Abstract
Resistance to linezolid (LZD) occurs through mutations in 23S rRNA and ribosomal proteins L3 and L4 or through methylation of 23S rRNA by Cfr. Here we report novel L3 mutations, ΔSer145/His146Tyr and ΔMet169-Gly174, co-occurring with cfr in LZD-resistant Staphylococcus aureus isolates recovered from a hospital outbreak in Madrid, Spain. LZD MIC values (16, 32, or 64 μg/ml) correlated with the presence and severity of the L3 mutation. All isolates had TR-700 (torezolid) MIC values of ≤2 μg/ml.
Linezolid (LZD) resistance was first associated with mutations in the domain V region of 23S rRNA genes (G2576T) (7, 30). Over time, a variety of 23S rRNA mutations have been identified, and these remain the most commonly reported class of mutation leading to LZD resistance (5, 9, 10). In rare cases, mutations in ribosomal protein L4 have also been associated with LZD resistance (8, 17, 32). More recently, a variety of mutations in ribosomal protein L3 have also been identified in both laboratory and clinically derived staphylococci associated with reduced susceptibility to oxazolidinones (15-17). Cfr-based LZD resistance, however, is potentially more worrisome than mutation-based chromosomally encoded resistance mechanisms (25, 29). The cfr gene encodes a methyltransferase which confers LZD resistance via methylation of carbon-8 on 23S rRNA base A2503 (6). Cfr is generally plasmid borne and transposon associated and therefore likely to be horizontally transmitted (11). Strains carrying cfr are resistant not only to LZD but also to phenicols, lincosamides, pleuromutilins, and streptogramin A class antibiotics (19), as well as 16-membered ring macrolides (28). Thus, selective pressure due to the use of any of these drug classes may lead to the spread of this resistance determinant.
The emergence of cfr and identification of additional LZD resistance mechanisms, including L3 mutations, raise the potential for multiple mechanisms to occur within a single strain. Our previous work identified coupled 23S rRNA and L3 mutations in both a laboratory LZD serially passaged Staphylococcus aureus strain (17) and a clinical Staphylococcus epidermidis isolate (15). Another recent report documented a Spanish outbreak of LZDr strains (S. aureus, S. epidermidis, Enterococcus faecium, and Enterococcus faecalis) which included strains possessing cfr alone, 23S rRNA mutations alone, or 23S rRNA G2576T mutations in conjunction with cfr (2). Finally, an outbreak of LZDr S. epidermidis in Ohio was comprised of isolates possessing an L4 mutation in conjunction with either the cfr gene or mutations in 23S rRNA (1). These reports are the first to demonstrate the co-occurrence of cfr with any other LZD resistance mechanism; however, given the low fitness cost of Cfr methylation (12), strains with resistance due to multiple mechanisms may not be unexpected. Characterization of such isolates and the evaluation of antibacterial activities of clinically relevant oxazolidinones are thus of high interest.
Previous reports of S. aureus cfr-positive clinical isolates and laboratory-generated cfr-transformed S. aureus strains typically cite LZD MIC values in the range of 8 to 16 μg/ml (13, 14, 21, 29). Analysis of cfr-positive LZDr MRSA from a 2008 hospital outbreak in Madrid, Spain, identified 18 isolates: 1 environmental and 12 patient intensive care unit (ICU) isolates, 3 patient isolates from other wards, and 2 additional patient isolates predating the outbreak that were identified in a retrospective study (22). LZD MIC values for these strains were 8 (n = 4), 16 (n = 13), or 32 (n = 1) μg/ml (22), all of which are above the LZD breakpoint of 4 μg/ml. This study investigated whether additional oxazolidinone resistance mechanisms could account for the variability in LZD resistance levels among these clinical cfr isolates.
(Portions of this work were presented at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy [J. B. Locke, G. Morales, M. Hilgers, Kedar G. C., S. Rahawi, J. J. Picazo, K. J. Shaw and J. L. Stein, abstr. C1-1431], Boston, MA, 12 to 15 September 2010.)
MICs of clinically relevant oxazolidinones, LZD (ChemPacific, Inc., Baltimore, MD), TR-700 (torezolid, formerly known as DA-7157; Trius Therapeutics, Inc., San Diego, CA), and radezolid (RZD) (RX-1741; Medicilon, Chicago, IL), as well as tiamulin (TIA) (Wako Pure Chemical Industries, Ltd., Richmond, VA), chloramphenicol (CHL) (Sigma-Aldrich Corp., St. Louis, MO), and vancomycin (VAN) (Sigma), were determined via broth microdilution in accordance with CLSI guidelines as previously described (3, 17). Quality control of oxazolidinones was performed via nuclear magnetic resonance (NMR), liquid chromatography-mass spectrometry (LC-MS), and biological activity assays. MIC values reported for each strain/drug combination were determined in at least three independent experiments, all yielding identical results. LZD MIC values for a panel of the 6 representative strains included in this study fell into three groups: group 1 (16 μg/ml; n = 1), group 2 (32 μg/ml; n = 4), or group 3 (64 μg/ml; n = 1), which corresponded to TR-700 MIC values of 0.5, 1, or 2 μg/ml and radezolid MIC values of 4, 4, or 8 μg/ml, respectively (Table 1). Both TR-700 and LZD MIC values in this study were 2-fold higher than those originally reported for these strains (22). As expected, the cfr isolates were resistant to TIA and CHL (Table 1).
TABLE 1.
Characteristics of clinical and laboratory-derived S. aureus strains possessing L3 mutations and/or the cfr methyltransferase gene
| Origin | Strain(s) | Reference or source | PFGE type | Presence of cfr | L3 mutation(s)a | MIC (μg/ml)b |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LZD | TR-700 | RZD | TIA | CHL | VAN | ||||||
| Clinical | Group 1c | 22 | C | + | 16 | 0.5 | 4 | >64 | >64 | 2 | |
| Group 2d | 22 | A, B | + | ΔSer145/His146Tyr | 32 | 1 | 4 | >64 | >64 | 1 | |
| Group 3e | 22 | D | + | ΔMet169-Gly174 | 64 | 2 | 8 | >64 | >64 | 2 | |
| Laboratory | 29213f | ATCC | NAi | − | 2 | 0.5 | 1 | 0.5 | 8 | 1 | |
| 29213-1g | 17 | NA | − | Gly155Arg | 4 | 1 | 1 | 8 | 8 | 1 | |
| 29213-2g | 17 | NA | − | Gly155Arg/Met169Leu | 8 | 2 | 2 | 4 | 8 | 2 | |
| 29213-3g | 17 | NA | − | ΔPhe127-His146 | 8 | 2 | 2 | 4 | 8 | 2 | |
| 29213(p42262)h | This study | NA | + | 16 | 0.5 | 2 | >64 | >64 | 1 | ||
| 29213-1(p42262)h | This study | NA | + | Gly155Arg | 32 | 1 | 4 | >64 | >64 | 2 | |
| 29213-2(p42262)h | This study | NA | + | Gly155Arg/Met169Leu | 64 | 2 | 8 | >64 | >64 | 2 | |
| 29213-3(p42262)h | This study | NA | + | ΔPhe127-His146 | 64 | 2 | 8 | >64 | >64 | 2 | |
Ribosomal protein L3 mutations are given using staphylococcal numbering.
MICs were determined via broth microdilution (CLSI) (3).
Group 1 contained isolate 42262.
Group 2 isolates included 32289, P-978 (environmental), 42292, and 56351.
Group 3 contained isolate 51312.
ATCC 29213 is included as an LZDs control strain and is not isogenic to any of the clinical cfr strains in this study.
29213-1, -2, and -3 L3 mutants were selected in vitro with LZD and/or TR-700 in a previous study.
p42262 is a cfr-containing plasmid isolated from group 1 strain 42262 and is used to transform ATCC 29213 wild type and the three isogenic L3 mutant strains.
NA, not analyzed.
The presence of additional ribosomal mutations was assessed by sequencing the domain V region of all 23S rRNA alleles and the genes encoding L3 (rplC) and L4 (rplD), as previously described (17, 20). No ribosomal mutations were detected in the group 1 representative S. aureus isolate with an LZD MIC value of 16 μg/ml (42262; pulsed-field gel electrophoresis [PFGE] type C) (Table 1). The 4 group 2 isolates with LZD MIC values of 32 μg/ml (32289, 56351, P-978 [environmental isolate], and 42292; PFGE A or B) all possessed the ΔC434-C436 mutation in rplC, translating into a ΔSer145/His146Tyr deletion/transversion mutation in L3. Strain 51312 (PFGE D), the only isolate with an LZD MIC of 64 μg/ml (group 3), possessed a ΔA505-T522 in-frame deletion in rplC, resulting in a 6-amino-acid ΔMet169-Gly174 deletion in L3. The 2- to 4-fold TR-700 MIC shifts for strains with L3 mutations is consistent with previous reports of MIC shifts associated with L3 mutations (15, 17), as is the TR-700 MIC of 0.5 μg/ml against the cfr-only strain (18). Radezolid demonstrates smaller fold shifts than LZD against a variety of ribosomal mutations (27) and against Cfr methylation; however, the presence of the acetamide results in 2- to 4-fold MIC increases (14). Together these resistance mechanisms contribute to 4- to 8-fold-reduced susceptibility to radezolid versus strains possessing both Cfr and L3 resistance mechanisms.
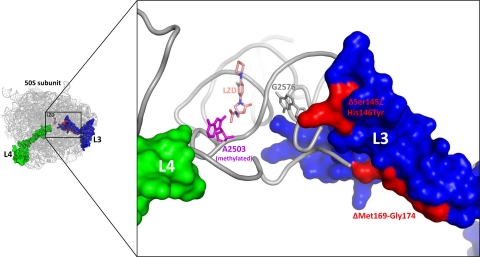
Reduced susceptibility to oxazolidinones has been documented in S. aureus strains possessing mutations in some of the residues involved in these L3 mutations, including ΔSer145 (15, 16), Met169Leu/Gly155Arg (17), and ΔPhe127-His146 (17). The precedence of similar L3 mutations in LZDr strains prompted us to investigate the newly identified mutants through analysis of the crystal structure of the Deinococcus radiodurans LZD-bound 50S subunit (PDB accession code 3DLL) (31). The ΔSer145/His146Tyr mutation abuts a cluster of 23S rRNA bases that include some known to influence those of the peptidyltransferase center (PTC) (e.g., G2576), therefore exerting its effect directly through a small set of intervening bases (Fig. 1). The ΔMet169-Gly174 mutation, although more distant from the PTC, involves the deletion of a significant strand of secondary structure (Fig. 1). In order to accommodate this dramatic deletion, regions of the L3 protein proximal to the PTC must necessarily undergo large rearrangements. These perturbations are likely propagated to the adjacent PTC, resulting in the observed reduction in oxazolidinone binding.
FIG. 1.
Structural analysis of L3 mutations and Cfr methylation in clinical LZDr strains. Ribosomal protein L3 mutations ΔSer145/His146Tyr and ΔMet169-Gly174 (both colored red) occur in close proximity to the LZD binding site in the PTC. 23S rRNA base A2503 (colored magenta) is modeled in a Cfr carbon-8 methylated state (the methyl group points toward the acetamide of LZD). For reference, 23S rRNA base G2576 (colored gray; the most common 23S rRNA base mutated in LZDr strains characterized to date) and ribosomal protein L4 (colored green; infrequent mutations in which are linked to LZD resistance) are shown. Images were generated with the PyMOL software program (4), using the coordinates of the D. radiodurans LZD-bound 50S subunit (31).
In an effort to recapitulate and validate the coupled Cfr plus L3 resistance trends observed in these clinical isolates, we generated a panel of isogenic comparator strains in the S. aureus ATCC 29213 background. The wild-type ATCC 29213 parent strain and three isogenic, laboratory-selected L3 mutants (17) having two different oxazolidinone susceptibility profiles were transformed with the p42262 cfr plasmid (isolated from clinical strain 42262) (23) via electroporation as previously described (24). Transformant MICs mirrored the oxazolidinone MICs observed in the clinical strains, suggesting that the Cfr and Cfr plus L3-based resistance mechanisms are the primary contributing factors to the observed LZD resistance levels in these clinical isolates. Based on the structural analyses and MIC trends from cfr-transformed 29213 L3 mutants, the difference in LZD resistance levels (i.e., 32 versus 64 μg/ml) among some of these clinical strains is likely due to differences in how severely each L3 mutation perturbs the PTC and thus reduces oxazolidinone binding affinity.
Novel mutations in ribosomal protein L3 were detected in each of the S. aureus cfr isolates examined with LZD MIC values of >16 μg/ml. TR-700 was the only oxazolidinone tested with MIC values of ≤2 μg/ml against all strains. Because the breakpoints for TR-700 and radezolid have not been determined, the clinical relevance of MIC values against these strains cannot yet be assessed. Enhanced activity of TR-700 is primarily due to reduced steric hindrance of the TR-700 C-5 hydroxymethyl substituent compared to the acetamide of LZD or radezolid in the presence of Cfr methylation (14, 26). This study is the first to document the co-occurrence of cfr in clinical S. aureus isolates possessing L3 mutations, following reports of cfr coupled with 23S rRNA (2) or L4 (1) mutation-based resistance mechanisms. Cfr methylation has now been shown to be compatible with each of the three documented classes of mutation-based staphylococcal resistance to LZD, thus highlighting the need for next-generation oxazolidinones to maintain activity against a variety of resistance mechanisms.
ADDENDUM IN PROOF
During the review process of this manuscript, work documenting the co-occurence of the cfr gene in clinical Staphylococcus aureus isolates possessing L3 mutations was published (R. E. Mendes, L. Deshpande, E. Rodriguez-Noriega, J. E. Ross, R. N. Jones, and R. Morfin-Otero, J. Clin. Microbiol. 48:3041-3043).
Acknowledgments
We thank Douglas Phillipson and Grayson Hough for quality control analysis of the oxazolidinones.
Gracia Morales was supported by a research contract with the Fundación para la Investigación Biomédica del HCSC.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Bonilla, H., J. P. Quinn, J. Siedel, H. Schmidt, M. Lescoe, S. P. McCurdy, M. M. Lemmon, M. D. Huband, L. Puzniak, T. Hart, K. Jafri, L. A. Brennan, and A. Tait-Kamradt. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-131. American Society for Microbiology, Washington, DC.
- 2.Cercenado, E., M. Marin, R. Insa, and E. Bouza. 2010. Abstr. 20th European Congress of Clinical Microbiology and Infectious Diseases, abstr. P979. European Society of Clinical Microbiology and Infectious Diseases, Munich, Germany.
- 3.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, CLSI Document M7-A7, 7th ed., vol. 26, no. 2. CLSI, Wayne, PA.
- 4.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA.
- 5.Farrell, D. J., R. E. Mendes, J. E. Ross, and R. N. Jones. 2009. Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn. Microbiol. Infect. Dis. 65:392-403. [DOI] [PubMed] [Google Scholar]
- 6.Giessing, A. M., S. S. Jensen, A. Rasmussen, L. H. Hansen, A. Gondela, K. Long, B. Vester, and F. Kirpekar. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzales, R. D., P. C. Schreckenberger, M. B. Graham, S. Kelkar, K. DenBesten, and J. P. Quinn. 2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 8.Holzel, C. S., K. S. Harms, K. Schwaiger, and J. Bauer. 2010. Resistance to linezolid in a porcine Clostridium perfringens strain carrying a mutation in the rplD gene encoding the ribosomal protein L4. Antimicrob. Agents Chemother. 54:1351-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, R. N., S. Kohno, Y. Ono, J. E. Ross, and K. Yanagihara. 2009. ZAAPS International Surveillance Program (2007) for linezolid resistance: results from 5591 Gram-positive clinical isolates in 23 countries. Diagn. Microbiol. Infect. Dis. 64:191-201. [DOI] [PubMed] [Google Scholar]
- 10.Jones, R. N., J. E. Ross, J. M. Bell, U. Utsuki, I. Fumiaki, I. Kobayashi, and J. D. Turnidge. 2009. Zyvox annual appraisal of potency and spectrum program: linezolid surveillance program results for 2008. Diagn. Microbiol. Infect. Dis. 65:404-413. [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg, C., F. M. Aarestrup, and S. Schwarz. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaMarre, J. M., T. Zhu, J. B. Locke, K. J. Shaw, and A. S. Mankin. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1346. American Society for Micriobiology, Washington, DC.
- 13.Lawrence, L., P. Danese, J. DeVito, F. Franceschi, and J. Sutcliffe. 2008. In vitro activities of the Rx-01 oxazolidinones against hospital and community pathogens. Antimicrob. Agents Chemother. 52:1653-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locke, J. B., J. Finn, M. Hilgers, G. Morales, S. Rahawi, K. G. C., J. J. Picazo, W. Im, K. J. Shaw, and J. L. Stein. 2010. Structure-activity relationships of diverse oxazolidinones for linezolid-resistant Staphylococcus aureus strains possessing the cfr methyltransferase gene or ribosomal mutations. Antimicrob. Agents Chemother. 54:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locke, J. B., M. Hilgers, and K. J. Shaw. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53:5275-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locke, J. B., M. Hilgers, and K. J. Shaw. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1349. American Society for Microbiology, Washington, DC.
- 17.Locke, J. B., M. Hilgers, and K. J. Shaw. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke, J. B., A. S. Mankin, J. LaMarre, V. Brown-Driver, and K. J. Shaw. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1364b. American Society for Microbiology, Washington, DC.
- 19.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, and H. S. Gold. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 21.Mendes, R. E., L. M. Deshpande, M. Castanheira, J. Dipersio, M. Saubolle, and R. N. Jones. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morales, G., and J. J. Picazo. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-808. American Society for Microbiology, Washington, DC.
- 23.Morales, G., J. J. Picazo, E. Baos, F. J. Candel, A. Arribi, B. Pelaez, R. Andrade, M. A. de la Torre, J. Fereres, and M. Sanchez-Garcia. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821-825. [DOI] [PubMed] [Google Scholar]
- 24.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz, S., C. Werckenthin, and C. Kehrenberg. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw, K. J., S. Poppe, R. Schaadt, V. Brown-Driver, J. Finn, C. M. Pillar, D. Shinabarger, and G. Zurenko. 2008. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob. Agents Chemother. 52:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skripkin, E., T. S. McConnell, J. Devito, L. Lawrence, J. A. Ippolito, E. M. Duffy, J. Sutcliffe, and F. Franceschi. 2008. Rx-01: a new family of oxazolidinones that overcomes ribosomal-based linezolid resistance. Antimicrob. Agents Chemother. 52:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, L. K., and A. S. Mankin. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toh, S. M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 38:207-208. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, D. N., F. Schluenzen, J. M. Harms, A. L. Starosta, S. R. Connell, and P. Fucini. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U. S. A. 105:13339-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolter, N., A. M. Smith, D. J. Farrell, W. Schaffner, M. Moore, C. G. Whitney, J. H. Jorgensen, and K. P. Klugman. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]