Abstract
Imatinib, a BCR-Abl inhibitor, is a successful front-line treatment for chronic myelogenous leukemia (CML). Despite the success of imatinib, multiple mechanisms of resistance remain a problem, including overexpression of Lyn kinase (Lyn) and Bcl-2 family antiapoptotic proteins. Profiling micro-RNA (miRNA) expression in a model of Lyn-mediated imatinib-resistant CML (MYL-R) identified approximately 30 miRNAs whose expression differed >2-fold compared with drug-sensitive MYL cells. In particular, the expression of the miR181 family (a–d) was significantly reduced (∼11- to 25-fold) in MYL-R cells. Incubation of MYL-R cells with a Lyn inhibitor (dasatinib) or nucleofection with Lyn-targeting short interfering RNA increased miR181b and miR181d expression. A similar Lyn-dependent regulation of miR181b and miR181d was observed in imatinib-resistant K562 CML cells. Sequence analysis of potential targets for miR181 regulation predicted myeloid cell leukemia-1 (Mcl-1), a Bcl-2 family member whose expression is increased in MYL-R cells and drug-resistant leukemias. Inhibition of Lyn or rescue of miR181b expression reduced Mcl-1 expression in the MYL-R cells. To further investigate the mechanism of Mcl-1 repression by miR181, a luciferase reporter construct incorporating the Mcl-1 3′-untranslated region was tested. Overexpression of miR181b reduced luciferase activity, whereas these effects were ablated by the mutation of the seed region of the miR181 target site. Finally, stimulation of Lyn expression by 1,25-dihydroxyvitamin D3 treatment in HL-60 cells, a cell model of acute myelogenous leukemia, decreased miR181b expression and increased Mcl-1 expression. In summary, our results suggest that Lyn-dependent regulation of miR181 is a novel mechanism of regulating Mcl-1 expression and cell survival.
Introduction
The discovery and application of the BCR-Abl inhibitor imatinib has been a major hallmark in the development of kinase inhibitors for cancer chemotherapy. However, despite the success of imatinib for the treatment of chronic myelogenous leukemia (CML) and other cancers, multiple mechanisms of imatinib resistance have been identified. These include BCR-Abl mutations that prevent imatinib binding (i.e., T315I) (Hochhaus et al., 2002; Yamamoto et al., 2004), BCR-Abl overexpression (Hochhaus et al., 2002), and increased expression and activity of Src family kinases (SFKs) or other prosurvival proteins (Illmer et al., 2004; Li, 2008). Lyn kinase (Lyn) has been implicated in imatinib resistance in both CML cells and patient samples. Overexpression of Lyn, the most abundant SFK in hematopoietic cells, may contribute to drug resistance through increased signal transducer and activator of transcription 5 phosphorylation, Bcl-2 expression, and other prosurvival responses (Donato et al., 2003; Dai et al., 2004; Nam et al., 2007). Inhibition of Lyn with SFK inhibitors reduced prosurvival signaling and reversed imatinib resistance in CML cells (Ito et al., 2007; Nam et al., 2007). Moreover, the dual-specificity BCR-Abl/SFK inhibitors (i.e., dasatinib, sorafenib, nilotinib) effectively treat patients who are nonresponsive to imatinib therapy (Li, 2008; Wu et al., 2008).
MicroRNAs (miRNAs) are small (22–24 nucleotides) noncoding RNA molecules that are key regulators of protein expression through their targeted binding to specific mRNAs. By forming a double-stranded RNA duplex with target mRNAs in the RNA-induced silencing complex, miRNAs trigger the degradation of the mRNA transcript or directly inhibit protein translation (Ambros, 2001). More than 700 miRNAs have been described in humans, and patterns of deletion, down-regulation, or up-regulation of specific miRNAs have been characterized in B-cell chronic lymphocytic leukemias, acute myelogenous leukemia (AML), and CML (Calin et al., 2005; Venturini et al., 2007; Dixon-McIver et al., 2008). Recent studies have demonstrated the importance of miR181 (a–d) expression in AML and chronic lymphocytic leukemia. MiR181a is involved in hematopoietic differentiation (Chen et al., 2004), and loss of miR181 strongly correlates with a common AML morphological subtype (Debernardi et al., 2007). Moreover, high miR181 (a–d) expression is prognostic for the achievement of complete remission and event-free survival in patients with AML (Marcucci et al., 2008; Schwind et al., 2009).
Bcl-2 family members are important prosurvival regulators of apoptosis that have been implicated in the promotion of drug resistance in cell models of leukemia (Shangary and Johnson, 2003; Dai et al., 2004; Bagrintseva et al., 2005). Myeloid cell leukemia-1 (Mcl-1) is a Bcl-2 family protein shown to correlate with leukemic relapse in AML and has been directly linked to resistance to chemotherapy (Kaufmann et al., 1998). In addition, Mcl-1 was recently implicated in AML survival in response to FLT-3 internal tandem duplications, a common mechanism of resistance in AML that involves the activation of Lyn (Okamoto et al., 2007; Breitenbuecher et al., 2009). Mcl-1 functions at the mitochondria by sequestering the proapoptotic BH3-only proteins Bim and Noxa, thereby preventing cytochrome c release and cell death (Warr and Shore, 2008). Mechanisms to inhibit Mcl-1 function include ubiquitination and degradation directed by the MULE/LASU1 E3-ligase (Zhong et al., 2005), and targeting of Mcl-1 mRNA for degradation by miRNA has been described previously (Mott et al., 2007; Chen et al., 2009; Crawford et al., 2009).
In this study, we describe a novel mechanism by which Lyn may confer multidrug resistance in a cell model of CML by repressing the expression of miR181. As our data show, miR181b directly represses Mcl-1 expression, and the Lyn-dependent loss of miR181 results in enhanced Mcl-1 levels and increased drug resistance. Thus, this is the first demonstration, to our knowledge, of Mcl-1 as a bona fide target of miR181 and suggests that the regulation of miR181 by Lyn in drug-resistant cells may contribute to this important antiapoptotic mechanism.
Materials and Methods
Cell Culture and Reagents.
MYL and MYL-R human CML cells were a generous gift from Dr. Hideo Tanaka (Department of Hematology and Oncology, Hiroshima University, Hiroshima, Japan). K562-R cells were obtained from Dr. Steven Grant [Massey Cancer Center, Virginia Commonwealth University, Richmond, VA; K562-R(1)]. A separate isolate of imatinib-resistant K562 cells was kindly provided by Dr. Nicholas Donato [Department of Internal Medicine, University of Michigan; K562-R(2)]. HL-60 cells were obtained from the University of North Carolina Tissue Culture Facility (Chapel Hill, NC). Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Norcross, GA) and 1% antibiotic/antimycotic (Invitrogen). The imatinib-resistant cells (MYL-R, K562R) were not continuously cultured in the presence of imatinib; however, imatinib resistance and Lyn overexpression was routinely checked and found to be stably maintained. HEK293T cells were a generous gift from Dr. Channing Der (Department of Pharmacology, University of North Carolina, Chapel Hill, NC) and were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic. Cell cultures were maintained and incubated with drug at 37°C in a 5% CO2 humidified atmosphere. Imatinib and dasatinib were purchased from LC Laboratories (Woburn, MA). SMARTpool siRNA was purchased from Thermo Fisher Scientific (Waltham, MA). 1,25-Dihydroxyvitamin D3 (1,25-D3) was purchased from Enzo Life Sciences International (Plymouth Meeting, PA).
Nucleofection.
Plasmids and siRNA were incorporated into cells using a nucleofection system (Amaxa Biosystems, Gaithersburg, MD; >90% transfection efficiency with >90% cell viability; data not shown). In brief, 1.0 to 1.5 × 106 cells were washed with phosphate-buffered saline and resuspended in nucleofection solution (Mirus Bio LLC, Madison, WI). Cells were nucleofected using the T-16 nucleofector program and added to 4.5 ml of nutrient-rich media. Cells were cultured in a 5% CO2 humidified atmosphere at 37°C for 24 h before experimentation.
Plasmid Constructs and Site-Directed Mutagenesis.
The mature human miR181b sequence was cloned into the SDSA 3.0 expression plasmid using the BglII and Xho1 restriction sites. After incorporation into cells, this plasmid expresses miR181b concomitantly with GFP. The Mcl-1 3′-UTR luciferase expression construct was a generous gift from Dr. Serge Nana-Sinkam (Division of Pulmonary, Allergy, Critical Care and Sleep Medicine, Ohio State University, Columbus, OH). This is a psicheck-2 plasmid vector that incorporates the 3′-UTR of Mcl-1 downstream of the luciferase gene (Crawford et al., 2009). Site-directed mutagenesis of the miR181b binding site was performed using the following primers: forward, 5′-CCATTTAAAAATAGGTATGAATAAGATGACTAAGATACTAATGGGGAAGAACTGCCCTG-3′; and reverse, 5′-CAGGGCAGTTCTTCCCCATTAGTATCTTAGTCATCTTATTCATACCTATTTTTAAAT GG-3′. The reaction mix included 1-ng template DNA, dNTPs, and forward and reverse primers. Touchdown PCR was performed on a Thermocycler (Eppendorf North America, New York, NY); after an initial incubation at 95°C for 2 min, the following steps were cycled 18 times: 95°C for 50 s, 70→55°C (each cycle was performed with a temperature 1° lower) for 50 s, and 72°C for 14 min. The PCR product was treated with Dpn1, and DH5α Escherichia coli (Invitrogen) were transformed according to the manufacturer's instructions. Mutation was confirmed by sequencing (Genome Analysis Facility, University of North Carolina, Chapel Hill, NC).
Caspase-3 Activity Assay.
To determine caspase-3 activity, cells (6 × 104/well) were incubated with drug or vehicle in a 96-well plate. Plates were then centrifuged for 5 min to pellet the cells. Cells were lysed on ice with buffer containing 250 mM HEPES, pH 7.4, 25 mM CHAPS, and 25 mM dithiothreitol. The nonionic detergent disrupts the plasma membrane and releases the cytosolic proteins without denaturing the caspase protein. The activity of caspase-3 in these samples was determined using the Caspase-3 Fluorimetric Assay Kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Fluorescence was measured using a FLUOstar Galaxy plate reader (BMG Labtech, Durham, NC) with a 360-nm excitation filter and a 460-nm emission filter.
qRT-PCR-Based miRNA Expression Profiling.
Total RNA was extracted from 40.0 × 106 MYL and MYL-R cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. miRNA enrichment was then performed using the Small RNA Isolation Kit (SABiosciences, Frederick, MD), and the miRNA purity was assessed using an Agilent Bioanalyzer Chip (Genomics and Bioinformatics Core, University of North Carolina, Chapel Hill, MD). Finally, 100-ng miRNA was 3′-polyadenylated and converted to cDNA using the First-Stand Synthesis Kit (SABiosciences) according to the manufacturer's instructions.
Approximately 100-ng cDNA was added to the Human Cancer RT2 miRNA qPCR Array (SABiosciences; ∼1.0 ng/well), and qRT-PCR was performed on an ABI 7500 Fast Real-Time Thermocycler (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Melting-curve analysis of the resulting transcript was determined. Analysis of the qRT-PCR data were performed using the RT2 miRNA PCR Array Data Analysis program (available at http://www.sabiosciences.com/pcr/arrayanalysis.php).
qRT-PCR Analysis of miR181 Expression.
MiRNA 181 expression was measured using the Taqman MicroRNA Assay (Applied Biosystems; miR181b and miR181d) according to the manufacturer's instructions with slight modification. In brief, total RNA was extracted from cells using TRIzol reagent. One microgram of RNA was added to the following reaction mix: RT buffer, 1 mM dNTPs, U6 small nuclear RNA RT primers (Applied Biosystems), and RT primers against miR181b. Twenty-five units of the Multiscribe RT enzyme (Applied Biosystems) was added, and the reverse-transcription reaction was performed on a PCR thermocycler. Twenty-five nanograms of the resulting cDNA was added to a qPCR mix containing the Taqman Fast Universal PCR Master Mix (Applied Biosystem) and the PCR primer/probe reaction mix against either miR181b or U6 small nuclear RNA. Quantitative PCR was performed on an ABI 7500 Real-Time System, and data were analyzed using the 7500 Fast software (Applied Biosystems).
Western Blot Analysis.
Cells were collected, washed twice with phosphate-buffered saline, and lysed using radioimmunoprecipitation assay buffer (without SDS; 150 mM NaCl, 9.1 mM Na2HPO4, 1.7 mM NaH2PO4, 1% NP-40, and 0.5% deoxycholic acid, pH 7.4) supplemented with protease and phosphatase inhibitors (150 μM Na3VO4, 0.25 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 10 nM microcystin). The lysate was centrifuged, and the protein concentration was determined using Bradford reagent (Thermo Fisher Scientific). Sample buffer (2×: 0.5 M Tris, 20% glycerol, 10% β-mercaptoethanol, and 0.002 μg/ml bromphenol blue, pH 6.8) was added to an equal volume of total protein (30–50 μg), and the samples were separated by SDS-polyacrylamide gel electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane (Millipore Bioscience Research Reagents, Billerica, MA). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline supplemented with Tween 20 (TBST; 9.9 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20, pH 8.0) for 1 h at room temperature. Primary antibodies against Lyn (Santa Cruz Biotechnology; Santa Cruz, CA), Mcl-1 (Santa Cruz Biotechnology), GFP (Sigma), SFK pY416 (Cell Signaling Technology, Danvers, MA), and SFK pY527 (Cell Signaling Technology) were diluted in 1% bovine serum albumin/TBST and applied to a membrane overnight at 4°C or for 1 h at room temperature. After the primary antibody incubation, membranes were washed three times for 5 min in TBST. Secondary antibodies (horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin; Santa Cruz Biotechnology) diluted in 1% gelatin/TBST were then applied to the membrane for 1 h at room temperature. β-Actin (Santa Cruz Biotechnology) or α-tubulin (Sigma-Aldrich) expression was measured as a loading control. Membranes were developed using enhanced chemiluminescence (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) with exposure to autoradiography film. Densitometry was performed using a FluorChem FC2 imager (Cell Biosciences, Santa Clara, CA).
Transfection and Luciferase Activity Assay.
HEK293T cells (3 × 105/well) were plated in poly(l-lysine)-treated six-well plates. The next day, cells were transfected using Lipofectamine (Invitrogen) according to the manufacturer's instructions.
Twenty-four hours after transfection, cells were lysed at room temperature with radioimmunoprecipitation assay buffer (without SDS). The lysate was centrifuged for 10 min at 10,000 rpm. Ten-microliter aliquots of each sample was applied to a 96-well plate, and 150 μl of the luciferase assay reaction mix (25 mM glycyl glycine, 15 mM MgSO4, 15 mM KPO4, and 4 mM EGTA, pH 7.8) containing 0.1 mg/ml luciferin (Sigma) was added to each well. Luciferase activity was measured on a Pherastar luminometer (BMG Labtech). Western blot analysis of GFP was performed to ensure equivalent transfection, and β-actin expression was determined as a loading control.
Statistics.
Data were analyzed using analysis of variance and t tests where appropriate using Prism 4 software (GraphPad Software Inc., San Diego, CA).
Results
Lyn Kinase Overexpression in MYL-R Cells Confers Drug Resistance.
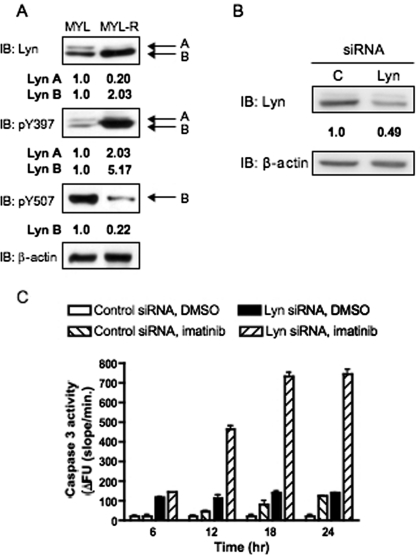
Previous studies have demonstrated that Lyn is important for MYL-R survival (Ito et al., 2007). To compare Lyn expression and activity, lysates of MYL and MYL-R cells were Western-blotted with antibodies that recognize total Lyn, active Lyn [P-Y397 (P-Y416 Src)], or inactive Lyn [P-Y507 (P-Y527 Src)]. As shown in Fig. 1A, the Lyn B splice form (53 kDa) was significantly overexpressed and phosphorylated on the activation loop (Tyr397), indicating increased activity in MYL-R cells. Likewise, the Lyn A splice form (56 kDa), although reduced in expression, was also more highly phosphorylated on this residue in the MYL-R cells (Fig. 1A). Loss of the inactivating Tyr507 phosphorylation was observed only for Lyn B, suggesting the elevated activity state of this kinase in MYL-R cells. It is noteworthy that the increase of Lyn activity was confirmed in these cells using a novel peptide-based fluorescent Lyn biosensor (Wang et al., 2010a).
Fig. 1.
Lyn kinase confers imatinib resistance in MYL-R cells. A, 2 × 106 cells were lysed and Western-blotted using the indicated antibodies (pY507, inhibitory phosphorylation site; pY397, autophosphorylation site). β-Actin expression was similarly measured as a loading control, and densitometry was performed. Representative data are shown. B and C, 1 × 106 MYL-R cells were nucleofected with siRNA against Lyn or nontargeting control siRNA. B, 48 h later, cells were lysed, and Western blot analysis was performed using antibodies against Lyn and β-actin, and densitometry was performed. Representative data are shown. C, cells were plated (6 × 105/well) in a 96-well plate and treated with imatinib (1.0 μM) or DMSO for the indicated time. Cells were then lysed, and caspase-3 activity was measured. Data represent the mean ± S.E.M. of triplicate samples.
Lyn hyperactivation has been attributed to imatinib resistance in MYL-R cells (Ito et al., 2007). In addition, incubation of MYL-R cells with gemcitabine, Ara-C, or adaphostin failed to activate caspase-3 in these cells, demonstrating that MYL-R are highly antiapoptotic (E. I. Zimmerman and L. M. Graves, unpublished results). To investigate the importance of Lyn in cell survival, MYL-R cells were transfected with Lyn-directed siRNA or nontargeting control siRNA and then exposed to 1 μM imatinib. This treatment resulted in a partial knockdown of Lyn and a significant increase in caspase 3 activation (Fig. 1, B and C). However, the magnitude of caspase activation was less than in MYL cells exposed to the same dose of imatinib (data not shown); this may be due to incomplete silencing of Lyn expression (Fig. 1B). In addition, pretreatment with 5-(4-chlorophenyl)-7-(dimethylethyl)pyrazolo[3,4-d]pyrimidine (PP2), a Lyn and SFK inhibitor, significantly elevated caspase activity in MYL-R cells treated with imatinib, an effect not observed after pretreatment with the inactive inhibitor analog, 1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (PP3) (data not shown).
MiR181 Expression Is Reduced in MYL-R Cells.
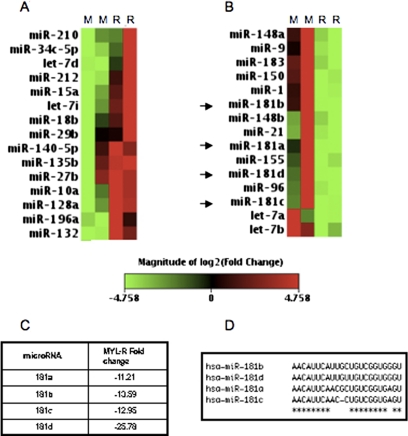
MiRNAs are small (20–22 nucleotides) noncoding RNA regulators of protein expression that have been implicated in the progression and survival of numerous cancers (Hammond, 2006). To identify miRNAs involved in antiapoptosis, MYL and MYL-R cells were profiled for miRNA expression using the Human Cancer RT2 miRNA qPCR Array (SA Biosciences). This qRT-PCR-based assay analyzed the expression of 88 known human miRNAs associated previously with cancer. Duplicate experiments demonstrated that 15 miRNAs had a >2-fold increase in expression in MYL-R cells relative to MYL cells (Fig. 2A) and that 15 miRNAs showed a >2-fold decrease in relative expression (Fig. 2B). It is noteworthy that we observed a strong down-regulation of the miR181 family (a–d) of miRNAs in MYL-R cells (Fig. 2, B and C). This family of miRNAs is highly conserved (Fig. 2D), and loss of miR181 expression is prognostic for aggressive AML (Marcucci et al., 2008; Schwind et al., 2009). To confirm the array results, individual qRT-PCR of miR181b and miR181d was performed; the results of these analyses showed that the expression of both miRNAs was reduced approximately 10-fold in MYL-R cells (Supplemental Fig. 1).
Fig. 2.
Loss of miR181 expression in MYL-R cells. A and B, miRNA expression was profiled in MYL and MYL-R cells by qRT-PCR using the SuperArray cancer array. The heat map depicts 15 miRNAs that had a >2-fold increase (A) or decrease (B) in expression in MYL-R cells compared with MYL cells. Arrows denote miR181 family members. Data represent duplicate experiments performed with triplicate samples. C, the fold change in miR181 expression in MYL-R cells in comparison to MYL cells. D, alignment of human miR181 miRNAs (5′→3′) using ClustalW software (http://www.ebi.ac.uk/Tools/clustalw2/index.html). An asterisk denotes a conserved nucleotide.
Lyn Inhibition Increases miR181 Expression in MYL-R Cells.
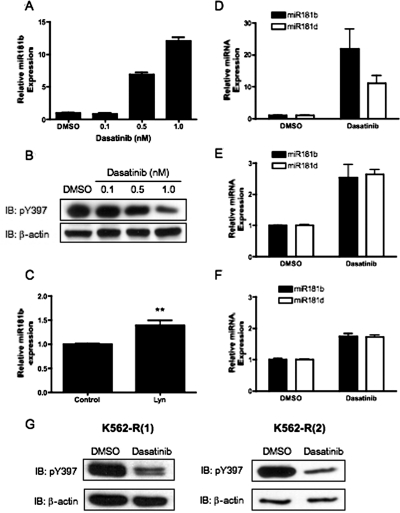
To determine whether hyperactivation of Lyn regulated miR181 expression in MYL-R cells, cells were treated with dasatinib, and miRNA expression was determined by qRT-PCR. As shown in Fig. 3, dasatinib treatment inhibited Lyn and increased miR181b expression in MYL-R cells in a dose-dependent manner (Fig. 3, A and B). Dasatinib (1 nM) treatment produced minimal effects on cell viability (∼15–20% cell loss), as determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt assay and cell proliferation (data not shown). In addition, nucleofection of MYL-R cells with Lyn-targeted siRNA significantly elevated miR181b expression compared with control siRNA-treated cells (Fig. 3C). K562-R cells are a model of imatinib-resistant CML, and overexpression of Lyn has been attributed to the mechanism of resistance in these cells (Donato et al., 2003; Dai et al., 2004). To further determine whether Lyn regulates miR181 expression, we tested two independently subcloned imatinib-resistant K562-R cell lines, termed K562-R(1) and K562-R(2). Similar to MYL-R cells, dasatinib treatment of K562-R cells inhibited Lyn activity and increased miR181b expression (Fig. 3E-G). In addition, dasatinib treatment increased miR181d expression in each imatinib-resistant cell line (Fig. 3, D–F). These data suggest that hyperactivation of Lyn represses miR181 expression in imatinib-resistant CML.
Fig. 3.
Lyn kinase inhibits miR181 expression. A and B, 2 × 106 MYL-R cells were treated for 24 h with the indicated dose of dasatinib or DMSO. A, total RNA was extracted from 1 × 106 cells, and miR181b expression was measured. Data represent the mean ± S.E.M. of triplicate samples. B, 1 × 106 cells were lysed, and Western blot analysis was performed using the indicated antibodies. Representative data are shown. C, 1 × 106 MYL-R cells were nucleofected with siRNA against Lyn or nontargeting control siRNA. Forty-eight hours later, RNA was extracted, and miR181b expression was determined. Data represent the mean ± S.E.M. of duplicate experiments performed with triplicate samples (**, p = 0.0036). D to G, 2 × 106 cells were treated for 24 h with 1 nM dasatinib or DMSO. D to F, total RNA was extracted from 1 × 106 MYL-R (D), K562-R(1) (E), and K562-R(2) cells (F), and miR181b and miR181d expression was measured. Data represent the mean ± S.E.M. of duplicate experiments performed with triplicate samples. G, 1 × 106 cells were lysed, and Western blot analysis was performed using the indicated antibodies. Representative data are shown.
Mcl-1 Is a Target of miR181b.
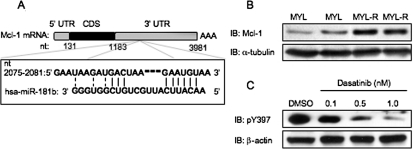
To investigate the potential significance of miR181 to cell survival, we sought to identify protein targets for miR181. Public database bioinformatic algorithms (available at http://www.targetscan.org) predict a binding site for miR181 on the 3′-UTR of Mcl-1, a Bcl-2 family member and antiapoptotic protein (Fig. 4A) (Warr and Shore, 2008). Mcl-1 expression inversely correlates with leukemia chemosensitivity (Kaufmann et al., 1998), and Western blot analysis determined that Mcl-1 expression was higher in MYL-R cells (Fig. 4B). Inhibition of Lyn with dasatinib decreased Mcl-1 expression in a dose-dependent manner, demonstrating the importance of Lyn activity in regulating Mcl-1 expression (Fig. 4C).
Fig. 4.
Mcl-1, a predicted target of miR181, is overexpressed in MYL-R cells. A, diagram of the predicted miR181 binding site in the Mcl-1 3′-UTR (available at http://www.targetscan.org). B, 2 × 106 cells were lysed, and Western blot analysis was performed with the indicated antibodies. Samples were loaded in duplicate. Representative data are shown. C, 2 × 106 MYL-R cells were treated for 24 h with the indicated dose of dasatinib or DMSO. Cells were lysed, and Western blotted with the indicated antibodies. Representative data are shown.
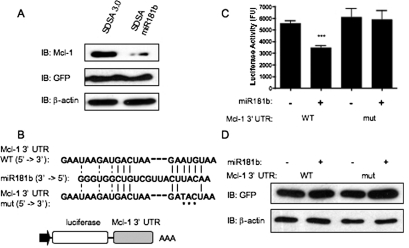
To determine whether miR181b regulated Mcl-1 expression, MYL-R cells were nucleofected with an miR181b-expressing plasmid vector (SDSA-miR181b) or empty vector (SDSA 3.0) control. Expression of GFP confirmed nucleofection and plasmid processing (Fig. 5A). In addition, qRT-PCR confirmed miR181b expression in these experiments (data not shown). As shown in Fig. 5, overexpression of miR181b in MYL-R cells significantly decreased Mcl-1 expression in comparison to the empty vector control (Fig. 5A).
Fig. 5.
miR181b inhibits Mcl-1 expression. A, 1 × 106 MYL-R cells were nucleofected with the SDSA 3.0 miR181b vector or empty vector (SDSA 3.0). Forty-eight hours later, cells were lysed, and Western blot analysis was performed with the indicated antibodies. Representative data are shown. B, diagram of luciferase constructs containing the Mcl-1 3′-UTR. Alignment of miR181b with the WT and mutant Mcl-1 3′-UTR sequences. An asterisk denotes a nucleotide mutation in the predicted miR181b binding site. C, HEK293T cells were cotransfected with the indicated SDSA 3.0 and psicheck Mcl-1 3′-UTR vectors. Twenty-four hours later, cells were lysed, and luciferase activity was measured. Data represent the mean ± S.E.M. of duplicate experiments performed with triplicate samples (***, p < 0.001). E, Western blot analysis was performed with the indicated antibodies. Representative data are shown.
miRNAs regulate protein expression by binding to the 3′-UTR of target mRNA and initiate mRNA degradation or inhibit translational processing (Ambros, 2001). To determine whether miR181-dependent inhibition of Mcl-1 expression was due to a direct interaction, HEK293T cells were cotransfected with the miR181b construct or empty vector and a luciferase-reporter construct containing the 3′-UTR of Mcl-1 mRNA (Fig. 5B). Using this approach, we observed that overexpression of miR181b significantly inhibited luciferase activity compared with the vector control cells (Fig. 5C). Because the miRNA “seed region” is critical for the recognition of miRNA targets, complementary nucleotides within this region were mutated in the 3′-UTR of the Mcl-1 luciferase reporter (Fig. 5B). These mutations ablated the miR181b-dependent repression of luciferase activity (Fig. 5C), whereas mutation of nucleotides outside the seed region failed to affect this process (data not shown). These data suggest that miR181b directly interacts with the 3′-UTR of Mcl-1 mRNA to inhibit Mcl-1 protein expression, and nucleotides within the seed region of this binding site are important to mediate this effect.
Lyn Regulates miR181b and Mcl-1 Expression in AML.
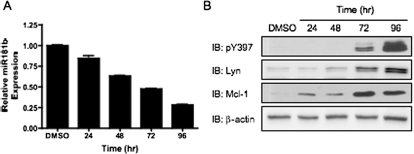
The HL-60 cell line is a commonly used cell model for the study of AML. It was shown previously that 1,25-D3 treatment increased the expression of Lyn in these cells (Wang et al., 2000). To determine whether manipulation of Lyn expression in HL-60 cells affected miR181b and Mcl-1 expression, we treated these cells with 1,25-D3 for up to 96 h. A strong increase in active Lyn expression was observed by Western blotting these samples. In agreement with our data obtained with the MYL-R and K562R cells, the increase in Lyn expression correlated with repression of miR181b and increased Mcl-1 expression in a time-dependent manner (Fig. 6, A and B). Thus, these results indicate that Lyn can affect Mcl-1 expression through modulation of miR181b expression in an AML cell line.
Fig. 6.
Enhancement of Lyn expression in HL-60 cells decreases miR181b expression and increases Mcl-1 expression. A and B, 1 × 106 HL-60 cells were treated with 10 ng/μl 1,25-D3 for the indicated time. A, total RNA was extracted from cells, and miR181b expression was measured. Data represent the mean ± S.E.M. of triplicate samples. B, cells were lysed, and Western blot analysis was performed using the indicated antibodies. Representative data are shown.
Discussion
These studies describe three important findings: 1, that the hyperactivation of Lyn suppresses the expression of miR181; 2, that miR181b represses the expression of a key antiapoptotic protein, Mcl-1; and 3, loss of miR181 upon Lyn activation may represent a novel mechanism of drug resistance in leukemia. This mechanism was observed in cell models of CML and AML, and the specific role of Lyn was confirmed by both dasatinib treatment and siRNA against Lyn. Considerable evidence now supports the role of Lyn as a “compensatory oncogene” in imatinib-resistant CML and specific subtypes of AML. Lyn is overexpressed in drug-resistant cell lines and patient samples (Donato et al., 2003; Dai et al., 2004; Wu et al., 2008) and activated in response to Flt3 activation or Flt3-internal tandem duplication mutation (Okamoto et al., 2007). Using a model of Lyn-dependent CML, we observed strong repression of the miR181 family of miRNAs. Low miR181 expression has previously been associated with poor prognosis in AML, whereas high miR181 (a–d) expression is prognostic for event-free survival in patients with AML (Marcucci et al., 2008; Schwind et al., 2009).
Although our studies primarily focused on miR181b, this family of miRNAs is highly conserved. Previous studies suggest Bcl-2 as a target for repression by miR181 microRNAs (Neilson et al., 2007; Chen et al., 2010). Comparison of the potential 3′-UTR targeting sequence of Bcl-2 with that of Mcl-1 demonstrates a remarkable homology between these sequences, which includes conservation within the seed sequence. The seed sequence is a key region of complementary necessary for the targeting of miRNAs (Wang et al., 2010b). Our results demonstrated that mutation of the nucleotides complementary to the miR181 seed sequence in the 3′-UTR of Mcl-1 ablated the effects of miR181b. Whether miR181 directly targets Bcl-2 has yet to be confirmed; however, this is would establish the importance of the miR181 family in the regulation of mitochondria-mediated apoptosis.
Previous studies have identified multiple miRNAs that are altered in drug-resistant cancer, suggesting that some miRNAs have tumor suppressive effects, whereas others regulate cell survival (Hummel et al., 2010). A recent study profiled miRNAs from patients with imatinib-resistant CML who did not have BCR/Abl mutations (San José-Enériz et al., 2009). These authors identified 19 miRNAs that were differentially expressed between drug-resistant patients and drug-responders. In agreement with their results, we observed down-regulation of the expression of miR183 and two members of the Let7 family (Let 7a, Let7b). However, in contrast to studies reporting decreased miR10a expression in drug-resistant CML, we observed increased miR10a expression in the MYL-R cells. Many of the other miRNAs reported by San José-Enériz et al. (2009) were either unchanged or not present on our arrays.
Previous studies have identified miRNAs that target Mcl-1 (Mott et al., 2007; Chen et al., 2009; Crawford et al., 2009), and we measured the expression of these miRNAs in our cell lines; however, the expression of these miRNAs was not implicated in the mediation of Mcl-1 regulation in MYL and MYL-R cells, suggesting that they may contribute to Mcl-1 expression in a cell type-specific manner. In addition to the miR181 family, we observed many unique differences in miRNA expression between MYL and MYL-R cells (miR10a, miR128a, miR132, miR150, miR155, miR183, miR196a, miR212). It is possible that some of these miRNAs may play a role in the mediation of drug resistance, and this will be a focus of future research.
Chen et al. (2010) demonstrated that miR181a overexpression sensitized cells to radiation. Moreover, Studzinski and colleagues have demonstrated that 1,25-D3 treatment of HL-60 cells not only increases Lyn expression, but also decreases miR181 expression and increases Mcl-1 expression (Wang and Studzinski, 1997; Wang et al., 2000, 2009). This work was published independently over the course of a decade. Our study is in agreement with their data, and, importantly, we believe our study provides further mechanistic insight to these observations. Finally, Studzinski and colleagues determined that 1,25-D3 treatment decreases sensitivity to drugs that induce apoptosis (Xu et al., 1993); this observation is in agreement with our data, which suggest that Lyn-dependent loss of miR181 may contribute to the development of imatinib resistance.
Although the mechanism of Lyn repression of miR181 is not known, miRNA expression can be regulated by both genetic and epigenetic mechanisms. For instance, the role of BCR-Abl in the silencing of miR328 in CML through a mitogen-activated protein kinase-dependent manner was shown recently (Eiring et al., 2010). Furthermore, activating mutations of the c-Kit kinase induce a MYC-dependent repression of miR29b in AML (Liu et al., 2010). In addition to transcription factor (CCAAT-enhancer-binding protein α, Myc, etc.)-dependent mechanisms, kinases may regulate miRNA expression through post-transcriptional events, such as that observed with LIN28 (Newman et al., 2008; Khusial et al., 2009). Future studies will aim to determine the mechanism of Lyn-dependent effects on miR181 expression.
Supplementary Material
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the American Heart Association [Grant 09PRE2200044]; and the National Institutes of Health National Institute of General Medical Sciences [Grant T32-GM008581-10.].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.066258.
- CML
- chronic myelogenous leukemia
- miRNA
- micro-RNA
- MYL-R
- Lyn-mediated imatinib-resistant chronic myelogenous leukemia
- siRNA
- short interfering RNA
- UTR
- untranslated region
- SFK
- Src family kinase
- AML
- acute myelogenous leukemia
- Mcl-1
- myeloid cell leukemia-1
- HEK
- human embryonic kidney
- GFP
- green fluorescent protein
- PCR
- polymerase chain reaction
- CHAPS
- 3-[(3-cholamidopropyl) dimethylammonio]propanesulfonate
- qRT-PCR
- quantitative reverse transcriptase-polymerase chain reaction
- TBST
- Tris-buffered saline/Tween 20
- PP2
- 5-(4-chlorophenyl)-7-(dimethylethyl)pyrazolo[3,4-d]pyrimidine
- DMSO
- dimethyl sulfoxide
- 1,25-D3
- 1,25-dihydroxyvitamin D3
- PP3
- 1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine.
References
- Ambros V. (2001) microRNAs: tiny regulators with great potential. Cell 107:823–826 [DOI] [PubMed] [Google Scholar]
- Bagrintseva K, Geisenhof S, Kern R, Eichenlaub S, Reindl C, Ellwart JW, Hiddemann W, Spiekermann K. (2005) FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L). Blood 105:3679–3685 [DOI] [PubMed] [Google Scholar]
- Breitenbuecher F, Markova B, Kasper S, Carius B, Stauder T, Böhmer FD, Masson K, Rönnstrand L, Huber C, Kindler T, et al. (2009) A novel molecular mechanism of primary resistance to FLT3-kinase inhibitors in AML. Blood 113:4063–4073 [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. (2005) A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353:1793–1801 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86 [DOI] [PubMed] [Google Scholar]
- Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. (2010) MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep 23:997–1003 [DOI] [PubMed] [Google Scholar]
- Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, Huang DD, Ding J, Shen F, et al. (2009) The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol 50:358–369 [DOI] [PubMed] [Google Scholar]
- Crawford M, Batte K, Yu L, Wu X, Nuovo GJ, Marsh CB, Otterson GA, Nana-Sinkam SP. (2009) MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun 388:483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Rahmani M, Corey SJ, Dent P, Grant S. (2004) A Bcr/Abl-independent, Lyn-dependent form of imatinib mesylate (STI-571) resistance is associated with altered expression of Bcl-2. J Biol Chem 279:34227–34239 [DOI] [PubMed] [Google Scholar]
- Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. (2007) MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 21:912–916 [DOI] [PubMed] [Google Scholar]
- Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. (2008) Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 3:e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, Talpaz M. (2003) BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood 101:690–698 [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140:652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. (2006) MicroRNAs as oncogenes. Curr Opin Genet Dev 16:4–9 [DOI] [PubMed] [Google Scholar]
- Hochhaus A, Kreil S, Corbin AS, La Rosée P, Müller MC, Lahaye T, Hanfstein B, Schoch C, Cross NC, Berger U, et al. (2002) Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia 16:2190–2196 [DOI] [PubMed] [Google Scholar]
- Hummel R, Hussey DJ, Haier J. (2010) MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer 46:298–311 [DOI] [PubMed] [Google Scholar]
- Illmer T, Schaich M, Platzbecker U, Freiberg-Richter J, Oelschlägel U, von Bonin M, Pursche S, Bergemann T, Ehninger G, Schleyer E. (2004) P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia 18:401–408 [DOI] [PubMed] [Google Scholar]
- Ito T, Tanaka H, Kimura A. (2007) Establishment and characterization of a novel imatinib-sensitive chronic myeloid leukemia cell line MYL, and an imatinib-resistant subline MYL-R showing overexpression of Lyn. Eur J Haematol 78:417–431 [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD, Reed JC. (1998) Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood 91:991–1000 [PubMed] [Google Scholar]
- Khusial PR, Vadla B, Goldberg GS. (2009) Src regulates the expression of Lin28: implications for cell growth, adhesion, and communication. Cell Commun Adhes 15:407–409 [DOI] [PubMed] [Google Scholar]
- Li S. (2008) Src-family kinases in the development and therapy of Philadelphia chromosome-positive chronic myeloid leukemia and acute lymphoblastic leukemia. Leuk Lymphoma 49:19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, et al. (2010) Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 17:333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, et al. (2008) MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358:1919–1928 [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. (2007) mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26:6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Williams A, Vultur A, List A, Bhalla K, Smith D, Lee FY, Jove R. (2007) Dasatinib (BMS-354825) inhibits Stat5 signaling associated with apoptosis in chronic myelogenous leukemia cells. Mol Cancer Ther 6:1400–1405 [DOI] [PubMed] [Google Scholar]
- Neilson JR, Zheng GX, Burge CB, Sharp PA. (2007) Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev 21:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Hayakawa F, Miyata Y, Watamoto K, Emi N, Abe A, Kiyoi H, Towatari M, Naoe T. (2007) Lyn is an important component of the signal transduction pathway specific to FLT3/ITD and can be a therapeutic target in the treatment of AML with FLT3/ITD. Leukemia 21:403–410 [DOI] [PubMed] [Google Scholar]
- San José-Enériz E, Román-Gómez J, Jiménez-Velasco A, Garate L, Martin V, Cordeu L, Vilas-Zornoza A, Rodríguez-Otero P, Calasanz MJ, Prósper F, et al. (2009) MicroRNA expression profiling in imatinib-resistant chronic myeloid leukemia patients without clinically significant ABL1-mutations. Mol Cancer 8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwind SGM, Maharry K, Radmacher MD, Whitman SP, Paschka P, Mrózek K, Kolitz JE, Larson RE, Bloomfield CD. (2009) MicroRNA 181a (miR-181a) expression as a prognosticator in cytogenetically normal acute myeloid leukemia (CN AML) (Abstract). J Clin Oncol 27:7001 [Google Scholar]
- Shangary S, Johnson DE. (2003) Recent advances in the development of anticancer agents targeting cell death inhibitors in the Bcl-2 protein family. Leukemia 17:1470–1481 [DOI] [PubMed] [Google Scholar]
- Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M, Scherr M. (2007) Expression of the miR-17–92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood 109:4399–4405 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zimmerman EI, Toutchkine A, Martin TD, Graves LM, Lawrence D. (2010a) Multicolor monitoring of dysregulated protein kinases in chronic myelogenous leukemia. ACS Chem Biol doi: 10.1021/cb100099h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Studzinski GP, Chen F, Coffman FD, Harrison LE. (2000) p53/56(lyn) antisense shifts the 1,25-dihydroxyvitamin D3-induced G1/S block in HL60 cells to S phase. J Cell Physiol 183:238–246 [DOI] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Xie K, Jennings MH, Hu Y, Stromberg AJ, Nelson PT. (2010b) Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol 7:373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gocek E, Liu CG, Studzinski GP. (2009) MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle 8:736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Studzinski GP. (1997) Antiapoptotic action of 1,25-dihydroxyvitamin D3 is associated with increased mitochondrial MCL-1 and RAF-1 proteins and reduced release of cytochrome c. Exp Cell Res 235:210–217 [DOI] [PubMed] [Google Scholar]
- Warr MR, Shore GC. (2008) Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med 8:138–147 [DOI] [PubMed] [Google Scholar]
- Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG, Darnay BG, Lamothe B, Sun H, Talpaz M, et al. (2008) Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst 100:926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HM, Tepper CG, Jones JB, Fernandez CE, Studzinski GP. (1993) 1,25-Dihydroxyvitamin D3 protects HL60 cells against apoptosis but down-regulates the expression of the bcl-2 gene. Exp Cell Res 209:367–374 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Kurosu T, Kakihana K, Mizuchi D, Miura O. (2004) The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem Biophys Res Commun 319:1272–1275 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121:1085–1095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.