Abstract
Sirtuins are NAD+-dependent protein deacylases that are conserved in all domains of life and are involved in diverse cellular processes, including control of gene expression and central metabolism. Eukaryotic sirtuins have N-terminal extensions that have been linked to protein multimerization and cellular localization. Here the first evidence of sirtuin isoforms in bacteria is reported. The enterobacterium Salmonella enterica synthesizes two isoforms of CobB sirtuin, a shorter 236-amino-acid isoform (here CobBS) and a longer 273-amino-acid isoform (here CobBL). The N-terminal 37-amino-acid extension of CobBL is amphipathic, containing 18 basic amino acids (12 of which are Arg) and 13 hydrophobic ones; both isoforms were active in vivo and in vitro. Northern blot and transcription start site analyses revealed that cobB is primarily expressed as two monocistronic cobB mRNAs from two transcription start sites, one of which was mapped within the neighboring ycfX gene and the other of which was located within cobB. Additionally, a low-abundance ycfX-cobB bicistronic mRNA was observed which could encode up to three proteins (YcfX, CobBL, and CobBS). CobBL isoforms are common within the family Enterobacteriaceae, but species of the genus Erwinia (including the plant pathogen Erwinia amylovora) encode only the CobBL isoform. The CobBL isoform from E. amylovora restored growth of as S. enterica cobB mutant strain on low acetate.
Homologues of the yeast silence information regulator 2 protein (ySir2p, also known as sirtuin) are Zn(II)-containing, NAD+-dependent deacylases present in cells of all domains of life (21, 24, 35). In eukaryotes, sirtuin function impacts gene expression, metabolism, cancer, stress responses, and life span control (5, 18, 22, 29, 38). Ongoing work on the regulation of sirtuin activity levels has shown that sirtuins are regulated at the transcriptional, posttranscriptional, and posttranslational levels (22, 34).
In eukaryotes, some sirtuins contain N-terminal extensions of the enzymatic core, and these regions can affect the binding of interacting partners (11, 20), facilitate oligomerization of sirtuins (46), mediate interactions with other sirtuin forms (8, 27), and direct cellular localization (8, 31). Thus, these regions seem to contribute to the regulation of sirtuin function in the eukaryotic cell. It is not known whether sirtuin isoforms exist in bacteria or archaea or, if they do, what their functions may be.
Bacterial sirtuins play roles in short-chain fatty acid metabolism in S. enterica serovar Typhimurium LT2 (here S. enterica) and Bacillus subtilis (16, 17, 38) and in the catabolism of aromatic and allicyclic acids in Rhodopseudomonas palustris (10). In S. enterica, the protein acetyltransferase Pat acetylates acetyl coenzyme A (acetyl-CoA) synthetase (Acs) and propionylates propionyl-CoA synthetase (PrpE) (17, 37). In both cases, Pat acylates a conserved active-site lysine within the adenylation domain, effectively inactivating the enzyme by preventing the formation of the acetyl-AMP or propionyl-AMP intermediate (17, 36). Deacylation of propionylated PrpE (PrpEPr) or acetylated Acs (AcsAc) by the CobB sirtuin of S. enterica reactivates both enzymes, allowing cells to grow on propionate or low levels of acetate, respectively (17, 38).
The role of sirtuins in S. enterica central metabolism was recently expanded by data that suggest that a large number of central metabolic enzymes in S. enterica may be modified by reversible lysine acetylation (44). In addition to cellular metabolism, sirtuins have been linked to bacterial chemotaxis through the ability to deacetylate the response regulator CheY in Escherichia coli (26).
Though the role of bacterial sirtuins is expanding, little is known about the transcriptional regulation of the genes that encode these enzymes. Wang et al. reported the transcriptional profile of the cobB gene of S. enterica (44), but the signals that induce cobB expression and the location of the cobB promoter elements were not identified.
Based on the annotated cobB sequence of S. enterica, we sought to determine whether the CobB sirtuin contains an N-terminal extension to the sirtuin catalytic core, as predicted by bioinformatic analysis of the region. We also determined the transcription start site from which cobB mRNA is synthesized. Here, we provide evidence that S. enterica synthesizes two biologically active isoforms of CobB, one form of which contains a cationic 37-residue N-terminal extension. We used in vitro and in vivo approaches to demonstrate that each CobB isoform was active. We provide evidence that cobB is transcribed from multiple promoters and propose that these alternate transcripts may allow S. enterica to differentially regulate CobB isoform synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in nutrient broth (NB), no-carbon essential (NCE) minimal medium (3), or 3-(N-morpholino)propanesulfonic acid (MOPS) minimal medium (28). For Western blot assays, cells were grown using NCE minimal medium. For growth curves when acetate was the sole carbon source, MOPS minimal medium was supplemented with MgSO4 (1 mM), l-methionine (0.5 mM), and acetate (10 mM, pH 7.0). For growth on glycerol, MOPS minimal medium was supplemented with MgSO4 (1 mM), trace minerals (1×), dicyanocobinamide (15 nM), 5,6-dimethylbenzamidazole (125 μM), and glycerol (22 mM). When used, ampicillin was at 100 μg/ml. Cells were grown at 37°C with shaking. For growth curves, cell density was measured using a microtiter plate and a microtiter plate reader (Bio-Tek Instruments). Doubling times were calculated using the Prism 4 software package (GraphPad, La Jolla, CA).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Referencea |
|---|---|---|
| S. enterica serovar Typhimurium LT2 strains | ||
| JE6583 | metE205 ara-9 | Laboratory collection |
| JE7088 | ΔmetE2702 ara-9 | Laboratory collection |
| Derivatives of JE6583 | ||
| JE10806 | ΔycfX128 | |
| JE11228 | ΔcobB1375 | |
| Derivatives of JE7088 | ||
| JE12939 | ΔcobB1375 ΔcobT1380 | Laboratory collection |
| JE12972 | ΔcobB1375 ΔcobT1380/pBAD30 | |
| JE12973 | ΔcobB1375 ΔcobT1380/pCOBB8 (or pCobBL/S) | |
| JE12974 | ΔcobB1375 ΔcobT1380/pCOBB19 (or pCobBL) | |
| JE12975 | ΔcobB1375 ΔcobT1380/pCOBB24 (or pCobBS) | |
| JE13302 | ΔcobB1375 ΔcobT1380/pCOBB93 (or pEaCobBL) | |
| E. coli JE6090 | C41(λDE3) | Laboratory collection |
| Plasmids | ||
| pBAD30 | ParaBAD expression vector, bla+ | 19 |
| pTEV6 | N-terminal, rTEV-cleavable MBP fusion overexpression vector, bla+ | 30 |
| pCOBB8 (or pCobBL/S) | cobB+ cloned into pBAD30, bla+ | 38 |
| pCOBB19 (or pCobBL) | cobB1372 allele (CobBM37A M38A) in pBAD30, bla+ | |
| pCOBB24 (or pCobBS) | cobB1373 allele (CobBM1A) in pBAD30, bla | |
| pCOBB93 (or pEaCobBL) | E. amylovora cobB+ allele in pBAD30, bla+ | |
| pCOBB71 | CobBS ORF in pTEV6 | |
| pCOBB72 | CobBL ORF in pTEV6 |
Unless otherwise indicated, all strains were constructed during the course of this work.
Strain construction.
Gene deletions in S. enterica were constructed by using the phage lambda Red recombinase system as described previously (12). Notably, the ycfX deletion was specifically designed to delete the 5′ portion (nucleotides [nt] 4 to 300) of the ycfX gene without disrupting putative cobB regulatory elements that exist in the 3′ region of the ycfX open reading frame (ORF).
Molecular techniques.
DNA manipulations were performed using standard techniques (4, 14). Restriction endonucleases were purchased from Fermentas. DNA was amplified using PfuUltra II Fusion DNA polymerase (Stratagene), and site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids were isolated using the Wizard Plus SV Miniprep kit (Promega), and PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega). DNA sequencing was performed using BigDye (ABI PRISM) protocols, and sequencing reactions were resolved at the University of Wisconsin—Madison Biotechnology Center. The sequences of the oligonucleotide primers used are listed in Table 2.
TABLE 2.
Primers and DNA probe used in this study
| Name | Sequencea |
|---|---|
| cobB deletion primers | |
| cobB(L) Wanner 5′ | GGTGCTGCTTTTTTACATCTTACCGACTAATCAAAAAAAGAGGTTGTTATGGTGTAGGCTGGAGCTGCTTC |
| cobB(L) Wanner 3′ | ACGTAACGTGAAATGTAGGCCGGATAAGGCGTTACCGGGCAAACAGCACTACATATGAATATCCTCCTTAG |
| ycfX deletion primers | |
| ycfX1 Wanner 5′ | GAGTATTGAGCGGCCAGTAATCGCGGAATATAAAAACAAGGACGGTGATGGTGTAGGCTGGAGCTGCTTC |
| ycfX1 Wanner 3′ | ATCATCCCAGGCTTCGGACAAGGCAAAACAGTTAGCGTCATTGTCCAGACGCATATGAATATCCTCCTTAG |
| Cloning primers | |
| SacI + 5′cobB_Eamy | CATGAGCTCCGCTTTGCTAAGGCGCG |
| SphI + 3′cobB_Eamy | ACTGCATGCTAAAGCGGGTTGGCCGTCG |
| KpnI + CobBL 5′ pTEV6 | GTAGGTACCATGCAGTCGCGTCGGTTTCATCG |
| KpnI + CobBS 5′ pTEV6 | ATAGGTACCATGGAAAACCCAAGAGTATTAGTCC |
| SalI + CobB 3′ pTEV6 | GATGTCGACCAGCACTACAGCCCTTTCAGG |
| Site-directed mutagenesis primers | |
| SeCobB M1A F | CAAAAAAAGAGGTTGTTGCGCAGTCGCGTCG |
| SeCobB M1A R | CGACGCGACTGCGCAACAACCTCTTTTTTTG |
| SeCobB M37A, M38A F | GACAGAGTGGTGCCGGAAGCGGCGGAAAACCCAAGAG |
| SeCobB M37A, M38A R | CTCTTGGGTTTTCCGCCGCTTCCGGCACCACTCTGTC |
| 5′ RACE analysis primers | |
| CobB_499_RT | CGCCGTTCCATTCCAGAATCTGG |
| CobB_435_RT | ATGAATGATGTTGCGATTGC |
| CobB_325_RT | GATAAGCTTCCGCATTGGGTTGTATTTCC |
| Northern blot probes | |
| cobB_Northern_98-mer | CGGATAGACATGACCGGACGTGCCAATGGCGATAAAAATATCCGCCATCGACAGCGCCATATAAATTTCATCCATGCCAAGCGGCATCTCGCCAAACC |
| ycfX_Northern_96-mer | TCTAAAAAGGCGCTATAGCTGGTATGGGGCGTGGGAACCCGTTTTTCCGAGTGCAGCCGCCGCGTTGAGTCAAATACGCCTAACGCAATGTTTGTT |
Bold type indicates restriction sites.
Cloning of S. enterica and Erwinia amylovora cobB genes.
Cloning of S. enterica cobB has been described previously (38). Site-directed mutagenesis of start codon M1 and M37/M38 yielded cobB alleles cobB1372 (encodes CobBM37A M38A or CobBL) on plasmid pCOBB19 (also referred to as pCobBL) and cobB1373 (encodes CobBM1A or CobBS) on plasmid pCOBB24 (also referred to as pCobBS).
The E. amylovora cobB gene was amplified from purified genomic DNA (a kind gift from N. Perna). The 1-kb fragment containing E. amylovora cobB+ was digested with SacI and SphI and ligated into pBAD30 (19), which had been digested with the same enzymes. The resulting 6-kb plasmid was called pCOBB93 or pEaCobBL.
Protein purification.
S. enterica cobB was amplified from JE6583 genomic DNA using primers to add a 5′ KpnI site and a 3′ SalI site to the amplification product. For CobBL overproduction, nt 1 to 822 of the cobB allele were amplified, and for CobBS overproduction, nt 109 to 822 of the cobB allele were amplified. Amplified fragments were digested with KpnI and SalI and subsequently ligated into plasmid pTEV6 (30) with the same enzymes. The resulting plasmids, pCOBB71 and pCOBB72, direct the synthesis of CobBS and CobBL, respectively, with recombinant tobacco etch virus (rTEV) protease-cleavable N-terminal maltose-binding protein (MBP) tags.
Overproduction and purification methods were the same for both CobBL and CobBS. Plasmids pCOBB71 and pCOBB72 were moved into E. coli strain C41(λDE3). The resulting strains were grown overnight and subcultured 1:100 (vol/vol) into 2 liters of super broth (13) containing ampicillin (100 μg/ml). Cultures were grown with shaking at 37°C to an A600 of ∼0.7, and MBP-H6-CobB synthesis was induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM). The culture was grown overnight at 37°C. Cells were harvested at 8,000 × g for 15 min at 4°C in a Beckman Coulter Avanti J-2 XPI centrifuge fitted with a JLA-8.1000 rotor. Cell pellets were resuspended in 30 ml cold His-Bind buffer (buffer A, containing sodium phosphate buffer [20 mM, pH 7.5], NaCl [500 mM], and imidazole [20 mM]). Cells were placed on ice and lysed by sonication for 2 min (2-s pulse followed by 2 s of cooling) at level 7 in a model 550 sonic dismembrator (Fisher). The extract was cleared by centrifugation at 4°C for 30 min at 43,367 × g. Clarified cell extract was loaded onto a 5-ml HisTrap FF column (GE Healthcare) connected to a computer-controlled ÄKTA fast protein liquid chromatography (FPLC) system. Unbound proteins were eluted off the column by extensive washing with buffer A. A 65-ml wash step with 95% buffer A and 5% buffer B (sodium phosphate buffer [20 mM, pH 7.5], NaCl [500 mM], imidazole [500 mM]) was applied to the column prior to a 65-ml linear gradient of 5 to 100% buffer B. All fractions containing MBP-H6-CobB were combined. rTEV protease (33) was added to MBP-H6-CobB, and samples were dialyzed at room temperature for 3 h in buffer C (sodium phosphate buffer [20 mM, pH 7.5], NaCl [500 mM]) containing 1 mM dithiothreitol. CobB-rTEV mixtures were then dialyzed at 4°C against buffer C for 3 h and again against buffer C containing imidazole (20 mM) for 12 h. After cleavage and dialysis, protein mixtures were passed over the 5-ml HisTrap column following the protocol described above. Protein that did not bind to the column was analyzed by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) (23). Fractions containing CobBL or CobBS were pooled. CobBL and CobBS were stored in HEPES buffer (20 mM) containing NaCl (100 mM) and 10% glycerol. CobB concentrations were determined by measuring absorbance at 280 nm. The molar extinction coefficient used to calculate CobBL and CobBS concentrations was 22,460 M−1 cm−1.
Western blot analysis.
A 200-ml overnight culture grown in NB was washed twice with 1× NCE minimal medium. Cells were then inoculated into either NB or NCE minimal medium containing l-methionine (0.5 mM) and MgSO4 (1 mM) and supplemented with acetate (50 mM), glycerol (50 mM), or glucose (50 mM) to an optical density at 650 nm (OD650) of 0.1. For Western blot assays of citrate-grown cultures, cultures grown overnight in NB were inoculated to an OD650 of 0.025 into NCE minimal medium containing l-methionine (0.5 mM) and MgSO4 (1 mM) and supplemented with citrate (50 mM) without washing. When cultures reached mid-log or stationary phase, they were placed on ice for 10 min to arrest growth and then centrifuged at 8,000 × g for 15 min at 4°C in a Beckman Coulter Avanti J-25I centrifuge fitted with a JLA-16.250 rotor. Cells were resuspended in HEPES buffer (50 mM, pH 7.5) containing phenylmethylsulfonyl fluoride (1 mM) and EDTA (10 mM). Cells were lysed by sonication on ice using a microtip at 50% duty for 1 min. Cell lysates were spun at 16,000 × g for 10 min at 4°C in an Eppendorf 5415D microcentrifuge to remove unbroken cells and cellular debris. Proteins in cellular extracts were quantified by absorbance at 280 nm. Fifty or 400 μg of cellular protein was resolved using SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore). Rabbit polyclonal CobB antiserum (Laboratory Animal Resources, University of Wisconsin—Madison) was used to detect isoforms of CobB. The CobB signal was visualized using alkaline phosphatase-conjugated goat anti-rabbit antibody (Pierce) and the one-step nitroblue tetrazolium—5-bromo-4-chloro-3′-indolylphosphate (NBT-BCIP) substrate according to the manufacturer's instructions.
CobB isoform abundance was quantified as described previously (43). Briefly, 2-fold dilutions of total cellular lysates (12.5 μg to 1.6 mg) prepared as described above were separated by SDS-PAGE using 1.5-mm-thick 12% polyacrylamide gels. Proteins were transferred and detected as described above. Band intensity was quantified using the TotalLab 1D gel analysis software v2003 (Nonlinear Dynamics, Durham, NC), and values within the linear range of detection were used to determine the relative CobB isoform levels. The values reported were in arbitrary units.
In vitro deacetylation assays.
Conditions for in vitro acetylation and deacetylation of Acs have been described previously (37). In the assays described here, we used homogeneous CobBL, CobBS, Pat, and Acs from S. enterica (17, 36, 37). Acs was radiolabeled using Pat protein acetyltransferase (37) and [1-14C]acetyl-CoA in a final volume of 1 ml. Excess [1-14C]acetyl-CoA was removed by buffer exchange; NAD+ was added to the AcsAc reaction mixture to a final concentration of 1 mM, and deacetylation reactions were initiated by the addition of CobBL or CobBS. Samples were periodically removed from the reaction mixture and quenched with SDS-PAGE loading buffer. Samples were resolved by SDS-PAGE and subjected to phosphorimaging analysis to assess the acetylation state of Acs after incubation with CobBS or CobBL.
Oligomeric state analysis of CobBL and CobBS.
Gel filtration was performed using a HiPrep 26/60 Sephacryl S-100 high-resolution column (GE Healthcare) connected to a computer-controlled ÄKTA FPLC system. The column was equilibrated with sodium phosphate buffer (50 mM, pH 7.5) containing NaCl (150 mM). CobBL (0.8 mg) or CobBS (1 to 2 mg) was applied to the column, which was developed isocratically at a rate of 2 ml min−1. Molecular weight calibrations were performed using the ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) components of the Bio-Rad gel filtration standards. The above standards were supplemented with bovine serum albumin (66.4 kDa; Promega) and DNase I (31 kDa; Sigma).
RNA isolation.
For Northern blot analysis, RNA was isolated as described previously (1). S. enterica cultures were grown in NB to an OD650 of 0.8 with shaking at 37°C. A 520-μl volume of cells was added to 8× lysis buffer and phenol-water (3.75:1; Invitrogen), and the mixture was incubated at 65°C with shaking at 500 rpm for 20 min. Lysates were centrifuged at 16,000 × g for 10 min at room temperature in an Eppendorf 5415D microcentrifuge, and the aqueous phase was extracted twice with 700 μl phenol-chloroform-isoamyl alcohol (25:24:1; Ambion). RNA was precipitated with 1.3 ml ethanol (100%) at −80°C for at least 30 min. RNA was pelleted by centrifugation at 4°C for 10 min as described above. The pellet was washed with 500 μl ethanol (75%) and centrifuged again for 10 min at 4°C. RNA pellets were allowed to dry at room temperature, and RNA was resuspended in Tris-EDTA buffer, pH 8.0 (Ambion).
To perform 5′ rapid amplification of cDNA ends (5′ RACE) analysis, RNA was isolated using the RNA Protect reagent and the RNeasy Mini kit (Qiagen). Cells were grown to an OD650 of 0.7, and RNA was isolated according to the manufacturer's protocols. RNA was resuspended in RNase-free water.
Northern blot analysis.
RNA (21 μg in 5.5 μl) was mixed with 1 μl MOPS buffer (10×), 3.5 μl formaldehyde, and 10 μl formamide (100%). RNA samples were incubated at 65°C for 15 min to denature the RNA. Samples were incubated on ice for 5 min, and 3 μl RNA agarose gel loading buffer (glycerol [50%], EDTA [1 mM], bromophenol blue [0.25%], xylene cyanol [0.25%]) was added. Samples were separated on a denaturing agarose gel (MOPS [1×], formaldehyde [2.2 M], agarose [1%]) with a Millennium marker (Ambion) run at 45 V for approximately 3 h. Gels were prepared for transfer as described previously (6).
RNA was transferred to 0.2-μm Hybond N+ membrane (GE Healthcare) by capillary transfer for 16 h. RNA was linked to the membrane by baking for 2 h at 80°C. Membranes were stained for 45 s for total RNA with methylene blue (0.03%) in sodium acetate (0.3 M, pH 5.2). Membranes were then destained for 2 min with diethyl pyrocarbonate-treated water. Membranes were prehybridized with Ultrahyb (Ambion) at 42°C for at least 30 min. The membrane was probed overnight with a 32P-radiolabeled single-stranded DNA probe (Table 2). Membranes were washed as described previously (6) and subjected to phosphorimager analysis to visualize the relative mobilities of the cobB and ycfX transcripts. The phosphorimager and methylene blue-stained membrane images were overlaid using Adobe Photoshop CS2 (Adobe Systems, Inc.) to determine the relative mobilities of the cobB and ycfX transcripts.
5′ RACE protocol.
5′ RACE was performed using the Version 2 5′ RACE system from Invitrogen according to the manufacturer's instructions. Nested, cobB-specific primers were used to synthesize the cDNA and for the subsequent amplification reactions. After the second round of amplification, the PCR products were separated by agarose gel electrophoresis and gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Fragments were sequenced using a cobB-specific primer to determine the 5′ end of the cobB transcript using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's protocol. Reaction mixtures were resolved and analyzed at the University of Wisconsin Biotechnology Center.
RESULTS AND DISCUSSION
Multiple start codons of sirtuins are conserved in Enterobacteriaceae.
An alignment of cobB homologues from representative members of the family Enterobacteriaceae suggested that most of these bacteria might synthesize two isoforms of CobB, a short (CobBS) and a long (CobBL) isoform (Fig. 1). The one exception we found was the plant pathogen E. amylovora, whose genome sequence appeared to direct the synthesis of only one form, CobBL; this feature was conserved within all sequenced Erwinia species (data not shown). Notably, all of the N-terminal extensions of the putative CobBL isoforms synthesized by enterobacteria were amphipathic in nature, with many conserved basic residues (Arg and Lys, highlighted in gray), and in several instances, additional Arg residues were present in the extension but were not conserved in all genera. The extended peptide of the S. enterica CobBL isoform contained 17 basic amino acids (12 Arg, 2 Lys, 2 Gln, and 1 Asn; 17/37 = 46%) and 11 hydrophobic amino acids (4 Phe, 4 Leu, 2 Val, and 1 Ile; 11/37 = 30%).
FIG. 1.
CobBL and the positively charged N-terminal region are conserved among the members of the family Enterobacteriaceae. The N-terminal regions of CobB homologues from the family Enterobacteriaceae were aligned. The CobBL and CobBS starting methionines are highlighted in black. Conserved, positively charged residues (Arg and Lys) are highlighted in gray.
S. enterica synthesizes short and long isoforms of CobB sirtuin.
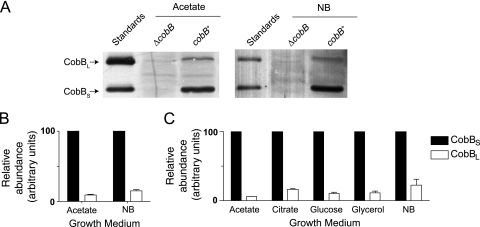
The cobB gene of S. enterica contains two putative translation initiation sites within the first 38 codons (M1 and M38, respectively; Fig. 1). Although another potential start codon, M37, lies juxtaposed to M38, we considered M38 to be the biologically relevant start codon due to its conservation in other enterobacteria (Fig. 1). To determine whether different CobB isoforms are synthesized from the putative M1 and M38 start codons, we grew S. enterica in rich medium (NB) or in NCE minimal medium with acetate (50 mM) as the sole source of carbon and energy. Under all of these conditions, S. enterica expressed two isoforms of CobB which matched the predicted size of purified CobB standards (Fig. 2A). Samples were serially diluted, and densitometry was used to quantify the relative abundance of each isoform on Western blot assays. As shown in Fig. 2B, the shorter, 236-amino-acid CobBS isoform (start at M38) was expressed at levels approximately 7- or 11-fold higher than the longer 273-amino-acid CobBL isoform (start at M1) on NB and acetate, respectively. Previous work demonstrated that cobB is translated most highly at the mid-log and late-log stages of growth (44). Therefore, we assessed relative CobBL and CobBS levels during mid-log phase in S. enterica grown in NB or minimal medium supplemented with acetate, citrate, glucose, or glycerol (50 mM) (see Fig. S1 in the supplemental material). Under these conditions, CobBL levels were about 5- to 20-fold higher than CobBS levels (Fig. 1C). The expression of these two CobB isoforms at differing levels suggested a previously unknown level of regulation of the CobB sirtuin in S. enterica.
FIG. 2.
S. enterica synthesizes two isoforms of CobB. Western blot analysis was performed using rabbit anti-CobB antiserum on whole-cell lysates of wild-type and ΔcobB mutant strains grown to stationary phase on NB and 50 mM acetate (A). The purified SeCobBL (31.1 kDa) and SeCobBS (26.2 kDa) proteins were included as molecular mass standards. Relative quantities of SeCobBL and SeCobBS were determined for cells grown to stationary phase (B) or mid-log phase (C) in NB or minimum medium supplemented with acetate, citrate, glucose, or glycerol (50 mM each). Quantities were normalized to SeCobBS levels, which were assigned a value of 100 arbitrary units.
The CobBL and CobBS isoforms deacetylate AcsAc in vitro.
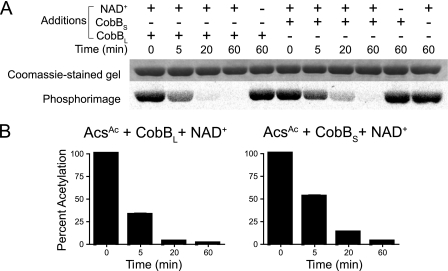
CobB sirtuin is the only annotated NAD+-dependent deacetylase of S. enterica. Previous in vitro work with CobBS demonstrated that this isoform efficiently deacetylates acetylated acetyl-CoA synthetase (AcsAc) in vitro (36). However, prior to this work, the ability of CobBL to perform this reaction had not been assessed. To assess the deacetylase activity of CobBL, S. enterica Acs was acetylated with radiolabeled acetyl-CoA, and the product of the reaction was used as the substrate for in vitro deacetylation activity assays. The decreasing amount of radiolabel associated with Acs over time showed that both CobBL and CobBS deacetylated AcsAc in vitro in a NAD+-dependent manner (Fig. 3).
FIG. 3.
SeCobBL and SeCobBS deacetylate AcsAc in vitro. SeCobBL or SeCobBS was incubated with NAD+ and AcsAc that was previously acetylated by Pat and [1-14C]acetyl-CoA. Samples were removed as a function of time and quenched with gel loading buffer. Proteins were resolved by SDS-PAGE and analyzed by phosphorimaging (A). The amount of radiolabeled AcsAc remaining at each time point was quantified by densitometry (B).
CobBL and CobBS deacetylate AcsAc in vivo.
To determine whether CobBL and CobBS are active in vivo, we used sirtuin-deficient S. enterica strain JE12939 (ΔcobB ΔcobT). To assess the activity of the two CobB isoforms in vivo, we relied on the role of CobB during growth on 10 mM acetate (38). Under such conditions, acetate activation depends on Acs activity, which is regulated by reversible Nɛ-Lys acetylation (36). The lack of cobB function in strain JE12939 prevents deacetylation and reactivation of AcsAc (36, 38). Therefore, growth of strain JE12939 on 10 mM acetate would only occur if CobBL or CobBS were active, that is, if AcsAc were deacetylated.
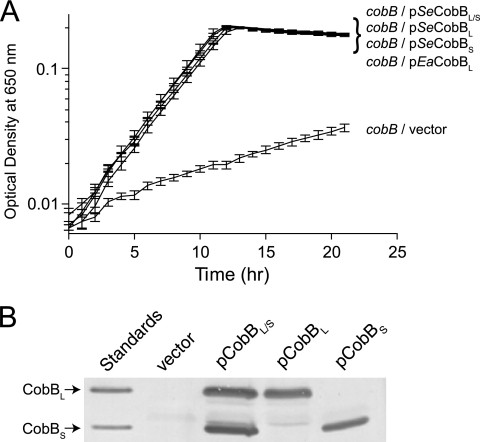
To assess CobBS and CobBL activity in vivo, we introduced plasmids that encode arabinose-inducible alleles cobB1372 (CobBM37A M38A = CobBL) and cobB1373 (CobBM1A = CobBS) into strain JE12939 (ΔmetE ΔcobB ΔcobT). In Fig. 4A, we show that CobBL and CobBS restored growth of strain JE12939 on 10 mM acetate equally well. Western blot analysis confirmed that each plasmid expressed the expected CobB isoform (Fig. 4B). As expected, strain JE12939 failed to grow in the absence of CobB, a phenotype that was corrected by the introduction of the cobB+ allele (positive control). Together, the data support the conclusion that CobBL and CobBS are functional in vivo.
FIG. 4.
SeCobBL, SeCobBS, and EaCobBL proteins restore growth of an S. enterica cobB mutant strain on low acetate. (A) Growth behavior of S. enterica on MOPS minimal medium supplemented with acetate (10 mM). Growth experiments were performed at 37°C using a microtiter plate and a microtiter plate reader (Bio-Tek Instruments). (B) CobB Western blot analysis of S. enterica cobB strains overproducing SeCobBL, SeCobBS, or CobBL/S during growth on acetate (10 mM). SeCobBL is CobBM37A M38A, encoded by cobB1372; SeCobBS is CobBM1A, encoded by cobB1373.
To determine whether the ability of CobBL to deacetylate AcsAc was conserved among the members of the family Enterobacteriaceae, we asked whether the E. amylovora cobB gene could restore growth of S. enterica strain JE12939 on 10 mM acetate. As shown in Fig. 4, the E. amylovora CobBL (EaCobBL) protein supported growth of S. enterica strain JE12939 on 10 mM acetate.
The CobBL and CobBS isoforms have phosphoribosyltransferase activity.
It has been known for some time that CobB compensates for the lack of the nicotinate mononucleotide:5,6-dimethylbenzimidazole (DMB) phosphoribosyltransferase (CobT) enzyme during the synthesis of coenzyme B12 from its precursor cobinamide (40, 41). It was not known, however, whether both isoforms of CobB could compensate for CobT. Thus, we determined whether S. enterica CobBL (SeCobBL), SeCobBS, and EaCobBL can restore adenosylcobalamin biosynthesis in a strain that carries a chromosomal deletion of the cobT gene. To ensure that the plasmid was the only source of CobB protein, we also deleted the chromosomal copy of cobB. Thus, the S. enterica JE12939 strain (ΔmetE ΔcobB ΔcobT) used in this analysis was a cobalamin auxotroph because it cannot synthesize adenosylcobalamin from the precursors cobinamide and DMB. Hence, growth of strain JE12939 on glycerol-cobinamide-DMB would only occur if SeCobBL, SeCobBS, or EaCobBL could activate DMB. Derivatives of strain JE12929 that synthesized SeCobBL, SeCobBS, or EaCobBL grew on minimal medium supplemented with glycerol (22 mM), dicyanocobinamide (15 nM), and DMB (125 μM) at the following very similar doubling times: JE12939/pSeCobBL, 1.2 ± 0.05 h; JE12939/pSeCobBS, 1.1 ± 0.12 h; JE12939/pEaCobBL, 1.5 ± 0.04 h; JE12939/vector, 13.3 ± 0.04 h. The above data show that all of the forms of CobB tested activated DMB and restored adenosylcobalamin biosynthesis in an S. enterica strain lacking the CobT enzyme. Results from Western blot analysis confirmed that the plasmids used in this experiment expressed the expected CobB isoform (see Fig. S2 in the supplemental material).
Initial characterization of the CobBL N terminus.
Previous work with the yeast sirtuin Hst2 demonstrated that an N-terminal extension to the core sirtuin domain could mediate sirtuin oligomerization (46). Gel filtration analysis of purified SeCobBL and SeCobBS demonstrated that both isoforms exist as monomers under the conditions tested (see Fig. S3 in the supplemental material). Thus, the N-terminal extension of SeCobBL does not mediate sirtuin oligomerization under the conditions tested.
Amphipathic helices have been demonstrated to allow the passage of small peptides through membranes (15). Because the N terminus of CobB is amphipathic in composition, we used secondary-structure prediction software to determine if this structure is capable of forming an amphipathic helix. The software package JPred3 (9) predicts that residues 6 to 28 form an alpha helix. However, a helical wheel projection of the predicted helix (see Fig. S4 in the supplemental material) suggested that this region of SeCobBL had charged residues evenly dispersed around the central axis of the helix and would not exist as a canonical amphipathic helix in which one face along the axis is hydrophobic while the other face is hydrophilic (32, 39).
In S. enterica, cobB is transcribed from two promoters independent of the neighboring ycfX gene.
To gain insights into the mechanisms that control the synthesis of CobB isoforms in S. enterica, we investigated whether the cell synthesized ≥1 cobB mRNA. We note that, as currently ascribed, cobB is located 18 nt 3′ of ycfX, a homolog of the nagK gene that in E. coli encodes N-acetylglucosamine kinase (42). The proximity of cobB to ycfX suggested that cobB and ycfX might constitute an operon. In fact, information available online describes nagK (ycfX) cobB as an operon (http://regulondb.ccg.unam.mx/gene?term=ECK120003173&organism=ECK&format=jsp). Although Uehara and Park suggested that a promoter for cobB lies within nagK, the idea was not pursued (42).
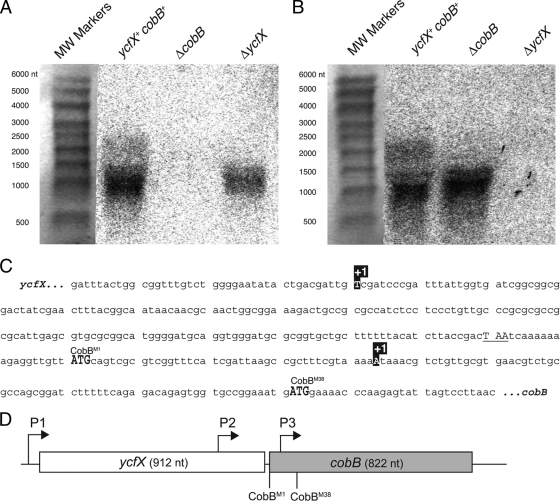
We used Northern blot analysis to determine the length of the cobB transcript in S. enterica. If cobB (822 bp) and ycfX (912 bp) were cotranscribed, we expected a >1.7-kb transcript. Using a cobB-specific probe, we identified a dominant transcript approximately 1,000 nt in size, which suggested that the transcript spanned only cobB (Fig. 5A). Noteworthy was the small amount of signal appearing around 1,800 nt that was detected using cobB-specific and ycfX-specific probes (Fig. 5A and B) but was absent in either the ΔycfX or the ΔcobB mutant strain, which suggested that a small amount of CobB was synthesized from a ycfX-cobB transcript.
FIG. 5.
The cobB gene has two promoters independent of the ycfX promoter. Northern blot assays of wild-type (cobB+ ycfX+) and ΔcobB and ΔycfX mutant strains probing for cobB (A) or ycfX (B) transcripts. RNA Millennium markers were used as molecular weight standards. The phosphorimage was overlaid over the total RNA-stained membrane to align the lanes. (C) Identities of the cobB transcription start sites within the ycfX and cobB ORFs. The transcription start sites are designated +1. The CobB translation start sites are highlighted in gray. The ycfX stop codon is underlined. (D) Model of cobB promoters P1, P2, and P3 (drawn to scale). Assignment of promoters was based on Northern blot analysis (P1) or Northern blot analysis and transcription start site data (P2 and P3). The CobBL start codon is designated CobBM1. The CobBS start codon is designated CobBM38.
We used 5′ RACE analysis to determine the transcription start sites of cobB. PCR amplification of the 5′ RACE products revealed two transcripts of different lengths. Although we refer to the 5′ mRNA ends identified from independent replicates as transcription start sites, the 5′ RACE protocol used does not distinguish between transcription start sites and mRNA processing sites. The sequences of these 5′ RACE products revealed a transcription site 5′ of each translation start site (Fig. 5C). One transcription start site was located within ycfX (nt 731), while the other was located within cobB (nt 44). The precise cobB transcription start site located within ycfX was somewhat ambiguous because the 5′ RACE protocol resulted in a poly(G) tail on the 5′ end of the transcript. It is possible that the G at the −1 site relative to the upstream cobB promoter could be the +1 nucleotide of the upstream cobB transcription start site.
A schematic of the cobB promoters is shown in Fig. 5D. Promoters were assigned based on the Northern blot analysis (for the ycfX-cobB promoter, P1) or Northern blot and 5′ RACE analyses (for the two promoters initiating monocistronic cobB mRNAs at P2 and P3). A small amount of cobB mRNA is transcribed from P1, and the P1-derived transcript contains the entire cobB ORF. The P2-derived transcript encompasses both of the ORFs that encode CobBL and CobBS. CobBL is likely synthesized from the P2-derived transcript, but it is unclear whether or not CobBS is synthesized from this transcript. Promoter P3 exists within the cobB ORF. The P3-derived transcript only spans the ORF that encodes CobBS, suggesting that only CobBS can be synthesized from this transcript. By separating the expression of each cobB isoform with independent transcription start sites, S. enterica could differentially regulate each transcription start site and thus each CobB isoform.
Notably, when the cobB ORF is expressed from the PBAD promoter, levels of CobBL and CobBS seem to be equalized (Fig. 4B) relative to when the cobB allele is expressed from the chromosome (Fig. 2A). The cobB complementation vector pCobBWT lacks the cobB P1 and P2 promoters but does contain the vector-based PBAD promoter and cobB P3. Therefore, CobBL/CobBS ratios seem to be regulated, in part, by the copy number of the cobB allele and/or the identity of the promoters driving cobB expression.
Contributions from this work.
This is the first report of sirtuin isoforms in bacteria or archaea. The existence of sirtuin isoforms in a single bacterium suggests that these enzymes have more than one physiological role. We have shown that S. enterica synthesizes two biologically active isoforms of the CobB sirtuin deacetylase, which are synthesized from two sets of in-frame start codons. Long and short CobB sirtuin isoforms appear to be common among the members of the family Enterobacteriaceae, with the notable exception of the members of the genus Erwinia, which appear to synthesize only the long sirtuin isoform. Our data show that the CobBL isoform of E. amylovora is biologically active (Fig. 4). In S. enterica, the two CobB isoforms do not appear to be the result of processing of CobBL into CobBS; instead, the isoforms are synthesized from alternative start codons. Examination of cobB gene expression revealed an additional level of regulation in which multiple cobB mRNAs are synthesized from transcription start sites located 5′ of each cobB translation initiation site (Fig. 5).
What roles could multiple isoforms of CobB be playing in S. enterica?
Unique roles for CobBL and CobBS are not immediately obvious. It is striking that, among the bacterial CobBL isoforms we analyzed (Fig. 1), on average, 37% of the residues in the CobBL extension are Lys or Arg, with 78% of the positively charged residues being Arg. The structure of CobB from the enterobacterium E. coli has been determined (Protein Data Bank code 1S5P) (47); however, since the structure was solved using CobBS, it is not clear what the structure of the N terminus of CobBL is or how it could be interacting with other macromolecules in the bacterial cell. Since E. amylovora is only capable of synthesizing CobBL, it seem that this isoform is the biologically relevant isoform for this organism and is capable of carrying out all necessary CobBL functions in E. amylovora.
Although both isoforms of CobB can deacetylate AcsAc, AcsAc may not be the only physiologically relevant substrate for both isoforms. Recent work suggests that approximately 191 proteins in S. enterica are modified by acetylation (44), and putative protein partners of CobB in E. coli have been identified using a multiprotein complex purification procedure (7). The N-terminal region of CobB may be important for mediating interaction with some of these putative acetylated protein substrates. Alternatively, the positively charged N terminus of CobB may also be needed to interact with other molecules or structures in the cell. Nonribosomal, Arg-rich peptides with amino acid compositions similar to that of the N terminus of CobBL have been implicated in protein-RNA interactions (2, 25, 45). Thus, the N terminus of CobBL could bind nucleic acids or target the isoform to another negatively charged molecule of the cell. Thus, many questions remain unanswered. For example, (i) why are the levels of CobBL and CobBS different; (ii) does the positively charged N terminus of CobBL affect its activity, localization, or substrate specificity; and (iii) are there any physiological conditions under which either isoform is essential? These questions need to be addressed as we broaden our understanding of the physiological role of bacterial sirtuins.
Supplementary Material
Acknowledgments
This work was supported by PHS grant R01-GM062203 to J.C.E.-S. A.C.T. was supported in part by the University of Wisconsin—Madison Molecular Biosciences Training Grant (MBTG; 5 T32 GM007215-35) from the National Institutes of Health.
V. Starai constructed plasmids pCOBB19 and pCOBB24. We thank N. Perna of the University of Wisconsin—Madison Genetics Department for her gift of E. amylovora DNA.
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 2.Bayer, T. S., L. N. Booth, S. M. Knudsen, and A. D. Ellington. 2005. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11:1848-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch, K. D., and B. Grossman. 1995. Digestion of DNA with restriction endonucleases, p. 3.1.1-3.1.21. In F. M. Ausubel, R. E. R. Brent, D. D. Kingston, J. G. Moore, J. A. Seidman, A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, NY.
- 5.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 6.Brown, T., K. Mackey, and T. Du. 2004. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.1-4.1.19. In F. M. Ausubel, R. E. R. Brent, D. D. Kingston, J. G. Moore, J. A. Seidman, A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, NY. [DOI] [PubMed]
- 7.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 8.Cockell, M. M., S. Perrod, and S. M. Gasser. 2000. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154:1069-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosby, H. A., E. K. Heiniger, C. S. Harwood, and J. C. Escalante-Semerena. 2010. Reversible N(E)-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol. 76:874-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuperus, G., R. Shafaatian, and D. Shore. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19:2641-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbing, K., and R. Brent. 2002. Media preparation and bacteriological tools, p. 1.1.3. In F. M. Ausubel, R. E. R. Brent, D. D. Kingston, J. G. Moore, J. A. Seidman, A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, NY. [DOI] [PubMed]
- 14.Elion, E. A., P. Marina, and L. Yu. 2007. Constructing recombinant DNA molecules by PCR, p. 3.17.1-3.17.12. In F. M. Ausubel, R. E. R. Brent, D. D. Kingston, J. G. Moore, J. A. Seidman, A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, NY. [DOI] [PubMed]
- 15.Futaki, S., T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda, and Y. Sugiura. 2001. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 276:5836-58340. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, J. G., and J. C. Escalante-Semerena. 2009. In Bacillus subtilis, the sirtuin protein deacetylase encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl-CoA synthetase. J. Bacteriol. 191:1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity, J., J. G. Gardner, W. Hawse, C. Wolberger, and J. C. Escalante-Semerena. 2007. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282:30239-30245. [DOI] [PubMed] [Google Scholar]
- 18.Guarente, L. 2007. Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72:483-488. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou, Z., J. R. Danzer, L. Mendoza, M. E. Bose, U. Muller, B. Williams, and C. A. Fox. 2009. Phylogenetic conservation and homology modeling help reveal a novel domain within the budding yeast heterochromatin protein Sir1. Mol. Cell. Biol. 29:687-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 22.Kwon, H. S., and M. Ott. 2008. The ups and downs of SIRT1. Trends Biochem. Sci. 33:517-525. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278:685-690. [DOI] [PubMed] [Google Scholar]
- 25.Lazinski, D., E. Grzadzielska, and A. Das. 1989. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59:207-218. [DOI] [PubMed] [Google Scholar]
- 26.Li, R., J. Gu, Y. Y. Chen, C. L. Xiao, L. W. Wang, Z. P. Zhang, L. J. Bi, H. P. Wei, X. D. Wang, J. Y. Deng, and X. E. Zhang. 2010. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 76:1162-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. U. S. A. 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD(+)-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 30.Rocco, C. J., K. L. Dennison, V. A. Klenchin, I. Rayment, and J. C. Escalante-Semerena. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segrest, J. P., H. De Loof, J. G. Dohlman, C. G. Brouillette, and G. M. Anantharamaiah. 1990. Amphipathic helix motif: classes and properties. Proteins 8:103-117. [DOI] [PubMed] [Google Scholar]
- 33.Shih, Y. P., H. C. Wu, S. M. Hu, T. F. Wang, and A. H. Wang. 2005. Self-cleavage of fusion protein in vivo using TEV protease to yield native protein. Protein Sci. 14:936-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, B. C., W. C. Hallows, and J. M. Denu. 2008. Mechanisms and molecular probes of sirtuins. Chem. Biol. Interact. 15:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U. S. A. 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 37.Starai, V. J., and J. C. Escalante-Semerena. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340:1005-1012. [DOI] [PubMed] [Google Scholar]
- 38.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 40.Trzebiatowski, J. R., G. A. O'Toole, and J. C. Escalante-Semerena. 1994. The cobT gene of Salmonella typhimurium encodes the NaMN:5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-alpha-d-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 176:3568-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsang, A. W., and J. C. Escalante-Semerena. 1998. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 273:31788-31794. [DOI] [PubMed] [Google Scholar]
- 42.Uehara, T., and J. T. Park. 2004. The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 186:7273-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venembre, P., N. Seta, A. Boutten, M. Dehoux, M. Aubier, and G. Durand. 1994. Comparison of enhanced chemiluminescence and colorimetric techniques for the immuno-detection of alpha 1-antitrypsin. Clin. Chim. Acta 227:175-184. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Q., Y. Zhang, C. Yang, H. Xiong, Y. Lin, J. Yao, H. Li, L. Xie, W. Zhao, Y. Yao, Z. B. Ning, R. Zeng, Y. Xiong, K. L. Guan, S. Zhao, and G. P. Zhao. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss, M. A., and N. Narayana. 1998. RNA recognition by arginine-rich peptide motifs. Biopolymers 48:167-180. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, K., X. Chai, A. Clements, and R. Marmorstein. 2003. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 10:864-871. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, K., X. Chai, and R. Marmorstein. 2004. Structure and substrate binding properties of CobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 337:731-741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.