Abstract
3-Methylindole (3MI) is a preferential pneumotoxicant found in cigarette smoke. A number of lung-expressed human cytochrome P450 enzymes, including 1A1, 2F1, and 2A13, catalyze the metabolism of 3MI to reactive intermediates that fragment DNA – measured with the Comet assay to assess DNA damage – in a cytochrome P450-dependent manner in primary normal human lung cells in culture, but the mutagenesis of 3MI has been controversial. In the present study, the mutagenic potential of 3MI was compared to the prototypical cigarette smoke carcinogens benzo(a)pyrene (B(a)P) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). 3MI, B(a)P, and NNK were incubated with the Salmonella typhimurium strain TA98, which is known to detect the most common subtype of cigarette smoke-induced mutagenicity, frameshift mutations in DNA, and with Salmonella typhimurium strain TA100 which detects base pair substitution mutants, with five sources of P450-mediated bioactivation: rat liver S9, human lung microsomes, recombinant CYP2A13, purified CYP2F3, and recombinant CYP1A1. Only B(a)P was mutagenic in TA100 and it was bioactivated by human lung microsomes and rat liver S9 sources of P450s. However with the TA98 strain, CYP1A1, CYP2A13, CYP2F3, and human lung microsomes bioactivated 3MI to highly mutagenic intermediates, whereas neither human nor rat liver S9 sub-cellular fractions formed mutagenic intermediates from 3MI. Quantitative western blot analysis verified that all three respiratory enzymes were present in human lung microsomes in widely varying amounts. These results indicate that metabolism of 3MI by human lung-expressed cytochrome P450 enzymes, but not hepatic P450s, elicits equivalent or higher mutagenicity than the prototype cigarette smoke mutagens B(a)P and NNK and indicates that 3MI is a likely human pulmonary carcinogen.
Introduction
Cigarette smoke has been reported to contain well over 4000 chemicals, at least 81 of which have been classified as carcinogens by the International Agency for Research on Cancer (1), including the known carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), which is present in cigarettes at a range of concentrations up to 500 ng per cigarette (2), and benzo(a)pyrene (B(a)P), which is present in the range of 3.36 to 40 ng per cigarette (3, 4). Several studies have examined cigarette smoke condensates and shown mutagenic activity in the TA98 Salmonella reverse mutation assay (5, 6). At least 90% of established human carcinogens are mutagenic in this assay, and very few non-carcinogenic compounds display false positives (7).
3-Methylindole (3MI) is formed by the pyrolysis of tryptophan during the burning of tobacco and is found in cigarette smoke at concentrations ranging from 0.4 to 1.7 μg per cigarette and up to 16 μg per cigarette when the cigarette smoke was analyzed from its particulate matter fractions (4, 8). These values are far higher than the reported levels of either NNK or B(a)P. Low micromolar concentrations (in the range of 0.1 to 10 μM) of 3MI have been previously demonstrated by our laboratory to induce markers of carcinogenesis, including DNA damage and p53 nuclear localization (9). Previous mutagenesis work by another group indicated that 3MI was not mutagenic in a TA100 Salmonella reverse mutation assay using rat liver S9 (10). However, the TA100 assay detects base pair mutagens, whereas another bacterial mutagenesis assay, the TA98 assay, detects frameshift mutations, which are the predominate DNA alterations associated with tobacco smoke-induced mutagenesis (6).
Work in our laboratory indicated that 3MI was not mutagenic to the TA98 strain with rat liver S9 fractions (9). However, in the same study, we demonstrated that 3MI was mutagenic in the presence of purified CYP2F3, the goat lung isoform of human CYP2F1, both of which have been demonstrated by our laboratory to preferentially catalyze the formation of the reactive intermediate, 3MEIN (11).
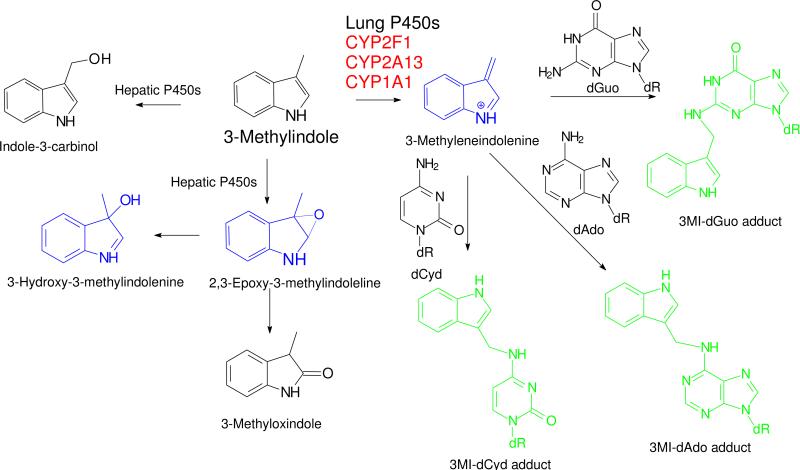
Bioactivation of 3MI to at least three electrophilic intermediates (Scheme 1) by a plethora of P450 enzymes has been extensively documented (11-17). However, 3MEIN, the dehydrogenated intermediate that reacts with glutathione (16, 18, 19), deoxynucleosides (20), DNA (9, 10, 20), and proteins (17, 21, 22), is only produced by a limited number of P450 enzymes (11), almost all of which are selectively expressed in respiratory tissues. CYP2F1 exclusively produces 3MEIN (11), and CYP1A1 and CYP2A13 (12) efficiently dehydrogenate 3MI to 3MEIN, and oxygenate 3MI to indole-3-carbinol and 3-methyloxindole. The extrahepatic CYP1B1 and the major hepatic P450s CYP2A6, CYP2C19, CYP2D6, CYP2B6, CYP3A4/5, and CYP2E1 do not form 3MEIN (11). Incubations of lung or liver microsomes with pure deoxynucleosides produced only the deoxyguanosine, deoxyadenine, and deoxycytidine adducts of 3MEIN shown in Scheme 1. No adduct of deoxythymidine or adducts of any deoxynucleoside with the other putative electrophilic intermediates were identified (20). The same adducts were identified from microsomal incubations with DNA, after isolation and hydrolysis of the adducted DNA, and we have identified the adducts in lung tissues of mice treated with inhaled 3MI (unpublished studies). These extensive previous studies provide robust evidence that CYP1A1. CYP2F1, and CYP2A13 efficiently catalyze the formation of 3MEIN, and this intermediate appears to react avidly with the exocyclic amine groups of DNA bases. Therefore, 3MEIN appears to be the most likely mutagenic intermediate.
Scheme 1. Illustration of 3MI Metabolism, Putative Bioactivation, and Nucleoside Adducts.
The dehydrogenated intermediate, 3-methyleneindolenine, is efficiently and selectively produced by the lung P450 enzymes CYP2F1, CYP2A13, and CYP1A1. Three putative electrophilic intermediates of 3MI bioactivation are shown in blue. Three nucleoside adducts (shown in green) of 3-methyleneindolenine with DNA have been previously identified (20), are produced from hydrolysis of 3MI-alkylated DNA, and are putative markers of the mutagenic activity of 3MI. Nucleoside adducts of the 2,3-epoxide or 3-hydroxy-3-methylindolenine have not been identified. R = deoxyribose
The previous results from our laboratory provided impetus for our hypothesis that hepatic S9 fractions lack the specific cytochrome P450 enzymes necessary to cause 3MI-mediated mutagenesis, but that 3MI-mediated mutagenesis is preferentially catalyzed by a small number of selectively expressed pulmonary P450 enzymes. Therefore, in the present study the mutagenic activity of 3MI, catalyzed by P450 enzymes in human lung microsomes, or the specific pulmonary enzymes known to catalyze the metabolism of 3MI to reactive intermediates (CYP1A1 (11), CYP2A13 (12), and CYP2F3) was compared to the mutagenic activities of the known carcinogens NNK and B(a)P.
Materials and Methods
Microbial Mutagenesis (Ames Assay)
Bacterial mutagenesis assays were conducted using a modified standard reverse mutagenesis plate test with Salmonella (23), as previously published (9). Briefly, 100 μL of a log-phase bacterial culture of either Salmonella typhimurium strain TA98 or TA100 was preincubated for 30 min at 37°C in a shaking incubator with the test compound (vehicle controls were treated with 5 μL of DMSO, and test groups were treated with increasing concentrations of either B[a]P, 3MI, or NNK dissolved in DMSO. For inhibitory assays, test compounds were incubated in the presence or absence of 1 μM 8-MOP or 0.5 mM ABT), 450 μL of 0.1 M phosphate buffer (50 mM Na2HPO4-7H2O, 0.04% H3PO4), 50 pmol of recombinant cytochrome P450 1A1 (CYP1A1 Supersomes from BD Gentest, Woburn, MA), 2F3 [expressed and purified, as previously published (24, 25)], recombinant 2A13 in Sf9 cell microsomes (expressed as previously published, (26)), 2 mg Aroclor 1254–induced rat liver S9 (Celsis, Baltimore, MD), or 0.5 mg human lung microsomes isolated from non-smokers (Celsis, Baltimore, MD) in the presence or absence of 18 mM NADPH. Recombinant enzymes were preincubated with human cytochrome P450 reductase microsomes (BD Biosciences, San Jose, CA) at a 1:3 ratio for 10 minutes, as previously published (25). Following the 30-min preincubation period, 2.5 ml of molten top agar containing 0.5 mM histidine and 0.5 mM biotin was added to each reaction mixture, then poured onto minimal growth agar plates, and incubated upside down for 48 h at 37°C. Test plates containing only the individual cytochrome P450 enzymes, TA98, or DMSO were used as controls. Masked colony counting was performed using a Fisher bacterial colony counter.
Quantitative Western Blot Analysis of Cytochrome P450 Expression in Human Lung Microsomes
30 μg human lung microsomal protein and 15 μL Laemmli buffer containing 5% β-mercaptoethanol were heated for 5 minutes at 90°C. For CYP1A1 and CYP2A13, 0.01, 0.05, 0.1, 0.5, and 1 pmol of recombinant CYP1A1 (BD Gentest) or recombinant CYP2A13 (26) were also mixed with 15 μL Laemmli buffer containing 5% β-mercaptoethanol and were heated for 5 minutes at 90°C to run as standards. Samples (and standards in the case of CYP1A1 and CYP2A13) were then fractionated by NuPAGE 4-12% Bis-Tris polyacrylamide gel (Invitrogen) electrophoresis at 200 V for 50 minutes. Following fractionation, proteins were transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% nonfat milk in 1 X PBST (1 X phosphate buffered saline containing 0.1% Tween-20) for 1 hour at room temperature. The membrane was subsequently incubated overnight at 4°C with primary anti-CYP1A1, anti-CYP2F1, or anti-CYP2A6/13 antibody (1A1 and 2F1 antibodies were purchased from Santa Cruz Biotechnologies, Santa Cruz, CA). Methods for the production and use of the polyclonal antibody that was used to detect CYP2A6/13 were previously published (27). The polyclonal antibody used for our studies was raised in rabbits to the mouse CYP2A5 purified enzyme, and it recognizes both CYP2A6 and CYP2A13 from human lung with equal reactivity (28). Therefore, we refer to it as a polyclonal CYP2A6/13 antibody. The antibody was used previously (28) to determine enzyme expression in 117 human lung microsomal samples, where high resolution SDS-polyacrylamide gel electrophoresis of affinity purified human lung CYP2A6 and CYP2A13 enzymes were separated by their mobility differences and identified and quantitated by western blot/densitometry analysis. The CYP2A6/13 antibody was used in our studies at a 1:200 dilution in the blocking buffer. After the overnight incubation, the membrane was rinsed twice with 1 X PBST and twice with 1 X PBS containing 1% NP-40 and was then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Sigma), at a 1:1000 dilution in 1 X PBST for 1 h at room temperature. After the secondary antibody was removed, the membrane was washed three times with 1 X PBST. For detection of the loading control, GAPDH, membranes were stripped with 0.2 M NaOH for 5 minutes at room temperature, rinsed once with 1 X PBS, blocked as indicated above, and incubated with primary anti-GAPDH antibody (ABCAM, Cambridge, MA) at a 1:5000 dilution in blocking buffer at 4°C overnight. Membranes were then washed and incubated with secondary antibody as indicated above. Protein was detected using Western Blot Luminol Reagent (Santa Cruz Biotechnologies) and visualized using a Kodak Image Station 400.
Statistical analysis
All experiments were repeated in triplicate, and data were reported as mean ± standard deviation. The difference between control and treated cells was tested using one-way analysis of variance (ANOVA) and differences were considered significant with a probability of p ≤ 0.05. Post-hoc analysis of the differences between 3MI- or NNK-treated cells and other treatment groups were calculated using student's t-test and differences were considered significant with a probability of p ≤ 0.05. The statistically significant differences between 3MI-treated cells and either NNK- or BaP-treated cells at each of the four concentrations was evaluated with the two-tailed Students t test.
Results
Ability of 3MI, NNK, and B(a)P to Cause Base Pair Substitution Mutations with Human Lung Microsomes and Rat Liver S9
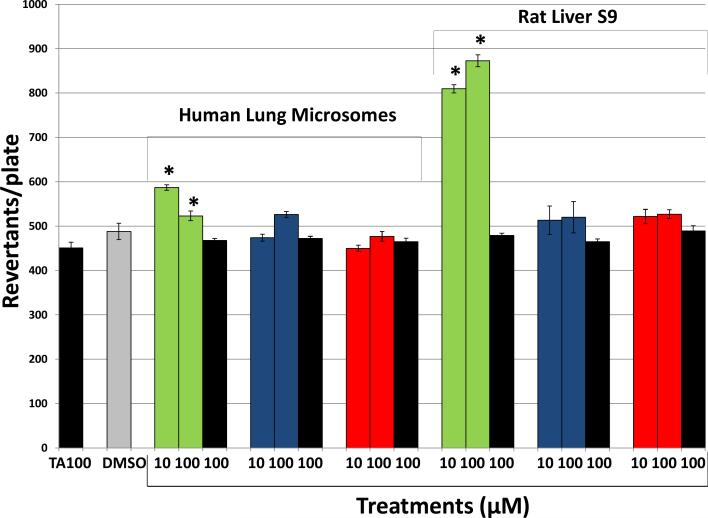
To confirm the results of the study conducted by Reddy et al. (10), mutagenicity of 3MI, NNK, and B(a)P was assessed using the Salmonella typhimurium strain TA100 and either 0.5 mg human lung microsomes or 2 mg rat liver S9 fractions, both in the presence and absence of NADPH, to determine cytochrome P450-dependence. Only B(a)P demonstrated a statistically significant increase in mutagenicity in the presence of NADPH with either human lung microsomes or rat liver S9 (Figure 1). B(a)P was not mutagenic in the absence of NADPH, indicating that the observed mutagenesis was cytochrome P450-dependent. Both 3MI and NNK failed to increase mutagenesis at any concentration in the presence of any activating system, indicating that these compounds do not cause base pair substitution mutations, and in the case of 3MI, confirming the report by Reddy et al. (10) Concentrations lower than those depicted failed to demonstrate significant mutagenesis with any of the test compounds (data not shown). Higher amounts of human lung microsomal proteins (2.0 mg per incubation) did not bioactivate the three chemicals to a greater extent (data not shown).
Figure 1. B(a)P Causes Base Pair Substitution Mutations (Salmonella typhimurium strain TA100), but NNK and 3MI do not.
Incubation of 10 and 100 μM B(a)P in the presence of NADPH (green bars) with either 0.5 mg human lung microsomes (pooled sample from 10 donors with an EROD activity = 0.9 pmol/mg/min) or 2 mg rat liver S9 caused significant increases in the number of revertants per plate, whereas NNK (blue bars) and 3MI (red bars) failed to cause mutagenesis at any concentration. In the absence of cytochrome P450 activation (no NADPH - black bars), the number of revertants per plate was not significantly different from the vehicle-treated control (DMSO - gray bar), indicating that B(a)P-mediated mutagenicity was cytochrome P450-dependent. * indicates p < 0.05 compared to DMSO-treated control. Revertants/plate represents the average number of colonies counted from 3 plates per treatment group from duplicate experiments.
Ability of 3MI, NNK, and B(a)P to Cause Frameshift Mutations with Human Lung Microsomes
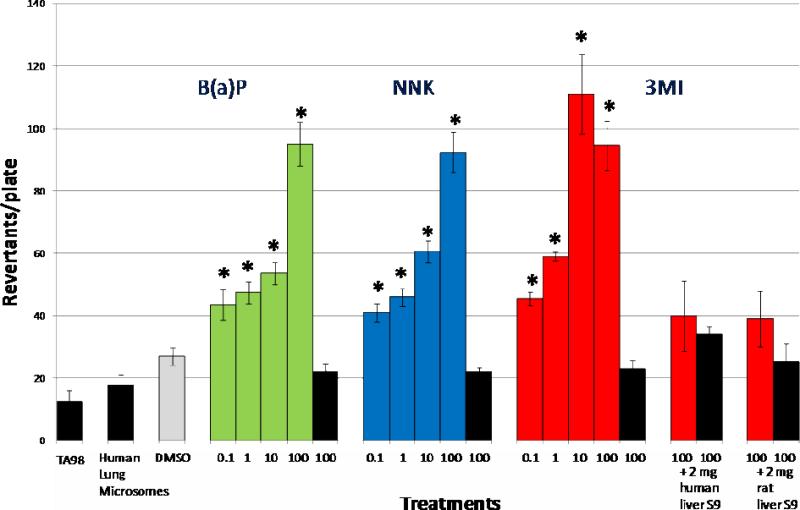
Mutagenicity of increasing concentrations of 3MI, NNK, and B(a)P was assessed with Salmonella typhimurium strain TA98 and 0.5 mg of human lung microsomes both in the presence and absence of NADPH to determine cytochrome P450 dependence. Each chemical demonstrated a statistically significant increase in mutagenicity in the presence of NADPH as compared to DMSO-treated control with as little as 0.1 μM (Figure 2). For NNK and B(a)P, mutagenicity increased in a concentration-dependent manner up through the 100 μM concentration. 3MI, on the other hand, showed increasing mutagenicity up through the 10 μM concentration, but lower revertants/plate with the highest concentration (100 μM). However, at this high concentration, the density of the bacterial background “lawn” appeared to decrease, probably because 3MI was cytotoxic. The cytotoxicity of 3MI to Salmonella typhimurium and to hepatocytes has been reported (10). 3MI was significantly more mutagenic than NNK was at the 0.1 μM (p=0.05), 1.0 μM (p=0.002), and 10 μM (p=0.007) concentrations, although not at the 100 μM (p=0.8) concentration. 3MI was also more mutagenic than BaP was at the 1.0 μM (p=0.006) and 10 μM (p=0.005) concentrations, although not at the 0.1 μM (p=0.1) or 100 μM (p=0.9) concentrations. Neither incubation of 3MI with Salmonella typhimurium strain TA98 and 2 mg human liver S9, or 2 mg rat liver S9 was mutagenic, indicating that the cytochrome P450 enzymes that catalyze the formation of mutagenic intermediates of 3MI are preferentially expressed in the lung. None of the chemicals was mutagenic in the absence of NADPH, indicating that cytochrome P450 activity is required for mutagenesis to occur with these compounds. Incubation with concentrations lower than 0.1 μM failed to produce mutagenicity with any compound (data not shown). This experiment was repeated using microsomes from a different batch of donors and using pooled human lung S9 and the results were nearly identical to those reported in Figure 2 (data not shown).
Figure 2. 3MI is the Most Mutagenic Chemical with Human Lung Microsomes.
Incubation of μM concentrations of benzo(a)pyrene (B(a)P - green bars), 4-(methylnitrosamino)- 1-(3-pyridyl)-1-butanone (NNK - blue bars), and 3-methylindole (3MI - red bars) with Salmonella typhimurium strain TA98 in the presence of 0.5 mg of human lung microsomes (pooled sample from 10 donors with an EROD activity = 0.9 pmol/mg/min) with NADPH indicated that 3MI was the most potent cigarette smoke mutagen. The analysis was repeated with a second batch of human lung microsomes from 10 different donors, and the results were essentially identical. In the absence of cytochrome P450 activation (no NADPH - black bars), the number of revertants per plate was not significantly different from the vehicle-treated control (DMSO - gray bar), indicating that mutagenicity was cytochrome P450-dependent. Neither incubations with 2 mg human liver S9 fractions, nor incubations with 2 mg rat liver S9 fractions, mutated the bacteria. * indicates p < 0.05 compared to DMSO-treated control. Revertants/plate represents the average number of colonies counted from 3 plates per treatment group from duplicate experiments.
Contribution of Individual Cytochrome P450 Enzymes to 3MI-, NNK-, and B(a)P-Mediated Mutagenicity
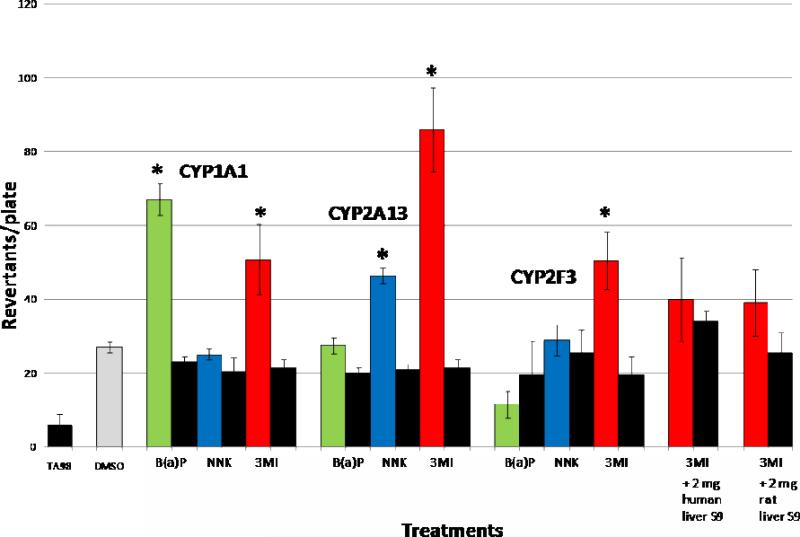
Incubation of 10 μM concentrations of 3MI and B(a)P with Salmonella typhimurium strain TA98 and 50 pmol of 1A1 Supersomes in the presence of NADPH produced statistically significant increases in mutagenicity as compared to DMSO-treated control, whereas incubation of the same concentration of NNK did not result in an increased number of revertants (Figure 3). Incubation of 10 μM concentrations of 3MI and NNK with Salmonella typhimurium strain TA98 and 50 pmol of recombinant 2A13 in the presence of NADPH produced statistically significant increases in the number of revertants per plate, whereas incubation with the same concentration of B(a)P did not. Only 10 μM 3MI produced a significant increase in the number of mutants when incubated with Salmonella typhimurium strain TA98 and 50 pmol of purified 2F3 in the presence of NADPH. Incubation with Salmonella typhimurium strain TA98 and both human and rat liver S9 failed to produce 3MI-mediated mutagenesis in the presence of NADPH, confirming that the lung expressed P450 enzymes required for 3MI-mediated mutagenesis are not expressed in sufficient amounts in liver to catalyze the formation of mutagenic intermediates. None of the chemicals exhibited mutagenicity in the absence of NADPH.
Figure 3. Lung P450s Selectively Bioactivate B(a)P, NNK, and 3MI to Mutate Bacteria.
Incubation of 10 μM B(a)P (green bars), NNK (blue bars), or 3MI (red bars) with Salmonella typhimurium strain TA98 in the presence of NADPH and 50 pmol of CYP1A1 supersomes, recombinant CYP2A13, and recombinant CYP2F3 indicated that all three lung cytochrome P450 enzymes bioactivate 3MI, with CYP2A13 causing the highest levels of mutagenicity. In the absence of NADPH (black bars), mutagenesis did not occur. Neither incubations with 2 mg human liver S9 fractions, nor incubations with 2 mg rat liver S9 fractions, mutated the bacteria, indicating specific metabolism by lung cytochrome P450 enzymes. * indicates p < 0.05 compared to DMSO-treated control. Revertants/plate represents the average number of colonies counted from 3 plates per treatment group from duplicate experiments.
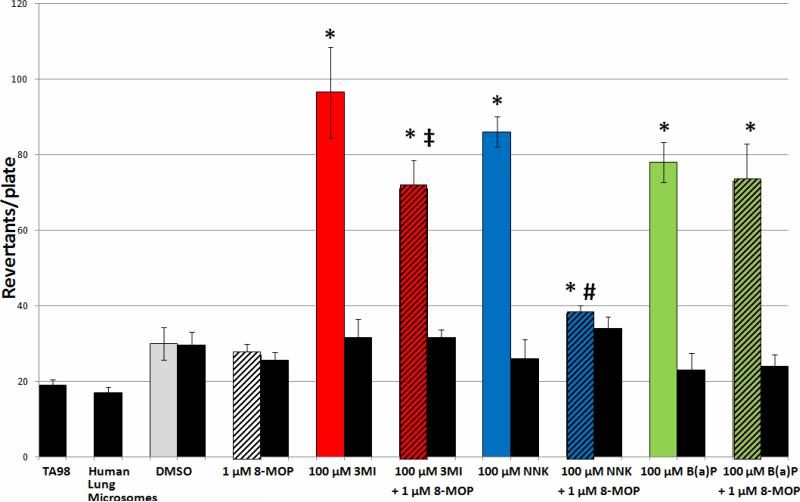
Contribution of 2A13 to the Mutagenicity Observed with Human Lung Microsomes
Incubation of Salmonella typhimurium strain TA98 and 0.5 mg of human lung microsomes with 100 μM 3MI in combination with 1 μM 8-MOP (the concentration previously demonstrated to inhibit CYP2A13 activity (29)) decreased the mutagenicity of 3MI by approximately 30% in the presence of NADPH (Figure 4). Interestingly, the mutagenesis response observed with 100 μM NNK in combination with 8-MOP, although still significantly elevated above DMSO-treated control, was much more attenuated than that observed with 3MI and 8-MOP, indicating that 2A13 plays a more significant role in NNK-mediated mutagenesis than that of 3MI-mediated mutagenesis. B(a)P-mediated mutagenesis, on the other hand, remained unaffected by 8-MOP co-incubation, confirming that its metabolism is not catalyzed by 2A13. Incubation of 8-MOP alone with Salmonella typhimurium strain TA98 and 0.5 mg of human lung microsomes in the presence of NADPH was not mutagenic, and none of the chemicals or combinations of chemicals was mutagenic in the absence of NADPH.
Figure 4. Inhibition of CYP2A13 Activity Inhibits 3MI- and NNK-Mediated Mutagenicity with Human Lung Microsomes, but Does Not Affect B(a)P-Mediated Mutagenicity.
Incubation of the inhibitor of CYP2A13 activity, 1 μM 8-methoxypsoralen (8-MOP), in combination with 100 μM 3MI (red bar with black stripes) and 100 μM NNK (blue bar with black stripes) with Salmonella typhimurium strain TA98 in the presence of 0.5 mg of human lung microsomes (pooled sample from 10 donors with an EROD activity = 0.9 pmol/mg/min) with NADPH significantly decreased mutagenicity as compared to either 3MI (red bar) or NNK (blue bar) incubation alone. However, mutagenicity remained significantly increased above DMSO treated control (gray bar), indicating that CYP2A13 was responsible for some, but not all, of the mutagenicity induced by these compounds. Inhibition of CYP2A13 had no effect on B(a)P-mediated mutagenicity. In the absence of cytochrome P450 activity (no NADPH - black bars) no mutagenesis occurred. 8-MOP incubation alone (white bar with black stripes) had no effect on mutagenesis. * indicates p < 0.05 compared to DMSO-treated control, ‡ indicates p < 0.05 as compared to 3MI treatment, # indicates p < 0.05 compared to NNK treatment. Revertants/plate represents the average number of colonies counted from 3 plates per treatment group from duplicate experiments.
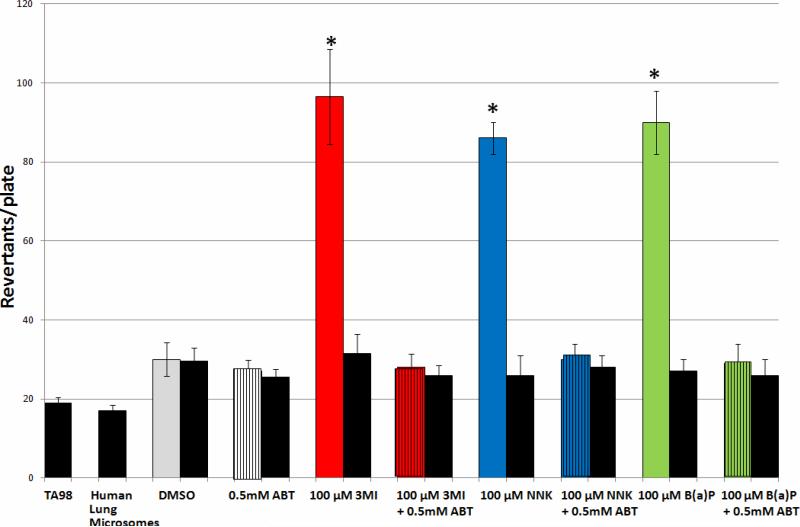
Dependence of 3MI-, NNK-, and B(a)P-Mediated Mutagenicity Upon Cytochrome P450 Activity with Human Lung Microsomes
Incubation of Salmonella typhimurium strain TA98 and 0.5 mg of human lung microsomes with 100 μM concentrations of 3MI, NNK, and B(a)P in combination with 0.5 mM ABT (the concentration previously determined to inhibit 3MEIN formation in human lung cells (30)) in the presence of NADPH completely inhibited the mutagenesis observed with treatment with each of the compounds alone (Figure 5), confirming that cytochrome P450-mediated metabolism of these compounds is necessary for mutagenesis to occur. ABT itself did not prove to be mutagenic, nor were any of the compounds mutagenic in the absence of NADPH.
Figure 5. CYP450 Activity is Required for 3MI, NNK, and B(a)P-Mediated Mutagenicity with Human Lung Microsomes.
Incubation with the suicide substrate inhibitor of virtually all cytochrome P450 activity, 0.5 mM 1-aminobenzotriazole (ABT), in combination with 3MI (red bar with black stripes), NNK (blue bar with black stripes), and B(a)P (green bar with black stripes) with Salmonella typhimurium strain TA98 in the presence of 0.5 mg of human lung microsomes (pooled sample from 10 donors with an EROD activity = 0.9 pmol/mg/min) in the presence of NADPH decreased mutagenesis levels until they were no longer significantly elevated above either DMSO control (gray bars) or treatments in the absence of NADPH (black bars), indicating that mutagenicity of all three compounds was dependent upon cytochrome P450 activity. Treatment with ABT alone (white bar with black stripes) had no effect on mutagenesis. * indicates p < 0.05 compared to DMSO-treated control. Revertants/plate represents the average number of colonies counted from 3 plates per treatment group from duplicate experiments.
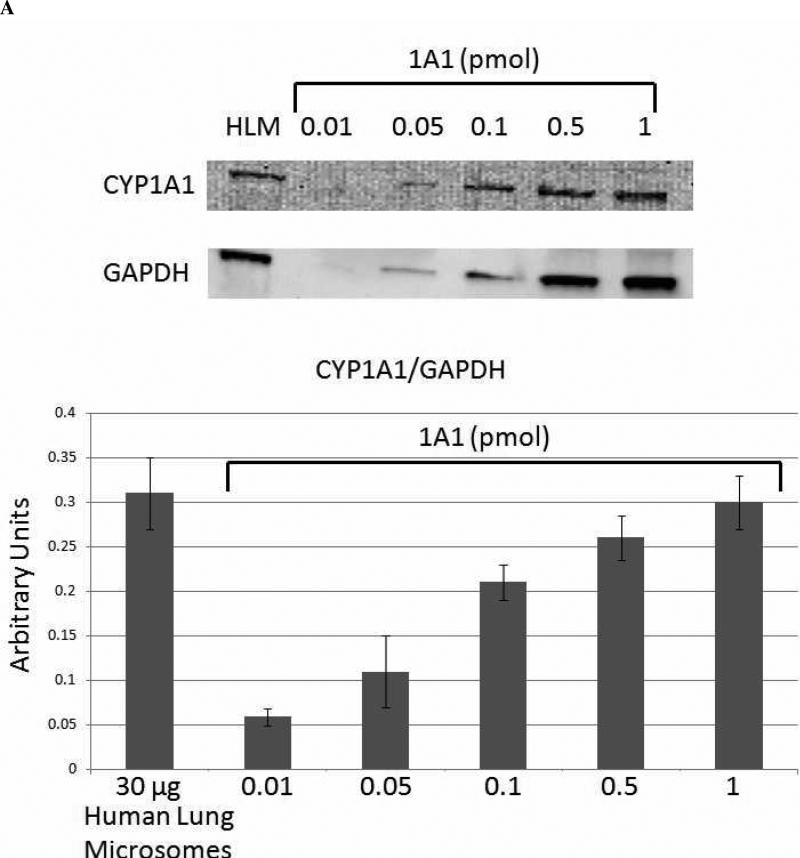
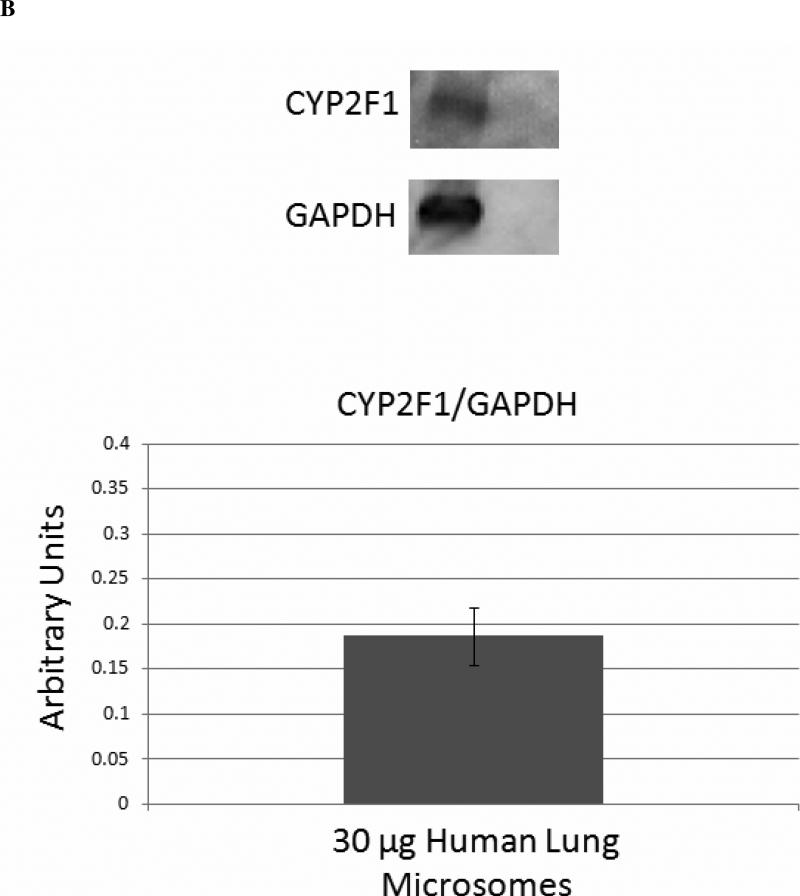
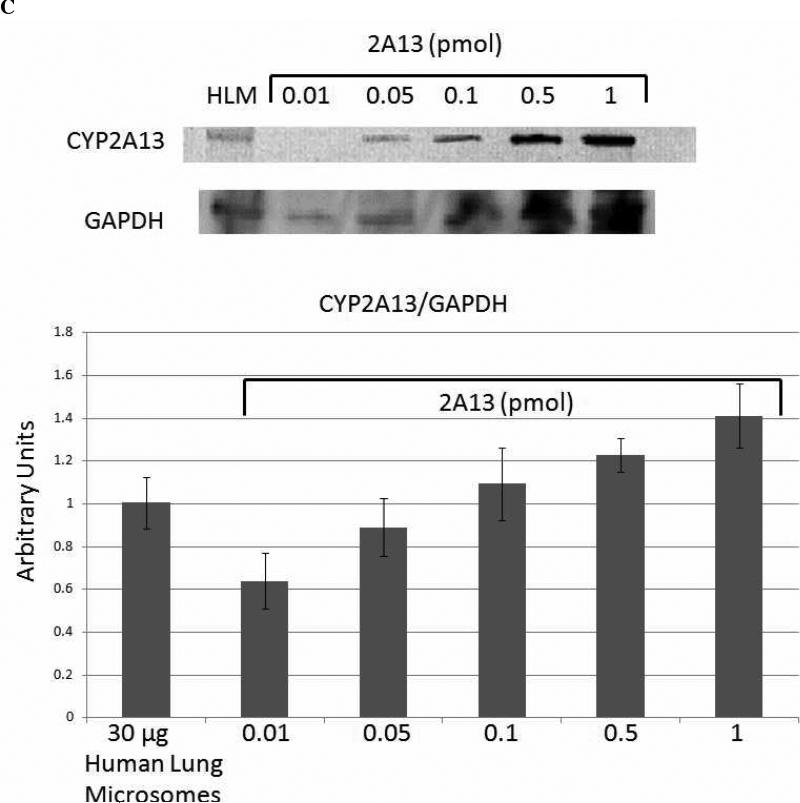
Quantitation of Cytochrome P450 Expression in Human Lung Microsomes
Quantitative western blot analysis was performed to determine the relative expression levels of CYP1A1 (Figure 6A) and CYP2A13 (Figure 6C), as extrapolated from standard curves. The results for the expression of CYP2A13 most likely represent the total expression of CYP2A6 and CYP2A13, because the antibody does not distinguish between the two highly homologous proteins, and they are both expressed in human lung microsomes (28). The human lung microsomes were pooled from 10 non-smoking donors, so it is reasonable to expect that both CYP2A6 and CYP2A13 would be detected, even though the expression of each enzyme is highly variable in human lung microsomal samples (28). 1A1 expression appeared to be nearly ten times higher than 2A6/13 expression in these pooled microsomes. Although we lacked a standard for 2F1 expression, and were thus unable to quantify its expression levels, the mobility of the band was correct, and our previous studies (30, 31) have demonstrated the expression and specificity of the antibody for CYP2F enzymes. Therefore, these results confirmed the expression of CYP2F1 in these human lung microsomes (Figure 6B).
Figure 6. Quantitative Analysis of Cytochrome P450 Expression in Human Lung Microsomes.
Densitometric analysis of quantitative western blots indicated that CYP1A1, CYP2F1, and CYP2A13 were all expressed in the pooled human lung microsomal samples. The results shown for CYP2A13 are most likely for the total expression of CYP2A6 and CYP2A13, because the antibody used does not differentiate between these two highly homologous enzymes, and they are both expressed in human lung microsomal samples (28). CYP1A1 showed the highest levels of expression in the range of 0.5 - 1 pmol CYP1A1/30 μg microsomes, whereas CYP2A13 expression was approximately one order of magnitude lower at 0.05 - 0.1 pmol CYP2A13/30 μg microsomes.
Discussion
Although the lung is the target organ for cigarette smoke-induced toxicity, the vast majority of mutagenesis research has been conducted using hepatic activating systems. This study represents the first examination of 3MI-mediated mutagenicity to focus on extrahepatic cytochrome P450 enzymes and pulmonary activating systems. We demonstrate that, although both human and rat hepatic S9 fail to catalyze 3MI-mediated mutagenicity (Figures 2 and 3), human lung microsomes (Figure 2), recombinant CYP1A1, CYP2A13 and CYP2F3 (Figure 3) all catalyze extensive 3MI-mediated frameshift mutations. The fact that cytochrome P450 activity is necessary for mutagenesis to occur (on the basis of the results from treatment groups without NADPH for all figures, as well as the ABT results in Figure 5) confirms our previous conclusions that 3MI must be bioactivated by cytochrome P450 enzymes. It is unlikely that 3MI is mutagenic in and of itself.
Bioactivation of 3MI to the highly reactive 3MEIN intermediate (Scheme 1) by specific lung, not liver, enzymes has been documented, and production of 3MEIN correlated to the toxicity of 3MI in rodents and in animal and human lung cells (11-13, 17, 30, 32-34). In addition, incubations of lung or liver microsomes with the four individual deoxynucleosides produced only the three adducts shown in Scheme 1. No adducts were detected with the other putative electrophiles (20). Thus, it seems reasonable to conclude that 3MEIN is the most likely DNA-modifying intermediate that produces the striking mutagenic response that we report here.
The present study confirms previous reports that B(a)P causes both frameshift and base pair substitution mutations and demonstrates for the first time that 3MI only causes frameshift mutations, similar to NNK, which does not cause base pair substitution mutations with TA100 (35). The potential carcinogenicity of 3MI is indicated by the observation that 3MI behaves so similarly to what has long been believed to be the most abundant and most potent carcinogen in cigarette smoke, NNK (36). In fact, in the current study, 3MI was an even more potent mutagen than NNK was, when tested at equal concentrations (0.1, 1.0, or 10 μM) with human lung microsomes (Figure 2). The potent mutagenic potential of 3MI in human lung, taken together with the fact that 3MI has been detected in cigarette smoke at higher levels than NNK, compels us to ascertain whether this chemical makes a significant contribution to cigarette smoke-induced lung cancer.
Smoking increases the risk of all types of lung cancers, but particularly the three major histological types: small cell lung carcinoma, squamous cell carcinoma, and adenocarcinoma. The specific cellular origins of these three tumor types have not been unequivocally established, but these tumors are generally believed to begin in the respiratory epithelium, especially in bronchiolar and alveolar epithelial cells (37). Both 2F1 and 2A13 are predominantly expressed in airway epithelial, not parenchymal cells, in human lungs (28, 38-40). CYP2F1 is induced by 3MI metabolites in primary human bronchial epithelial cells (31). Only CYP2W1 and CYP4A1 (from an analysis of human lung tissues of all 57 human P450 genes) have higher ratios of bronchiolar to parenchymal tissue transcription (41). Thus, epithelial cells of the respiratory tract not only contain sufficient amounts of several P450 enzymes that produce 3MEIN, including an enzyme (CTP2F1) that is induced by 3MI, but these cells are probably also the pre-neoplastic cells that give rise to the major tumor types from smoking.
This study provides evidence that 3MI is more mutagenic than is either NNK or B(a)P (Figure 2), the prototype cigarette smoke carcinogens (36), in human lung microsomes. Quantitative western blot analysis revealed that all three of the enzymes examined in this study are expressed in the human lung microsomes used to conduct these assays. 3MI is bioactivated to reactive electrophiles by 1A1 (11), 2F1 (11, 33, 42), and 2A13 (12); NNK is metabolized to reactive intermediates by 2A13 (28) and, to a much lesser extent, by 2F1 (43), and B(a)P is metabolized to its carcinogenic intermediate, benzo(a)pyrene diol epoxide, by 1A1 (44). Thus, the presence of all three enzymes that bioactivate these chemicals in human lung microsomal fractions is consistent with the assumption that the enzymes contribute extensively to the mutagenicity that was observed in our study. Since 3MI is present at such higher concentrations than NNK or B(a)P is in cigarette smoke, it is reasonable to conclude that 3MI may contribute significantly to mutagenesis in the lungs of human smokers, to an extent even greater than the mutagenesis contributed by either NNK or B(a)P, two prototype lung carcinogens. A logical extension of this reasoning is to hypothesize that 3MI exposures from cigarette smoke could be more tumorigenic than are exposures to B(a)P or NNK. Obviously, confirmation of such a vivid theory requires considerable additional experimental and epidemiological studies.
Previous studies have indicated a chemopreventive effect of 8-MOP in a mouse model of lung adenoma (45) and have gone on to attribute this effect to the inhibition of CYP2A enzymes (46). Since recent work has identified CYP2A13 as such an efficient catalyst of the dehydrogenation of 3MI (12), we investigated the ability of 8-MOP to block 3MI-mediated mutagenesis in human lung microsomes. Although 8-MOP blocked nearly all of the NNK-mediated mutagenesis, we were surprised to observe that it only blocked approximately 1/3 of 3MI-mediated mutagenesis (Figure 4). However, after examining the P450 expression in the human lung microsomes (Figure 6), we believe that this is likely due to the fact that CYP2A protein expression is at least ten times lower than 1A1 expression in the samples examined. The amounts of CYP2A13 protein present in these microsomal preparations were probably even lower, given the fact that both CYP2A6 and CYP2A13 are detected by the anti-CYP2A antibody used. Nonetheless, it is reasonable to expect that 3MI-mediated mutagenesis will be more efficient in individuals with relatively high levels of CYP2A13 expression than in those with low levels of CYP2A13 expression (28), and that, in a given individual, 3MI-mediated mutagenesis will be more efficient in parts of the lung where CYP2A13 protein is more abundant, i.e., in airway epithelial cells (40), than in other lung cell types. Recent extensive quantitation of P450 transcription in bronchial and peripheral former smoker human lung tissues confirmed the robust localization (at least 100-fold higher) of CYP2F1 and CYP2A13 in bronchial airways, rather than in pulmonary parenchymal tissues (41). Very little CYP1A1 transcription was found in any lung section of these former smokers. Thus, it is quite probable that bioactivation and mutagenesis of 3MI by CYP2F1 and/or CYP2A13 would be pervasive if airway cells were examined.
In summary, we have demonstrated that 3MI is more mutagenic in human lung microsomes than the prototypical pneumotoxicants NNK and B(a)P and that this effect is likely due to the fact that the three main enzymes responsible for the dehydrogenation of 3MI are specifically expressed in human lung microsomes. We have confirmed that 3MI-mediated mutagenesis is cytochrome P450-dependent and is partially inhibited by blocking the activity of a known metabolizer of 3MI. This research indicates that 3MI is a likely human lung carcinogen and additional investigations into the mechanisms of the mutagenic effects of 3MI in smokers’ lung are warranted.
Acknowledgements
The authors would like to thank David Martin and Evan Makin for their assistance with the bacteria assays. This work was supported by the National Heart, Lung, and Blood Institute (HL13645 to G.S.Y.) and the National Cancer Institute (CA092596 to X.D.).
The abbreviations used are
- 3MI
3-methylindole
- P450
cytochrome P450
- B(a)P
benzo(a)pyrene
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- 3MEIN
3-methyleneindolenine
- S9
supernatant fraction from 9,000 × g centrifugation
- 8-MOP
8-methoxypsoralen
- ABT
1-aminobenzotriazole
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
References
- 1.Smith CJ, Perfetti TA, Garg R, Hansch C. IARC carcinogens reported in cigarette mainstream smoke and their calculated log P values. Food Chem Toxicol. 2003;41:807–817. doi: 10.1016/s0278-6915(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 2.Gray N, Boyle P. The case of the disappearing nitrosamines: a potentially global phenomenon. Tob Control. 2004;13:13–16. doi: 10.1136/tc.2003.005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiserman MJ, Rickert WS. Carcinogens in tobacco smoke: benzo[a]pyrene from Canadian cigarettes and cigarette tobacco. Am J Public Health. 1992;82:1023–1026. doi: 10.2105/ajph.82.7.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann D, Hoffmann I, El-Bayoumy K. The less harmful cigarette: a controversial issue. a tribute to Ernst L. Wynder. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 5.DeMarini DM, Gudi R, Szkudlinska A, Rao M, Recio L, Kehl M, Kirby PE, Polzin G, Richter PA. Genotoxicity of 10 cigarette smoke condensates in four test systems: comparisons between assays and condensates. Mutat Res. 2008;650:15–29. doi: 10.1016/j.mrgentox.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Maertens RM, White PA, Rickert W, Levasseur G, Douglas GR, Bellier PV, McNamee JP, Thuppal V, Walker M, Desjardins S. The genotoxicity of mainstream and sidestream marijuana and tobacco smoke condensates. Chem Res Toxicol. 2009;22:1406–1414. doi: 10.1021/tx9000286. [DOI] [PubMed] [Google Scholar]
- 7.McCann J, Spingarn NE, Kobori J, Ames BN. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci U S A. 1975;72:979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wynder EL, Hoffman D. Tobacco and Tobacco Smoke: Studies in Experimental Carcinogenesis. Academic Press; New York, NY: 1967. [Google Scholar]
- 9.Weems JM, Cutler NS, Moore C, Nichols WK, Martin D, Makin E, Lamb JG, Yost GS. 3-Methylindole is mutagenic and a possible pulmonary carcinogen. Toxicol Sci. 2009;112:59–67. doi: 10.1093/toxsci/kfp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy MV, Storer RD, Laws GM, Armstrong MJ, Barnum JE, Gara JP, McKnight CG, Skopek TR, Sina JF, DeLuca JG, Galloway SM. Genotoxicity of naturally occurring indole compounds: correlation between covalent DNA binding and other genotoxicity tests. Environ Mol Mutagen. 2002;40:1–17. doi: 10.1002/em.10088. [DOI] [PubMed] [Google Scholar]
- 11.Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- 12.D'Agostino J, Zhuo X, Shadid M, Morgan DG, Zhang X, Humphreys WG, Shu YZ, Yost GS, Ding X. The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab Dispos. 2009;37:2018–2027. doi: 10.1124/dmd.109.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skiles GL, Yost GS. Mechanistic studies on the cytochrome P450-catalyzed dehydrogenation of 3-methylindole. Chem Res Toxicol. 1996;9:291–297. doi: 10.1021/tx9501187. [DOI] [PubMed] [Google Scholar]
- 14.Skordos KW, Laycock JD, Yost GS. Thioether adducts of a new imine reactive intermediate of the pneumotoxin 3-methylindole. Chem Res Toxicol. 1998;11:1326–1331. doi: 10.1021/tx9801209. [DOI] [PubMed] [Google Scholar]
- 15.Skordos KW, Skiles GL, Laycock JD, Lanza DL, Yost GS. Evidence supporting the formation of 2,3-epoxy-3-methylindoline: a reactive intermediate of the pneumotoxin 3-methylindole. Chem Res Toxicol. 1998;11:741–749. doi: 10.1021/tx9702087. [DOI] [PubMed] [Google Scholar]
- 16.Yan Z, Easterwood LM, Maher N, Torres R, Huebert N, Yost GS. Metabolism and bioactivation of 3-methylindole by human liver microsomes. Chem Res Toxicol. 2007;20:140–148. doi: 10.1021/tx060239e. [DOI] [PubMed] [Google Scholar]
- 17.Yost GS. Mechanisms of 3-methylindole pneumotoxicity. Chem Res Toxicol. 1989;2:273–279. doi: 10.1021/tx00011a001. [DOI] [PubMed] [Google Scholar]
- 18.Skiles GL, Smith DJ, Appleton ML, Carlson JR, Yost GS. Isolation of a mercapturate adduct produced subsequent to glutathione conjugation of bioactivated 3-methylindole. Toxicology and applied pharmacology. 1991;108:531–537. doi: 10.1016/0041-008x(91)90099-z. [DOI] [PubMed] [Google Scholar]
- 19.Nocerini MR, Yost GS, Carlson JR, Liberato DJ, Breeze RG. Structure of the glutathione adduct of activated 3-methylindole indicates that an imine methide is the electrophilic intermediate. Drug Metab Dispos. 1985;13:690–694. [PubMed] [Google Scholar]
- 20.Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. Detection and characterization of DNA adducts of 3-methylindole. Chem Res Toxicol. 2001;14:1014–1024. doi: 10.1021/tx0100237. [DOI] [PubMed] [Google Scholar]
- 21.Ruangyuttikarn W, Skiles GL, Yost GS. Identification of a cysteinyl adduct of oxidized 3-methylindole from goat lung and human liver microsomal proteins. Chem Res Toxicol. 1992;5:713–719. doi: 10.1021/tx00029a019. [DOI] [PubMed] [Google Scholar]
- 22.Thornton-Manning JR, Nichols WK, Manning BW, Skiles GL, Yost GS. Metabolism and bioactivation of 3-methylindole by Clara cells, alveolar macrophages, and subcellular fractions from rabbit lungs. Toxicology and applied pharmacology. 1993;122:182–190. doi: 10.1006/taap.1993.1186. [DOI] [PubMed] [Google Scholar]
- 23.McCann J, Choi E, Yamasaki E, Ames BN. Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc Natl Acad Sci U S A. 1975;72:5135–5139. doi: 10.1073/pnas.72.12.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kartha JS, Skordos KW, Sun H, Hall C, Easterwood LM, Reilly CA, Johnson EF, Yost GS. Single mutations change CYP2F3 from a dehydrogenase of 3-methylindole to an oxygenase. Biochemistry. 2008;47:9756–9770. doi: 10.1021/bi8005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kartha JS, Yost GS. Mechanism-based inactivation of lung-selective cytochrome P450 CYP2F enzymes. Drug Metab Dispos. 2008;36:155–162. doi: 10.1124/dmd.107.017897. [DOI] [PubMed] [Google Scholar]
- 26.Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- 27.Gu J, Zhang QY, Genter MB, Lipinskas TW, Negishi M, Nebert DW, Ding X. Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J Pharmacol Exp Ther. 1998;285:1287–1295. [PubMed] [Google Scholar]
- 28.Zhang X, D'Agostino J, Wu H, Zhang QY, von Weymarn L, Murphy SE, Ding X. CYP2A13: variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J Pharmacol Exp Ther. 2007;323:570–578. doi: 10.1124/jpet.107.127068. [DOI] [PubMed] [Google Scholar]
- 29.von Weymarn LB, Zhang QY, Ding X, Hollenberg PF. Effects of 8-methoxypsoralen on cytochrome P450 2A13. Carcinogenesis. 2005;26:621–629. doi: 10.1093/carcin/bgh348. [DOI] [PubMed] [Google Scholar]
- 30.Nichols WK, Mehta R, Skordos K, Mace K, Pfeifer AM, Carr BA, Minko T, Burchiel SW, Yost GS. 3-methylindole-induced toxicity to human bronchial epithelial cell lines. Toxicol Sci. 2003;71:229–236. doi: 10.1093/toxsci/71.2.229. [DOI] [PubMed] [Google Scholar]
- 31.Weems JM, Yost GS. 3-Methylindole metabolites induce lung CYP1A1 and CYP2F1 enzymes by AhR and non-AhR mechanisms, respectively. Chem Res Toxicol. 2010;23:696–704. doi: 10.1021/tx9004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowland AM, Yost GS. Selected Pneumotoxic Agents, Comprehensive Toxicology McQueen, C. A., Ed. In: Yost GS, editor. Respiratory Toxicology. 2nd Edition ed. Elsevier Science; Oxford: 2010. pp. 511–547. [Google Scholar]
- 33.Lanza DL, Code E, Crespi CL, Gonzalez FJ, Yost GS. Specific dehydrogenation of 3-methylindole and epoxidation of naphthalene by recombinant human CYP2F1 expressed in lymphoblastoid cells. Drug Metab Dispos. 1999;27:798–803. [PubMed] [Google Scholar]
- 34.Huijzer JC, Adams JD, Jr., Yost GS. Decreased pneumotoxicity of deuterated 3-methylindole: bioactivation requires methyl C-H bond breakage. Toxicology and applied pharmacology. 1987;90:60–68. doi: 10.1016/0041-008x(87)90306-1. [DOI] [PubMed] [Google Scholar]
- 35.Yim SH, Hee SS. Bacterial mutagenicity of some tobacco aromatic nitrogen bases and their mixtures. Mutat Res. 2001;492:13–27. doi: 10.1016/s1383-5718(01)00152-8. [DOI] [PubMed] [Google Scholar]
- 36.Hecht SS. Tobacco smoke carcinogens and lung cancer. Journal of the National Cancer Institute. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 37.Wistuba II, Mao L, Gazdar AF. Smoking molecular damage in bronchial epithelium. Oncogene. 2002;21:7298–7306. doi: 10.1038/sj.onc.1205806. [DOI] [PubMed] [Google Scholar]
- 38.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 39.Carr BA, Wan J, Hines RN, Yost GS. Characterization of the human lung CYP2F1 gene and identification of a novel lung-specific binding motif. The Journal of biological chemistry. 2003;278:15473–15483. doi: 10.1074/jbc.M300319200. [DOI] [PubMed] [Google Scholar]
- 40.Zhu LR, Thomas PE, Lu G, Reuhl KR, Yang GY, Wang LD, Wang SL, Yang CS, He XY, Hong JY. CYP2A13 in human respiratory tissues and lung cancers: an immunohistochemical study with a new peptide-specific antibody. Drug Metab Dispos. 2006;34:1672–1676. doi: 10.1124/dmd.106.011049. [DOI] [PubMed] [Google Scholar]
- 41.Leclerc J, Tournel G, Courcot-Ngoubo Ngangue E, Pottier N, Lafitte JJ, Jaillard S, Mensier E, Lhermitte M, Broly F, Lo-Guidice JM. Profiling gene expression of whole cytochrome P450 superfamily in human bronchial and peripheral lung tissues: Differential expression in non-small cell lung cancers. Biochimie. 2010;92:292–306. doi: 10.1016/j.biochi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Thornton-Manning J, Appleton ML, Gonzalez FJ, Yost GS. Metabolism of 3-methylindole by vaccinia-expressed P450 enzymes: correlation of 3-methyleneindolenine formation and protein-binding. J Pharmacol Exp Ther. 1996;276:21–29. [PubMed] [Google Scholar]
- 43.Smith TJ, Stoner GD, Yang CS. Activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in human lung microsomes by cytochromes P450, lipoxygenase, and hydroperoxides. Cancer Res. 1995;55:5566–5573. [PubMed] [Google Scholar]
- 44.Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers’ lung: comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–1977. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi H, Saoo K, Yokohira M, Ikeda M, Maeta H, Miyazaki M, Yamazaki H, Kamataki T, Imaida K. Pretreatment with 8-methoxypsoralen, a potent human CYP2A6 inhibitor, strongly inhibits lung tumorigenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in female A/J mice. Cancer Res. 2003;63:7581–7583. [PubMed] [Google Scholar]
- 46.Miyazaki M, Yamazaki H, Takeuchi H, Saoo K, Yokohira M, Masumura K, Nohmi T, Funae Y, Imaida K, Kamataki T. Mechanisms of chemopreventive effects of 8-methoxypsoralen against 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced mouse lung adenomas. Carcinogenesis. 2005;26:1947–1955. doi: 10.1093/carcin/bgi156. [DOI] [PubMed] [Google Scholar]