Abstract
Here, we report the sequencing and analysis of eight complete mitochondrial genomes of chimpanzees (Pan troglodytes) from each of the three established subspecies (P. t. troglodytes, P. t. schweinfurthii and P. t. verus) and the proposed fourth subspecies (P. t. ellioti). Our population genetic analyses are consistent with neutral patterns of evolution that have been shaped by demography. The high levels of mtDNA diversity in western chimpanzees are unlike those seen at nuclear loci, which may reflect a demographic history of greater female to male effective population sizes possibly owing to the characteristics of the founding population. By using relaxed-clock methods, we have inferred a timetree of chimpanzee species and subspecies. The absolute divergence times vary based on the methods and calibration used, but relative divergence times show extensive uniformity. Overall, mtDNA produces consistently older times than those known from nuclear markers, a discrepancy that is reduced significantly by explicitly accounting for chimpanzee population structures in time estimation. Assuming the human–chimpanzee split to be between 7 and 5 Ma, chimpanzee time estimates are 2.1–1.5, 1.1–0.76 and 0.25–0.18 Ma for the chimpanzee/bonobo, western/(eastern + central) and eastern/central chimpanzee divergences, respectively.
Keywords: mitochondrial genome, Pan troglodytes, Pan paniscus, divergence time
1. Introduction
Mitochondrial DNA has long been used to investigate questions about primate taxonomy and demography (e.g. Morin et al. 1994; Horai et al. 1995; Gagneux et al. 1999; Schrago & Russo 2003; Eriksson et al. 2004; Raaum et al. 2005). The ability to sequence the complete mtDNA genome relatively quickly and inexpensively has resulted in a number of studies in humans that investigate population history (Ingman et al. 2000; Maca-Meyer et al. 2001; Herrnstadt et al. 2002; Ingman & Gyllensten 2003; Macaulay et al. 2005; Thangaraj et al. 2005; Sun et al. 2006) and selection (Nachman et al. 1996; Mishmar et al. 2003; Elson et al. 2004). However, the application of complete mtDNA sequence data to questions about population history and selection within other species has not been common.
Chimpanzees (Pan troglodytes) and bonobos (Pan paniscus) are our sister species, and studies of genetic variation in these species shed light on their evolutionary histories as well as serve as a comparison to our own history. Both paleoanthropological and genetic studies indicate that that the human and chimpanzee + bonobo lineages diverged 6.5–4.2 Ma (Sarich & Wilson 1973; White et al. 1994; Chen & Li 2001; Glazko & Nei 2003; Kumar et al. 2005), while chimpanzees and bonobos diverged more recently with estimates ranging from 2.5 to 0.8 Ma (Horai et al. 1992; Kaessmann et al. 1999; Stone et al. 2002; Yu et al. 2003; Fisher et al. 2004; Won & Hey 2005; Caswell et al. 2008). Genetic data, as well as some morphological data, suggest strong population structuring within chimpanzees that correlates with subspecies boundaries, and this structure appears to be demarcated by river and habitat boundaries and reinforced by dispersal patterns (Gagneux et al. 2001; Guy et al. 2003; Lockwood et al. 2004; Gonder et al. 2006; Becquet et al. 2007).
Currently, three subspecies, distributed across the central part of Africa, are recognized within chimpanzees. Pan t. schweinfurthii is the easternmost subspecies, located in Tanzania, Burundi, Rwanda, Uganda and the Democratic Republic of Congo. The central subspecies, P. t. troglodytes, is found in Congo, Gabon, the Central African Republic, Equatorial Guinea and Cameroon, and P. t. verus, the westernmost subspecies, is found in Senegal, Guinea-Bissau, Guinea, Sierra Leone, Liberia, Mali and Ivory Coast. MtDNA research has also suggested a potential fourth subspecies, P. t. ellioti (Oates et al. 2009), formerly known as P. t. vellerosus, in Nigeria (Gonder et al. 1997, 2006; Gagneux et al. 1999), although the limited Y-chromosome evidence has failed to support this claim (Stone et al. 2002).
Demographic inferences about chimpanzee subspecies are limited but mostly indicate a larger effective population size in the central subspecies, and an initial split between the western and central/eastern subspecies (Deinard & Kidd 1999; Kaessmann et al. 1999; Stone et al. 2002; Fisher et al. 2004; Won & Hey 2005; Becquet et al. 2007). There has also been a distinction between haploid markers (both mtDNA and Y-chromosome studies) that have shown high levels of structure (corresponding to subspecies designations) and autosomal nuclear data that have suggested significant gene flow, differences in male and female effective population sizes and/or incomplete sorting of lineages. On the one hand, this has led to some researchers nominating new subspecies based on results from haploid markers (Gonder et al. 1997; Gonder Disotell & Oates 2006), while others have proposed elimination of subspecies designations altogether based on the nuclear data (Fisher et al. 2006). In addition, some experts suggest that the western and Nigerian chimpanzees should form one subspecies (P. t. vellerosus), while the central and eastern chimpanzees should belong to a second subspecies (P. t. troglodytes; Gonder Disotell & Oates 2006). More recently, a large autosomal microsatellite dataset has supported substructure within chimpanzees that corresponds to the subspecies designations (Becquet et al. 2007).
Comparisons of levels of intraspecific variability have important implications for understanding the evolution of hominoid genomes and clarifying the demographic history of contemporary populations of humans and great apes (Stone & Verrelli 2006). Because of the different signals regarding population history as well as the conflicting estimates of divergence times based on different loci, a better sampling of markers and populations is needed. In this study, we report complete mtDNA sequences in eight chimpanzees including individuals from all of the three currently recognized subspecies as well as an individual with a P. t. ellioti mtDNA haplotype to assess the neutrality of evolutionary patterns of the mtDNA genome and to examine intraspecific diversity. A major emphasis of our mtDNA analysis is to investigate why the timing of divergence between chimpanzees and bonobos and among the subspecies of chimpanzees that have been reported from previous mtDNA studies are much older than those obtained using the nuclear DNA (e.g. Horai et al. 1995; Stone et al. 2002; Fisher et al. 2004; Won & Hey 2005).
2. Material and methods
(a). Materials
The complete mitochondrial genomes from seven wild-born chimpanzees (four P. troglodytes verus, two P. t. schweinfurthii and one P. t. ellioti) and one captive born chimpanzee (haplotype corresponds to P. t. troglodytes) were sequenced for this study. When unknown, subspecies status was determined based on a comparison of the mtDNA HVI region sequence to those from chimpanzees of known subspecies (Morin et al. 1994; Gonder et al. 1997; Stone et al. 2002). Two P. t. verus samples, Pt115 and Pt120 (ISIS no. 2738 and 2216), and the P. t. ellioti sample, Pt114 (ISIS no. 2412), were from the New Iberia Primate Center. The remaining two P. t. verus samples, Pt82 and Pt105 (North American regional studbook for the chimpanzee no. 341 and ISIS no. 2435), were from the Riverside Zoo in Scottsbluff, NE, and the Southwest Foundation for Biomedical Research, respectively. The two P. t. schweinfurthii samples, Pt96 and Pt161 (ISIS no. 3020 and 925), and the P. t. troglodytes sample, Pt13 (ISIS no. 4441), were from the Primate Foundation of Arizona. Whole blood samples (5–10 ml) were taken during routine veterinary examinations, and DNA was isolated using a standard phenol/chloroform-based extraction (Sambrook & Russell 2001).
(b). Polymerase chain reaction and nucleotide sequencing
The complete mtDNA genome was amplified in 28 overlapping segments using the polymerase chain reaction (PCR) in a PTC-200 thermal cycler (MJ Research). PCR primers for each segment are listed in the electronic supplementary material. For most segments, PCR conditions were: 94°C for 5 min (94°C, 30 s; annealing temperature specified in table S1 in the electronic supplementary material, 30 s; 72°C, 30 s) for 35 cycles, followed by a single final extension of 72°C for 5 min. A touchdown PCR protocol was used for segments amplified with primers L846 and H1620, L4589 and H5276 and L14110 and H14900. For these segments, PCR conditions were: 94°C for 5 min (94°C, 30 s; 10°C above annealing temperature specified in table S1 in the electronic supplementary material minus 0.5°C per cycle, 30 s; 72°C, 30 s) for 20 cycles, (94°C, 30 sec; annealing temperature specified in table S1 in the electronic supplementary material, 30 s; 72°C, 30 s) for another 20 cycles, then 72°C for 5 min. PCR products were purified with the QIAquick Purification Kit (Qiagen) and sequenced in two directions, using the BigDye terminator cycle sequencing kit v. 3.1 (Applied Biosystems) and an Applied Biosystems 3730 capillary sequencer. Sequence trace files were assembled using the SeqMan program (DNAStar), and then manually checked and aligned.
MtDNA insertions into the nuclear genome (numts) can complicate analyses and invalidate conclusions if they are mistakenly amplified in the place of authentic mtDNA sequence (Bensasson et al. 2001; Thalmann et al. 2004). Although there was no direct evidence that numts had been amplified here (i.e. there were no ‘heterozygous’ positions and no nucleotide mismatches between overlapping fragments), we checked the authenticity of our sequences with the long-range PCR method described by Thalmann et al. (2004) using Platinum Taq HiFi (Invitrogen). Briefly, for each chimpanzee individual, we amplified two large fragments, A (approx. 9 kb) and B (approx. 10 kb), which overlap each other at both ends to cover the entire mtDNA genome using primers listed in table S1 in the electronic supplementary material. Five microlitres of the PCR product was electrophoresed through a 2 per cent NuSieve GTG low-melting agarose (Cambrex) gel. Bands were excised and melted in 100 µl Molecular Biology Reagent Water (Sigma) at 55°C for 1 h, and then vortexed thoroughly. Four microlitres of this mixture was then used to amplify and sequence three of our original segments, using the methods specified above. One of these segments (primers L15255 and H107, 1214 bp) was amplified from both fragments A and B. Additionally, we amplified a 687 bp segment (L4589 and H5276) from fragment A and an 862 bp segment (L8333 and H9195) from fragment B. The resulting sequences were compared with each other and with the original sequence for each individual. There were no observed discrepancies, supporting the authenticity of the complete mtDNA genome sequences generated for these eight individuals. These data were submitted to GenBank and are listed under accession numbers GU112738–GU112745.
(c). Data analysis
We compared our sequences with those from previous studies, including two P. t. verus (Jenny, GenBank accession number X93335 and the chimpanzee mtDNA reference sequence no. NC 001643), one P. paniscus (no. NC 001644), one gorilla (Gorilla gorilla, no. NC 001645), one orangutan from each of the two subspecies (Pongo pygmaeus, no. NC 001646 and Pongo pygmaeus abelii, no. NC002083), one gibbon (Hylobates lar, no. NC 002082), the Cambridge human reference sequence (Homo sapiens, no. AC000021) and 53 additional humans (Anderson et al. 1981; Horai et al. 1992, 1995; Arnason et al. 1996; Xu & Arnason 1996; Andrews et al. 1999; Ingman et al. 2000). The hypervariable region or D-loop is non-coding and was excluded from most analyses because it is known to have a very different mutational pattern.
Two estimates of diversity were calculated for each locus: π is based on the average number of nucleotide differences per site between two sequences randomly drawn from a sample and θs is based on the sample size-corrected proportion of segregating sites (Watterson 1975; Nei 1987). The Jukes & Cantor (1969) correction was applied to all sequence comparisons involving interspecific variation and divergence (Lynch & Crease 1990). Under equilibrium conditions with respect to mutation and drift, both π and θs estimate the neutral parameters: 2Neμ for mtDNA, where Ne is the effective population size and μ is the neutral mutation rate. Tajima's D-statistic was calculated to test for deviations from neutral frequency distribution (Tajima 1989). The measures of diversity and tests of neutrality were performed with the program DnaSP 4.0 (Rozas et al. 2003).
The mutation rate for the complete genome, excluding the D-loop region, was calculated from the data as follows: mutation rate per site per year = (k/2Tsplit) × l, where k is the mean genetic distance, l the length of the sequence and Tsplit the time in years since the human and chimpanzee divergence. Tsplit is assumed to be 6 Ma. The mean genetic distance was estimated between the 10 chimpanzees examined in this study and the 53 humans from Ingman et al. (2000) using the Tamura–Nei distance (Tamura & Nei 1993) with the evolutionary rates among sites modelled using the gamma distribution in MEGA 4 (Tamura et al. 2007). The shape parameter for the gamma distribution was calculated using ModelTest as noted below.
ModelTest (Posada & Crandall 1998) was used to select the most appropriate evolutionary model for each dataset. For the 13 protein-coding genes, the general time-reversible (GTR) model with a proportion of the sites invariable (I) and gamma-distributed rates among sites (Γ) was selected by the AIC criterion. For the complete genome without the D-loop, the GTR + Γ model was selected by the AIC. Phylogenetic analyses were then performed using PAUP* (Swofford 2003) using the model and parameters estimated by ModelTest. Ten random addition sequences with TBR branch swapping were used to obtain the maximum-likelihood estimates of the phylogenies. Bootstrap analyses were subsequently performed using 100 bootstrap replicates and TBR searches for each replicate for each dataset.
We calculated divergence times between species and subspecies using two relaxed-clock methods: MultiDivTime (MDT) and BEAST (Thorne & Kishino 2002; Drummond & Rambaut 2007). In these methods, a Markov Chain Monte Carlo (MCMC) procedure is used within a Bayesian analysis framework to estimate the posterior distributions of evolutionary rates and divergence times, given priors on phylogenetic relationships and calibration nodes. These analyses were performed using DNA sequence alignment of the complete mitochondrial genome, except the D-loop region, and for 13 protein-coding genes separately. The protein-coding genes were analysed at amino acid and DNA sequence level as a super-alignment. We also analysed the fourfold (4F) degenerate sites by themselves, as their evolutionary patterns are expected to be the closest to the strict neutrality (Kumar et al. 2005).
In MDT, branch lengths of the amino acid dataset (16 taxa, 3772 sites) were estimated with the mitochondrial mammal (mtmam.dat) model of substitution, while for nucleotides (genome and 4F sites) the F84 + gamma model was used within the PAML program package (Yang 2007). Initial number of sites analysed for whole genome and 4F degenerate sites were 15 514 and 1843, respectively. All sites containing gaps and ambiguous nucleotides were excluded from the analyses. Other parameters used in MDT were: 10 000 sampling of the Markov chain, with sampling frequency every 100, burn-in of 100 000, and bigtime was set to 50 Ma. The root to tip distance (rttm) was set at 25 Ma with identical standard deviation (rttmsd).
The same data sets were analyzed also in BEAST with an uncorrelated lognormal relaxed clock. Substitution model used were the mtREV + gamma + invariant sites model for amino acids and the GTR + gamma + invariant sites for nucleotides. We also separated models for population and species divergences by using two populations based on the geographical distribution of the western and eastern + central chimpanzee subspecies (two populations plus one species model). The remaining divergences were estimated under a Yule speciation process. The length of the chain for each analysis was adjusted to the dataset in order to obtain effective sample sizes above 200 for all parameters.
The same prior information on times (lower and upper bounds) was used for the two molecular clock methods. These included the times of the following species divergences: gorilla versus chimpanzee + human divergence (10–6.5 Ma) and the chimpanzee and human divergence (6.5–4.2 Ma) times defined as uniform distributions. These calibrations were used together or separately depending on the dataset and the hypothesis to test. MDT and BEAST yielded divergence times and their 95% credibility intervals (CrIs).
In addition to species and subspecies divergences, BEAST also produces the age of the most recent common ancestor (TMRCA) based on the population sample included. We compared these estimates with those obtained using the GeneTree program, which simulates a coalescent process including time information conditional on a specific haplotype tree with a given value of θ (Griffiths & Tavaré 1994; Bahlo & Griffiths 2000). We estimated θ for each dataset by GeneTree using the maximum-likelihood method. Indels and the D-loop were not included in the analyses, and constant population sizes and panmixia were assumed. Simulation results are based on 10 million replicate runs. To calculate the TMRCA in years, a chimpanzee generation time of 15 years was used and the divergence between chimpanzees and humans was set at 6 Ma.
3. Results
The complete mtDNA genomes (16 541 bp) of 10 chimpanzees contain 695 segregating sites (table 1). We also observe insertion/deletion (indel) polymorphisms in both the 12S rRNA gene and the D-loop. In the 12S rRNA gene, an insertion of a guanine (G) after nucleotide position 138 was present in all sequences except the chimpanzee reference sequence. This G is also present in humans (Anderson et al. 1981; Ingman et al. 2000) where it is found in stem 7 of the secondary structure model of Neefs et al. (1993). In addition, a length polymorphism was found in the 12S rRNA gene in the loop between stems 23 and 24. Here, at nucleotide positions 378–384, an unstable run of cytocines resulted in significant variation with at least 6–15 cytocines present. This region was difficult to PCR and produced a heteroplasmic sequence. In the D-loop, a total of 12 indels was found. Because it is difficult to properly incorporate indels into models of sequence evolution for estimating divergence times, these were removed from further analyses.
Table 1.
Diversity statistics for mtDNA. Length is excluding gaps. *Significance: p < 0.05.
| taxa | n | length | s | π (%) | θ (%) | Tajima's D |
|---|---|---|---|---|---|---|
| complete genome | ||||||
| P. troglodytes | 10 | 16 543 | 695 | 1.48 | 1. 49 | −0.16 |
| P. t. schweinfurthii | 2 | 16 556 | 38 | 0.23 | 0.23 | |
| P. t. s. and P. t. t. | 3 | 16 556 | 134 | 0.54 | 0.54 | |
| P. t. verus (w/Nigeria) | 7 | 16 548 | 413 | 0.95 | 1.02 | −0.49 |
| P. t. verus | 6 | 16 548 | 320 | 0.84 | 0.85 | −0.1 |
| H. sapiensa | 53 | 16 553 | 657 | 0.37 | 0.87 | −2.12* |
| without D-loop | ||||||
| P. troglodytes | 10 | 15 441 | 544 | 1.23 | 1.25 | −0.16 |
| P. t. schweinfurthii | 2 | 15 443 | 26 | 0.17 | 0.17 | |
| P. t. s. and P. t. t. | 3 | 15 443 | 95 | 0.41 | 0.41 | |
| P. t. verus (w/Nigeria) | 7 | 15 441 | 298 | 0.72 | 0.79 | −0.59 |
| P. t. verus | 6 | 15 441 | 224 | 0.62 | 0.64 | −0.17 |
| H. sapiensa | 53 | 15 430 | 516 | 0.29 | 0.74 | −2.23* |
Non-synonymous polymorphisms were found in all 13 protein-coding genes. On average, chimpanzee sequences contained 28.8 non-synonymous differences when compared with 8.91 observed in humans. The estimates of synonymous and non-synonymous substitution diversities for each mitochondrial gene as well as comparative data from humans are shown in table 2. When examining silent and replacement sites separately (table 2), Tajima's test rejected strict neutrality at a 5 per cent significance level in chimpanzees only for non-synonymous sites at CO3 and ND4L. Humans show many significantly negative Tajima's D-statistic for both synonymous and non-synonymous sites. When data from only P. t. verus (without Pt114) was examined, three genes (ND1, CO2 and ND6) showed significantly negative Tajima's D-values at replacement sites. When Pt114 was included in P. t. verus, only replacement sites at CO3 produced a significantly negative value (data not shown).
Table 2.
Population diversity estimates and tests of neutrality at mtDNA protein-coding genes in humans (n = 53, Ingman et al. 2000) and chimpanzees (n = 10). *p < 0.05.
| taxa and genes | lengtha | replacement |
silent |
||||
|---|---|---|---|---|---|---|---|
| S | πb | TDc | S | π | TD | ||
| Pan troglodytes | |||||||
| ND1 | 954 | 3 | 0.0016 | 0.13 | 41 | 0.0539 | −0.27 |
| ND2 | 1041 | 16 | 0.0066 | −0.46 | 33 | 0.046 | 0.18 |
| CO1 | 1539 | 4 | 0.0016 | 1.32 | 42 | 0.042 | 0.51 |
| CO2 | 681 | 2 | 0.0013 | −0.03 | 14 | 0.026 | −0.46 |
| ATP8 | 204 | 4 | 0.0097 | 0.23 | 4 | 0.025 | −0.45 |
| ATP6 | 678 | 7 | 0.0054 | 0.39 | 25 | 0.051 | 0.11 |
| CO3 | 783 | 6 | 0.0021 | −1.8* | 28 | 0.058 | 0.68 |
| ND3 | 345 | 5 | 0.0055 | −0.83 | 10 | 0.045 | 0.41 |
| ND4L | 294 | 2 | 0.0019 | −1.4* | 9 | 0.048 | 0.81 |
| ND4 | 1377 | 12 | 0.0032 | −1 | 46 | 0.045 | −0.05 |
| ND5 | 1809 | 17 | 0.0046 | 0.23 | 63 | 0.05 | 0.07 |
| ND6 | 522 | 3 | 0.0022 | 0.54 | 16 | 0.045 | 0.33 |
| CYTB | 1140 | 8 | 0.0035 | 0.11 | 54 | 0.069 | 0.18 |
| all genes | 11 316 | 88 | 0.0034 | −0.34 | 385 | 0.049 | 0.14 |
| Homo sapiens | |||||||
| ND1 | 954 | 8 | 0.0007 | −1.93* | 19 | 0.0087 | −1.49* |
| ND2 | 1041 | 13 | 0.0012 | −2.04* | 29 | 0.0071 | −2.31* |
| CO1 | 1539 | 8 | 0.0008 | −1.32 | 40 | 0.0083 | −2.15* |
| CO2 | 681 | 3 | 0.0007 | −1.11 | 19 | 0.0067 | −2.32* |
| ATP8 | 204 | 2 | 0.0014 | −0.96 | 6 | 0.0158 | −1.18 |
| ATP6 | 678 | 8 | 0.002 | −1.17 | 22 | 0.0096 | −2.08* |
| CO3 | 783 | 4 | 0.0003 | −1.73* | 25 | 0.0105 | −2.01* |
| ND3 | 345 | 3 | 0.0028 | 0.12 | 10 | 0.0108 | −1.67* |
| ND4L | 294 | 1 | 0.0002 | −1.32* | 9 | 0.015 | −1.09 |
| ND4 | 1377 | 12 | 0.0007 | −2.15* | 43 | 0.0116 | −1.89* |
| ND5 | 1809 | 23 | 0.0015 | −1.95 | 51 | 0.0095 | −2.12* |
| ND6 | 522 | 5 | 0.0013 | −1.31 | 20 | 0.0118 | −2.01* |
| CYTB | 1140 | 16 | 0.0011 | −2.2* | 26 | 0.0099 | −1.66* |
| all genes | 11 316 | 106 | 0.0011 | −2.19* | 318 | 0.0096 | −2.17* |
aLength does not include the stop codon, for ‘all genes’, ATP8 and ND4L were truncated so that overlapping regions with other genes were not counted twice.
bNucleotide diversity.
cTajima's D-test.
Our analysis shows that chimpanzees harbour approximately four times more nucleotide diversity (π) than humans (table 1), while θs is 1.7 times greater. Within chimpanzees, P. t. verus exhibited the most variation; however, multiple samples of the central subspecies were not included in this study and only two P. t. schweinfurthii were sampled. Despite the population structure within chimpanzees, Tajima's test of the complete genome and also of only the protein-coding genes did not show a significant departure from neutrality, while it is significantly negative in humans (tables 1 and 2). These results support the assumption of constant population size in chimpanzees used in the GeneTree analysis.
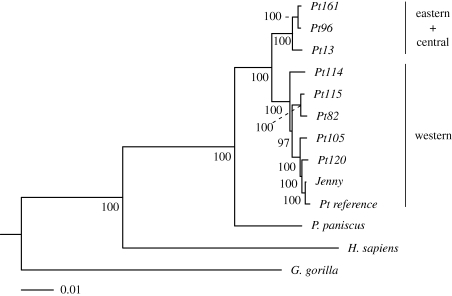
For a reduced dataset including only the chimpanzees, the best evolutionary model selected by ModelTest using both likelihood ratio tests and the AIC was the GTR + Γ model with the shape parameter in the gamma distribution, α, estimated to be 0.066. PAUP* was then used to estimate the ML tree and bootstrap support. Using the entire dataset, the phylogenetic analyses of both the complete genome without the D-loop and using only the protein-coding genes produced similar results (figure 1). Bootstrap support was high (greater than 97%) for all nodes. The western chimpanzee sequences, Pt82, Pt105, Pt115 and Pt120, the chimpanzee mtDNA reference sequence and the sequence for the chimpanzee Jenny cluster together with the Nigerian sequence, Pt114, as their most closely related taxon. The eastern chimpanzees, Pt96 and Pt161, cluster with the central chimpanzee sequence, Pt13.
Figure 1.
Maximum-likelihood phylogeny of chimpanzees, human and gorilla. Full mitochondrial genome (excluding the D-loop) and 13 mitochondrial protein-coding genes gave similar results. The geographical location of the chimpanzee populations is shown. Bootstrap values are shown near each node.
We first inferred a time tree of chimpanzee and bonobo evolution by using the MDT software. In this case, we used two primary time constrains: 10.0–6.5 Ma for gorilla/(human + chimpanzee) divergence and 6.5–4.2 for human/chimpanzee divergence. Using this calibration for full mitochondrial genome (excluding the D-loop), only amino acid alignments of 13 protein-coding genes and only 4F degenerate sites produced similar estimates (table 3). The chimpanzee/bonobo split was estimated to be 3.2–2.3 Ma. The time estimates for western/(eastern + central) and eastern/central splits were dated to 1.68–1.13 and 0.46–0.34 Ma, respectively (table 3). Interestingly, MDT analysis yielded a human/chimpanzee divergence date of approximately 5.7 Ma, which would be considered young by some experts. Therefore, we re-estimated divergence times in MDT by assuming the human/chimpanzee divergence to be 6.5 and found that all the resulting time estimates increased proportionally (table 3).
Table 3.
Time estimates and 95% CrIs (in million years) for major divergences in the phylogeny obtained using MDT and BEAST for three datasets (mitochondrial genome, amino acids, and 4F degenerate sites). MDT and BEAST coalescent model do not model population structure within chimpanzees, which, instead, is accounted for in BEAST 2 + 1 model. MDT divergence times in parenthesis for the 4F degenerate sites are obtained with the human/chimpanzee calibration fixed at 6.5 Ma. All other times are estimated using gorilla/human + chimpanzee (10.0–6.5 Ma) and human/chimpanzee (6.5–4.2 Ma).
| Mt genomea |
amino acidsb |
4Fc |
||||
|---|---|---|---|---|---|---|
| time | 95% CrI | time | 95% CrI | time | 95% CrI | |
| MDT | ||||||
| human/chimpanzee | 5.69 | 4.76–6.46 | 5.73 | 4.72–6.46 | 5.65 (6.5) | 4.49–6.46 |
| chimpanzee/bonobo | 2.33 | 1.75–3.14 | 3.17 | 2.23–4.24 | 2.81 (3.35) | 1.68–4.19 |
| western/eastern + central | 1.13 | 0.80–1.70 | 1.5 | 0.86–2.47 | 1.68 (2.04) | 0.84–3 |
| Nigerian/western | 0.55 | 0.37–0.86 | 0.78 | 0.4–1.48 | 0.811 (1) | 0.35–1.66 |
| eastern/central | 0.34 | 0.22–0.58 | 0.43 | 0.16–0.93 | 0.458 (0.57) | 0.7–1.11 |
| BEAST (coalescent model) | ||||||
| human/chimpanzee | 5.48 | 4.63–6.5 | 5.43 | 4.46–6.5 | 5.28 | 4.24–6.35 |
| chimpanzee/bonobo | 2.02 | 1.53–2.57 | 2.79 | 1.88–3.72 | 1.79 | 1.23–2.42 |
| western/eastern + central | 0.96 | 0.70–1.22 | 1.13 | 0.84–1.85 | 0.92 | 0.62–1.27 |
| Nigerian/western | 0.48 | 0.35–0.61 | 0.72 | 0.45–1.03 | 0.42 | 0.27–0.60 |
| eastern/central | 0.27 | 0.18–0.37 | 0.35 | 0.15–0.58 | 0.21 | 0.11–0.33 |
| BEAST (2 + 1 model)d | ||||||
| human/chimpanzee | 4.94 | 4.2–5.68 | 4.87 | 4.2–5.73 | 4.7 | 4.2–5.43 |
| chimpanzee/bonobo | 1.76 | 1.35–2.19 | 2.28 | 1.5–3.16 | 1.49 | 1.05–1.99 |
| western/eastern + central | 0.83 | 0.64–1.04 | 1.05 | 0.67–1.45 | 0.76 | 0.51–1.01 |
| Nigerian/western | 0.41 | 0.32–0.52 | 0.57 | 0.36–0.80 | 0.35 | 0.23–0.47 |
| eastern/central | 0.23 | 0.16–0.31 | 0.27 | 0.1–0.44 | 0.18 | 0.095–0.27 |
aMitochondrial genome excluding the D-loop.
bThirteen mitochondrial protein-coding genes.
c4F degenerate sites of the 13 protein-coding genes.
dTwo populations (western and eastern + central) and one speciation model (Yule).
Because MDT assumes that evolutionary rates among lineages are autocorrelated, we also estimated divergence times using the BEAST software, which allows rates among lineages to vary independently. For genomic DNA and amino acid sequence analysis, BEAST analyses produced estimates very similar to those from MDT for the same sequences (table 3). However, the BEAST analysis of 4F degenerate sites yielded much younger estimates (table 3). The above analyses, however, did not take into account the population structure of chimpanzees. In order to incorporate this biological reality, we conducted BEAST analysis by assuming that each P. t. subspecies constitutes a population (western subspecies and eastern + central subspecies). This leads to a two populations plus one speciation model (2 + 1 model), which yields younger divergence times than those obtained from MDT or from other BEAST analyses (table 3).
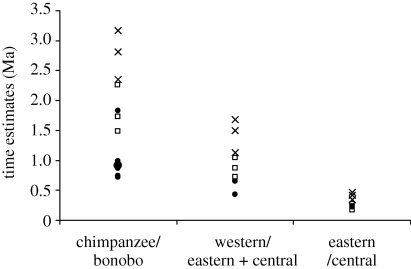
Although the use of 2 + 1 model in BEAST makes results younger compared with the estimates obtained without considering population structure within chimpanzees, the time estimates based on amino acid sequences are 50 per cent older than those from the analysis of DNA sequences (genome as well as 4F degenerate sites). Kumar et al. (2005) have previously shown that amino acid time estimates are often older and not preferred for relatively recent divergences. Overall, the 2 + 1 model produces time estimates that are more similar to those reported from nuclear autosomal loci (figure 2). Based on the 2 + 1 BEAST analysis, the average divergence times within the chimpanzees across three datasets are 2.28–1.49, 1.05–0.76 and 0.27–0.18 Ma for the chimpanzee/bonobo, western/(eastern + central) and the eastern/central divergences, respectively (table 3). These results highlight the need for modelling populations correctly while estimating times of closely related species and subspecies divergences when using relaxed-clock methods.
Figure 2.
Compilation of estimated divergence times between chimpanzees and bonobos and within chimpanzees from literature (autosomal loci only, filled circle) and this study (MDT, cross; BEAST (2 + 1), square).
The time to the MRCA (TMRCA) for each chimpanzee population in the BEAST analysis was 0.35 and 0.18 (table 3). We compared these estimates of TMRCA with those obtained from GeneTree, which employs a coalescence process and estimates of diversity to generate time estimates. The estimate of sequence diversity for the complete genome, excluding the D-loop, was 1.38 × 10−8 substitutions per site per year assuming a chimpanzee–human divergence time of 6 Ma. Using this estimate and applying GeneTree separately to each subspecies (western and eastern + central), we obtained TMRCA of 202 000 (±14 000) and 180 000 (±19 000) years, respectively. These results were similar to those found by BEAST in the 4F dataset.
4. Discussion
Studies of diversity and divergence times in chimpanzees, and primates in general, have frequently used mtDNA. Similar to some previous studies (Morin et al. 1994; Gagneux et al. 1999), our new data indicate that diversity within chimpanzees is greatest in the western subspecies (π = 0.94% for all or 0.84% without the Nigerian individual), although the sample size for the eastern/central subspecies (π = 0.54%) is rather small (n = 3). This pattern of diversity is contrary to those found at nuclear loci where diversity estimates are higher in the central subspecies. For example, analyses of NRY data showed that central chimpanzees had higher levels of diversity, because five different haplotypes were observed when compared with only one observed in a much larger sample of western chimpanzees (Stone et al. 2002). In a survey of approximately 10 kb at Xq13.3 from 30 chimpanzees (17 P. t. verus, 12 P. t. troglodytes and 1 P. t. schweinfurthii), Kaessmann et al. (1999) found that P. t. troglodytes were the most diverse. The mean pairwise sequence diversity at this locus among chimpanzees was 0.13 per cent, which is about four times greater than that in humans (0.037%). We also found a similar difference.
Kaessmann et al. (1999) also noted that the subspecies were not monophyletic for X-chromosome haplotypes (one western and one central chimpanzee shared the same haplotype). However, Verrelli et al. (2006) found that haplotypes were not shared between subspecies for the X-chromosome locus G6PD. At this locus, diversity was also greatest among central chimpanzees in a survey of 56 chimpanzees (6 P. t. troglodytes and 48 P. t. verus). Because the X-chromosome has three times the effective population size (Ne) of mtDNA, it should take three times as long as mtDNA to achieve monophyly. So it is not surprising that some of the X-chromosome lineages are not completely sorted among the subspecies given their likely divergence times.
Autosomal sequences also indicate higher diversity in central chimpanzees followed by eastern and western chimpanzees (Deinard & Kidd 1999; Yu et al. 2003; Fisher et al. 2004, 2006). Fisher et al. (2004) found that central chimpanzees are 2.0–2.5 times more diverse than western chimps and worldwide samples of humans. Central chimpanzees also show a relatively high proportion of rare alleles that could be the result of an old bottleneck or fine-scale population structure. Won & Hey (2005) found evidence for one-way gene flow from P. t. verus to P. t. troglodytes and suggest that this may be the result of interactions between the Nigerian chimpanzees and the central subspecies.
The distinct pattern of higher mtDNA diversity and lower diversity at other loci found in western chimpanzees suggests that the founding population of this subspecies may have been skewed with a larger number of females and a smaller group of closely related males. This pattern is perhaps not surprising given the philopatric mating patterns of chimpanzees where females disperse at adolescence, and males remain within their natal group (Nishida 1979; Pusey 1979; Wrangham 1979; Goodall 1986; Pusey & Packer 1986; Langergraber et al. 2007; Inoue et al. 2008). However, the pattern of higher mtDNA diversity/lower nuclear diversity in western chimpanzees compared with central + eastern chimpanzees indicate that the finding and/or subsequent demography of the western subspecies was somehow different from that found in the other subspecies. Genetic studies confirm that the males within many chimpanzee groups are typically more closely related than females; however, this is not true in all groups, particularly those where habitat fragmentation, disease or poaching affect group demography (Morin et al. 1994; Vigilant et al. 2001; Lukas et al. 2005; Inoue et al. 2008). The mtDNA data generated in this study indicate that chimpanzee female effective population size has been large (Ne = approx. 36 000 for all chimpanzees and approx. 20 000 for P. t. verus) with no evidence of population bottleneck or expansion.
MtDNA data may also be affected by selection, and several studies suggest that many mtDNA protein polymorphisms are slightly deleterious in humans (e.g. Nachman et al. 1996; Rand & Kann 1996; Kivisild et al. 2006). In this study, we also found evidence for a significantly negative value of Tajima's D for most mitochondrial protein-coding genes in humans (and for the genome as a whole) which can be indicative of population expansion and/or selection. However, in these chimpanzee data, a significant departure from neutrality was only found for CO3 and ND4L at replacement sites and there was no evidence for a departure from neutrality for the genome as a whole (table 2).
In molecular phylogenetics analysis of the complete mtDNA of chimpanzees, bonobos, humans and other great apes, human lineages are the most recent closest relative of Pan, in agreement with the scores of previous studies. A more controversial issue in chimpanzee evolutionary genomics has been the timing of divergence between chimpanzees and bonobos and among the subspecies of chimpanzees owing to the discrepancy between nuclear and mitochondrial results. The results from our mtDNA analysis provide partial resolution for this discrepancy. We find that the lack of consideration of population substructure of the chimpanzee subspecies when using mtDNA is a major cause of this difference. Therefore, we consider the time estimates from the use of two populations + one speciation model for analysing 4F degenerate sites data in BEAST to be the least biased and most appropriate for estimating chimpanzee divergence times. However, the absolute divergence times are strongly influenced by the calibration points used. For example, when using two calibration ranges (10.0–6.5 and 6.5–4.2 for gorilla/(human + chimpanzee) and human/chimpanzee splits, respectively), we obtain chimpanzee/bonobo divergence of 1.49 Ma. However, the human/chimpanzee divergence predicted by BEAST in this case is only 4.70 Ma, which is significantly lower than the current expectation.
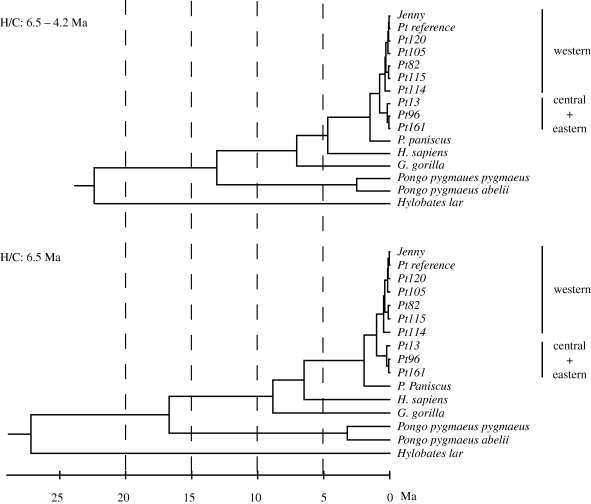
By constraining the human/chimpanzee divergence to be 6.5, the chimpanzee/bonobo time increases to 1.94 Ma. Interestingly, the ratio of the estimates of human/chimpanzee and chimpanzee/bonobo divergences is very similar (0.30–0.32) in different BEAST analyses under 2 + 1 model (table 4). Therefore, the estimate of chimpanzee/bonobo divergence times scales proportionally with the human/chimpanzee calibration. In this case, mtDNA analyses suggest a range of 2.09–1.49 Ma for chimpanzee/bonobo divergence, because the best estimates of human/chimpanzee divergence are in the range of 7–5 Ma (Kumar et al. 2005; Hedges et al. 2006; figure 3).
Table 4.
Divergence times (in million years) and ratios among major divergences for 4F degenerate sites from BEAST 2 + 1 model. The human/chimpanzee (H/C) time is used as the reference in calculating the time ratios. The gorilla/human + chimpanzee divergence was used as calibration (10.0–6.5 Ma) in all the estimations. The human/chimpanzee calibration was used either as a range (6.5–4.2 Ma) or fixed (6.5 Ma). Absolute times with H/C at 5 or at 7 Ma were scaled according to the ratios calculated.
| BEAST (2 + 1)a |
||||||
|---|---|---|---|---|---|---|
| absolute times (4F) |
time ratios (4F) |
absolute times |
||||
| H/C = (4.2–6.5) | H/C = 6.5 | H/C = (4.2–6.5) | H/C = 6.5 | H/C = 5 | H/C = 7 | |
| human/chimpanzee | 4.70 | 6.50 | — | — | 5.00 | 7.00 |
| chimpanzee/bonobo | 1.49 | 1.94 | 32% | 30% | 1.49 | 2.09 |
| western/(eastern + central) | 0.76 | 0.98 | 16% | 15% | 0.76 | 1.06 |
| eastern/central | 0.18 | 0.23 | 4% | 4% | 0.18 | 0.25 |
aTwo populations (western and eastern + central) and one speciation model (Yule).
Figure 3.
Timetrees obtained from BEAST (2 + 1 model) from 4F degenerate sites. The time scale is in million years (Ma). Both trees used the gorilla/human + chimpanzee calibration (10.0–6.5 Ma). Additionally, the upper tree used the human/chimpanzee (H/C) calibration range of 6.5–4.2 Ma while the lower tree fixed it at 6.5 Ma.
These mtDNA estimates for chimpanzee/bonobo are consistent with the range of 1.8–0.93 Ma based on the earlier analysis of Y-chromosomes, non-coding and non-repetitive genomic segments and X-chromosomes (Xq13.3; Kaessmann et al. 1999; Stone et al. 2002; Yu et al. 2003). However, more recent nuclear genome analyses have typically yielded a much younger date for this divergence (0.93–0.79 Ma). For example, Fisher et al. (2004) estimated a divergence time of 0.80 Ma for this divergence using a moment estimator method that examines the numbers of segregating sites at particular frequencies (Wakeley & Hey 1997). Won & Hey (2005) obtained an estimate of 0.89–0.86 Ma from multiple datasets (Deinard & Kidd 1999; Kaessmann et al. 1999; Stone et al. 2002; Yu et al. 2003) using an isolation with migration (IM) model, as did Becquet & Przeworski (2007) who estimated a split of 0.92–0.79 Ma from two datasets using an MCMC method to estimate the parameters of an IM model. Recently, Hey (2010) estimated a split of 0.68–1.54 Ma using a multi-population IM model, while Wegmann & Excoffier (2010) used an approximate Bayesian computation approach to estimate a divergence time of 1.6 Ma. These previous studies and our analysis show how the time estimates are sensitive not only to the choice of dataset, but also to the models used to describe the chimpanzee's population structure.
Within chimpanzees, the western and central + eastern clades diverged between 1.06–0.76 Ma according to our mtDNA analyses. This is younger than previous estimates from mtDNA (1.6–1.3 Ma; Morin et al. 1994), but older than other estimates based on nuclear loci. Fisher et al. (2004) examined 5.4 kb of autosomal sequence in 14 central and 16 western chimpanzees, and they propose a time of 0.65–0.43 Ma for the divergence of western and central + eastern groups. Won & Hey (2005) also calculated a divergence time estimate of 0.43 Ma for the central and western subspecies, while Becquet & Przeworski (2007) obtained much younger time estimates (0.28 Ma for western/eastern split and 0.44 Ma western/central divergence). More recent estimates range from 0.34–0.91 Ma (Caswell et al. 2008; Wegmann & Excoffier 2010). Therefore, nuclear DNA time estimates are again much younger than those indicated by mtDNA.
The eastern and central subspecies of chimpanzee are estimated to have diverged recently. Although these two subspecies do not appear to share either mtDNA or NRY haplotypes, these haplotypes are not monophyletic (Gagneux et al. 2001; Stone et al. 2002). From the limited number of sequences in this study, we estimate the divergence of eastern and central subspecies to be approximately 0.25–0.18 Ma. The Nigerian lineage appears to have diverged significantly earlier (approx. 0.4 Ma). Even though the single Nigerian lineage potentially diverged much earlier than its closest relatives, there is yet no evidence from Y-chromosome and autosomal STR markers to elevate the Nigerian chimpanzees to a separate subspecies (Stone et al. 2002; Becquet et al. 2007).
The consistent discrepancy among the different divergence estimates, both within chimpanzees and between chimpanzees and bonobos, are probably due to the different population histories reflected by different parts of the genome. The older divergence times based on mtDNA data may reflect a demographic history of greater female effective population size in Pan compared with the male effective population size (Stone et al. 2002; Eriksson et al. 2006).
Despite the wealth of information gleaned from complete mtDNA genome sequences to provide insight into the maternal history and patterning of humans (e.g. Ingman et al. 2000; Macaulay et al. 2005), such population genetic data have not been available for other primates. In chimpanzees, complete mitochondrial DNA sequences have previously been published for only one of the three subspecies (Horai et al. 1995; Arnason et al. 1996). Analysis of these sequences along with eight additional sequences representing all of the recognized subspecies, including one individual from the proposed subspecies P. t. ellioti, add to the picture of diversity and population history in this species; however, these data also illustrate the need for additional sampling of chimpanzees throughout their range. In addition, mtDNA data may be affected by certain demographic scenarios (sex-biased dispersal and bottlenecks), as suggested in this study, as well as selection, and thus may not provide an accurate time scale of evolutionary or population events unless population structure is considered (Nachman et al. 1996; Ballard & Rand 2005). Much of the primate diversity and taxonomic data published to date relies solely on mtDNA data. These studies should be eyed with caution until additional data are available.
Acknowledgements
We thank Cecil Lewis, Vinod Swarna and Brian Verrelli for helpful discussions and comments pertaining to this study and/or manuscript. This study would not have been possible without the samples generously provided by the New Iberia Primate Center (supported by an NCRR grant no. U42 RR015087 to the University of Louisiana at Lafayette, New Iberia Research Center), the Primate Foundation of Arizona, the Southwest Foundation for Biomedical Research and Riverside Zoo, Scottsbluff, Nebraska. This research was supported by the National Science Foundation to A.S. (BCS-0073871), the National Institutes of Health to S.K. (HG002096) and the Arizona State University.
Footnotes
One contribution of 14 to a Discussion Meeting Issue ‘The first four million years of human evolution’.
References
- Anderson S., et al. 1981Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (doi:10.1038/290457a0) [DOI] [PubMed] [Google Scholar]
- Andrews R. M., Kubacka I., Chinnery P. F., Lightowlers R. N., Turnbull D. M., Howell N.1999Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147. [DOI] [PubMed] [Google Scholar]
- Arnason U., Gullberg A., Xu X.1996A complete mitochondrial DNA molecule of the white-handed gibbon, Hylobates lar, and comparison among individual mitochondrial genes of all hominoid genera. Hereditas 124, 185–189 (doi:10.1111/j.1601-5223.1996.00185.x) [Google Scholar]
- Bahlo M., Griffiths R. C.2000Inference from genetrees in a subdivided population. Theor. Popul. Biol. 57, 79–95 (doi:10.1006/tpbi.1999.1447) [DOI] [PubMed] [Google Scholar]
- Ballard J. W. O., Rand D. M.2005The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36, 621–642 (doi:10.1146/annurev.ecolsys.36.091704.175513) [Google Scholar]
- Becquet C., Przeworski M.2007A new approach to estimate parameters of speciation models with application to apes. Genome Res. 17, 1505–1519 (doi:10.1101/gr.6409707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C., Patterson N., Stone A. C., Przeworski M., Reich D.2007Genetic structure of chimpanzee populations. PLoS Genet. 3, e66 (doi:10.1371/journal.pgen.0030066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D., Zhang D., Hartl D. L., Hewitt G. M.2001Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol. Evol. 16, 314–321 (doi:10.1016/S0169-5347(01)02151-6) [DOI] [PubMed] [Google Scholar]
- Caswell J. L., Mallick S., Richter D. L., Neubauer J., Schirmer C., Gnerre S., Reich D.2008Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 4, e1000057 (doi:10.1371/journal.pgen.1000057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.-C., Li H.2001Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68, 444–456 (doi:10.1086/318206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinard A., Kidd K.1999Evolution of a HOXB6 intergenic region within the great apes and humans. J. Hum. Evol. 36, 687–703 (doi:10.1006/jhev.1999.0298) [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson J. L., Turnbull D. M., Howell N.2004Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet. 74, 229–238 (doi:10.1086/381505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J., Hohmann G., Boesch C., Vigilant L.2004Rivers influence the population genetic structure of bonobos (Pan paniscus). Mol. Ecol. 13, 3425–3435 (doi:10.1111/j.1365-294X.2004.02332.x) [DOI] [PubMed] [Google Scholar]
- Eriksson J., Siedel H., Lukas D., Kayser M., Erler A., Hashimoto C., Hohmann G., Boesch C., Vigilant L.2006Y-chromosome analysis confirms highly sex-biased dispersal and suggests a low male effective population size in bonobos (Pan paniscus). Mol. Ecol. 15, 939–949 (doi:10.1111/j.1365-294X.2006.02845.x) [DOI] [PubMed] [Google Scholar]
- Fischer A., Wiebe V., Paabo S., Przeworski M.2004Evidence for a complex demographic history of chimpanzees. Mol. Biol. Evol. 21, 799–808 (doi:10.1093/molbev/msh083) [DOI] [PubMed] [Google Scholar]
- Fischer A., Pollack J., Thalmann O., Nickel B., Paabo S.2006Demographic history and genetic differentiation in apes. Curr. Biol. 16, 1133–1138 (doi:10.1016/j.cub.2006.04.033) [DOI] [PubMed] [Google Scholar]
- Gagneux P., et al. 1999Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc. Natl Acad. Sci. USA 96, 5077–5082 (doi:10.1073/pnas.96.9.5077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P., Gonder M. K., Goldberg T. L., Morin P. A.2001Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Phil. Trans. R. Soc. Lond. B 356, 889–897 (doi:10.1098/rstb.2001.0865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G. V., Nei M.2003Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20, 424–434 (doi:10.1093/molbev/msg050) [DOI] [PubMed] [Google Scholar]
- Gonder M. K., Oates J. F., Disotell T. R., Forstner M. R. J., Morales J. C., Melnick D. J.1997A new west African chimpanzee subspecies? Nature 388, 337 (doi:10.1038/41005) [DOI] [PubMed] [Google Scholar]
- Gonder M. K., Disotell T. R., Oates J. F.2006New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. Int. J. Primatol. 27, 1103–1127 (doi:10.1007/s10764-006-9063-y) [Google Scholar]
- Goodall J.1986The chimpanzees of Gombe: patterns of behavior. Cambridge, UK: Harvard University Press [Google Scholar]
- Griffiths R. C., Tavaré S.1994Simulating probability distributions in the coalescent. Theor. Popul. Biol. 46, 131–159 (doi:10.1006/tpbi.1994.1023) [Google Scholar]
- Guy F., Brunet M., Schmittbuhl M., Viriot L.2003New approaches in hominoid taxonomy: morphometrics. Am. J. Phys. Anthropol. 121, 198–218 (doi:10.1002/ajpa.10261) [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Dudley J., Kumar S.2006TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (doi:10.1093/bioinformatics/btl505) [DOI] [PubMed] [Google Scholar]
- Herrnstadt C., et al. 2002Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 70, 1152–1171 (doi:10.1086/339933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J.2010The divergence of chimpanzee species and subspecies as revealed in multi-population isolation-with-migration analyses. Mol. Biol. Evol. 27, 921–933 (doi:10.1093/molbev/msp298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai S., Satta Y., Hayasaka K., Kondo R., Inoue T., Ishida T., Hayashi S., Takahata N.1992Man's place in the hominoidea revealed by mitochondrial DNA genealogy. J. Mol. Evol. 35, 32–43 (doi:10.1007/BF00160258) [DOI] [PubMed] [Google Scholar]
- Horai S., Hayasaka K., Kondo R., Tsugane K., Takahata N.1995Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc. Natl Acad. Sci. USA 92, 532–536 (doi:10.1073/pnas.92.2.532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M., Gyllensten U.2003Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res. 13, 1600–1606 (doi:10.1101/gr.686603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M., Kaessmann H., Pääbo S., Gyllensten U.2000Mitochondrial genome variation and the origin of modern humans. Nature 408, 708–713 (doi:10.1038/35047064) [DOI] [PubMed] [Google Scholar]
- Inoue E., Inoue-Murayama M., Vigilant L., Takenaka O., Nishida T.2008Relatedness in wild chimpanzees: influence of paternity, male philopatry, and demographic factors. Am. J. Phys. Anthropol. 137, 256–262 (doi:10.1002/ajpa.20865) [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Cantor C. R.1969Evolution of protein molecules. In Mammalian protein metabolism (ed. Munro H. N.), pp. 21–132 New York, NY: Academic Press [Google Scholar]
- Kaessmann H., Wiebe V., Pääbo S.1999Extensive nuclear DNA sequence diversity among chimpanzees. Science 286, 1159–1162 (doi:10.1126/science.286.5442.1159) [DOI] [PubMed] [Google Scholar]
- Kivisild T., et al. 2006The role of selection in the evolution of human mitochondrial genomes. Genetics 172, 373–387 (doi:10.1534/genetics.105.043901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Filipski A., Swarna V., Walker A., Hedges S. B.2005Placing confidence limits on the molecular age of the human–chimpanzee divergence. Proc. Natl Acad. Sci. USA 102, 18 842–18 847 (doi:10.1073/pnas.0509585102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber K. E., Siedel H., Mitani J. C., Wrangham R. W., Reynolds V., Hunt K., Vigilant L.2007The genetic signature of sex-biased migration in patrilocal chimpanzees and humans. PLoS ONE 2, e973 (doi:10.1371/journal.pone.0000973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood C. A., Kimbel W. H., Lynch J. M.2004Morphometrics and hominoid phylogeny: support for a chimpanzee–human clade and differentiation among great ape subspecies. Proc. Natl Acad. Sci. USA 101, 4356–4360 (doi:10.1073/pnas.0306235101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas D., Reynolds V., Boesch C., Vigilant L.2005To what extent does living in a group mean living with kin? Mol. Ecol. 14, 2181–2196 (doi:10.1111/j.1365-294X.2005.02560.x) [DOI] [PubMed] [Google Scholar]
- Lynch M., Crease T. J.1990The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 7, 377–394 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N., Gonzalez A. M., Larruga J. M., Flores C., Cabrera V. M.2001Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2, 13 (doi:10.1186/1471-2156-2-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V., et al. 2005Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308, 1034–1036 (doi:10.1126/science.1109792) [DOI] [PubMed] [Google Scholar]
- Mishmar D., et al. 2003Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 100, 171–176 (doi:10.1073/pnas.0136972100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. A., Moore J. J., Chakraborty R., Jin L., Goodall J., Woodruff D. S.1994Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265, 1193–1201 (doi:10.1126/science.7915048) [DOI] [PubMed] [Google Scholar]
- Nachman M. W., Brown W. M., Stoneking M., Aquadro C. F.1996Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R.1993Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21, 3025–3049 (doi:10.1093/nar/21.13.3025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M.1987Molecular evolutionary genetics. New York, NY: Columbia University Press [Google Scholar]
- Nishida T.1979The social structure of chimpanzees of the Mahale Mountains. In The great apes (eds Hamburg D. A., McCown E. R.), pp. 73–121 Menlo Park, CA: Benjamin/Cummings [Google Scholar]
- Oates J. F., Groves C. P., Jenkins P. D.2009The type locality of Pan troglodytes vellerosus (Gray 1862), and implications for the nomenclature of west African chimpanzees. Primates 50, 78–80 (doi:10.1007/s10329-008-0116-z) [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A.1998ModelTest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- Pusey A.1979Intercommunity transfer of chimpanzees in Gombe National Park. In The great apes (eds Hamburg D. A., McCown E. R.), pp. 465–479 Menlo Park, CA: Benjamin/Cummings [Google Scholar]
- Pusey A. E., Packer C.1986Dispersal and philopatry. In Primate societies (eds Smuts B. B., Cheney D. L., Seyfarth R. M., Wrangham R. W., Struhsaker T. T.), pp. 250–266 Chicago, IL: University of Chicago Press [Google Scholar]
- Raaum R. L., Sterner K. N., Noviello C. M., Stewart C. B., Disotell T. R.2005Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 48, 237–257 (doi:10.1016/j.jhevol.2004.11.007) [DOI] [PubMed] [Google Scholar]
- Rand D. M., Kann L. M.1996Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 13, 735–748 [DOI] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R.2003DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497 (doi:10.1093/bioinformatics/btg359) [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.2001Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sarich V. M., Wilson A. C.1973Generation time and genomic evolution in primates. Science 179, 1144–1147 (doi:10.1126/science.179.4078.1144) [DOI] [PubMed] [Google Scholar]
- Schrago C. G., Russo C. A.2003Timing the origin of New World monkeys. Mol. Biol. Evol. 20, 1620–1625 (doi:10.1093/molbev/msg172) [DOI] [PubMed] [Google Scholar]
- Stone A. C., Verrelli B. C.2006Focusing on comparative ape population genetics in the post-genomic age. Curr. Opin. Genet. Dev. 16, 586–591 (doi:10.1016/j.gde.2006.09.003) [DOI] [PubMed] [Google Scholar]
- Stone A. C., Griffiths R. C., Zegura S. L., Hammer M. F.2002High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc. Natl Acad. Sci. USA 99, 43–48 (doi:10.1073/pnas.012364999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., et al. 2006The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol. Biol. Evol. 23, 683–690 (doi:10.1093/molbev/msj078) [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2003PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinaur Associates [Google Scholar]
- Tajima F.1989Statistical method for testing the neutral muation hypothesis by DNA polymorphism. Genetics 123, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M.1993Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Thalmann O., Hebler J., Poinar H. N., Paabo S., Vigilant L.2004Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol. Ecol. 13, 321–335 (doi:10.1046/j.1365-294X.2003.02070.x) [DOI] [PubMed] [Google Scholar]
- Thangaraj K., Chaubey G., Kivisild T., Reddy A. G., Singh V. K., Rasalkar A. A., Singh L.2005Reconstructing the origin of Andaman Islanders. Science 308, 996 (doi:10.1126/science.1109987) [DOI] [PubMed] [Google Scholar]
- Thorne J. L., Kishino H.2002Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 51, 689–702 (doi:10.1080/10635150290102456) [DOI] [PubMed] [Google Scholar]
- Verrelli B. C., Tishkoff S. A., Stone A. C., Touchman J. W.2006Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol. Biol. Evol. 23, 1592–1601 (doi:10.1093/molbev/msl024) [DOI] [PubMed] [Google Scholar]
- Vigilant L., Hofreiter M., Siedel H., Boesch C.2001Paternity and relatedness in wild chimpanzee communities. Proc. Natl Acad. Sci. USA 98, 12 890–12 895 (doi:10.1073/pnas.231320498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., Hey J.1997Estimating ancestral population parameters. Genetics 145, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A.1975On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (doi:10.1016/0040-5809(75)90020-9) [DOI] [PubMed] [Google Scholar]
- Wegmann D., Excoffier L.2010Bayesian inference of the demographic history of chimpanzees. Mol. Biol. Evol. 27, 1425–1435 (doi:10.1093/molbev/msq028) [DOI] [PubMed] [Google Scholar]
- White T. D., Suwa G., Asfaw B.1994Australopithicus ramidus, a new species of early hominid from Aramis, Ethiopia. Nature 371, 306–312 (doi:10.1038/371306a0) [DOI] [PubMed] [Google Scholar]
- Won Y. J., Hey J.2005Divergence population genetics of chimpanzees. Mol. Biol. Evol. 22, 297–307 (doi:10.1093/molbev/msi017) [DOI] [PubMed] [Google Scholar]
- Wrangham R. W.1979Sex differences in chimpanzee dispersion. In The great apes (eds Hamburg D. A., McCown E. R.), pp. 481–489 Menlo Park, CA: Benjamin/Cummings [Google Scholar]
- Xu X., Arnason U.1996The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J. Mol. Evol. 43, 431–437 (doi:10.1007/BF02337514) [DOI] [PubMed] [Google Scholar]
- Yang Z.2007PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- Yu N., Jensen-Seaman M. I., Chemnick L., Kidd J. R., Deinard A. S., Ryder O., Kidd K. K., Li W. H.2003Low nucleotide diversity in chimpanzees and bonobos. Genetics 164, 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]