Abstract
Addition of fucose (Fuc) to glycoprotein N-linked glycans or in O-linkage directly to Ser/Thr residues modulates specific cell–cell interactions and cell signaling events. Vertebrates and invertebrates add Fuc in α6-linkage to the reducing terminal N-acetylglucosamine residue of N-glycans. In Drosophila and other invertebrates, Fuc can also be added in α3-linkage to the same residue. These difucosylated N-glycans are recognized by anti-horseradish peroxidase (anti-HRP) antisera, providing a well-established marker for insect neural tissue. To understand the mechanisms and consequences of tissue-specific glycan expression, we identified a single α3-fucosyltransferase (FucTA) that produces the anti-HRP epitope in Drosophila embryos. FucTA transcripts are temporally and spatially restricted to cells that express the anti-HRP epitope and are missing in a mutant that lacks neural α3-fucosylation. Transgenic expression of FucTA, but not of any other candidate α3-fucosyltransferase, rescues the anti-HRP epitope in the embryonic nervous system of this mutant. Mass spectrometric characterization of the N-glycans of Drosophila embryos overexpressing FucTA confirms that this enzyme is indeed responsible for the biosynthesis of difucosylated glycans in vivo. Whereas ectopic expression of FucTA in the larval wing disc produces mild wing notching, the heterochronic, pan-neural expression of FucTA in early differentiating neurons generates neurogenic and cell migration phenotypes; this latter effect is associated with reduced GDP-Fuc levels in the embryo and indicates that the diversion of fucosylation resources towards fucosylation of N-glycans has an impact on developmental signaling associated with O-fucosylation.
Keywords: anti-HRP, Drosophila, fucosyltransferase, FucTA, N-glycans
Introduction
Cells in developing tissues express glycans that are unique for their transient state of differentiation and, ultimately, for their fully differentiated identity. Linked to protein or lipid, many of these carbohydrate structures are essential for specific cellular and developmental functions, including cell–cell recognition, cell adhesion, signaling, and protein targeting. Mechanisms that control tissue- and cell-specific glycosylation are, nevertheless, largely unknown, but studying glycans with restricted expression patterns presents opportunities for uncovering relevant pathways through genetic analysis. Fucose (Fuc) is frequently one of the monosaccharide components of cellular glycans that participate in carbohydrate-mediated recognition and signaling events (Becker and Lowe 2003). For instance, Fuc-containing epitopes drive selectin-mediated lymphocyte homing in mammals, and a defect in the Golgi GDP-Fuc transporter results in human leukocyte adhesion deficiency (Lübke et al. 2001; Lühn et al. 2001; Noda et al. 2003; Moriwaki et al. 2007). Furthermore, the addition of core α6-linked Fuc to N-linked glycans of immunoglobulins inversely correlates with the efficiency of antibody-dependent cellular cytotoxicity (Shinkawa et al. 2003), and Notch-based signaling requires the direct modification of the Notch polypeptide with O-linked Fuc residues (Okajima and Irvine 2002; Stanley 2007).
Species-specific glycosylation in invertebrates often results in the production of glycan structures that are immunogenic in vertebrates. One example is the presence of core α3-Fuc, in the context of a GlcNAcβ4(Fucα3)GlcNAcβ1-Asn motif (Figure 1). This structure is a target for IgE from patients allergic to plant foods and pollens (Fötisch and Vieths 2001; Pöltl et al. 2007) as well as of antisera raised against plant and insect glycoproteins (Prenner et al. 1992; Wilson et al. 1998; Jin et al. 2006). A well-known example of the latter is anti-horseradish peroxidase (anti-HRP), an antiserum raised against the plant glycoprotein HRP. This antiserum not only recognizes plant glycoproteins modified with core α3-Fuc but also specific glycans of many invertebrate species and is used as a reagent to stain the neural tissue of insects, including Drosophila melanogaster (Jan and Jan 1982; Snow et al. 1987; Kurosaka et al. 1991), as well as of other Ecdysozoa (Haase et al. 2001), including the nematode Caenorhabditis elegans (Siddiqui and Culotti 2007). In the nematode, this cross-reaction is eliminated in a mutant lacking an α3-fucosyltransferase (Paschinger et al. 2004).
Fig. 1.
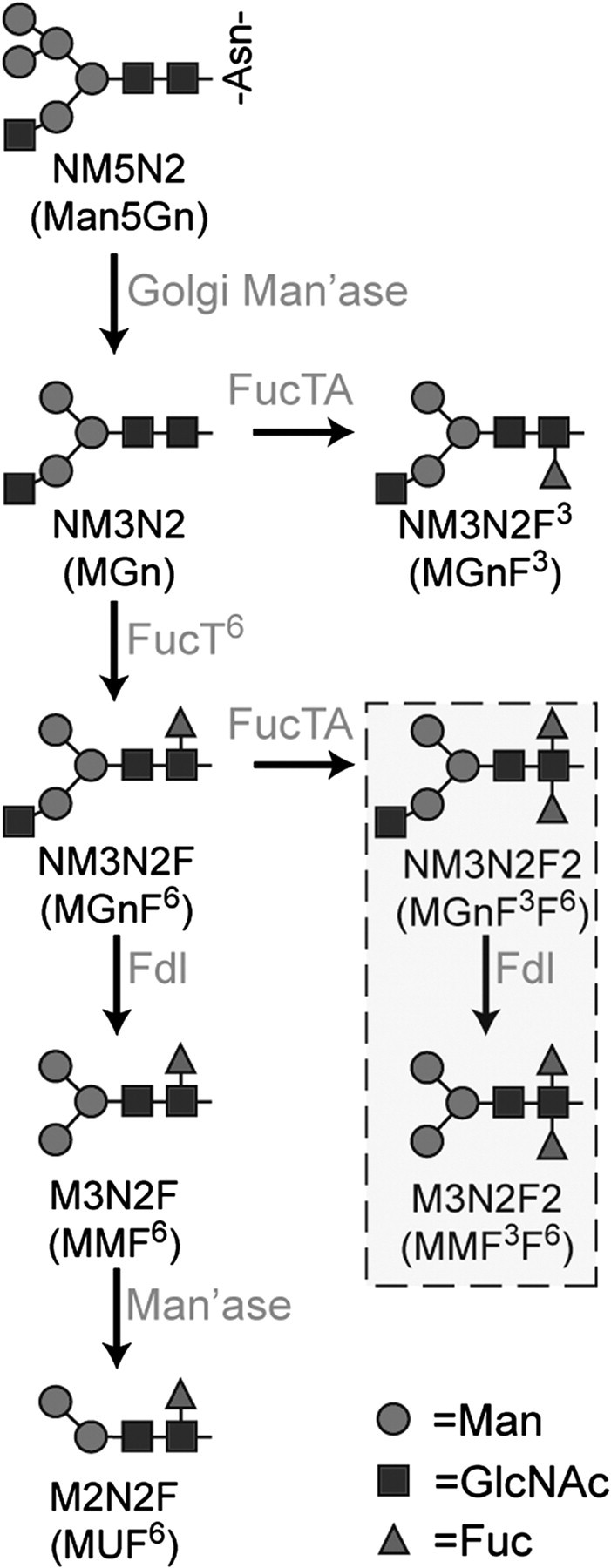
Pathways for N-linked glycan fucosylation in Drosophila. N-acetylglucosaminyltransferase I (GlcNAc-TI) adds N-acetylglucosamine to the high-mannose glycan Man5GlcNAc2 on glycoproteins transported to the Golgi, resulting in glycans with the composition Man5GlcNAc3. This octasaccharide is substrate for the production of all hybrid, complex, and paucimannose N-glycans in the Drosophila embryo. Two shorthand nomenclatures are in general use to describe glycan structures that derive from Man5GlcNAc3, also known as NM5N2 (Man5Gn; alternative names are in parentheses). Following trimming by Golgi mannosidases (Golgi Man’ase), the NM3N2 (MGn) glycan and its monofucosylated derivative NM3N2F6 (MGnF6) are substrates for production of the HRP epitopes, NM3N2F3 (MGnF3) and NM3N2F2 (MGnF3F6), which are found in very low prevalence in the embryo. The major pathway for HRP epitope synthesis is initiated by addition of fucose (Fuc) in α6-linkage to NM3N2 (MGn) by Fucosyltransferase 6 (FucT6), followed by addition of α3-linked Fuc, which is catalyzed by Fucosyltransferase A (FucTA) in vitro. FucTA prefers substrates already containing α6-linked Fuc; thus, most α3-fucosylated glycans are difucosylated. The hexosaminidase known as “Fused lobes” (Fdl) removes non-reducing terminal GlcNAc to produce paucimannose glycans such as M3N2F6 (MMF6) or M3N2F2 (MMF3F6). An unidentified mannosidase activity (Man’ase) removes an additional Man residue from the M3N2F6 (MMF6) structure to generate M2N2F6 (MUF6). The boxed glycans are a subset of the HRP epitopes that have been detected in Drosophila embryos, of which M3N2F2 (MMF3F6) is the most prevalent. Graphical representations of glycans are consistent with the nomenclature of the Consortium for Functional Glycomics.
We have previously demonstrated the presence of core α3-fucosylated N-glycans in Drosophila (Fabini et al. 2001; Aoki et al. 2007) as well as of a relevant α3-fucosyltransferase, designated FucTA, which can synthesize the epitope recognized by anti-HRP (hereafter referred to as the “HRP epitope”) in vitro (Fabini et al. 2001). Furthermore, knock-down of FucTA by RNA interference in a Drosophila neural cell line reduced HRP epitope expression (Rendić et al. 2006). Based on these and other studies of glycan processing in insects (Paschinger et al. 2005; Léonard et al. 2006; Sarkar et al. 2006; Shah et al. 2008), the biosynthetic pathway required for HRP epitope expression can be proposed (Figure 1). However, there are four α3-fucosyltransferase homologues in Drosophila (FucTA, B, C, D), and evidence has been lacking as to which of these is responsible in vivo for the tissue-specific expression of the HRP epitope. In the course of determining which of the aforementioned four candidate α3-fucosyltransferase homologues might synthesize the HRP epitope in vivo, we observed that FucTA is the only homologue responsible for neural-specific production of this N-linked epitope. Furthermore, unexpected neurogenesis, glial migration, and wing formation phenotypes were observed when FucTA expression was manipulated, generally consistent with previously described effects of reduced Notch O-glycosylation (Okajima and Irvine 2002; Edenfeld et al. 2007). The developmental phenotypes engendered by imbalanced consumption of GDP-Fuc highlight the importance of regulated glycan expression.
Results
FucTA expression coincides with the presence of the HRP epitope
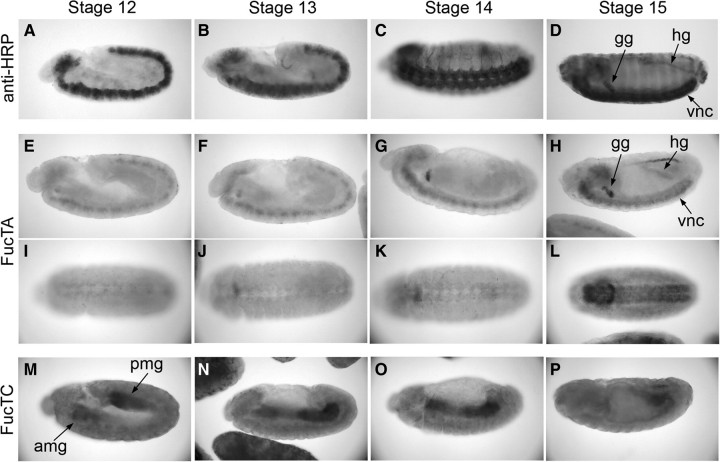
Previous studies have shown that anti-HRP staining first appears in differentiating neurons of Drosophila embryos during stage 11 (Jan and Jan 1982). As neurogenesis gives way to differentiation and progeny acquire terminal identities, HRP epitope expression increases across the nervous system. In addition to the ventral nerve cord, anti-HRP reactivity is also observed in the peripheral nervous system as well as in the garland gland (gg) and hindgut (hg) by stage 14 (Figure 2A–D). To determine which of the four candidate α3-fucosyltransferases (FucTA, B, C, or D) share a similar temporal and spatial expression patterns as the HRP epitope, single-stranded RNA probes for each enzyme were synthesized and applied to wild-type embryos. Only the FucTA probe hybridized to neural tissue, to the garland gland, and to hindgut, spatially and temporally coincident with anti-HRP staining in the wild-type embryo (Figure 2E–L). FucTA transcripts are weakly detected in the neurectoderm as early as stage 11 and increase throughout embryonic development. In contrast, FucTC transcripts were present in the developing anterior and posterior midgut until fusion of the anlage after which expression was decreased (Figure 2M–P). Consistent with the inability of reverse transcription–polymerase chain reaction (PCR) to detect FucTB and FucTD transcripts in embryos (Rendić et al. 2006), specific hybridization patterns were not observed with the FucTB and FucTD probes (Figure S1). These results clearly show that the spatial and temporal expression patterns of FucTA, but of no other α3-fucosyltransferase homologue, in wild-type embryos mimic the expression pattern of the HRP epitope.
Fig. 2.
Tissue-specific expression of α3-fucosyltransferase homologues in the Drosophila embryo. Wild-type embryos (Oregon R) were probed with either anti-HRP or with single-stranded, digoxigenin-labeled RNA probes specific for FucTA or FucTC transcripts. (A–D) Oligosaccharides recognized by staining of wild-type embryos with anti-HRP are first expressed late during stage 11 (not shown in this figure) in differentiating neurons as they delaminate from the ventral neurectoderm. Through stages 12 and 13, anti-HRP staining tracks with continuing elaboration of the ventral nerve cord (vnc) during germband retraction. By stage 14, anti-HRP positive axons are visible in the lateral ectoderm, and elements of the peripheral nervous system begin to acquire the epitope. The only non-neural embryonic tissues that also stain with anti-HRP antibody, the garland gland (gg) and hindgut (hg), also become strongly stained by stage 14. (E–L) FucTA transcripts are expressed in the same pattern as the HRP epitope. Lateral (E–H) and ventral views (I–L) of embryos at progressively older stages demonstrate that FucTA and HRP epitope are both detected in the ventral nerve cord, garland gland, and hindgut. (M–P) The FucTC probe detected transcript in the developing anterior and posterior midguts (amg, pmg; lateral views); hybridization to midgut was evident from the first appearance of the anlage at stage 12 through the completion of fusion by late stage 14. Expression of FucTC was consistently higher at the anterior and posterior ends of the midgut than at the leading edges of its growth towards the mid-body. Transcripts were not detected with probes for FucTB and FucTD (Figure S1). All embryos are oriented with their anterior ends to the left.
Neural expression of FucTA rescues α3-fucosylation in TM3 embryos
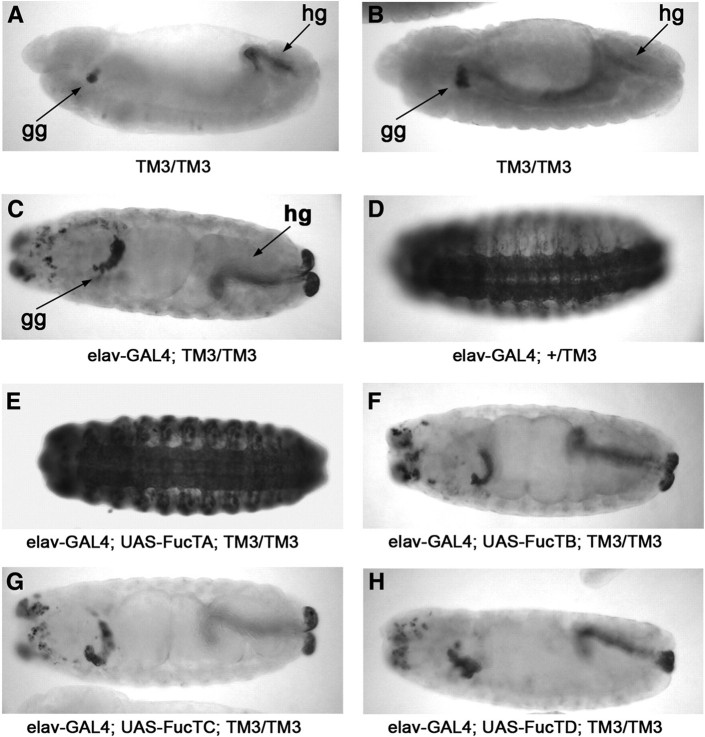
Tissue-specific regulation of fucosyltransferase activity is revealed in embryos bearing chromosomal aberrations that affect HRP epitope expression. Embryos homozygous for the TM3 balancer chromosome fail to express the HRP epitope in the nerve cord but retain expression in the garland gland and hindgut (Snow et al. 1987; Seppo et al. 2003). Consistent with the loss of neural HRP epitopes (Figure 3A), FucTA transcripts in TM3/TM3 embryos are detected only in the garland gland and hindgut (Figure 3B). Thus, this aberration provides a useful background for validating the assignment of FucTA as the only fucosyltransferase capable of synthesizing the epitope in neural tissue. Multiple UAS-FucT transgenic lines were generated for each of the α3-fucosyltransferase homologues, and transgenic chromosomes were crossed into the TM3-lacZ background and mated to a neural-specific GAL4 driver line (elav-GAL4). Expression of the HRP epitope was only restored in progeny embryos carrying the UAS-FucTA chromosome (Figure 3C–H). The amounts of transcript per cell, detected by in situ hybridization signals, were comparable for each of the elav-GAL4; UAS-FucT lines tested for rescue (Figure S2).
Fig. 3.
FucTA synthesizes the HRP epitope in embryos. (A–B, lateral views, late stage 13) Embryos homozygous for the TM3 balancer chromosome express the HRP epitope (A, anti-HRP staining) and transcript (B, in situ hybridization) for FucTA in the garland gland (gg) and the hindgut (hg) but not in neural tissue. (C–H, all ventral views except for H, which is lateral, stages 14–15). Each of the four candidate α3-fucosyltransferases (FucTA, B, C, and D) was expressed as a UAS-transgene in neural cells under control of the neural-specific elav-GAL4 driver. In homozygous TM3 embryos, the elav-GAL4 driver by itself does not rescue HRP epitope expression (C). Introduction of a single copy of a wild-type third chromosome generates normal expression of the epitope (D). Only FucTA (E), not FucTB, C, nor D (F–H), rescued neural expression of the HRP epitope in the TM3 homozygous background.
Increased α3-fucosylation induces neurogenic and cell migration phenotypes
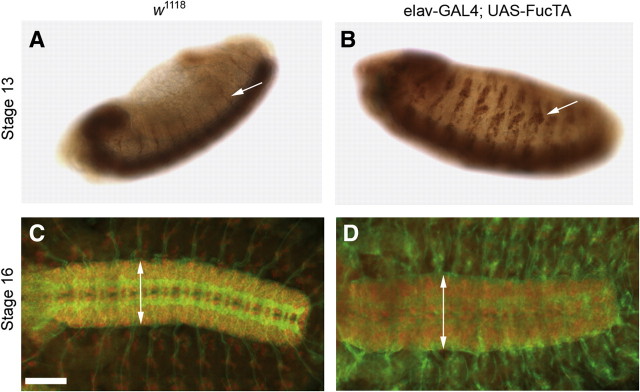
Anti-HRP staining of elav-GAL4; UAS-FucTA embryos reveals clusters of peripheral sensory neurons larger than expected for the developmental stage of the embryo. The development of enlarged peripheral sensory complexes is first apparent in stage 13 embryos (Figure 4A–B) and, in part, reflects the fact that the elav promoter drives transcription at stages that precede the normal appearance of the HRP epitope. Early heterochronic induction of the enzyme produces premature accumulation of the glycan. However, peripheral cluster augmentation is also apparent by in situ hybridization for FucTA transcript, but not for other FucT transcripts in elav-GAL4; UAS-FucTB, C, or D embryos (Figure S2). Therefore, enlarged peripheral neural clusters result specifically from the expression of the FucTA enzyme. In older embryos, the Notch-like neurogenic impact of increased HRP epitope expression continues to be evident in the expanded dimensions of the ventral nerve cord (Figure 4C–D).
Fig. 4.
Increased α3-fucosylation induces neurogenic phenotypes. (A–B, anti-HRP staining with DAB precipitate) In stage 13 wild-type embryos (A), anti-HRP staining in the lateral ectoderm is limited to the axonal processes of motor neurons that have grown out from the ventral nerve cord (arrow in A). In elav-GAL4; UAS-FucTA embryos of the same age (B), prominent clusters of peripheral neural cells express the epitope (arrow in B). (C–D, immunofluorescence, anti-HRP in green, anti-ELAV in red). By stage 16, the ventral nerve cord of wild-type embryos (C) is narrower than the ventral nerve cord in elav-GAL4; UAS-FucTA embryos (D). Double-headed arrows are equivalent to 92 and 115 μm in C and D, respectively. Scale bar corresponds to 60 μm in C and D.
Increased N-linked core fucosylation also induces aberrant glial cell migration. Glial cell number was reduced in the ventral nerve cord but increased along peripheral migratory routes in elav-GAL4; UAS-FucTA embryos. The wild-type distribution of repo-positive glial nuclei at the lateral edge of the ventral nerve cord was significantly attenuated in elav-GAL4; UAS-FucTA embryos (Figure 5A–D, arrowheads in C, D), and extremely fine, thread-like, repo-positive nuclear profiles were observed extending towards nerve exit sites (arrows in Figure 5D), indicating that α3-fucosylation enhances active glial migration along efferent neural pathways. The decrease in ventral nerve cord glia was associated with an increase in the number of repo-positive glial nuclei in peripheral tissue (Figure 5E–F). These extranumerary peripheral glia were detected along migratory routes previously described to be Notch dependent (Edenfeld et al. 2007; Silies et al. 2007).
Fig. 5.
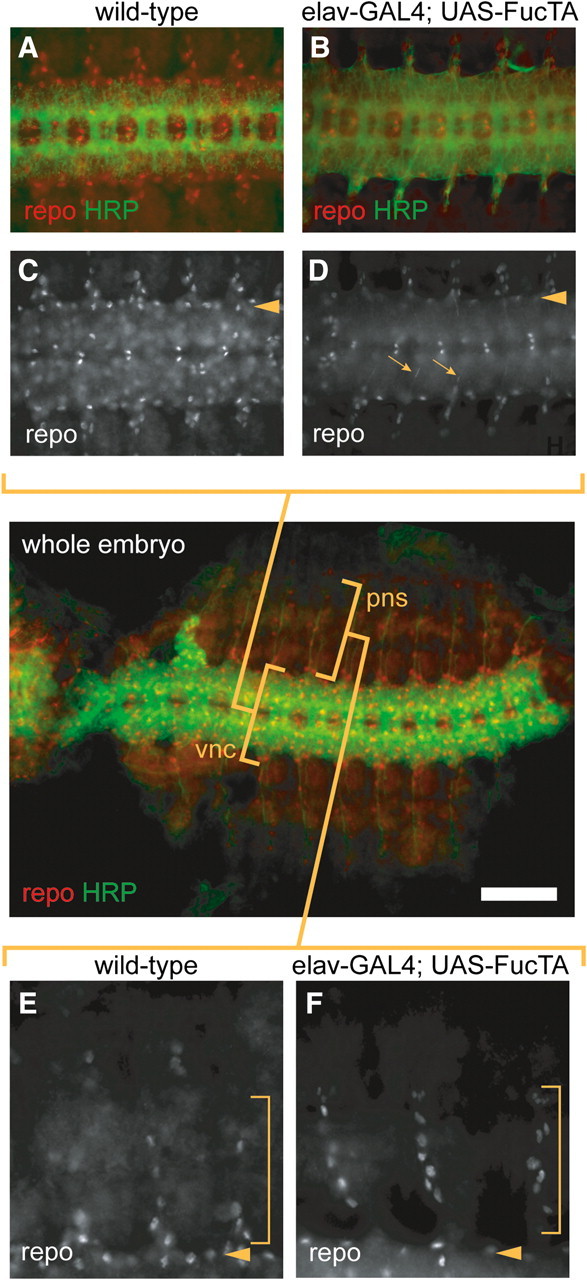
Increased α3-fucosylation affects cell migration. A low magnification image of a dissected, wild-type whole embryo at stage 14 is presented in the middle panel to provide spatial orientation for the higher magnification images in panels A–F. The low magnification image provides an overview of the distribution of repo-positive glial cells in relation to the HRP epitope in the ventral nerve cord (vnc) and peripheral nervous system (pns). (A–D) Higher magnification images of the ventral nerve cord of wild-type (A, C) or elav-GAL4; UAS-FucTA (B, D) embryos at stage 14, double-stained to visualize glial nuclei (anti-repo, red) and HRP epitope expression (green). The nuclei of repo-positive glial cells are broadly distributed throughout the nerve cord in wild-type (C) embryos but also delineate the lateral limit of the ventral nerve cord (arrowhead). The prevalence of the lateral glial population is reduced by increased α3-fucosylation (D, arrowhead), and elongated profiles of repo-positive glial nuclei (D, arrows) indicate glial migration along efferent neural pathways in elav-GAL4; UAS-FucTA embryos. (E–F) Migration of glial cells into peripheral tissues is visualized by anti-repo staining. Three segments of the ventral and ventrolateral ectoderm (stage 14) of wild-type (E) or elav-GAL4; UAS-FucTA embryos (F) are shown; the lateral edge of the ventral nerve cord is indicated (arrowhead). Within the bracketed region of the ventral and ventrolateral ectoderm, significantly more glial nuclei are detected in embryos with increased α3-fucosylation. Scale bar represents 60 μm in the whole embryo panel, 80 μm in A, B and 150 μm in C, D.
The expression of FucTA is limiting for α3-fucosylation in many different cell types
Non-neural ectodermal cells do not express the HRP epitope during normal embryonic development. However, when FucTA expression is driven with en-GAL4, HRP epitope expression in the early embryo is detected in a segmental pattern consistent with engrailed (En) expression (Figure 6A). The en-GAL4 driver also induces ectopic expression of the HRP epitope in the posterior compartment of the wing imaginal disc in UAS-FucTA larvae (Figure 6B–C). In embryonic and larval imaginal tissues that are normally devoid of the HRP epitope, FucTA expression is sufficient to drive the production of glycans bearing α3-linked Fuc. Therefore, availability of nucleotide sugar precursor (GDP-Fuc), nucleotide sugar transporter activity, and appropriately processed core glycan acceptor substrates on glycoprotein targets all exist within a broad range of cell types at levels that can support α3-fucosylation.
Fig. 6.
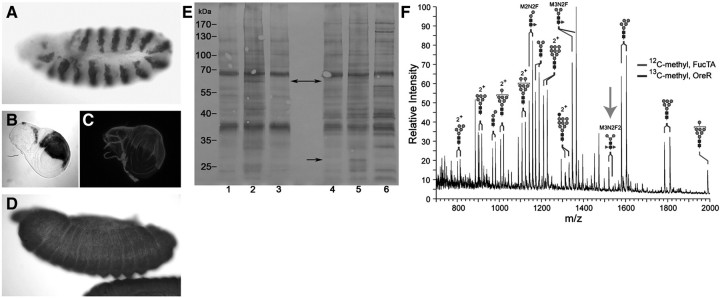
FucTA is limiting for production of the HRP epitope and of α3-fucosylated glycans in multiple cell types. (A–C) en-GAL4-driven FucTA expression generates HRP epitope in cells that do not normally express the glycan. In embryos of the genotype en-GAL4; UAS-FucTA, anti-HRP staining is detected in the expected engrailed pattern of segmentally repeated ectodermal cells (A, DAB precipitate, stage 11). Wing discs harvested from larvae of the same genotype express HRP epitope in the posterior compartment (B, en-GAL4; UAS-lacZ, X-gal staining; C, en-GAL4; UAS-FucTA, anti-HRP immunofluorescence). (D, late stage 13) A single UAS-FucTA transgenic line, designated UFA5, constitutively expresses the enzyme in the absence of a GAL4 driver. HRP epitope is produced throughout the ectoderm and amnioserosa at all stages of embryonic development in UFA5 embryos (DAB precipitate). (E) Equal amounts of proteins of female (lanes 1–3) and male (lanes 4–6) adult flies were probed with anti-HRP on a western blot. Extracts from UFA5 line (lanes 2 and 5) show an overall higher intensity when compared to wild-type flies (OreR, lanes 1 and 4; w1118, lanes 3 and 6), a result in line with anti-HRP stainings of UFA5 embryos. Although similar to a great extent to wild-type flies, both UFA5 males and females show a slightly extended anti-HRP binding pattern (see arrows for examples). (F) N-linked glycans were prepared from wild-type (OreR) and UAS-FucTA (UFA5) embryos, differentially permethylated with either [12C]-methyl iodide (UFA5) or [13C]-methyl iodide (OreR) and mixed together based on equal protein amounts before analysis by NSI-MS. Differential mass shifts, resulting from incorporation of 12C or 13C isotopes, allow direct comparison of glycan abundance between two preparations in the same full MS analysis. Ectopic expression of FucTA brings the M3N2F2 (MMF3F6) glycan significantly above threshold (arrow). The reduced prevalence of M3N2F (MMF6) and M2N2F (MUF6) in UFA5 is consistent with the diversion of precursor glycans towards the production of HRP epitopes.
One of the six UAS-FucTA lines generated by P-element insertion produced ectopic expression of the HRP epitope without introducing a GAL4 driver. In this constitutively expressing line, designated UFA5, the epitope is detected throughout the entire ectoderm and amnioserosa (Figure 6D). By probing equal amounts of protein extracts of UFA5 and wild type with anti-HRP in a western blotting experiment, an overall more intensive and a slightly different anti-HRP binding pattern is noticeable (Figure 6E). Expression of the epitope is broadly increased such that the core difucosylated M3N2F2 glycan, an HRP epitope (see Figure 1 for structures), is easily detected by nanospray ionization mass spectrometry (NSI-MS) in the total glycan profile of UFA5 embryos (Figure 6F). Direct comparison of differentially permethylated glycans prepared from control embryos (w1118) emphasizes the magnitude of the increased synthesis of this glycan. Additionally, decreased prevalence of glycans with only one Fuc residue (M3N2F and M2N2F) is consistent with the diversion of precursors (NM3N2 and NM3N2F) towards synthesis of difucosylated structures. The N-linked glycan profile of UFA5 embryos indicates that FucTA expression pulls oligosaccharide synthesis towards the production of difucosylated glycans at the expense of the major monofucosylated, paucimannose structures.
Increased α3-fucosylation in neural tissue affects nucleotide sugar levels but not global O-fucosylation
Since elav-GAL4-driven expression of FucTA generates Notch-like phenotypes in the embryonic nervous system, we hypothesized that increased α3-fucosylation of N-linked glycans might affect levels of GDP-Fuc, placing stress on the pathway for O-fucosylation of the Notch protein. Therefore, we measured nucleotide sugar levels in control (w1118) and elav-GAL4; UAS-FucTA embryos. Significant decreases were detected for both GDP-Fuc and for its precursor GDP-Man, while UDP-GlcNAc and UDP-Gal were not affected (Figure 7A). The impact of increased α3-fucosylation on nucleotide sugar levels was only evident in embryos overexpressing FucTA in the nervous system; changes in GDP-Fuc or GDP-Man were not detected in UFA5 embryos nor were Notch-like phenotypes detected in this background (data not shown); this divergence can be explained by the strong driving by elav in a subset of cells, whereas ectopic expression in UFA5 is due to a leakiness in a range of cells. Interestingly, direct characterization of the O-linked glycans expressed in elav-GAL4; UAS-FucTA and UFA5 embryos revealed that the prevalence of the only O-Fuc glycan detectable in Drosophila embryos (GlcNAcβ3(GlcAβ4)Fuc) was unchanged by expanded expression of N-linked α3-fucosylation, although the abundance of the core 1 disaccharide was significantly elevated (Figure 7B–D).
Fig. 7.
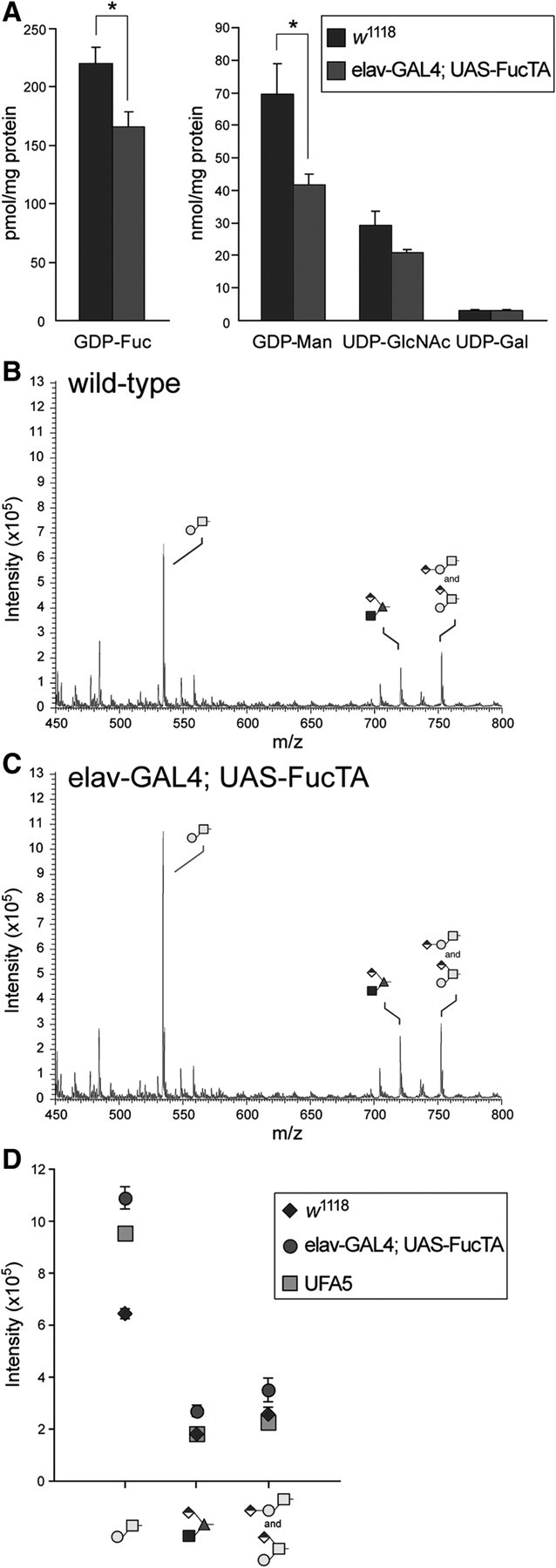
Increased α3-fucosylation in neural tissue reduces GDP-Fuc and GDP-Man but does not affect global O-fucosylation. (A) Nucleotide sugars were measured in extracts of wild-type (w1118) or FucTA transgenic embryos. Both GDP-Fuc and GDP-Man levels were reduced per milligram of protein in elav-GAL4; UAS-FucTA embryos, which exhibit notch-like neural phenotypes (* indicates P < 0.05, mean ± s.e.m. for n = 3 independent preparations of each genotype). Nucleotide sugars unrelated to fucosylation (UDP-GlcNAc and UDP-Gal) were unchanged in embryos with increased α3-fucosylation. O-linked glycans were prepared from wild-type (B, w1118) or from elav-GAL4; UAS-FucTA (C) embryos by reductive β-elimination. Glycans derived from equivalent amounts of embryonic protein were permethylated and analyzed by NSI-MS. Three major signals predominate the mass spectrum of O-linked glycans in the Drosophila embryo. The core 1 disaccharide (Galβ3GalNAc) is detected at m/z = 534. The only detected O-glycan containing Fuc was previously characterized as GlcNAcβ3(GlcAβ4)Fuc and is found at m/z = 722. Finally, the signal at m/z = 752 arises from an isobaric mixture of two structures, linear and branched isomers of the core 1 disaccharide bearing GlcA. (D) Increased α3-fucosylation of N-linked glycans does not affect the prevalence of the O-Fuc glycan but does result in increased prevalence of the core 1 disaccharide in elav-GAL4; UAS-FucTA embryos. The mean ± s.e.m. for three independent preparations is plotted for wild-type (w1118) and for elav-GAL4; UAS-FucTA. A single determination is shown for the UFA5 genotype, which expresses the HRP epitope constitutively in the embryonic ectoderm and amnioserosa (see Figure 6).
Increased α3-fucosylation in the posterior half of the wing disc alters compartment boundaries
Since HRP epitope expression is induced in the posterior compartment of wing discs harvested from en-GAL4; UAS-FucTA larvae (see Figure 6C), we assessed the impact of ectopic FucTA expression on the formation of compartment boundaries in the developing wing. The distribution of the wingless protein (wg) is a downstream reporter of Notch activation, which establishes the dorsoventral boundary of the wing disc. If the appearance of en-driven, ectopic HRP epitope negatively influences Notch signaling, reduced wg expression should be detected along the dorsoventral boundary in the posterior compartment. In en-GAL4; UAS-FucTA discs, the wg pattern is not completely eliminated but is disrupted in a graded manner (Figure 8A–H). Rather than extending across the entire dorsoventral boundary, wg expression is attenuated more severely at the posterior extreme of the boundary in the half disc where FucTA is expressed. Consistent with the partial loss of wg expression, mature wings from en-GAL4; UAS-FucTA adults exhibit notched phenotypes along their posterodistal margin, albeit with relatively low penetrance (Figure 8I–K and Table I). A more robust disruption is seen in the partial to complete loss of the anterior cross vein (Figure 8L–N and Table I). Previous fate mapping studies indicate that the progenitors of the anterior cross vein straddle the anterior/posterior compartment boundary, where they express En (Blair and Ralston 1997; Blair 2007). Significant cross vein defects and wing notching were not observed in wings of adults expressing another FucT (en-GAL4; UAS-FucTB, Table I) nor in the UFA5 line, which also lacked notch-like neural phenotypes and had unaltered nucleotide sugar levels.
Fig. 8.
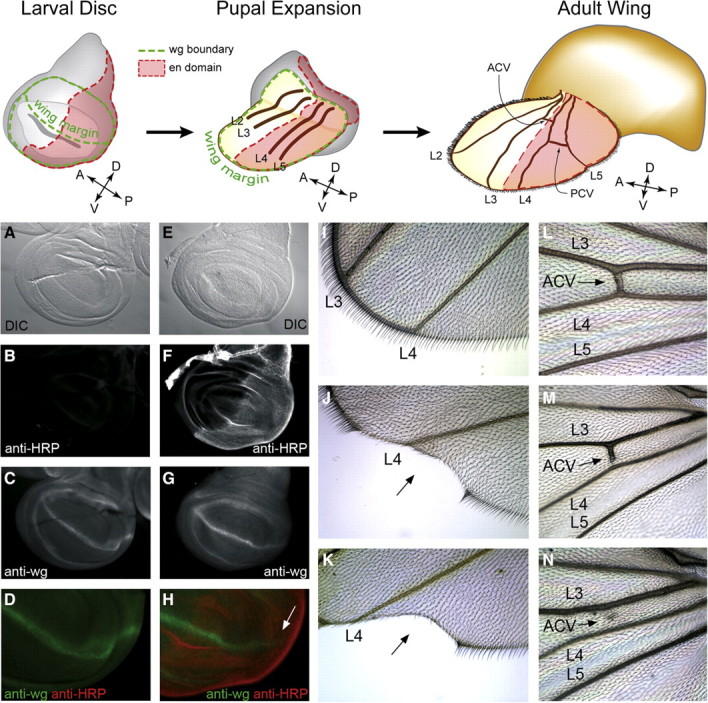
Ectopic α3-fucosylation in the posterior compartment of the wing disc alters Notch signaling and crossvein development. Reference figures across the top highlight the compartment boundaries that are established in the larval wing disc and illustrate their morphogenetic transformation into the adult wing. The posterior compartment of the wing disc is defined by the expression of engrailed (en, shown in red). In the adult wing, engrailed expressing cells contribute to the posterior half of the wing beginning at the intervein region between longitudinal veins L3 and L4. The anterior crossvein (ACV) derives from this region, while the posterior crossvein (PCV) arises from the intervein between L4 and L5. The dorsovental boundary in the larval wing disc prefigures the wing margin of the mature wing and is defined by expression of the wingless protein (wg, shown in green). Appropriate Notch signaling is essential for wg expression and the subsequent formation of the dorsoventral boundary. (A–D) Wild-type larval wing disc visualized by DIC (A), anti-HRP staining (B), anti-wg staining (C), and merge of anti-HRP/anti-wg (D). (E–H) Wing disc from en-GAL4; UAS-FucTA larva stained as in A–D. En-driven expression of FucTA attenuates wg expression along the dorsoventral boundary in the posterior compartment (arrow in H). (I–K) Appearance of the distal posterior wing margin in wild-type (I) and en-GAL4; UAS-FucTA (J, K) adults. With low penetrance (see Table I), the attenuation of wg expression observed in the larval wing disc results in wing notching (arrow in J, K). (L–N) The anterior crossvein (ACV) in wild-type wings interconnects L3 and L4 (L). In en-GAL4; UAS-FucTA wings, the ACV is frequently incomplete (M) and often completely missing (N).
Table I.
Penetrance of wing phenotypes induced by HRP epitope expression
| Genotype | Penetrance |
||
|---|---|---|---|
| Incomplete anterior crossvein (%) | Incomplete posterior crossvein (%) | Wing notch (%) | |
| w1118 (n = 59) | 14 | 0 | 0 |
| en-GAL4; UAS-FucTA (n = 60) | 48 | 3 | 8 |
| en-GAL4; UAS-FucTB (n = 60) | 7 | 0 | 0 |
Discussion
Fucosyltransferase A catalyzes α3-fucosylation in the Drosophila embryo
The use of anti-HRP antibody to stain Drosophila neural tissue was first reported nearly 30 years ago (Jan and Jan 1982; Paschinger et al. 2009). Since then, the Drosophila epitope has been convincingly demonstrated to consist of Fuc in α3-linkage to the reducing terminal GlcNAc residue of the N-linked core (Snow et al. 1987; Katz et al. 1988; Kurosaka et al. 1991). Drosophila glycans bearing α3-linked Fuc are greatly enriched in neural tissue but still account for only 1% of the total N-glycans in the whole organism (Fabini et al. 2001; North et al. 2006; Aoki et al. 2007). Many of the proteins that bear the HRP epitope are expressed in neural tissue, including a subset of the fasciclins (Snow et al. 1987), the Nervana ATPase (Sun and Salvaterra 1995), and a receptor tyrosine phosphatase (Desai et al. 1994). In some cases, these proteins are also expressed outside the nervous system where they are alternatively glycosylated.
Four candidate α3-fucosyltransferase genes (FucTA, B, C, and D) were previously identified in Drosophila. Of these four, only FucTA was capable of generating the HRP epitope when expressed in S2 cells, and only knock-down of FucTA decreased anti-HRP staining in a Drosophila neuronal cell line (Rendić et al. 2006). We now demonstrate that FucTA also meets all of the criteria for being the enzyme responsible for producing the epitope in the Drosophila embryo. FucTA is expressed coincident with the appearance of the epitope in the wild-type embryo and in a restricted distribution that correlates with residual, non-neural epitope production in a mutant that lacks the glycan in neural tissue. Re-expression of the enzyme in neural tissue rescues synthesis of the epitope in this mutant background. Furthermore, direct structural analysis of N-linked glycans by mass spectrometry verifies that the expected difucosylated glycan (M3N2F2, MMF3F6) is increased by ectopic expression of the FucTA enzyme. Finally, expression of the enzyme is sufficient to produce the epitope in a broad range of cell types, many of which do not normally generate α3-fucosylated glycans. Therefore, multiple lines of evidence support the assignment of FucTA as the physiologically relevant and limiting enzyme responsible for α3-fucosylation.
Neural phenotypes associated with altered α3-fucosylation
Mutations in three genes have been previously described that decrease α3-fucosylation. Loss of GlcNAcT-1 (Mgat1), the enzyme necessary for production of all hybrid, complex, and fucosylated N-glycans in Drosophila, eliminates α3-fucosylation and produces deficits in adult locomotion, life span, and larval brain development (Sarkar et al. 2006; Sarkar et al. 2010). The other two previously described mutations affect the expression of a more restricted set of N-linked glycans. One of these is neurally altered carbohydrate (nac), which is associated with a maternal-effect lethality, and aberrant wing, eye, and neural development at 18°C, as well as sensory axon misrouting at 25°C (Phillis et al. 1993; Whitlock 1993); the molecular basis for the nac phenotype, though, has not yet been proven. The third mutation that affects α3-fucosylation is carried on the TM3 balancer chromosome and eliminates neural expression of the HRP epitope in embryos, but does not affect non-neural, larval, or adult anti-HRP staining (Snow et al. 1987; Katz et al. 1988). The relevant defect on the TM3 chromosome was molecularly mapped to the tollo/toll-8 gene, and adults that carry a non-complementing deletion in combination with the TM3 chromosome exhibit a moderate, although unquantified, locomotion defect (Seppo et al. 2003). Glycan analysis indicates that the mutation carried on TM3 also affects the expression of other minor glycans, but the relative prevalence of the major monofucosylated structure (M3N2F) is unchanged (Aoki et al. 2007). For all three of these mutations, no direct mechanism has yet been proposed that links observed developmental and behavioral phenotypes with loss of specific glycans.
In the course of testing the four FucT candidates for rescue of α3-fucosylation, we observed phenotypes that evoked comparisons to previously described roles of Fuc in the modulation of Notch signaling. Mutations affecting OFUT1 (O-Fucosyltransferase 1), the enzyme which adds Fuc in O-linkage to the Notch protein, Gmd (GDP-Man dehydratase), necessary for the production of the GDP-Fuc donor substrate, Gfr (GDP-Fuc transporter of Golgi), or Efr (GDP-Fuc transporter of the ER) have been shown to enhance or phenocopy Notch signaling phenotypes in various contexts (Okajima and Irvine 2002; Sasamura et al. 2003; Ishikawa et al. 2005; Okajima et al. 2005; Ishikawa et al. 2010). These results, as well as those from studies of Notch signaling in vertebrates, have led to the well-tested hypothesis that appropriate O-linked fucosylation of Notch is essential for its normal signaling activity (Okajima et al. 2003; Stanley 2007; Vodovar and Schweisguth 2008), although other data suggest that inactive OFUT1 can act as a Notch-specific chaperone (Okajima et al. 2008; Stahl et al. 2008). In this study, manipulation of N-linked fucosylation produced enlarged peripheral sensory clusters, expansion of the ventral nerve cord, aberrant glial cell migration, and compartment-specific wing notching, each of which is consistent with altered Notch signaling.
Unexpected effects of N-linked fucosylation on Notch signaling
Notch is modified by O-Fuc, O-Glc, and O-GlcNAc but also carries N-glycans (Johansen et al. 1989; Luo and Haltiwanger 2005; Acar et al. 2008; Matsuura et al. 2008). Therefore, FucTA-dependent Notch signaling defects could result directly from altered N-linked glycosylation of Notch or of Notch’s glycoprotein ligands (Delta or Serrate). Consistent with this possibility, an existing Drosophila Notch mutation (Nts1) (Xu et al. 1992) is associated with a change from an N-linked glycosylation sequon with a highly favored amino acid composition (NGS) to a less favored one (NDS) (Koles et al. 2007). Alternatively, FucTA overexpression may indirectly affect Notch O-fucosylation through utilization of a disproportionate share of the intracellular GDP-Fuc pool. The amount of GDP-Fuc in insect cells (Tomiya et al. 2001) and in Drosophila embryos (Figure 7) is low, especially in comparison with other nucleotide sugars; in our measurements, GDP-Fuc levels are some 300-fold less than those of GDP-Man. Diversion of the donor substrate towards α3-fucosylation could cause reduced availability severe enough to impact O-fucosylation. However, despite our determination that GDP-Fuc and GDP-Man levels are reduced when FucTA is overexpressed in neural tissue, a coordinated decrease in global O-fucosylation is not detectable in whole embryos (Figure 7). Indeed, considering the complexity of the glycosylation pathway, it is remarkable, but difficult to explain, that both GDP-Man levels and mucin-type core 1 glycosylation are affected in the elav-GAL4; UAS-FucTA line. The only detectable O-Fuc glycan in Drosophila embryos accounts for 11% of the total O-linked glycan content; the high abundance of this single glycan implies that it is carried on proteins other than just Notch (Aoki et al. 2008). Therefore, characterization of global O-fucosylation is not likely to report critical alterations on low abundance proteins, such as Notch, nor in subsets of cells (e.g. neuronal) in embryos. Determining whether glycosylation of endogenous Notch is affected by FucTA overexpression presents a major analytical challenge but will ultimately be essential for assigning phenotypes to specific post-translational modifications.
The pleiotropy of aberrant N-linked fucosylation
The effects of altered α3-fucosylation are extremely well tolerated through development and maturity in Drosophila. The UFA5 UAS-FucTA line is viable and fertile despite the broad expansion of α3-fucosylation to embryonic ectoderm and amnioserosa. Other ectopic expressions of FucTA generate only non-lethal, moderately neurogenic effects and mild wing notching. Thus, a significant increase in the flux through an otherwise minor, tissue-restricted glycosylation pathway does not result in global pathology. Nevertheless, anti-HRP blots demonstrate that many glycoproteins carry α3-fucosylation in wild-type and in our UAS-FucTA lines (Figure 6E), consistent with the expectation that altered fucosylation might reveal Fuc-dependent, Notch-independent processes. Indeed, one phenotype that we observe in our en-GAL4-driven FucTA lines indicates that increased α3-fucosylation impacts development in a manner independent of Notch signaling. Specifically, in this case, ectopic expression of FucTA generates incomplete crossvein phenotypes predominantly in the anterior, rather than the posterior, crossvein of the wing. The differentiation of anterior crossvein cells from the developing wing epithelium is initiated by localized expression of the TGF-β homologues, decapentaplegic (dpp) and glass bottom boat (gbb), which activate bone morphogenetic protein (BMP) signaling in the presumptive crossvein cells. For the posterior crossvein, longitudinal veins serve as the nearby TGF-β/BMP source for establishing a local morphogen gradient that induces differentiation (Blair 2007). At the posterior crossvein, more so than at the anterior crossvein, Notch signaling spatially restricts BMP-induced differentiation of provein cells, thereby limiting the width of the crossvein (Huppert et al. 1997; Marcus 2001). Therefore, at least in this context, our data dissect aberrant fucosylation away from Notch signaling and suggest that altered glycosylation impacts a component of the TGF-β/BMP pathway that is required differentially in the morphogenesis of the anterior and posterior crossveins. Interestingly, both fucosylation of N-glycans and addition of O-linked GalNAc are essential for signaling through vertebrate TGF-β receptors, suggesting evolutionarily conserved contexts in which specific glycan processing has been selected (Schachter 2005; Wang et al. 2005; Herr et al. 2008).
Although core α3-fucosylation is not a feature of vertebrates, these organisms display a fucosylation repertoire which has been expanded to include a non-reducing terminal α3-Fuc on N-, O-, and lipid-linked glycans, structures that have not yet been described in Drosophila. In many cases, these vertebrate fucosylated glycans have important functions as recognition ligands and modulators of signaling receptors (Becker and Lowe 2003). Thus, over and above a significant quantity of α6-fucosylated N-linked glycans, vertebrates and invertebrates have superimposed other uses for Fuc, including modification of critical developmental signaling pathways. Regardless of the acceptor, the utilization of Fuc as a regulatory and recognition element in cell–cell interactions is a phenomenon that is shared across species.
Materials and methods
Fucosyltransferase constructs
The entire open reading frames of D. melanogaster α1,3-fucosyltransferase homologues were isolated by PCR using the pIZT/V5-His vectors encoding these enzymes (Rendić et al. 2006) or Canton S cDNA as templates; the following primer pairs were used: FucTA/5′/EcoRI (cggaattccatgcggcgtccgaag) with FucTA/3′/KpnI (ccggggtacctcagtcgtcgctggagtcg), FucTB/5′/XhoI (ccgctcgagataatcatgcgactggcac) with FucTB/3′/XbaI (gctctagattaagtatttgaactattactgc), FucTC/5′/EcoRI (cggaattcatgtatttagggagggttc) with FucTC/3′/KpnI (cggggtacctcacaaacgtattcggctttg), and FucTD/5′/EcoRI (cggattccaaaaggcaatgccgatagac) with FucTD/3′/KpnI (ccggggtacctcagaggaaggtggtggatc). The purified PCR fragments were cut with relevant restriction enzymes prior to ligation into the pUAST vector. Plasmid DNA was then used to inject w1118 embryos and create transformant lines by standard procedures (Brand et al. 1994). For each FucT construct, multiple insertion lines on all three chromosomes were generated. The FucT transformant results reported here were validated by repeating the rescue experiments with at least one other independent insertion line. The UFA5 line was the only line found to constitutively express FucTA in the absence of a GAL4 driver.
Drosophila lines
The TM3 balancer used in this study carries an embryonically expressed lacZ marker (D, ry/TM3-DZ, Pry + t7.2 = H22.7, Bloomington Stock Center, Bloomington, IN, USA), allowing the anti-HRP staining status of blue embryos to be correlated with rescue of the TM3 mutation (Seppo et al. 2003). Driver lines (en-GAL4, 2nd chromosome insertion; elav-GAL4, X-insertion) were obtained from the Bloomington Stock Center.
Anti-HRP staining, lacZ activity, and in situ hybridization
Embryo collections were dechorionated, fixed, devitellinized, stained with antibodies, and staged according to standard methods (Campos-Ortega and Hartenstein 1985; Patel 1994). Primary antibody dilutions were 1:1000 for rabbit anti-HRP (Jackson ImmunoResearch, West Grove, PA, USA) and 1:5 for anti-ELAV (Developmental Studies Hybridoma Bank, University of Iowa). Alexafluor-conjugated (Invitrogen, Carlsbad, CA, USA) or peroxidase-conjugated (Jackson ImmunoResearch) secondary antibodies were used at 1:500 dilutions. LacZ activity was detected in embryos as previously described (Klämbt et al. 1991). The distribution of four fucosyltransferase mRNAs (FucTA, B, C, or D) in embryos was visualized using single-stranded RNA probes labeled with digoxigenin (Tautz and Pfeifle 1989). The four probes demonstrated transcript-specific hybridization to the nerve cord in embryos collected from matings of an elav-GAL4 driver to UAS-FucTA, B, C, or D lines.
Glycan preparation and mass spectrometry
N-glycans and O-glycans were prepared from wild-type and FucTA transgenic lines and analyzed by NSI-MSn as previously described (Aoki et al. 2007; Aoki et al. 2008).
Extraction and analysis of sugar nucleotides
Overnight embryo collections taken from wild-type or FucTA transgenic parents (approximately 50 μL of packed embryos) were homogenized and extracted with chloroform:methanol:water::4:8:3 (v:v:v) as previously described for the preparation of embryo proteins for glycan analysis (Aoki et al. 2007). Following precipitation of proteins by centrifugation, the supernatant was dried under a nitrogen stream. Protein content was measured by bicinchoninic acid assay of the pellet to normalize sugar nucleotide quantification. The dried supernatant was dissolved in 200 μL of 50% methanol and applied onto BAKERBOND SPE Octadecyl C18 cartridge columns (100 mg, JT Baker, Phillipsburg, NJ). The columns were then washed with 3 mL of Milli-Q water. The flow-through and wash fractions were pooled and lyophilized. Dried material was resuspended in 40 mM phosphate buffer, pH 9.2 (120 μL buffer per 0.5 mg of protein), and filtered by centrifugation through 10 K molecular weight cutoff membranes (Millipore, Bedford, MA) before analysis by high-performance anion-exchange chromatography according to the method of Tomiya et al. (2001). Sugar nucleotides were detected by absorbance at 260 nm, identified by co-elution with standard, and quantified by comparison with known amounts of standard nucleotide sugars.
Western blotting
Gender separated, adult flies (OreR, w1118, UFA5; four flies per 150 μL lysis buffer) were mixed with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxyglycolate, 1% NP40, 10% glycerol, 0.4 mM ethylenediaminetetraacetic acid) supplemented with 1:100 dilution of Sigma Protease Inhibitor Cocktail (for use in purification of histidine-tagged proteins) and lysed using a Branson Sonifier 250 (50% duty cycle, 30% output, 2 min). Extracts were spun down for 30 mins at 21,000g and 4°C and supernatants were then mixed with an equal volume of 2 × SDS–polyacrylamide gel electrophoresis buffer, followed by a 5-min incubation at 95°C. After transfer to the membrane, Ponceau S staining was performed to verify the transfer efficiency of all samples. For probing the blot, 1:10,000 dilution of rabbit anti-HRP (Sigma, St Louis, MO, USA) as primary antibody and 1:2000 dilution of the secondary, alkaline phosphatase-conjugated antibody (Vector Laboratories, Burlingame, CA, USA) were used. The development was performed using SigmaFAST BCIP/NBT solution.
Supplementary Material
Acknowledgements
The authors are grateful for the excellent technical contributions of Denise Kerner, Mindy Porterfield, and Johan Koo, and to Dr. Katharina Paschinger for her comments on the manuscript.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- Fuc
fucose
- gg
garland gland
- hg
hindgut
- HRP
horseradish peroxidase
- NSI-MS
nanospray ionization mass spectrometry
- PCR
polymerase chain reaction
- SDS
sodium dodecyl sulfate
- wg
wingless protein
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by grants from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (P17681 and L314 to I.B.H.W) and from National Institutes of Health, National Institute of General Medical Sciences (NIH/NIGMS) (GM072839 to M.T.).
Conflict of interest statement
None declared.
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem. 2008;283:30385–30400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- Blair SS, Ralston A. Smoothened-mediated Hedgehog signalling is required for the maintenance of the anterior-posterior lineage restriction in the developing wing of Drosophila. Development. 1997;124:4053–4063. doi: 10.1242/dev.124.20.4053. [DOI] [PubMed] [Google Scholar]
- Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu Rev Cell Dev Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Drosophila melanogaster: Practical uses in cell and molecular biology. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. New York: Springer-Verlag; 1985. [Google Scholar]
- Desai CJ, Popova E, Zinn K. A Drosophila receptor tyrosine phosphatase expressed in the embryonic CNS and larval optic lobes is a member of the set of proteins bearing the ‘HRP’ carbohydrate epitope. J Neurosci. 1994;14:7272–7283. doi: 10.1523/JNEUROSCI.14-12-07272.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenfeld G, Altenhein B, Zierau A, Cleppien D, Krukkert K, Technau G, Klambt C. Notch and Numb are required for normal migration of peripheral glia in Drosophila. Dev Biol. 2007;301:27–37. doi: 10.1016/j.ydbio.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Fabini G, Freilinger A, Altmann F, Wilson IBH. Identification of core α1,3 fucosylated glycans and cloning of the requisite fucosyltransferase cDNA from Drosophila melanogaster. Potential basis of the neural anti-horseradish peroxidase epitope. J Biol Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- Fötisch K, Vieths S. N- and O-linked oligosaccharides of allergenic glycoproteins. Glycoconj J. 2001;18:373–390. doi: 10.1023/a:1014860030380. [DOI] [PubMed] [Google Scholar]
- Haase A, Stern M, Wachtler K, Bicker G. A tissue-specific marker of Ecdysozoa. Dev Genes Evol. 2001;211:428–433. doi: 10.1007/s004270100173. [DOI] [PubMed] [Google Scholar]
- Herr P, Korniychuk G, Yamamoto Y, Grubisic K, Oelgeschlager M. Regulation of TGF-β signalling by N-acetylgalactosaminyltransferase-like 1. Development. 2008;135:1813–1822. doi: 10.1242/dev.019323. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Higashi S, Ayukawa T, Sasamura T, Kitagawa M, Harigaya K, Aoki K, Ishida N, Sanai Y, Matsuno K. Notch deficiency implicated in the pathogenesis of congenital disorder of glycosylation IIc. Proc Natl Acad Sci USA. 2005;102:18532–18537. doi: 10.1073/pnas.0504115102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa HO, Ayukawa T, Nakayama M, Higashi S, Kamiyama S, Nishihara S, Aoki K, Ishida N, Sanai Y, Matsuno K. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. J Biol Chem. 2010;285:4122–4129. doi: 10.1074/jbc.M109.016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci USA. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Bencúrová M, Borth N, Ferko B, Jensen-Jarolim E, Altmann F, Hantusch B. Immunoglobulin G specifically binding plant N-glycans with high affinity could be generated in rabbits but not in mice. Glycobiology. 2006;16:349–357. doi: 10.1093/glycob/cwj071. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Fehon RG, Artavanis-Tsakonas S. The notch gene product is a glycoprotein expressed on the cell surface of both epidermal and neuronal precursor cells during Drosophila development. J Cell Biol. 1989;109:2427–2440. doi: 10.1083/jcb.109.5.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz F, Moats W, Jan YN. A carbohydrate epitope expressed uniquely on the cell surface of Drosophila neurons is altered in the mutant nac (neurally altered carbohydrate) EMBO J. 1988;7:3471–3477. doi: 10.1002/j.1460-2075.1988.tb03222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: A model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology. 2007;17:1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- Kurosaka A, Yano A, Itoh N, Kuroda Y, Nakagawa T, Kawasaki T. The structure of a neural specific carbohydrate epitope of horseradish peroxidase recognized by anti-horseradish peroxidase antiserum. J Biol Chem. 1991;266:4168–4172. [PubMed] [Google Scholar]
- Léonard R, Rendić D, Rabouille C, Wilson IBH, Preat T, Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- Lübke T, Marquardt T, Etzioni A, Hartmann E, von Figura K, Körner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- Lühn K, Wild MK, Eckhardt M, Gerardy-Schahn R, Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- Marcus JM. The development and evolution of crossveins in insect wings. J Anat. 2001;199:211–216. doi: 10.1046/j.1469-7580.2001.19910211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- Moriwaki K, Noda K, Nakagawa T, Asahi M, Yoshihara H, Taniguchi N, Hayashi N, Miyoshi E. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology. 2007;17:1311–1320. doi: 10.1093/glycob/cwm094. [DOI] [PubMed] [Google Scholar]
- Noda K, Miyoshi E, Gu J, Gao CX, Nakahara S, Kitada T, Honke K, Suzuki K, Yoshihara H, Yoshikawa K, et al. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003;63:6282–6289. [PubMed] [Google Scholar]
- North SJ, Koles K, Hembd C, Morris HR, Dell A, Panin VM, Haslam SM. Glycomic studies of Drosophila melanogaster embryos. Glycoconj J. 2006;23:345–354. doi: 10.1007/s10719-006-6693-4. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307:1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008;6:1. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K, Rendić D, Lochnit G, Jantsch V, Wilson IBH. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J Biol Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IBH. Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology. 2005;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Rendić D, Wilson IBH. Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj J. 2009;26:385–395. doi: 10.1007/s10719-008-9155-3. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg E, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. San Diego: Academic Press; 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- Phillis RW, Bramlage AT, Wotus C, Whittaker A, Gramates LS, Seppala D, Farahanchi F, Caruccio P, Murphey RK. Isolation of mutations affecting neural circuitry required for grooming behavior in Drosophila melanogaster. Genetics. 1993;133:581–592. doi: 10.1093/genetics/133.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöltl G, Ahrazem O, Paschinger K, Ibanez MD, Salcedo G, Wilson IBH. Molecular and immunological characterization of the glycosylated orange allergen Cit s 1. Glycobiology. 2007;17:220–230. doi: 10.1093/glycob/cwl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenner C, Mach L, Glössl J, März L. The antigenicity of the carbohydrate moiety of an insect glycoprotein, honey-bee (Apis mellifera) venom phospholipase A2. The role of α1,3-fucosylation of the asparagine-bound N-acetylglucosamine. Biochem J. 1992;284:377–380. doi: 10.1042/bj2840377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendić D, Linder A, Paschinger K, Borth N, Wilson IBH, Fabini G. Modulation of neural carbohydrate epitope expression in Drosophila melanogaster cells. J Biol Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- Sarkar M, Leventis PA, Silvescu CI, Reinhold VN, Schachter H, Boulianne GL. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. J Biol Chem. 2006;281:12776–12785. doi: 10.1074/jbc.M512769200. [DOI] [PubMed] [Google Scholar]
- Sarkar M, Iliadi KG, Leventis PA, Schachter H, Boulianne GL. Neuronal expression of Mgat1 rescues the shortened life span of Drosophila Mgat11 null mutants and increases life span. Proc Natl Acad Sci USA. 2010;107:9677–9682. doi: 10.1073/pnas.1004431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, et al. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- Schachter H. The search for glycan function: Fucosylation of the TGF-β1 receptor is required for receptor activation. Proc Natl Acad Sci USA. 2005;102:15721–15722. doi: 10.1073/pnas.0507659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppo A, Matani P, Sharrow M, Tiemeyer M. Induction of neuron-specific glycosylation by Tollo/Toll-8, a Drosophila Toll-like receptor expressed in non-neural cells. Development. 2003;130:1439–1448. doi: 10.1242/dev.00347. [DOI] [PubMed] [Google Scholar]
- Shah N, Kuntz DA, Rose DR. Golgi α-mannosidase II cleaves two sugars sequentially in the same catalytic site. Proc Natl Acad Sci USA. 2008;105:9570–9575. doi: 10.1073/pnas.0802206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Siddiqui SS, Culotti JG. Examination of neurons in wild type and mutants of Caenorhabditis elegans using antibodies to horseradish peroxidase. J Neurogenet. 2007;21:271–289. doi: 10.1080/01677060701693461. [DOI] [PubMed] [Google Scholar]
- Silies M, Edenfeld G, Engelen D, Stork T, Klambt C. Development of the peripheral glial cells in Drosophila. Neuron Glia Biol. 2007;3:35–43. doi: 10.1017/S1740925X07000622. [DOI] [PubMed] [Google Scholar]
- Snow PM, Patel NH, Harrelson AL, Goodman CS. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. J Neurosci. 1987;7:4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Salvaterra PM. Characterization of Nervana, a Drosophila melanogaster neuron-specific glycoprotein antigen recognized by anti-horseradish peroxidase antibodies. J Neurochem. 1995;65:434–443. doi: 10.1046/j.1471-4159.1995.65010434.x. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: Sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- Vodovar N, Schweisguth F. Functions of O-fucosyltransferase in Notch trafficking and signaling: Towards the end of a controversy? J Biol. 2008;7:7. doi: 10.1186/jbiol68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, et al. Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci USA. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock KE. Development of Drosophila wing sensory neurons in mutants with missing or modified cell surface molecules. Development. 1993;117:1251–1260. doi: 10.1242/dev.117.4.1251. [DOI] [PubMed] [Google Scholar]
- Wilson IBH, Harthill JE, Mullin NP, Ashford DA, Altmann F. Core α1,3-fucose is a key part of the epitope recognized by antibodies reacting against plant N-linked oligosaccharides and is present in a wide variety of plant extracts. Glycobiology. 1998;8:651–661. doi: 10.1093/glycob/8.7.651. [DOI] [PubMed] [Google Scholar]
- Xu T, Caron LA, Fehon RG, Artavanis-Tsakonas S. The involvement of the Notch locus in Drosophila oogenesis. Development. 1992;115:913–922. doi: 10.1242/dev.115.4.913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.