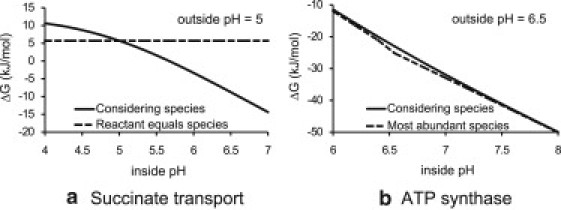
(a) The ΔG of succinate transport (see Eq. 8) is shown at varying pH of the inside compartment. Comparison of correctly considering the presence of species (solid line) versus assuming that the reactant concentration is equal to the transported species concentration (dashed line). Reactant concentrations (see Table 1) are kept constant, outside pH is 5, the membrane potential difference (ϕm) is 40 mV, and ionic strengths are kept 0.15 M in both compartments. (b) The ΔG of ATP hydrolysis is shown at varying pH of the inside compartment. Comparison of correctly considering the presence of species (solid line) versus assuming that the reactant concentration approximates the most abundant species concentration (dashed line). Reactant concentrations (see Table 1) are kept constant, the outside pH is 6.5, the membrane potential difference (ϕm) is −180 mV, and ionic strengths are kept 0.15 M in both compartments. See Section 3 in the Supporting Material for detailed examples of such calculations.
When the different species of the reactant succinate are not considered and Eq. 4 would be used with the reactant concentration, the value of the ΔG does not depend on the pH (Fig. 2 a, dashed line), because the reactant concentrations are independent of pH. This way of calculating the ΔG is sometimes applied for the analysis of genome-scale metabolic networks. The resulting ΔG is equal to the calculated ΔG that considers species, at an inside pH of 5. This is because, at pH 5, both compartments have an equal pH, which makes the relative abundance of the species equal in both compartments, and therefore the gradient of each species over the membrane equal to the total reactant concentration gradient over the membrane. From Fig. 2 a we can see that the effect of considering species on the calculated ΔG at an inside pH of 6.5 is ∼15 kJ/mol, which is a significant difference.
Another example is the synthesis of ATP. This is a process in which ADP is phosphorylated to ATP by the transport of protons over the mitochondrial membrane driven by their electrochemical potential. The biochemical process can be described as
| (10) |
Under concentrations found in the mitochondrial matrix, in vivo synthesis of ATP from ADP and Pi is thermodynamically unfavorable. The source of energy to drive this process is the transport of positively charged hydrogen ions down the electrochemical gradient across the mitochondrial inner membrane.
In Fig. 1 a, the ATP synthase reaction is shown schematically on the species level. From Eq. 10, we can define the stoichiometric coefficients that we can use in Eqs. 6 and 7,
| (11) |
where the indices represent ADP, Pi, , ATP, H2O, and , respectively. With the stoichiometric coefficients as defined above, and the data on values in Table 1, Eq. 6 can be used to calculate the ΔG of the transport/reaction process (note that when we define [H+] = 10−pH, then because a pH is defined as the hydrogen ion chemical activity, which includes the effects of ionic strength and temperature).
To calculate the ΔG using binding polynomials (Eq. 7), we must first define the chemical reaction in terms of reference species for each reactant. Here, we use as the reference species of each reactant the most unbound species (i.e., the species with the least bound ions). In terms of reference species, Eq. 10 becomes
| (12) |
Now we can define the stoichiometry of reacting protons,
| (13) |
with the order of indices equal to Eq. 11. With the additional information on the stoichiometry of the reacting protons, we can also calculate the ΔG according to Eq. 7. For simplicity, we consider only binding of protons, and ignore binding of other ions such as magnesium. In Section 3 in the Supporting Material, we provide a table with all the data that is used in the examples, and we show in detail how to calculate the ΔG for such transport/reaction processes and how to correct for ionic strength.
We obtain the same ΔG as the value calculated with Alberty's Legendre transform formalism (Eq. 6). In Fig. 2 b we show the ΔG as a function of inside pH. The ΔG that is calculated when considering the species (i.e., using Eqs. 6 or 7) is compared to assuming that the reactant concentration represents the most abundant species. In this case we can see that considering species does not have a significant effect on the calculated ΔG.
With Eqs. 6 and 7 we have a framework to calculate changes in Gibbs energies for reaction, transport, and transport/reaction processes. The example calculations show that in certain cases it is important to consider the various species that exist for each reactant. The difference in ΔG shown in Fig. 2 a is large at physiological conditions, and can even mean the difference between a positive or negative value of the ΔG. For the ATP synthase example, we see that there is only a minimal difference between correctly considering species and assuming that the reactant concentration represents the most abundant species concentration.
Conclusions
In this work, we have developed equations to correctly describe the thermodynamics of biochemical transport/reaction processes. Specifically, we have formulated two equivalent equations to calculate the change in Gibbs energy of a transport/reaction process based on two different ways of treating biochemical thermodynamics. We illustrate the need of these equations by showing that in some cases there is a significant difference between considering species and by assuming the reactant concentration represents a species concentration. Therefore, for the analysis of metabolic networks where many different reactions are analyzed, it is relevant to be able to consider species for the calculation of ΔG values by using metabolite concentrations. With the developed equations, thermodynamic analysis of metabolic networks spanning over multiple physical compartments can now be correctly described.
Acknowledgments
Funding is acknowledged from the Swiss Initiative in Systems Biology (SystemsX.ch project YeastX, to M.H.), Ecole Polytechnique Federale de Lausanne (to V.H.), Swiss National Science Foundation (to V.H.), and the National Institutes of Health (grant No. HL072011 to D.A.B.).
Footnotes
Anne Kümmel's present address is Novartis Institutes for BioMedical Research, Forum 1, Novartis Campus, 4056 Basel, Switzerland.
Supporting Material
References
- 1.Mavrovouniotis M.L. Identification of localized and distributed bottlenecks in metabolic pathways. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1993;1:275–283. [PubMed] [Google Scholar]
- 2.Mavrovouniotis M.L. Duality theory for thermodynamic bottlenecks in bioreaction pathways. Chem. Eng. Sci. 1996;51:1495–1507. [Google Scholar]
- 3.Pissarra P.D.N., Nielsen J. Thermodynamics of metabolic pathways for penicillin production: analysis of thermodynamic feasibility and free energy changes during fed-batch cultivation. Biotechnol. Prog. 1997;13:156–165. [Google Scholar]
- 4.Maskow T., von Stockar U. How reliable are thermodynamic feasibility statements of biochemical pathways? Biotechnol. Bioeng. 2005;92:223–230. doi: 10.1002/bit.20572. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka M., Okuno Y., Kanehisa M. Extraction of a thermodynamic property for biochemical reactions in the metabolic pathway. Genome Informatics. 2003;14:370–371. [Google Scholar]
- 6.Yang F., Qian H., Beard D.A. Ab initio prediction of thermodynamically feasible reaction directions from biochemical network stoichiometry. Metab. Eng. 2005;7:251–259. doi: 10.1016/j.ymben.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Kümmel A., Panke S., Heinemann M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics. 2006;7:512. doi: 10.1186/1471-2105-7-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry C.S., Jankowski M.D., Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys. J. 2006;90:1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holzhütter H.-G. The principle of flux minimization and its application to estimate stationary fluxes in metabolic networks. Eur. J. Biochem. 2004;271:2905–2922. doi: 10.1111/j.1432-1033.2004.04213.x. [DOI] [PubMed] [Google Scholar]
- 10.Price N.D., Thiele I., Palsson B.O. Candidate states of Helicobacter pylori's genome-scale metabolic network upon application of “loop law” thermodynamic constraints. Biophys. J. 2006;90:3919–3928. doi: 10.1529/biophysj.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian H., Beard D.A., Liang S.D. Stoichiometric network theory for nonequilibrium biochemical systems. Eur. J. Biochem. 2003;270:415–421. doi: 10.1046/j.1432-1033.2003.03357.x. [DOI] [PubMed] [Google Scholar]
- 12.Beard D.A., Liang S.D., Qian H. Energy balance for analysis of complex metabolic networks. Biophys. J. 2002;83:79–86. doi: 10.1016/S0006-3495(02)75150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beard D.A., Babson E., Qian H. Thermodynamic constraints for biochemical networks. J. Theor. Biol. 2004;228:327–333. doi: 10.1016/j.jtbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Price N.D., Famili I., Palsson B.Ø. Extreme pathways and Kirchhoff's second law. Biophys. J. 2002;83:2879–2882. doi: 10.1016/S0006-3495(02)75297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian H., Beard D.A. Metabolic futile cycles and their functions: a systems analysis of energy and control. Syst. Biol. (Stevenage) 2006;153:192–200. doi: 10.1049/ip-syb:20050086. [DOI] [PubMed] [Google Scholar]
- 16.Nigam, R., and S. Liang. 2005. Perturbing thermodynamically unfeasible metabolic networks. Brazilian Symposium on Bioinformatics, Unisinos, Brazil.
- 17.Nolan R.P., Fenley A.P., Lee K. Identification of distributed metabolic objectives in the hypermetabolic liver by flux and energy balance analysis. Metab. Eng. 2006;8:30–45. doi: 10.1016/j.ymben.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Fleming R.M.T., Thiele I., Nasheuer H.P. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophys. Chem. 2009;145:47–56. doi: 10.1016/j.bpc.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kümmel A., Panke S., Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100074. 2006.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F., Beard D.A. Thermodynamically based profiling of drug metabolism and drug-drug metabolic interactions: a case study of acetaminophen and ethanol toxic interaction. Biophys. Chem. 2006;120:121–134. doi: 10.1016/j.bpc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Beard D.A., Qian H. Relationship between thermodynamic driving force and one-way fluxes in reversible processes. PLoS ONE. 2007;2:e144. doi: 10.1371/journal.pone.0000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiechert W. The thermodynamic meaning of metabolic exchange fluxes. Biophys. J. 2007;93:2255–2264. doi: 10.1529/biophysj.106.099895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aledo J.C. Metabolic pathways: does the actual Gibbs free energy change affect the flux rate? Biochem. Mol. Biol. Educ. 2001;29:142–143. [Google Scholar]
- 24.Alberty R.A. Relations between biochemical thermodynamics and biochemical kinetics. Biophys. Chem. 2006;124:11–17. doi: 10.1016/j.bpc.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Beard D.A., Qian H. Thermodynamic-based computational profiling of cellular regulatory control in hepatocyte metabolism. Am. J. Physiol. Endocrinol. Metab. 2005;288:E633–E644. doi: 10.1152/ajpendo.00239.2004. [DOI] [PubMed] [Google Scholar]
- 26.Vinnakota K.C., Kemp M.L., Kushmerick M.J. Dynamics of muscle glycogenolysis modeled with pH time course computation and pH-dependent reaction equilibria and enzyme kinetics. Biophys. J. 2006;91:1264–1287. doi: 10.1529/biophysj.105.073296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard D.A. A biophysical model of the mitochondrial respiratory system and oxidative phosphorylation. PLOS Comput. Biol. 2005;1:e36. doi: 10.1371/journal.pcbi.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen J. Metabolic control analysis of biochemical pathways based on a thermokinetic description of reaction rates. Biochem. J. 1997;321:133–138. doi: 10.1042/bj3210133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F., Jeneson J.A.L., Beard D.A. Oxidative ATP synthesis in skeletal muscle is controlled by substrate feedback. Am. J. Physiol. Cell Physiol. 2007;292:C115–C124. doi: 10.1152/ajpcell.00237.2006. [DOI] [PubMed] [Google Scholar]
- 30.Büscher J.M., Czernik D., Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal. Chem. 2009;81:2135–2143. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- 31.Alberty R.A. Wiley-Interscience; Hoboken, NJ: 2003. Thermodynamics of Biochemical Reactions. [Google Scholar]
- 32.Beard D.A., Qian H. Cambridge University Press; London, UK: 2008. Chemical Biophysics: Quantitative Analysis of Cellular Systems. [Google Scholar]
- 33.Wyman J., Gill S.J. University Science Books; Mill Valley, CA: 1990. Binding and Linkage: Functional Chemistry of Biological Macromolecules. [Google Scholar]
- 34.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 35.Robinson A.J., Kunji E.R.S. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. USA. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolay K., Scheffers W.A., Kaptein R. In vivo 31P NMR studies on the role of the vacuole in phosphate metabolism in yeasts. Arch. Microbiol. 1983;134:270–275. doi: 10.1007/BF00407801. [DOI] [PubMed] [Google Scholar]
- 37.Fendt S.-M., Buescher J.M., Sauer U. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol. Syst. Biol. 2010;6:356. doi: 10.1038/msb.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


