Abstract
Latency-associated transcript (LAT) deletion mutants of herpes simplex virus type 1 (HSV-1) have reduced reactivation phenotypes. Thus, LAT plays an essential role in the latency-reactivation cycle of HSV-1. We have shown that LAT has antiapoptosis activity and demonstrated that the chimeric virus, dLAT-cpIAP, resulting from replacing LAT with the baculovirus antiapoptosis gene cpIAP, has a wild-type HSV-1 reactivation phenotype in mice and rabbits. Thus, LAT can be replaced by an alternative antiapoptosis gene, confirming that LAT’s antiapoptosis activity plays an important role in the mechanism by which LAT enhances the virus’ reactivation phenotype. However, because cpIAP interferes with both of the major apoptosis pathways, these studies did not address whether LAT’s proreactivation phenotype function was due to blocking the extrinsic (Fas-ligand–, caspase-8–, or caspase-10–dependent pathway) or the intrinsic (mitochondria-, caspase-9–dependent pathway) pathway, or whether both pathways must be blocked. Here we constructed an HSV-1 LAT(−) mutant that expresses cellular FLIP (cellular FLICE-like inhibitory protein) under control of the LAT promoter and in place of LAT nucleotides 76 to 1667. Mice were ocularly infected with this mutant, designated dLAT-FLIP, and the reactivation phenotype was determined using the trigeminal ganglia explant model. dLAT-FLIP had a reactivation phenotype similar to wild-type virus and significantly higher than the LAT(−) mutant dLAT2903. Thus, the LAT function responsible for enhancing the reactivation phenotype could be replaced with an antiapoptosis gene that primarily blocks the extrinsic signaling apoptosis pathway.
Keywords: Herpes simplex virus, latency, LAT, FLIP, antiapoptosis
Introduction
Herpes simplex virus type 1 (HSV-1) is widespread, with seropositivity estimates of 45% to 98% worldwide and 57% to 85% of adults in the United States (Brugha et al, 1997; Bunzli et al, 2004; Corey and Spear, 1986; Miller and Ship, 1977; Nahmias et al, 1990; Spruance, 1992; Xu et al, 2006; reviewed in Fatahzadeh and Schwartz, 2007). HSV-1 infects peripheral mucosal surfaces including the genitals, mouth, and eyes. It then travels up axons and establishes lifelong latent infections in sensory neurons in the host’s ganglia. The latent virus can reactivate sporadically and cause recurrent disease at the original site of infection. HSV-1–induced diseases, including genital lesions, cold sores in and around the mouth, corneal disease that can lead to impaired vision, and viral encephalitis, are all more likely to occur following viral reactivations rather than primary infection. This is probably because a primary infection can result in large numbers of recurrent episodes and also because some aspects of HSV-1–induced disease involve immunopathological responses to the virus that are less likely to occur in a naïve individual encountering the virus for the first time.
The HSV-1 latency-associated transcript (LAT) gene is the only viral gene whose transcript is readily and consistently detected during neuronal latency (Rock et al, 1987; Stevens et al, 1987). LAT-null mutants have a reduced reactivation phenotype, indicating that LAT provides a function that enhances the virus’ reactivation phenotype (Leib et al, 1989; Perng et al, 1994; Trousdale et al, 1991). Although the primary LAT transcript is approximately 8.3 kb long (Wagner et al, 1988b; Wechsler et al, 1988), the first 18% (1.5 kb) of LAT is sufficient to produce an apparently wild-type reactivation phenotype (Perng et al, 1996b). Thus, there is a LAT function that is located completely within the first 1.5 kb of LAT that enhances the reactivation phenotype.
In 2000 we reported that LAT has antiapoptosis activity and hypothesized that LAT’s antiapoptosis activity was the LAT function that enhanced the reactivation phenotype (Perng et al, 2000a). Although there was initially some controversy regarding LAT’s antiapoptosis activity (Thompson and Sawtell, 2001), a large body of data has now convincingly shown that LAT does in fact have antiapoptosis activity (Ahmed et al, 2002; Branco and Fraser, 2005; Carpenter et al, 2007; Henderson et al, 2002; Inman et al, 2001; Jin et al, 2003, 2005, 2007; Mott et al, 2003; Peng et al, 2004; Perng et al, 2002). In addition, we have shown that the first 1.5 kb of the primary LAT transcript has significant antiapoptosis activity (Inman et al, 2001; Jin et al, 2003). This is the same region to which we previously mapped LAT’s ability to enhance the HSV-1 reactivation phenotype (Perng et al, 1996b). Furthermore, we have shown that CJLAT and dLAT-cpIAP, chimeric viruses in which the first 1.5 kb of LAT was replaced with different alternative antiapoptosis genes, both have wild type–like reactivation phenotypes (Jin et al, 2005, 2007; Mott et al, 2003; Perng et al, 2002). Together, these findings confirm that LAT has antiapoptosis activity and that this antiapoptosis activity is sufficient to account for the wild-type reactivation phenotype.
Interestingly, we have found that LAT can block both major apoptotic pathways, namely the extrinsic pathway, also known as the caspase-8–dependent pathway or the TNF/Fas ligand pathway, and the intrinsic pathway, also known as the caspase-9–dependent pathway or the mitochondrial pathway (Henderson et al, 2004; Jin et al, 2003; Peng et al, 2004). This raises the question as to whether LAT’s ability to block the caspase-8–dependent pathway, the caspase-9–dependent pathway, or both pathways is critical for LAT’s ability to enhance the reactivation phenotype. The antiapoptosis activity genes used to replace LAT in CJLAT and dLAT-cpIAP can each block both major apoptotic pathways and therefore these mutants do not address this question (Jin et al, 2005; Perng et al, 2002). It is also possible that LAT inhibits a downstream step of apoptosis, for example activation (cleavage) of caspase-3, thus enabling it to inhibit both the intrinsic and extrinsic pathways. Defining which apoptotic pathway is blocked by LAT is important for understanding the molecular mechanism(s) by which LAT helps regulate latency and reactivation. Therefore, here we constructed and tested the mutant dLAT-FLIP (cellular FLICE-like inhibitory protein), which contains cellular FLIP (cFLIP) in place of LAT nucleotides (nts) 76 to 1667. FLIP, which is highly homologous to caspase-8, but is not catalytically active, can bind caspase-8 and FADD (Fas-associated death domain) (Irmler et al, 1997; Shu et al, 1997). Consequently, FLIP blocks the formation of DISC (death-induced signaling complex) (Krueger et al, 2001), which is formed after a death receptor is activated. DISC only contains initiator caspases, primarily caspase-8 or caspase-10 (Kischkel et al, 2001; Schmitz et al, 2000). Thus, FLIP primarily inhibits the initiation and signaling of death receptor–mediated apoptosis (Krueger et al, 2001; Scaffidi et al, 1999; Schmitz et al, 2000). If the crucial LAT function is to block a step that is only activated during the extrinsic pathway, expression of FLIP from the LAT promoter would effectively restore the wt reactivation phenotype. We report here that dLAT-FLIP had a reactivation phenotype significantly higher than that of the LAT(−) mutant dLAT2903 and at least as high as wild-type HSV-1. These results suggest that LAT can support the high wild-type reactivation phenotype by blocking early steps in the caspase-8–dependent pathway, for example inhibition of DISC formation.
Results
Construction and genomic structure of dLAT-FLIP
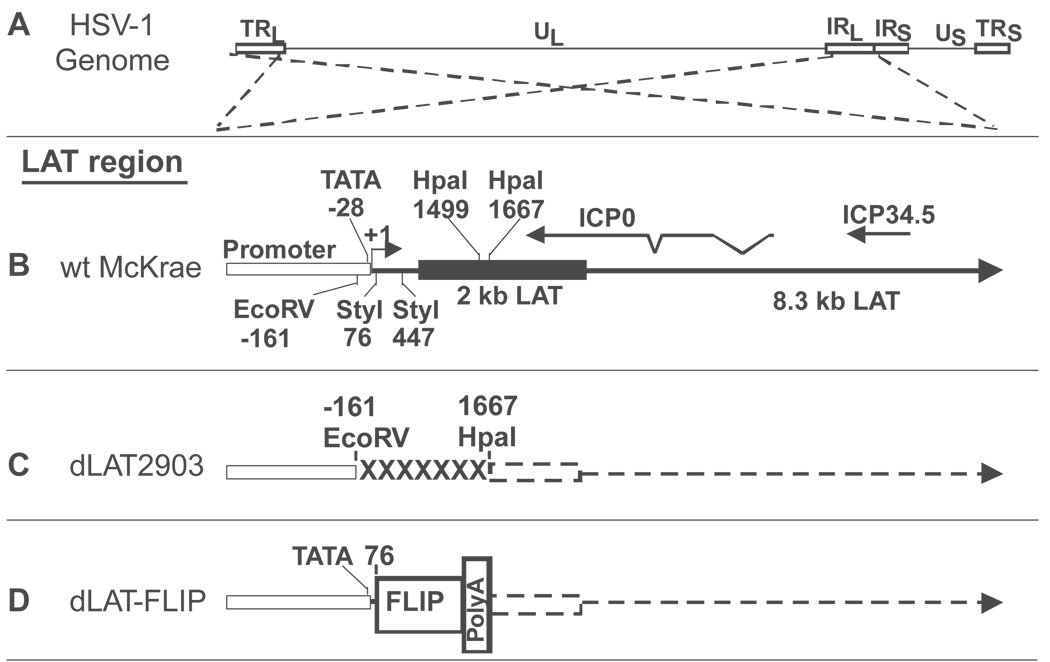
The genomic structure of wild-type HSV-1 strain McKrae, the LAT deletion mutant dLAT2903, which was derived from McKrae (Perng et al, 1994), and dLAT-FLIP, which was derived from dLAT2903, are shown schematically in Figure 1. Figure 1A shows the prototypic structure of the wild-type HSV-1 McKrae genome. The dashed lines originating from the viral long repeats (terminal long repeat [TLR] and internal long repeat [IRL]) indicate that the LAT region contained in these repeats are expanded in Figure 1B, C, and D to show the relative location and status of the LAT locus in the different viruses. The TLR and IRL are inverted relative to each other (i.e., the DNA sequences are in opposite orientations). This is indicated by the dashed lines crossing. Figure 1B shows a schematic of the LAT locus in wild-type McKrae. The locations of the ICP0 and ICP34.5 mRNAs are shown for reference. Restriction sites for EcoRV, StyI, and HpaI are also shown for reference. The primary LAT transcript is approximately 8.3 kb (Wagner et al, 1988a; Wechsler et al, 1988) (large arrow). A very stable and easily detected 2-kb LAT (black rectangle) is an intron derived by splicing of the primary LAT. The open rectangle indicates the relative location of the LAT promoter. The start of LAT transcription is indicated by the arrow at +1. Figure 1C shows the LAT region of dLAT2903. The construction and characterization of this mutant have been described previously (Perng et al, 1994). dLAT2903 is deleted for LAT nts −161 to +1677 in both copies of LAT. The deletion is indicated by “XXXXX”. dLAT2903 is missing key promoter elements including the TATA box, transcribes no detectable LAT RNA (indicated by dashed line), and is thus a true LAT-null mutant. dLAT2903 has a low reactivation phenotype compared to wt HSV-1 and its marker rescued virus (Perng et al, 1994). Figure 1D shows a schematic of the LAT locus in dLAT-FLIP. This mutant contains the complete LAT promoter (including nts −161 to +76 that are missing in dLAT2903) followed by the FLIP open reading frame (ORF) and a polyadenylation signal sequence (see Materials and Methods for more details). Both LAT loci in dLAT-FLIP therefore contain the FLIP ORF followed by a strong poly-A signal in place of LAT nts 76 to 1667, with the inserted region driven by the LAT promoter. We have previously shown that insertion of a poly-A site at this location decreases transcription of LAT downstream of an inserted ORF to undetectable levels (Jin et al, 2005). This is represented by the dashed arrow.
Figure 1.
Structure of the LAT region of dLAT-FLIP. (A) HSV-1 genomic structure. TRL and IRL indicate the viral long repeats (terminal and internal). IRS and TRS indicate the viral short repeats. UL and US indicate the long and short unique regions. The dashed lines indicate that the region of the TRL and IRL are expanded below with the TRL inverted relative to the IRL so that both identical regions can be represented by a single image in the subsequent schematics. (B) Wild-type (wt) McKrae. The LAT promoter is indicated by open rectangle. The solid black rectangle indicates the stable 2-kb LAT. The large arrow indicates the extent of the primary 8.3-kb LAT RNA. The relative locations of ICP0 and ICP34.5 are shown for reference, as are various restriction enzyme sites. LAT transcription starts at LAT nucleotide (nt) +1 (genomic nt 118,801 for the copy of LAT in the ILR). All nt numbers are shown relative LAT nt +1. The location of the LAT TATA box is shown at nt −28. (C) dLAT2903. dLAT2903 is deleted from LAT nt −161 to +1667 (XXXXXX). dLAT2903 is a true LAT-null mutant that is missing primary LAT promoter elements between −161 and +1. dLAT2903 is also deleted for a putative secondary LAT promoter, LAP2, located within the 5’ end of the primary LAT transcript prior to the start of the 2-kb LAT. This mutant therefore is not capable of expressing any LAT RNA (dashed lines) (Perng et al, 1994). (D) dLAT-FLIP contains the antiapoptosis gene FLIP followed by a poly-A signal inserted in place of LAT nts 76 to 1667. The entire LAT promoter is present. No LAT RNA is transcribed past the poly-A site.
The FLIP gene has no sequence homology to the HSV-1 LAT gene. In order to insert the FLIP ORF and poly-A signal site into the LAT locus by homologous recombination, FLIP was cloned into a plasmid that contains appropriate HSV-1 flanking sequences. The details of construction of this plasmid and additional details of constructing the mutant dLAT-FLIP can be found in Materials and Methods.
Confirmation of the dLAT-FLIP genomic structure by Southern blot analysis
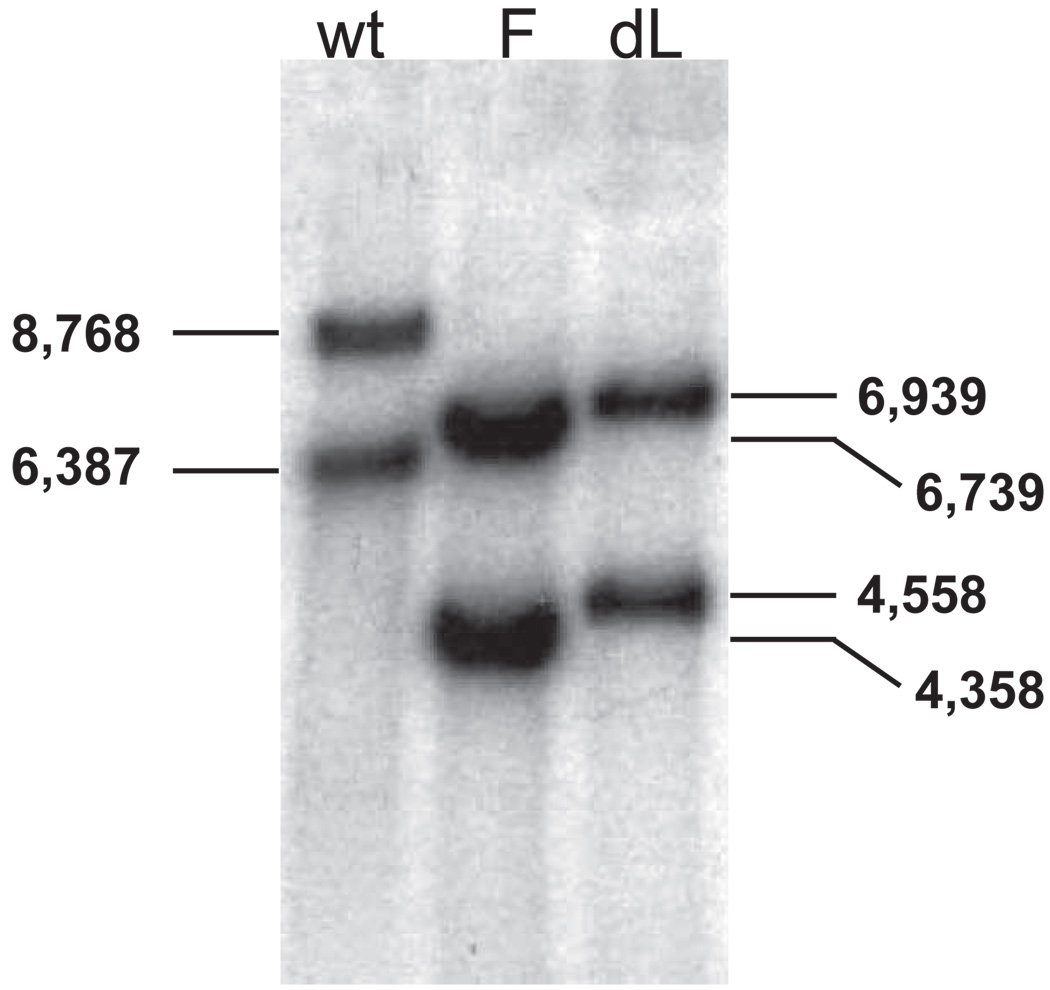
Viral DNAs were purified and digested with SalI. The resulting DNA fragments were separated on a 0.8% agarose gel, denatured, and transferred to a nylon membrane for Southern blot analysis (Figure 2). A SmaI-EcoRV DNA restriction fragment corresponding to LAT nts −864 to −161 was radiolabeled with α-32P-dCTP by nick translation and used as a probe. In wild-type McKrae, SalI digestion cuts both copies of LAT (one in each long repeat) at LAT nt 2102. It also cuts in the unique long region, the location of which differs near the internal long repeat compared to the terminal long repeat. The SalI site in the unique long region near the terminal long repeat is 8768 nts away from LAT nt 2102 in the terminal long repeat. The SalI site in the unique long region near the internal long repeat is 6387 nts away from LAT nt 2102 in the internal long repeat. Thus, SalI digestion of wild-type McKrae DNA produces a DNA fragment containing the 5’ end of LAT from the terminal long repeat that is larger than the LAT DNA fragment from the internal long repeat. This allows the status of both copies of LAT to be determined. In Figure 2, the wild-type LAT fragments are 8768 and 6387 bp, whereas the dLAT2903 LAT fragments are each 1829 nts shorter (6939 and 4558 bp) because there is an EcoRV (−161) to HpaI (1667) deletion in both copies of LAT. The FLIP insert in dLAT-FLIP contains a SalI site approximately 200 nts from its 5′ end that is not present in wt McKrae or dLAT2903. SalI cuts here instead of just over 400 nts downstream of the deletion in dLAT2903 at LAT nt 2102. The dLAT-FLIP SalI-SalI fragments are therefore predicted to be approximately 200 nts smaller (approximately 6739 and 4358 bp) than the corresponding restriction fragments from dLAT2903. These mobility of the dLAT-FLIP fragments are consistent with these sizes (Figure 2), confirming that the FLIP containing insert is present in both LAT regions of dLAT-FLIP.
Figure 2.
Southern hybridization analysis of dLAT-FLIP genomic structure. dLAT-FLIP (F), wt McKrae (wt), and dLAT2903 (dL) viral DNAs were purified, digested with SalI, separated using a 0.8% agarose gel, transferred to a membrane, and hybridized to a 32P-labeled SmaI-EcoRV (LAT nts −864 to −161) restriction fragment as described in the text. The numbers indicate the approximate size in base pairs of the restriction fragments.
Replication of dLAT-FLIP in tissue culture
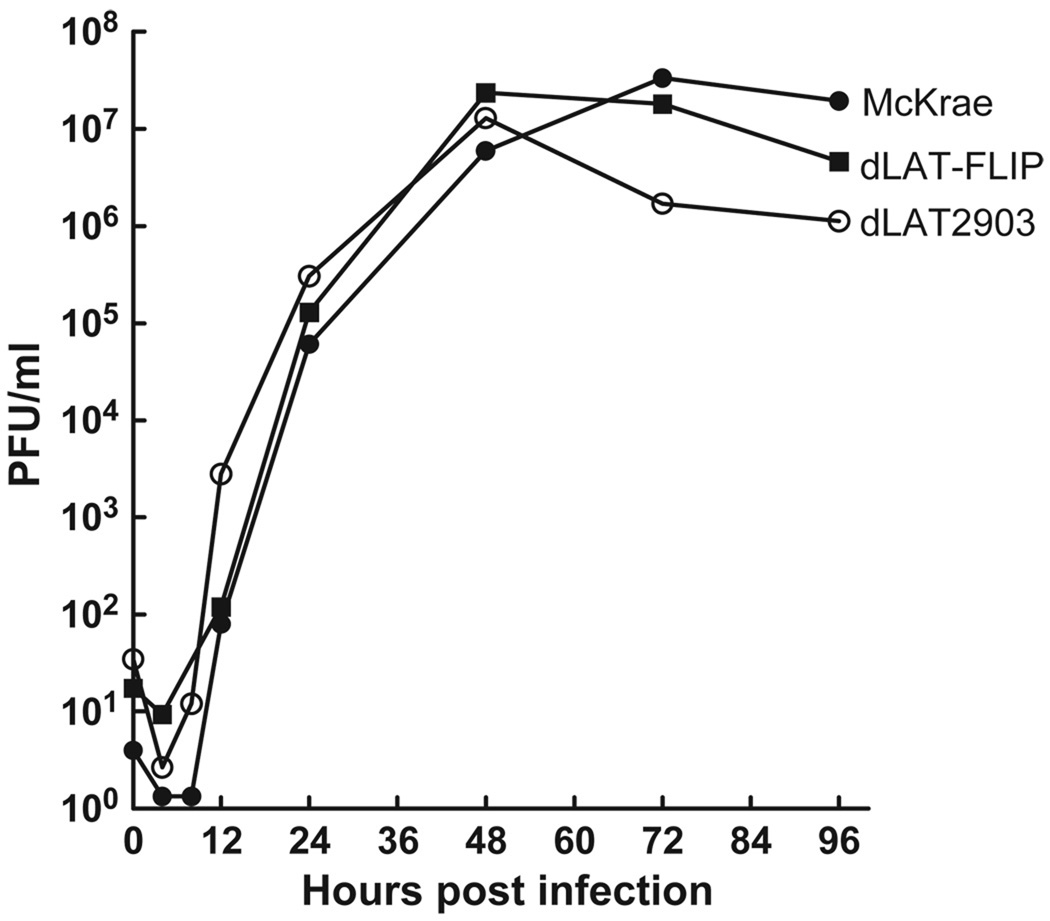
Rabbit skin (RS) cells were infected at an MOI of 0.01 with dLAT-FLIP, dLAT2903 (the immediate parental virus), or wild-type McKrae (the parental virus for dLAT2903) and replication was determined by plaque assays on RS cells as described in Materials and Methods. As expected from previous studies, wild-type McKrae and dLAT2903 replicated similarly and reached similar peak titers (Perng et al, 1994). Replication of dLAT-FLIP appeared similar to both dLAT2903 and wild-type virus (Figure 3). Thus, inserting the FLIP gene and the bovine growth hormone (BGH) poly-A signal sequence in place of LAT nts 76 to 1667 did not significantly alter virus replication in tissue culture. Normal virus replication of dLAT-FLIP also indicates that the ICP0 gene was functioning correctly in dLAT-FLIP.
Figure 3.
Replication of dLAT-FLIP in RS cells. Cell monolayers were infected with virus at a low MOI of 0.01. At the indicated times, infected cell monolayers with the culture medium were harvested, freeze-thawed to release virus, and total infectious virus was determined using standard plaque assays.
Replication of dLAT-FLIP in mouse eyes
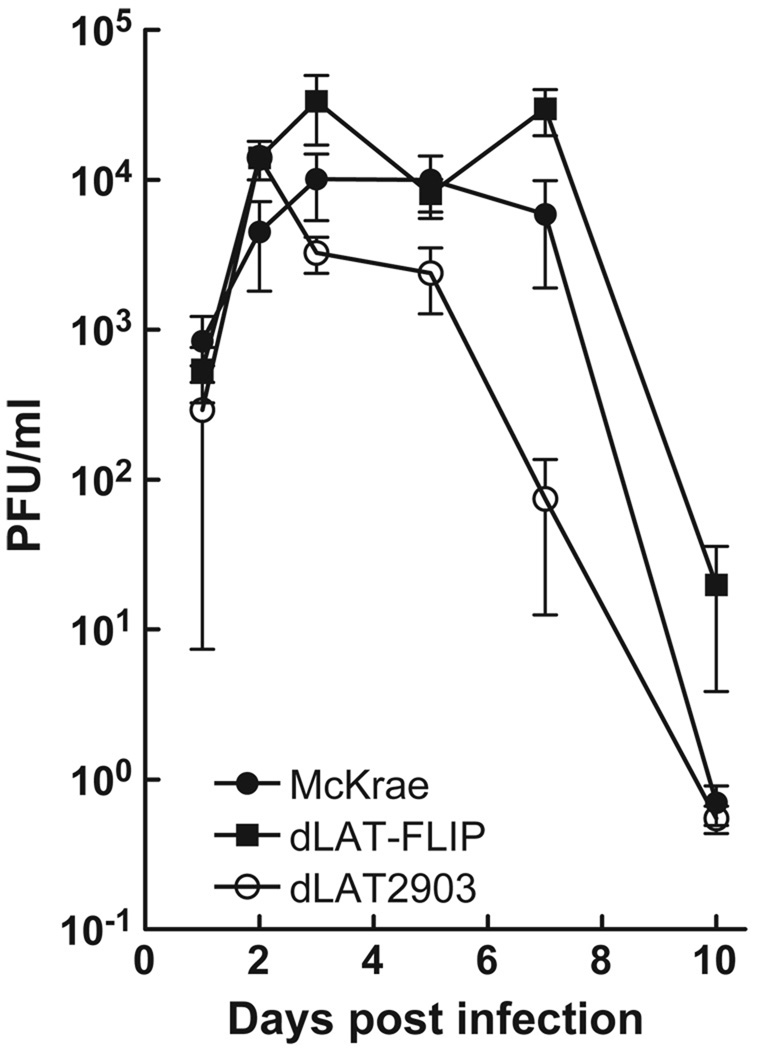
Eight- to 10-week-old Swiss Webster mice were infected with 2 × 105 plaque-forming units (PFU)/eye of dLAT-FLIP, dLAT2903, or wild-type McKrae as described in Materials and Methods. Tear swabs were collected from 10 eyes/group on the days indicated and the amount of infectious virus in each swab was determined by standard plaque assays on RS cell monolayers. Two independent experiments were performed and the combined results are shown (Figure 4; n = 20/group). There were no statistically significant differences between any of the groups on days 1, 2, 3, 5, or 10 post infection (analysis of variance [ANOVA]; P > .05). On day 7 post infection, there was no significant difference between wt and dLAT2903 (P > .05), but dLAT-FLIP was higher than the other groups (P < .05). This may be due to slower clearance of the virus because on day 7 post infection virus was still detected in 19/20 dLAT-FLIP, but only 11/20 McKrae and 9/20 dLAT2903 eyes.
Figure 4.
Replication of dLAT-FLIP in mouse eyes. Mice were ocularly infected with 2 × 105 PFU/eye. Tear swabs were collected on the indicated days. Each symbol represents the average titer for each group (n = 20). The results show the combined data from two independent experiments.
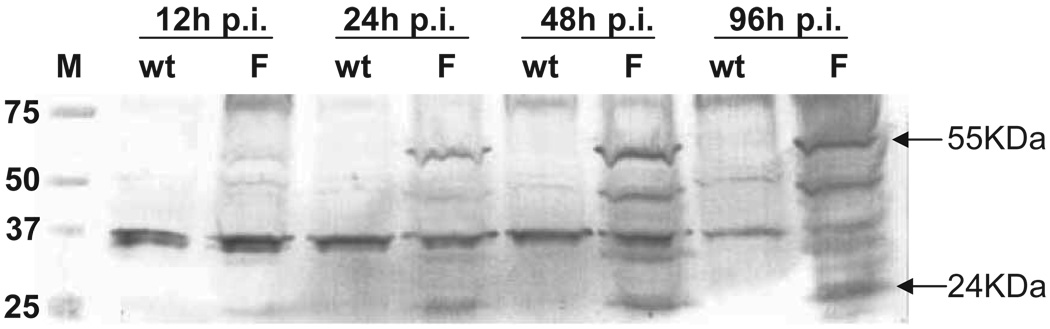
Expression of FLIP protein in dLAT-FLIP–infected cells
RS cells were infected at an MOI of 0.1 with either dLAT-FLIP or wt McKrae, harvested at the indicated times, and total cell extracts were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to a membrane, and Western analysis performed using a FLIP-specific antibody (Figure 5). The FLIP antibody recognized a band with an apparent mobility of about 55 kDa (arrow), which is the expected size for FLIP. A band with an apparent mobility of about 24 kDa was also recognized and this corresponds in size to a known processed product of FLIP. These bands were visualized in the dLAT-FLIP (F) lanes, but not in the wild-type McKrae (wt) lanes and were clearly visible by 24 h post infection (p.i.), despite the low MOI used. Several bands are also present in both the wt and F lanes and probably represent either cellular proteins (in the case of bands that have similar intensities at all time points) or HSV-1 proteins (in the case of bands whose intensity increased between 12 and 24 or 48 h p.i) that cross react with the anti-FLIP antibody. These results confirm that FLIP was expressed during productive tissue culture infection and that FLIP expression did not significantly impair viral replication in tissue culture.
Figure 5.
Western blot detection of FLIP protein expressed by dLAT-FLIP. RS cells were infected at an MOI of 0.1 with wild-type McKrae (wt) or dLAT-FLIP (F). Total cell extracts were harvested at the indicated times post infection, separated by SDS-PAGE, transferred to a membrane, and probed using rabbit FLIP-specific antibody (Anaspec). The antibody bound to the blots was visualized with anti-rabbit IgG conjugated to horseradish peroxidase (Chemicom). The arrows indicate the sizes of the two main forms of FLIP. Lane M = molecular weight markers in kDa.
Wild-type explant TG reactivation of dLAT-FLIP
In the mouse explant TG model, the decreased reactivation phenotype of LAT(−) viruses compared to wt viruses is sometimes only apparent when the time to the reactivation event is examined rather than just determining the percent of TG from which reactivated viruses is detected at some arbitrary late time point after explant (Perng et al, 2001b). Therefore the experiments reported here were performed by plating individual TG from latently infected mice in tissue culture medium, removing aliquots of the medium daily for 10 days and plating the medium on indicator cells (RS cells) to look for the presence of reactivated virus. This allows the experimenter to determine the day after explant on which reactivated virus is first present in the tissue culture medium. These data are then plotted as a survival curve and subjected to survival analysis (Kaplan-Meier). Because this analysis is appropriate for any kind of experiment where the result is expressed as the time to a well defined end point, we feel that this analysis is often preferable to Fisher’s exact test (or chi-square).
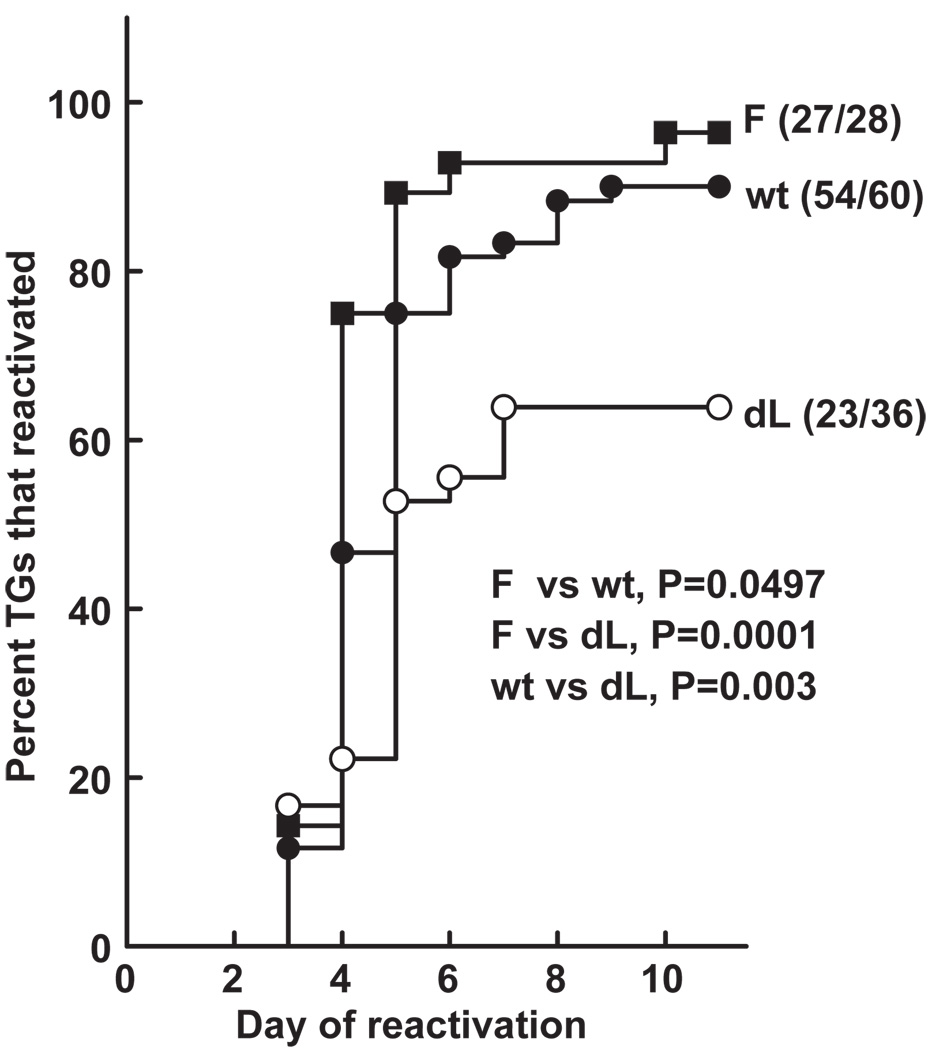
The results of three experiments all had the same trend. Thus, it was statistically valid to combine the data to achieve higher statistical power (Figure 6). Mice were infected with 2 × 105 PFU/eye of dLAT-FLIP, wt McKrae, or dLAT2903. On day 30 p.i., surviving mice were sacrificed, individual TG were cultured in tissue culture medium and the time to virus reactivation was determined. dLAT-FLIP reactivated more efficiently than dLAT2903 (P = .0001) and may have even reactivated more efficiently than wt McKrae (P = .0497). An additional analysis by the more typically used Fisher’s exact test, which does not take into account the time to reactivation, is shown in Table 1. Ninety-six percent of the TG from the latently infected dLAT-FLIP mice reactivated compared to 90% of the TG from wt infected mice (P = .42). Only 64% of the TG from dLAT2903-infected mice reactivated and this was significantly less than seen with dLAT-FLIP (P = .002). Thus, regardless of whether the mouse explanted TG reactivation data were analyzed by survival curve analysis or Fisher’s exact test, dLAT-FLIP appeared to reactivate at least as well as wild-type McKrae and significantly better than dLAT2903. This demonstrates that a LAT function involved in enhancing reactivation can be replaced by the alternative antiapoptosis gene FLIP, which mainly blocks the death receptor–mediated apoptosis pathway.
Figure 6.
Explant reactivation of dLAT-FLIP from mouse TG. Eight- to 10-week-old female Swiss Webster mice were infected with 2 × 105 PFU/eye of virus. On day 30 p.i. individual TG were explanted into tissue culture medium. Aliquots were removed daily and plated on indicator cells (RS cells) to look for the presence and time of first appearance of reactivated virus. The numbers next to each plot are the number of TG that reactivated (i.e., produced infectious virus)/the total number of TG in that group. F = dLAT-FLIP; wt = wild-type McKrae. P value calculated by Kaplan-Meier survival analysis. The combined results of three experiments are shown.
Table 1.
Reactivation analyzed by Fisher’s exact test
| dLAT-FLIP wt | McKrae | dLAT2903 | |
|---|---|---|---|
| No. reactivated TG/total | 27/28 (96%) | 54/60 (90%) | 23/36 (64%) |
| P versus dLAT2903 | .002 | .003 | |
| P versus wt McKrae | .42 | .003 |
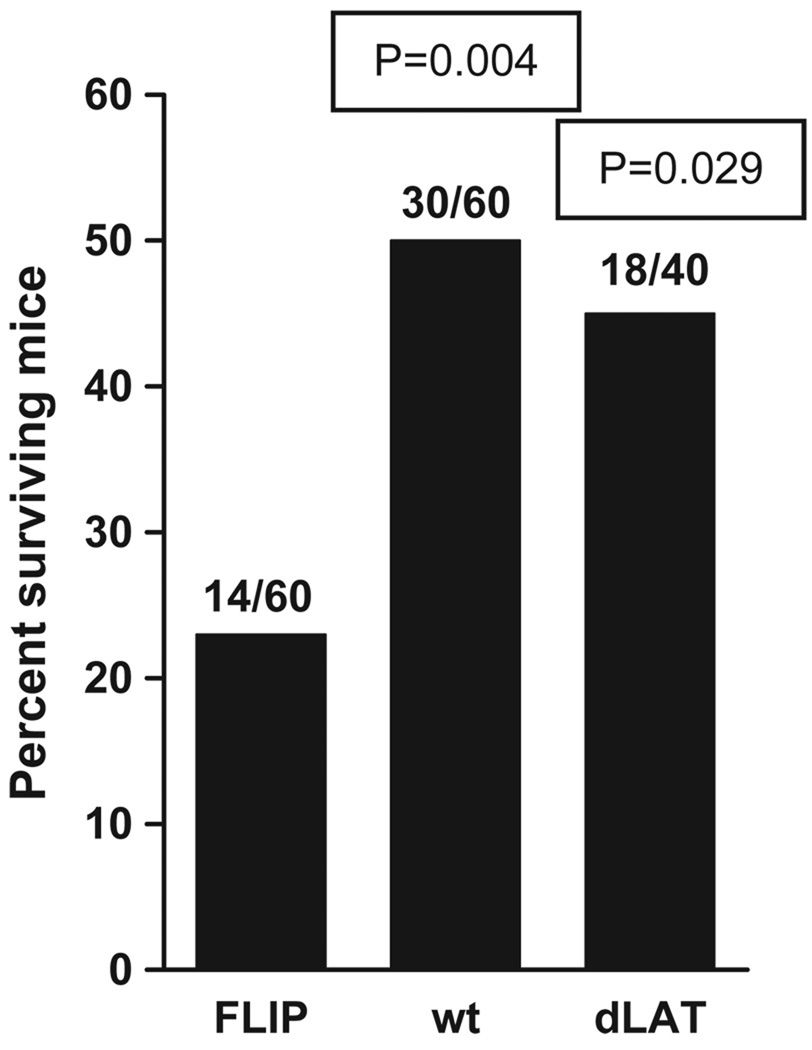
Decreased survival of mice ocularly infected with dLAT-FLIP
In the above studies, compared to both wild-type and dLAT2903-infected mice, fewer dLAT-FLIP–infected mice survived (Figure 7; P = .004 and P = .029), respectively. Thus, replacing LAT nts 76 to 1667 with the FLIP open reading frame followed by a poly-A signal appeared to increase the virulence of the virus compared to wild-type virus, which has those LAT nucleotides, and also compared to dLAT2903, which does not contain those LAT nucleotides. Interestingly, CJLAT, which like dLAT-FLIP contains an alternative antiapoptosis gene (the bovine herpes virus LR gene) in place of LAT, also has increased virulence in mice (Perng et al, 2002). In contrast, dLAT-cpIAP, which contains the alternative antiapoptosis gene cpIAP in place of LAT has decreased virulence in mice (Jin et al, 2005). We have previously shown that there is no connection between viral virulence and reactivation phenotype, because a LAT mutant with increased virulence has a reduced spontaneous reactivation (Perng et al, 1999), whereas a mutant that is completely avirulent has a wild-type reactivation phenotype (Perng et al, 1996a). Therefore it is unlikely that the wild-type reactivation phenotype of dLAT-FLIP is due to increased virulence. The reason for the difference in virulence of dLAT-FLIP, dLAT-cpIAP, and CJLAT is not yet understood.
Figure 7.
Survival of mice from the experiment shown in Figure 6. Numbers above each bar represent the number of mice the survived/total number of mice. P values are compared to dLAT-FLIP and were determined by the Fisher’s exact test.
Discussion
We report here the construction of dLAT-FLIP, an HSV-1 virus in which LAT nts 76 to 1667 were replaced by FLIP, a cellular gene that inhibits the extrinsic pathway of apoptosis. The recombinant or chimeric virus dLAT-FLIP had a reactivation phenotype that was at least as efficient as wild-type virus and significantly more efficient than the LAT-null mutant dLAT2903. These results provide additional confirmation for our hypothesis that LAT’s antiapoptosis activity is a key function involved in enhancing the reactivation phenotype.
It is important to note that the wild-type reactivation phenotype of dLAT-FLIP was not due to insertion of random nonviral DNA and/or a polyA signal in place of LAT nts 76 to 1667, because we previously showed that the mutant dLAT-EGFP, which is identical to dLAT-FLIP except that it contains the open reading frame (ORF) for enhanced green fluorescence protein (EGFP) in place of the FLIP ORF, has a low dLAT2903-like reactivation phenotype (Jin et al, 2005; Perng et al, 2000b). The wild-type reactivation phenotype of dLAT-FLIP was also not due to expression of LAT past LAT nt 1667, because we previously showed that dLAT1.5, a mutant with the same LAT deletion as dLAT-FLIP (LAT nts 76 to 1667), but without any foreign gene or poly-A signal to stop transcription, has a dLAT2903-like reactivation phenotype (Jin et al, 2005; Perng et al, 2001a). These mutants eliminate both the influence of potential read through transcription past the location of the inserted FLIP gene (LAT nt 1667) and the expression of a random foreign gene at the location of the inserted FLIP gene. Thus, in the case of dLAT-FLIP, rescue of the wild-type reactivation phenotype by inserting FLIP into dLAT2903 in place of the 5′ end of LAT must have been due to a function supplied by FLIP. Similarly, as we previously showed (Jin et al, 2005), rescue of the wild-type reactivation phenotype in dLAT-cpIAP must be due to a function supplied by cpIAP. Both FLIP and cpIAP are well-characterized antiapoptosis genes.
There are two major apoptosis pathways, the extrinsic apoptosis pathway also known as “the death receptor pathway” or “the caspase-8–mediated pathway,” and the intrinsic apoptosis pathway also known as “the mitochondrial pathway” or “the caspase-9–dependent pathway.” Very briefly, the caspase-8–mediated pathway is activated by ligation of cell surface receptor members of the tumor necrosis factor (TNF) receptor superfamily (Schulze-Osthoff et al, 1998), including TNF and Fas ligand (Nagata and Golstein, 1995). This leads to formation of the Fas-activated death domain (FADD) (Chinnaiyan et al, 1995), which leads to recruitment of procaspase-8 molecules into the death-inducing signaling complex (DISC). Here caspase-8 is activated by self cleavage (Boldin et al, 1996; Muzio et al, 1996), released, and then cleaves (activates) the main downstream caspase effector, caspase-3 (Stennicke et al, 1998).
Also very briefly, the caspase-9–dependent pathway is activated by signals that destabilize mitochondria and trigger release of cytochrome c from the mitochondria to the cytosol. A complex known as the apoptosome forms that includes cytochrome c, apoptosis protease activating factor-1 (Apaf-1), (d)ATP, and procaspase-9 (Zou et al, 1997; Zou et al, 1999). This leads to activation of caspase-9, which then cleaves (activates) caspase-3 (Li et al, 1997).
Interestingly, we have previously shown that in the absence of other viral genes LAT has the potential to significantly inhibit both major pathways of apoptosis (Jin et al, 2003; Peng et al, 2004). Although we have shown that LAT’s ability to block apoptosis is a key factor in LAT’s ability to enhance the virus’ reactivation phenotype, it is not yet known whether LAT’s ability to block the caspase-9 pathway, the caspase-8 pathway, or both together is required. One of our approaches to determining which apoptosis pathway is most important for LAT to block is to construct mutants in which LAT is replaced by various antiapoptosis genes with different abilities to block the major apoptosis pathways.
Similar to LAT, cpIAP can also block both major apoptosis pathways. cpIAP can block caspase-9 cleavage (Huang et al, 2000). It can also block apoptosis induced by fumonisin B1 (Jones et al, 2001), a chemical that activates the caspase-8 pathway by engaging the TNF receptor. Furthermore, cpIAP can block caspase-3 cleavage (Seshagiri and Miller, 1997), which should inhibit apoptosis induced through either of the major pathways. Because cpIAP and LAT both block apoptosis induced by both major pathways, the ability of cpIAP to replace LAT in dLAT-cpIAP and efficiently support a wild-type reactivation phenotype did not demonstrate which major apoptosis pathway LAT needs to block.
In contrast, FLIP blocks mainly the caspase-8 pathway. FLIP has sequence homology to caspase-8 and inhibits the caspase-8 pathway by being incorporated into DISC and preventing caspase-8 cleavage (Scaffidi et al, 1999). Thus, it was of interest to construct and test dLAT-FLIP to see if like dLAT-cpIAP, it would support a wild type–like reactivation phenotype. We reasoned as follows: (1) If LAT needs to efficiently block both the caspase-8 and caspase-9 pathways to exert its proreactivation phenotype effect, then dLAT-cpIAP, but not dLAT-FLIP, would have a wild type–like reactivation phenotype; (2) if LAT functions mainly by blocking the caspase-9 pathway, then again, dLAT-cpIAP, but not dLAT-FLIP, would have a wild type–like reactivation phenotype; and (3) if LAT functions mainly by blocking the caspase-8 pathway, then both dLAT-cpIAP and dLAT-FLIP would have wild type–like reactivation phenotypes. Thus, the results reported here support the hypothesis that LAT’s main affect on enhancing the reactivation phenotype is due to its ability to block the extrinsic or caspase-8 apoptotic pathway. Additional studies are underway to test the corollary hypothesis that a virus containing an antiapoptosis gene that only blocks the caspase-9 pathway in place of LAT will not fully support the wild type–like reactivation phenotype.
Previous studies aimed at mapping the regions of LAT that most efficiently block caspase-8– versus caspase-9–induced apoptosis, combined with studies mapping the region of LAT essential for supporting the reactivation phenotype, suggested that LAT’s ability to block the caspase-8 apoptotic pathway may be more important than LAT’s ability to block the caspase-9 apoptotic pathway. Thus, we previously found that expression of LAT nts 1 to 1499 inhibits caspase-8–induced apoptosis as efficiently as plasmids expressing LAT nts 1 to 2850 or 1 to 4658. In contrast, LAT nts 1 to 1499 only protect against caspase-9–induced apoptosis about 50% as efficiently as the larger LAT fragments (Peng et al, 2004). Because LAT nts 1 to 1499 can fully support a wild type–like reactivation phenotype (Perng et al, 1996b), we proposed at that time that LAT’s ability to block caspase-8–induced apoptosis might play a more significant role in enhancing the reactivation phenotype than LAT’s ability to block caspase-9–induced apoptosis (Peng et al, 2004). The ability of FLIP to efficiently replace LAT in dLAT-FLIP as reported here is consistent with this hypothesis. Although we think it is less likely, it is also possible that in dLAT-FLIP, cFLIP produces a “growth-enhancing” function outside of its antiapoptosis activity and that this somehow substitutes for LAT’s antiapoptosis function. It is also possible that LAT may have several different regions that inhibit apoptosis at several distinct steps in the apoptosis pathways.
Despite significant advances in recent years in our understanding of the role of LAT in the HSV-1 latency-reactivation cycle, including the now general consensus that LAT exerts a major effect on the reactivation phenotype via its antiapoptosis activity, many questions remain. Does LAT block apoptosis via a LAT-encoded protein, via micro-RNAs, via other RNA functions, via a combination of protein(s) and RNA(s)? Does LAT enhance the reactivation phenotype by mechanisms in addition to blocking apoptosis? Does LAT have additional functions not related to the reactivation phenotype? Regarding LAT’s ability to enhance the reactivation phenotype, does LAT function mainly at the time of establishment of latency, during maintenance of latency, during the reactivation step, or a combination of these times? Despite the apparent selective pressure to maintain the entire sequence of the primary 8.3-kb LAT transcript, studies in our laboratory have shown that the first 1.5 kb of the 8.3 kb primary LAT transcript is sufficient to support a wild type–like reactivation phenotype (Perng et al, 1996b). What then is the remainder of LAT doing? Likewise, do the stable 2-kb LAT and the 1.4- to 1.5-kb spliced RNA derived from the 2-kb LAT play a significant role in the reactivation phenotype or in some as yet to be determined phenotype?
Finally, how does LAT’s antiapoptosis activity enhance the reactivation phenotype? Some likely hypotheses include (1) LAT blocks viral-induced apoptosis in neurons, thereby increasing the number of latently infected neurons during establishment and/or maintenance of latency; (2) perhaps reactivation is triggered by insults that in the absence of LAT would induce apoptosis and kill the latently infected neuron before reactivation can be completed; (3) LAT may protect neurons against T cell–induced apoptosis. Recently it has been shown that during latency some neurons are surrounded by T cells, and it has been proposed that these T cells suppress reactivation (Khanna et al, 2003; Liu et al, 2001; Liu et al, 2000). Cytotoxic T cells kill target cells, in part by apoptosis. Thus, the high levels of LAT in some latently infected neurons may act to prevent elimination of these neurons by T cells. This type of LAT-related immune evasion could be important during establishment of, maintenance of, and/or reactivation from latency.
Whether LAT’s antiapoptosis activity functions by one or more of the above mechanisms or by a different mechanism, and whether LAT’s main effect on the reactivation phenotype occurs during establishment of latency, maintenance of latency, or reactivation from latency, is currently not known. It should be noted that, because the time in the latency-reactivation cycle at which LAT exerts its main influence is not yet known, we routinely talk about LAT mutants effecting the “reactivation phenotype” rather than just “reactivation.” The use of “reactivation phenotype” makes it clear that we are not assuming that LAT necessarily functions mostly in the reactivation process. Regardless, the results presented here show that a LAT function capable of supporting the wild-type reactivation phenotype can be replaced by FLIP, an antiapoptosis protein that is thought to block the caspase-8–dependent TNF/Fas ligand apoptotic pathway. This provides additional evidence that LAT’s antiapoptosis activity is directly or indirectly involved in the mechanism by which LAT enhances the reactivation phenotype. Although it is possible that cross-talk between the major apoptotic pathways can in some instances result in cFLIP interfering with both the extrinsic and intrinsic pathways, the studies reported here suggest that it may be more important for LAT to inhibit the extrinsic rather than the intrinsic apoptotic pathway.
Materials and methods
Cell lines
Rabbit skin (RS) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 µg/ml) (Sigma-Aldrich) at 37°C with 5% CO2 in a humidified incubator.
Viruses
All parental and mutant viruses were triple plaque purified and passaged only one or two times in rabbit skin (RS) cells prior to use. Wild-type McKrae (wt) and dLAT2903, have been previously described (Perng et al, 1994).
Virus growth curves
RS cell monolayers were infected at a multiplicity of infection (MOI) of 0.01. Virus was incubated with the cell monolayers for 1 h at 37°C and washed once with phosphate-buffered saline (PBS; 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, pH 7.4). The same amount of tissue culture medium (DMEM supplemented with 5% fetal bovine serum [FBS] and antibiotics as above) was added to each monolayer and cultures were incubated in a humidified, CO2 incubator at 37°C. At various times, monolayers were subjected to three freeze-thaw cycles, large cellular debris was removed by brief centrifugation, and the total amount of infectious virus was determined by standard plaque assays on RS cells. Each time point for each virus was done in quadruplicate.
Construction of dLAT-FLIP
Plasmid, pLAT5.6, was constructed by cloning the SwaI to MluI LAT locus (corresponding to 800 bp of the LAT promoter and the first 2850 bp of the primary 8.3-kb LAT from HSV-1 strain McKrae) into the BamHI site of pNEB193. The plasmid was digested with StyI and HpaI to remove a StyI-HpaI region corresponding to LAT nts 76 to 1667. BamHI linkers were added followed by ligation. The resulting plasmid pNEBLAT thus contains HSV-1 DNA corresponding to LAT nts −800 to +76, followed by a BamHI site, followed by LAT nts +1667 to +2850. A pcDNA3 mycFLIP plasmid was purchased from ScienceReagents (El Cajon, CA). This plasmid contains the long-form mRNA for the coding sequence of Homo sapiens FLICE-like inhibitory protein (cFLIP) inserted between the EcoRI and XhoI sites of pcDNA3. BglII linkers were added to the beginning of the cFLIP open reading frame (ORF) and the end of the bovine growth hormone (BGH) poly-A signal (nts 1018 to 1249 of pcDNA3) using the PCR-TOPO cloning method. The entire cFLIP ORF sequence and BGH poly-A signal sequence was isolated following BglII digestion and cloned into the BamHI restriction site of pNEBLAT between LAT nts 76 and 1667. The resulting plasmid, pNEBLAT-FLIP, was verified by DNA restriction digestion and DNA sequencing. It was then cotransfected with infectious dLAT2903 (Perng et al, 1994) genomic DNA into RS cells to generate the mutant dLAT-FLIP by homologous recombination as we previously described for construction of other HSV-1 mutants (Jin et al, 2005; Perng et al, 2001a, 2002, 1996c). Briefly, the cotransfection mix was plated on RS cells, individual viral plaques were analyzed by restriction digestion and Southern analysis, and plaques containing mixtures of dLAT2903 and dLAT-FLIP virus were repeatedly replaqued and analyzed until all the plaques appeared free of dLAT2903 virus. That plaque was then triple plaque purified and its structure confirmed as above. In the resulting chimeric virus, dLAT-FLIP, LAT nts 76 to 1667 are replaced by the complete cFLIP ORF followed by the BGH poly-A signal sequence, thus placing cFLIP under control of the intact LAT promoter.
Southern analysis
Briefly, viral DNA was digested with SalI, the restriction fragments were separated on a 0.8% agarose gel, transferred to Zeta paper, rinsed in 2 × SSC for 5 min, cross-linked to the membrane by ultraviolet (UV) light, and DNA-DNA hybridization performed with a LAT region specific 32P-labeled probe as we previously described (Perng et al, 1994, 2002).
Mice
Eight- to 10-week-old Swiss-Webster female mice (Jackson Labs) were used for all experiments. Viral infections were done without corneal scarification as we previously described (Perng et al, 1994).
Western blots
Total cell extracts were separated by 4% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was incubated with polyclonal rabbit anti-cFLIP antibody (Anaspec) at a 1:1000 dilution, washed, and the antibody bound to the blots was visualized with anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase diluted 1:2000 (Chemicom).
Statistical analysis
Analyses were performed using the personal computer program GraphPad Prism version 4.00 for Windows, GraphPad Software (San Diego, CA; www.graphpad.com).
Acknowledgments
This work was supported by Public Health Service grants EY13191, EY016663, P20RR15635, and R21AI069176, The Discovery Fund for Eye Research, The Henry L. Guenther Foundation, and Research to Prevent Blindness. Dr. Wechsler is an RPB Senior Scientific Investigator.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ahmed M, Lock M, Miller CG, Fraser NW. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J Virol. 2002;76:717–729. doi: 10.1128/JVI.76.2.717-729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- Branco FJ, Fraser NW. Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J Virol. 2005;79:9019–9025. doi: 10.1128/JVI.79.14.9019-9025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugha R, Keersmaekers K, Renton A, Meheus A. Genital herpes infection: a review. Int J Epidemiol. 1997;26:698–709. doi: 10.1093/ije/26.4.698. [DOI] [PubMed] [Google Scholar]
- Bunzli D, Wietlisbach V, Barazzoni F, Sahli R, Meylan PR. Seroepidemiology of Herpes simplex virus type 1 and 2 in Western and Southern Switzerland in adults aged 25–74 in 1992–93: a population-based study. BMC Infect Dis. 2004;4:10–22. doi: 10.1186/1471-2334-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D, Hsiang C, Brown DJ, Jin L, Osorio N, BenMohamed L, Jones C, Wechsler SL. Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis. Virology. 2007;369:12–18. doi: 10.1016/j.virol.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- Corey L, Spear PG. Infections with herpes simplex viruses (1) N Engl J Med. 1986;314:686–691. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- Fatahzadeh M, Schwartz RA. Human herpes simplex virus infections: epidemiology, pathogenesis, symptomatology, diagnosis, and management. J Am Acad Dermatol. 2007;57:737–763. doi: 10.1016/j.jaad.2007.06.027. quiz 764–766. [DOI] [PubMed] [Google Scholar]
- Henderson G, Peng W, Jin L, Perng GC, Nesburn AB, Wechsler SL, Jones C. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 latency-associated transcript. J NeuroVirol. 2002;8:103–111. doi: 10.1080/13550280290101085. [DOI] [PubMed] [Google Scholar]
- Henderson G, Perng GC, Nesburn AB, Wechsler SL, Jones C. The latency-related gene encoded by bovine herpesvirus 1 can suppress caspase 3 and caspase 9 cleavage during productive infection. J NeuroVirol. 2004;10:64–70. doi: 10.1080/13550280490261716. [DOI] [PubMed] [Google Scholar]
- Huang Q, Deveraux QL, Maeda S, Salvesen GS, Stennicke HR, Hammock BD, Reed JC. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc Natl Acad Sci U S A. 2000;97:1427–1432. doi: 10.1073/pnas.97.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman M, Perng G, Henderson G, Ghiasi H, Nesburn A, Wechsler S, Jones C. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild type spontaneous reactivation promotes cell survival in tissue culture. J Virol. 2001;75:3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, Rimoldi D, French LE, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- Jin L, Peng W, Perng GC, Brick DJ, Nesburn AB, Jones C, Wechsler SL. Identification of herpes simplex virus type 1 latency-associated transcript sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J Virol. 2003;77:6556–6561. doi: 10.1128/JVI.77.11.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Perng GC, Carpenter D, Mott KR, Osorio N, Naito J, Brick DJ, Jones C, Wechsler SL. Reactivation phenotype in rabbits of a herpes simplex virus type 1 mutant containing an unrelated antiapoptosis gene in place of latency-associated transcript. J NeuroVirol. 2007;13:78–84. doi: 10.1080/13550280601164333. [DOI] [PubMed] [Google Scholar]
- Jin L, Perng GC, Mott KR, Osorio N, Naito J, Brick DJ, Carpenter D, Jones C, Wechsler SL. A herpes simplex virus type 1 mutant expressing a baculovirus inhibitor of apoptosis gene in place of latency-associated transcript has a wild-type reactivation phenotype in the mouse. J Virol. 2005;79:12286–12295. doi: 10.1128/JVI.79.19.12286-12295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Ciacci-Zanella JR, Zhang Y, Henderson G, Dickman M. Analysis of fumonisin B1-induced apoptosis. Environ Health Perspect. 2001;109 Suppl 2:315–320. doi: 10.1289/ehp.01109s2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity. 2003;18:593–603. doi: 10.1016/s1074-7613(03)00112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, Gazdar A, Blenis J, Arnott D, Ashkenazi A. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol Cell Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu T, Khanna KM, Carriere BN, Hendricks RL. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J Virol. 2001;75:11178–11184. doi: 10.1128/JVI.75.22.11178-11184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Khanna KM, Chen X, Fink DJ, Hendricks RL. CD8(+) T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. J Exp Med. 2000;191:1459–1466. doi: 10.1084/jem.191.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MF, Ship II. A retrospective study of the prevalence and incidence of recurrent aphthous ulcers in a professional population, 1958–1971. Oral Surg Oral Med Oral Pathol. 1977;43:532–537. doi: 10.1016/0030-4220(77)90105-0. [DOI] [PubMed] [Google Scholar]
- Mott KR, Osorio N, Jin L, Brick DJ, Naito J, Cooper J, Henderson G, Inman M, Jones C, Wechsler SL, Perng GC. The bovine herpesvirus-1 LR ORF2 is critical for this gene’s ability to restore the high wild-type reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J Gen Virol. 2003;84:2975–2985. doi: 10.1099/vir.0.19421-0. [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- Nahmias AJ, Lee FK, Beckman-Nahmias S. Sero-epidemiological and -sociological patterns of herpes simplex virus infection in the world. Scand J Infect Dis Suppl. 1990;69:19–36. [PubMed] [Google Scholar]
- Peng W, Jin L, Henderson G, Perng GC, Brick DJ, Nesburn AB, Wechsler SL, Jones C. Mapping herpes simplex virus type 1 latency-associated transcript sequences that protect from apoptosis mediated by a plasmid expressing caspase-8. J Neuro Virol. 2004;10:260–265. doi: 10.1080/13550280490468690. [DOI] [PubMed] [Google Scholar]
- Perng G, Jones C, Ciacci-Zanella H, Henderson G, Yukht A, Slanina S, Hofman F, Ghiasi H, Nesburn A, Wechsler S. Virus induced neuronal apoptosis blocked by the herpes simplex virus latency associated transcript (LAT) Science. 2000a;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol. 1994;68:8045–8055. doi: 10.1128/jvi.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Esmaili D, Slanina SM, Yukht A, Ghiasi H, Osorio N, Mott KR, Maguen B, Jin L, Nesburn AB, Wechsler SL. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J Virol. 2001a;75:9018–9028. doi: 10.1128/JVI.75.19.9018-9028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. High-dose ocular infection with a herpes simplex virus type 1 ICP34.5 deletion mutant produces no corneal disease or neurovirulence yet results in wild-type levels of spontaneous reactivation. J Virol. 1996a;70:2883–2893. doi: 10.1128/jvi.70.5.2883-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996b;70:976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Maguen B, Jin L, Mott KR, Osorio N, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, Inman M, Henderson G, Jones C, Wechsler SL. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J Virol. 2002;76:1224–1235. doi: 10.1128/JVI.76.3.1224-1235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Slanina SM, Ghiasi H, Nesburn AB, Wechsler SL. A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. J Virol. 1996c;70:2014–2018. doi: 10.1128/jvi.70.3.2014-2018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Slanina SM, Ghiasi H, Nesburn AB, Wechsler SL. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. J Gen Virol. 2001b;82:1117–1122. doi: 10.1099/0022-1317-82-5-1117. [DOI] [PubMed] [Google Scholar]
- Perng GC, Slanina SM, Yuhkt A, Drolet BS, Keleher WJ, Ghiasi H, Nesburn AB, Wechsler SL. A herpes simplex virus type 1 latency associated transcript (LAT) mutant with increased virulence and reduced spontaneous reactivation. J Virol. 1999;73:920–929. doi: 10.1128/jvi.73.2.920-929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, Wechsler SL. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol. 2000b;74:1885–1891. doi: 10.1128/jvi.74.4.1885-1891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock DL, Nesburn AB, Ghiasi H, Ong J, Lewis TL, Lokensgard JR, Wechsler SL. Detection of latency- related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- Schmitz I, Kirchhoff S, Krammer PH. Regulation of death receptor-mediated apoptosis pathways. Int J Biochem Cell Biol. 2000;32:1123–1136. doi: 10.1016/s1357-2725(00)00048-0. [DOI] [PubMed] [Google Scholar]
- Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter ME. Apoptosis signaling by death receptors. Eur J Biochem. 1998;254:439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Miller LK. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc Natl Acad Sci U S A. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu HB, Halpin DR, Goeddel DV. Casper is a FADD- and caspase-related inducer of apoptosis. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- Spruance SL. The natural history of recurrent oral-facial herpes simplex virus infection. Semin Dermatol. 1992;11:200–206. [PubMed] [Google Scholar]
- Stennicke HR, Jurgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X, Zhou Q, Ellerby HM, Ellerby LM, Bredesen D, Green DR, Reed JC, Froelich CJ, Salvesen GS. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Thompson RL, Sawtell NM. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol. 2001;75:6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trousdale MD, Steiner I, Spivack JG, Deshmane SL, Brown SM, MacLean AR, Subak-Sharpe JH, Fraser NW. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol. 1991;65:6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EK, Devi-Rao G, Feldman LT, Dobson AT, Zhang YF, Flanagan WM, Stevens JG. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988a;62:1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EK, Flanagan WM, Devi-Rao G, Zhang YF, Hill JM, Anderson KP, Stevens JG. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol. 1988b;62:4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler SL, Nesburn AB, Watson R, Slanina SM, Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988;62:4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz LE. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]