Abstract
Messenger RNAs undergo 5' capping, splicing, 3'-end processing, and export before translation in the cytoplasm. It has become clear that these mRNA processing events are tightly coupled and have a profound effect on the fate of the resulting transcript. This processing is represented by modifications of the pre-mRNA and loading of various protein factors. The sum of protein factors that stay with the mRNA as a result of processing is modified over the life of the transcript, conferring significant regulation to its expression.
Transcription, capping, splicing, and RNA export are coupled. The interdependence of these activities profoundly affects the fate of mRNAs, providing additional layers of regulation and quality control.
Messenger RNA (mRNA) transcripts are extensively processed before export. 5′ capping, splicing, and 3′-end processing represent nuclear processes that are large determinants of the fate of a transcript. As mRNA processing events involve different cellular machinery (Fig. 1), RNA sequences, and have different consequences for target mRNAs, these processes were long seen to be independent of one another. It has become clear over the last decade, however, that these events are integrated and coordinated in space and time (Schroeder et al. 2000; reviewed in Bentley 2002; reviewed in Moore and Proudfoot 2009). Nuclear processing steps require a large set of proteins, many of which are loaded onto the transcript as a result of processing, adding a layer of regulatory information that can affect export, localization, translation, and stability of the transcript. Examples of such proteins include the exon-junction complex (EJC), left behind from splicing, and the THO/TREX complex, loaded during elongation. Indeed, the highly integrated nature of nuclear mRNA processing adds a new level of complexity to our picture of gene regulation. The availability of genetic fluorescent tags and sophisticated microscopy technology adds a dynamic component to this picture, providing spatial and temporal information and highlighting how nuclear structure might regulate gene expression (reviewed in Gorski et al. 2006; reviewed in Moore and Proudfoot 2009).
Figure 1.
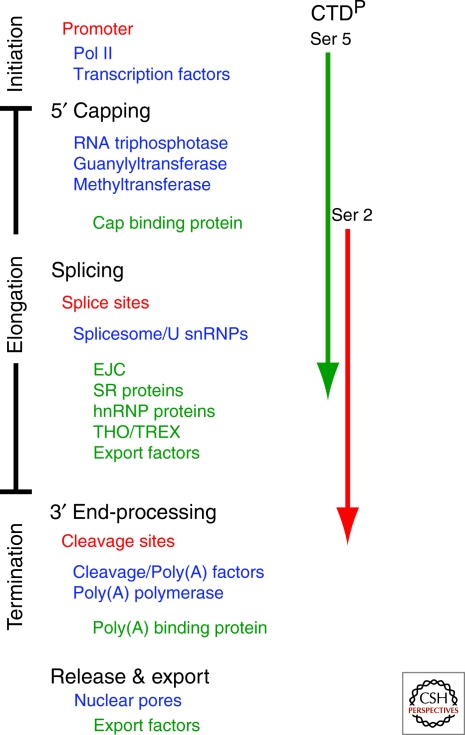
mRNA processing is tightly coupled to transcription. The phosphorylation state of the C-terminal domain (CTD) of RNA Polymerase II (Pol II) is given on the right in relation to major steps of transcription. Listed are functional processing sequences (red), components of processing machinery (blue) and factors that are loaded onto the transcript as a result of processing (green).
Transcription, the major contributor to RNA biogenesis, takes place under constraints of an anisotropic nuclear landscape that is highly structured (chromatin, distinct nuclear bodies, etc.) and dynamic (gene mobility, diffusive factors, genomic reorganization during cell cycle progression, etc.) (Yao et al. 2008). The availability of increasingly sensitive equipment and fluorescent markers has made it possible to intensively interrogate transcriptional dynamics (Fig. 2) (Becker et al. 2002; Janicki et al. 2004; reviewed in Shav-Tal et al. 2004b; Darzacq et al. 2007; Yao et al. 2007). Such approaches provide a new perspective on mRNA metabolism that has been mostly based on biochemical data. For instance, whereas preformed transcription complexes are sufficient for in vitro transcription, live cell imaging experiments show that different transcription factors show a wide range of dwell times at the promoter and suggest a link between transcription complex assembly dynamics and transcriptional output, consistent with a subunit assembly model for transcription complex recruitment (Gorski et al. 2008; reviewed in Darzacq et al. 2009). Modulation of transcriptional speed and processivity is suggested to be a way of regulating gene expression (Darzacq et al. 2007; reviewed in Core and Lis 2008; Core et al. 2008). These techniques have revealed highly inefficient transcription by RNA polymerase II (Pol II) and stochastic assembly of transcription complexes (Darzacq et al. 2007; Gorski et al. 2008), and both assembly and processivity can be regarded as rate limiting steps of in vivo transcription (reviewed in Core and Lis 2008). A novel way to look at gene expression has been recently presented by analyzing transcript amounts in individual cells on the single molecule level. Here the mean expression level and its variation are accessible, leading to a detailed understanding of variability in gene expression within a population (Zenklusen et al. 2008). Time lapse experiments add further information concerning the expression mode of individual genes, providing insights into differences between constitutive expression and bursts in different species (reviewed in Larson et al. 2009). These experimental approaches allow for in depth characterization of transcriptional dynamics (reviewed in Darzacq et al. 2009; reviewed in Larson et al. 2009), and are likely to provide greater insight into downstream processes.
Figure 2.
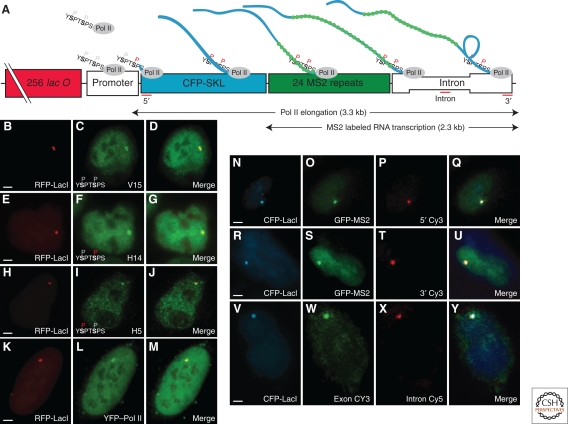
In vivo detection of transcription using fluorescence microscopy. Schematic shows a reporter cassette that is integrated as gene arrays into the genome. RFP-LacI labels reveal array locus and immunofluorescence shows that RNA Pol II with three distinct phosphorylation states is recruited to these active transcription sites (B–M). Similarly, the gene array, visualized by CFP-LacI, colocalizes with both MS2-tagged mRNAs, seen by GFP, and mRNA FISH probes targeting exonic and intronic regions (N–Y). (Reprinted, with permission, from Darzacq et al. 2007 [© 2007 Macmillan Publishers, Ltd.].)
TRANSCRIPTION SETS THE STAGE FOR PROCESSING
Transcription, particularly the carboxy-terminal domain (CTD) of RNA Pol II, contributes significantly to the integration of nuclear mRNA processing (Fig. 3). The CTD is an essential domain of the largest RNA Pol II subunit, composed of conserved YSPTSPS heptad repeats that are subject to reversible phosphorylation. It is well established that the CTD functions in transcription, and it has an equally important role in mRNA processing. The CTD interacts with a large number of protein factors, and among the protein domains shown to show preferential CTD binding are: CTD interacting domains (CIDs), WW domains, and FF domains (Verdecia et al. 2000; Smith et al. 2004; Noble et al. 2005). Serine 5 phosphorylation of the CTD occurs when RNA Pol II is at the 5′ end of the gene and is mediated by the TFIIH-associated kinase CDK7 (Kin28 in yeast) as transcription initiates. Serine 2 phosphorylation is mediated by PTEFb (CTDK1 in yeast) as the processive RNA Pol II elongates through the body of the gene (Komarnitsky et al. 2000; Peterlin and Price 2006). These phosphorylation marks are critical to the proper progression of transcription and are required for coordinating processing events (reviewed in Hirose and Manley 2000). Indeed, the CTD has been shown to bind over 100 different yeast proteins in its phosphorylated state (Phatnani et al. 2004). It can adopt different conformations depending on phosphorylation patterns, protein interactions, and proline isomerization via peptidyl-prolyl cis/trans isomerases (PPIases) (reviewed in Hirose and Ohkuma 2007). In this way, the CTD functions as a recruitment scaffold for different processing factors throughout transcription, thereby integrating processing events in time. Furthermore, changes in chromatin structure on transcriptional activation are likely to contribute to mRNA processing through gene positioning, or relocation at or near nuclear pores, and gene looping, or formation of DNA loops in which 5′ and 3′ ends make contact (reviewed in Moore and Proudfoot 2009). Recent evidence from studies using galactose genes in yeast showed that gene positioning can be a further regulatory step, and an interaction has been shown between transcription-dependent complexes (e.g TREX/TREX-2), chromatin remodeling complexes (e.g. SAGA), and nuclear pores (reviewed in Blobel 1985; Cabal et al. 2006; Klockner et al. 2009; reviewed in Moore and Proudfoot 2009). Export however, is likely to depend on successful maturation of the mRNA, and whereas in yeast gene gating to nuclear pores is suggested, mRNAs of other genes reach nuclear pores in a diffusive manner. Studies in mammalian cell systems have not supported gene gating, as fluorescence correlation spectroscopy (FCS), FRAP and single particle tracking consistently suggest diffusive behavior (Politz et al. 1998; Shav-Tal et al. 2004a; Grunwald et al. 2006; Politz et al. 2006; Braga et al. 2007; Siebrasse et al. 2008). In addition, high resolution studies of the nuclear periphery led to the conclusion that DNA is absent at the nuclear basket of nuclear pores (Schermelleh et al. 2008). Thus, transcription, and the associated machinery, serves to localize processing factors to the appropriate place on the nascent mRNA, stimulate and coordinate processing and possibly establish functionally significant chromatin conformations.
Figure 3.
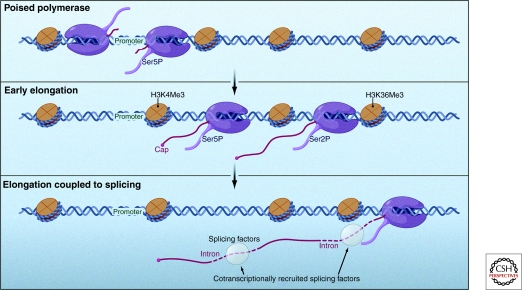
Depiction of several events that coincide with the switch from a promoter-engaged “poised” RNA polymerase to an elongating productive RNA polymerase (shown in purple). Histone modifications and changing CTD phosphorylation states are known to be associated with this switch. Furthermore, transcription is intimately coupled to mRNA processing events such as 5′ capping and splicing. (Reprinted, with permission, from Moore and Proudfoot 2009 [© 2009, Elsevier].)
CAPPING
The first processing event an mRNA undergoes is 5′-end capping which requires three enzymatic activities: RNA triphosphatase, guanylyltransferase and 7-methyltransferase (reviewed in Shuman 2001). Occurring early in transcription after RNA Pol II has transcribed the first 25–30 nucleotides, the RNA triphosphatase first acts on the terminal nucleotide to remove the γ-phosphate. The guanylyltransferase then transfers GMP from GTP to form GpppN, which is subsequently methylated. In mammals, a bifunctional capping enzyme includes both amino-terminal RNA triphosphate activity and carboxy-terminal guanylyltransferase activity. In yeast, the RNA triphosphatase (Cet1) and separate guanylyltransferase (Ceg1) form a heterodimeric capping enzyme. In both cases, a separate methyltransferase (Abd1 in yeast) is required to methylate the guanine at the N7 position.
TRANSCRIPTION AND CAPPING ARE TIGHTLY COUPLED
Mammalian capping enzyme binds directly to the elongating RNA Pol II with a phosphorylated CTD (termed RNA Pol II0) through its guanylyltransferase domain (Yue et al. 1997), thereby coupling capping to the early stage of transcription. Yeast capping enzyme subunits also bind directly and independently to RNA Pol II0, and this interaction is dependent on Kin28, the subunit of TFIIH responsible for serine 5 phosphorylation (Rodriguez et al. 2000). The loss of serine 5 phosphorylation during transcription correlates with the release of the capping enzyme, which is believed to occur before the nascent transcript is 500 nucleotides long (reviewed in Zorio and Bentley 2004). Ceg1 guanylylation activity is inhibited by the phosphorylated CTD but is restored and enhanced by Cet1, and this allosteric regulation may represent a means to temporally coordinate guanylylation and triphosphatase activities (Cho et al. 1998; Ho et al. 1998). In mammals, CTD binding to the guanylyltransferase has an allosteric affect, causing a twofold increase in affinity of guanylyltransferase for GTP (Ho and Shuman 1999). CTD phosphorylation that accompanies the transition from initiation to elongation has a clear impact on capping and allows communication between the transcriptional machinery and capping enzymes. It seems that capping also has a direct impact on transcription. Recent evidence indicates that capping enzymes can relieve transcriptional repression, suggesting an additional role in promoter clearance (Mandal et al. 2004). Temperature sensitive ceg1 yeast mutants are sensitive to drugs that disrupt elongation and show decreased transcription through promoter-proximal pause sites (Kim et al. 2004a). Furthermore, yeast capping enzyme subunits influence RNA Pol II occupancy at the 5′ end and may regulate transcription reinitiation (Myers et al. 2002; Schroeder et al. 2004). Taken together, it seems that transcription complexes are held at the promoter until capping occurs, after which the polymerase switches into an elongating mode.
CONSEQUENCES OF SUCCESSFUL CAPPING
Capping of transcripts confers stability. In yeast as well as mammals, capping helps protect the transcript from 5′→3′ exonucleases present in both the nucleus and cytoplasm (Hsu and Stevens 1993; Walther et al. 1998). The 5′→3′ degradation pathway involves deadenylation followed by rapid decapping by Dcp1/Dcp2, and degradation by the processive 5′→3′ exonuclease, Xrn1 (Hsu and Stevens 1993; Muhlrad et al. 1995). The cap is also important in mediating mRNA recruitment to ribosomes. The protein complex eIF4F recognizes the cap before translation, and facilitates circularization of mRNAs via an interaction with polyA-binding protein (PAB1), thereby aiding in translation reinitiation and enhancing protein synthesis (Tarun and Sachs 1996; Wakiyama et al. 2000). Transcripts engaged in translation are protected from degradation suggesting competition between translation and degradation (reviewed in Jacobson and Peltz 1996). Finally, depletion of CBC (cap-binding complex) from HeLa cell extracts inhibits the endonucleolytic cleavage step of 3′-end formation, reduces the stability of poly(A) cleavage complexes, and disrupts communication between 5′ and 3′ ends (Flaherty et al. 1997).
SPLICING
The precise removal of noncoding intervening sequences, or introns, from many pre-mRNAs is a required process for proper protein expression. In both yeast and mammals, this reaction is catalyzed by the spliceosome, consisting of the U1, U2, U4, U5 and U6 small nuclear RNPs (snRNPs) in conjunction with a large number of additional proteins (reviewed in Stark and Luhrmann 2006). Within the spliceosome, a series of RNA–RNA, RNA–protein, and protein–protein interactions is needed to identify and remove intronic regions and join exons, producing a mature transcript (reviewed in Collins and Guthrie 2000). The mature spliceosome carries out splicing through two transesterification reactions. First, the 2′-OH of a branch point nucleotide performs a nucleolytic attack on the first nucleotide of the intron, forming a lariat intermediate. Second, the 3′-OH from the free exon performs a nucleolytic attack on the last nucleotide of the intron, thereby joining exons and releasing the lariat intron. Intron identification relies on certain sequences, including the 5′ splice site, branch point (and downstream polypyrimidine tract) and 3′ splice site. In yeast, splice sites are easily identified, and although only 3% of genes contain single short introns, they account for more than 25% of cellular mRNAs (Ares et al. 1999; Lopez and Seraphin 1999; reviewed in Barrass and Beggs 2003). In mammals, however, splice sites are less clear, and many genes contains multiple introns that vary from a few hundred to hundreds of thousands of nucleotides (Lander et al. 2001). The presence of putative splice sites in higher eukaryotes does not necessarily lead to selection of these sites by the spliceosome. Flanking pre-mRNA regulatory elements, including intronic and exonic splicing enhancers or silencers, bind trans-acting regulatory factors that enhance or repress snRNP recruitment to splice sites. Generally, exonic splicing enhancers are bound by Serine/Arginine-rich (SR) proteins, whereas exonic splicing silencers are bound by heterogenous nuclear ribonucleoprotein (hnRNP) proteins (reviewed in Cartegni et al. 2002; reviewed in Singh and Valcarcel 2005). Therefore in higher eukaryotes, it is the cumulative effect of multiple factors that modulates splice site selection. As a result, 92%–94% of human transcripts are subject to alternative splicing, representing an important source of diversity in gene expression with serious implications for health and disease (reviewed in Nissim-Rafinia and Kerem 2005; Wang et al. 2008). Conversely, in yeast, SR proteins do not appear to have a significant role in splicing, consistent with the absence of exonic splicing enhancers (reviewed in Wahl et al. 2009).
The spliceosome is rich in proteins, containing approximately 125 different proteins (more than two-thirds of its mass), and spliceosome assembly is characterized by a remarkable exchange of components from one step to the next (reviewed in Wahl et al. 2009). Although RNA-RNA base pairing interactions are critical to the precise recognition of splice sites, they are generally weak and require additional proteins for enhanced stabilization. DExD/H-type RNA-dependent ATPases/helicases have long been implicated in rearrangements within the spliceosome, and many are conserved between yeast and humans. These proteins act at discrete stages of splicing including single-strand RNA translocation, strand annealing, and protein displacement (reviewed in Pyle 2008). Human spliceosomes also contain several PPIases that are absent in yeast, though the role of these proteins in splicing is not well understood. Similar to DExD/H-type RNA-dependent ATPases/helicases, they are recruited at discrete stages of splicing and are thought to be involved in at least one protein conformational switching event (reviewed in Wahl et al. 2009). Furthermore, post-translational modification of splicing factors and spliceosomal proteins may act as switches to allow fine tuning of the spliceosome (Bellare et al. 2008; reviewed in Wahl et al. 2009). The nature of interactions during such a tightly regulated protein-rich process is not very well documented and may be best studied using in vivo imaging techniques. For example, one recent study (Fig. 4) employed FRET-FLIM and revealed for the first time that different complexes of splicing factors show differential distributions in live cell nuclei (Ellis et al. 2008).
Figure 4.
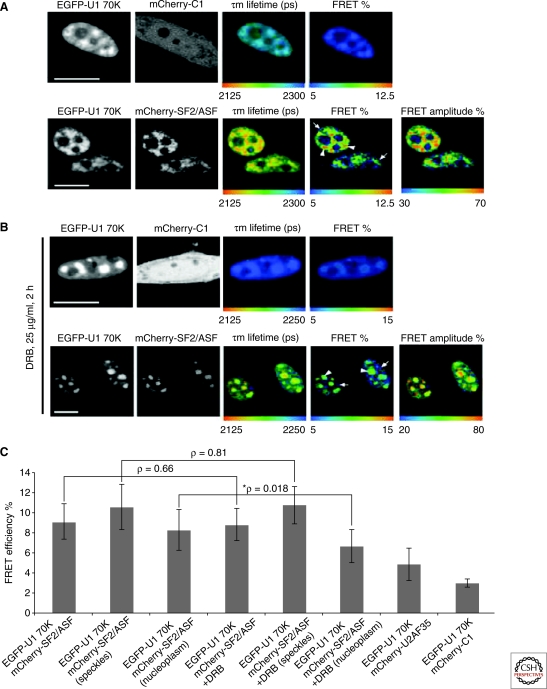
Spatial mapping of the interaction of U1 70K with SF2/ASF in vivo. Shown are confocal images of cells transfected with EGFP-U1 70K and cotransfected with either mCherry-C1 or mCherry-SF2/ASF. Mean fluorescence lifetime (in picoseconds) and percentages of FRET efficiency and FRET amplitude are shown (pseudocolor) in these same cells (A). The same experiment was repeated in the presence of 25 µg/ml DBR for 2 hours before imaging (B). FRET efficiencies calculated from FLIM measurements for the interaction of SF2/ASF with U1 70K (C). © Ellis et al., 2008. J. Cell Biol. 181: 921–934.
SPLICING LEAVES A MARK
Mature mRNAs are occupied by a number of different proteins that determine their fate in many ways, and several of these associations are splice-dependent. As mentioned, various proteins associate cotranscriptionally and accompany the packaged mRNA into the cytoplasm where they can direct localization, translation and decay. Shuttling SR proteins specifically serve as mRNP binding sites for export factors, and the phosphorylation state of these proteins confers export competence (reviewed in Huang and Steitz 2005; reviewed in Moore and Proudfoot 2009). The THO/TREX complex, which functions in transcription and export, associates with spliced mRNAs at the 5′-most exon (Cheng et al. 2006). THO/TREX recruitment is enhanced by splicing, and promotes rapid export (Valencia et al. 2008). In mammals, REF/Aly and UAP56 (homologs of yeast Yra1 and Sub2), are recruited as a consequence of splicing and have a role in aiding export. Perhaps the most studied splice-dependent mark is the exon junction complex, or EJC. EJCs are stably deposited 20–24 nucleotides upstream of exon-exon junctions late in splicing (Le Hir et al. 2000). Interestingly, spliced mRNAs appear to have greater translational efficiency than their cDNA counterparts (reviewed in Le Hir et al. 2003). Aside from their role in nonsense mediated decay, EJCs appear to directly enhance translation initiation. Although there are several proposed mechanisms by which EJCs do this, they ultimately serve to promote the pioneer round of translation (reviewed in Moore and Proudfoot 2009). Finally, a number of DEAD-box proteins have recently been identified as associating with mRNAs in a splice-dependent manner, and these are believed to influence many aspects of mRNA metabolism (Merz et al. 2007; reviewed in Rosner and Rinkevich 2007). Taken together, it is clear that spliced mRNAs carry with them numerous protein marks that indicate their splicing history and have important downstream effects.
LINKS BETWEEN TRANSCRIPTION AND SPLICING
Initial work focusing on the link between transcription and splicing suggested that splicing occurs cotranscriptionally and factors involved in splicing colocalize with transcription sites (Beyer and Osheim 1988; Zhang et al. 1994). Similar to capping, the CTD of RNA Pol II has an important role in splicing. CTD truncation causes inefficient splicing in mammalian cells and inhibition of colocalization of splicing factors with transcription sites (McCracken et al. 1997; Misteli and Spector 1999). Furthermore, RNA Pol II0 has been shown to physically associate with splicing factors that do not bind RNA Pol IIA (RNA Pol II with an unphosphorylated CTD), suggesting this interaction depends on the phosphorylation state of the CTD. Both anti-CTD antibodies and CTD peptides can inhibit splicing in vitro, and expression of phosphorylated CTD peptides has a similar effect on mammalian cells in vivo (Yuryev et al. 1996; Du and Warren 1997). RNA Pol II0 enhances splicing in vitro, whereas RNA Pol IIA has an inhibitory effect (Hirose et al. 1999). This is believed to result from RNA Pol II0-dependent stimulation of the early steps of spliceosome assembly, possibly by facilitating the binding of snRNPs to the nascent transcript. Therefore, it seems that CTD phosphorylation can act to recruit splicing factors to the nascent transcript to ensure rapid and accurate splicing. Transcription is also linked to splicing in ways independent of the CTD. Promoter identity and expression levels of certain SR proteins are known to affect alternative splicing (Cramer et al. 1997). Furthermore, in both yeast and mammals, disruption of RNA Pol II elongation markedly shifts the balance of alternatively spliced isoforms (de la Mata et al. 2003; Howe et al. 2003). This is consistent with a “first come first served” model in which elongation rate regulates splice site selection, as 5′ splice sites are more likely to pair with newly transcribed 3′ splice sites. Additionally, the transcriptional coactivator, p52, is known to interact with SF2/ASF and stimulates splicing (Ge et al. 1998), suggesting that transcriptional machinery can modulate splicing factor recruitment.
Finally, splicing can also have an impact on transcription. The presence of introns can confer increased transcriptional efficiency, possibly through increase initiation rates (Brinster et al. 1988). Recruitment of snRNPs by TAT-SF1, an elongation factor that associates with P-TEFb, enhances elongation and this effect is dependent on the presence of functional splice signals (Fong and Zhou 2001; Kameoka et al. 2004). Reminiscent of the effect of capping on transcription, certain splicing factors have also been shown to promote elongation (Lin et al. 2008).
LINKS BETWEEN CAPPING AND SPLICING
Capping also has a role in splicing. The 5′ cap structure increases splicing efficiency in mammalian cell extracts and in vivo (Konarska et al. 1984; Inoue et al. 1989). Depletion of CBC in cell extracts prevents spliceosome assembly at an early step in complex formation (Izaurralde et al. 1994). In yeast, CBC interacts with components of the earliest identified splicing complexes (Colot et al. 1996). Similarly, in mammals, successful capping and CBC recruitment is implicated in U1 small nuclear ribonucleoproteins (snRNP) recognition of the 5′ splice site, but this effect is specific to mRNAs with only one intron (Lewis et al. 1996).
QUALITY CONTROL OF SPLICING
Splicing defects can lead to potentially harmful protein variants. mRNAs are subject to quality control in the nucleus resulting in the prevention of export of splicing-defective transcripts. To date, several mutations in cis (conserved splice sites) and in trans (spliceosome components required for 1st or 2nd step catalysis) yield drastic reductions in mature mRNA without a corresponding increase in unspliced pre-mRNAs (reviewed in Staley and Guthrie 1998; Bousquet-Antonelli et al. 2000). Mature mRNA levels are largely restored in these same mutants when degradation is inhibited, suggesting that spliceosomes are able to act successfully on these substrates if they are not quickly destroyed. Thus, quality control acts on a number of different splice-defective pre-mRNAs, and degradation is in direct competition with productive splicing. This sort of kinetic competition is exemplified by the finding that decreased ATP hydrolysis of the DExD/H-type RNA-dependent ATPases, Prp16, leads to productive splicing of pre-mRNAs harboring mutant branch points that would normally be discarded (Burgess and Guthrie 1993). This same principal has been extended to other splice site mutations and members of the DExD/H-type RNA-dependent ATPases/helicases family (Mayas et al. 2006). By coupling spliceosomal rearrangements with irreversible ATP hydrolysis, these proteins ensure that splice-aberrant mRNAs which are unable to complete splicing within a time frame dictated by ATP hydrolysis rates, are discarded (reviewed in Villa et al. 2008). Quality control takes place in the nucleus, can act at numerous stages in the splicing process and serves to commit unspliced mRNAs to degradation pathways. One such pathway involves both 3′ → 5′ degradation by the exosome and 5′ → 3′ degradation by the nuclear exonuclease Rat1 (Bousquet-Antonelli et al. 2000). Other degradation pathways include Dbr1-mediated debranching of aberrant lariat-intermediates followed by export and cytoplasmic degradation, and Rnt1-mediated endonucleolytic cleavage of unspliced pre-mRNAs followed by nuclear degradation (Danin-Kreiselman et al. 2003; Hilleren and Parker 2003). An additional quality control mechanism involves spliceosome-dependent nuclear retention of unspliced transcripts. A number of different proteins seem to be involved in anchoring these transcripts at the nuclear side of the nuclear pore complex, and retention may be regulated by desumoylation (reviewed in Dziembowski et al. 2004; Galy et al. 2004; Palancade et al. 2005; Lewis et al. 2007). Furthermore, splice-defective mRNAs are also known to be retained at the site of transcription (Custodio et al. 1999; reviewed in Custodio and Carmo-Fonseca 2001). Thus, the cell has evolved various ways to ensure that unspliced transcripts do not leak into the cytoplasm. Finally, nonsense mediated decay (NMD) represents a highly specific form of quality control that extends to higher eukaryotes. The presence of an EJC downstream of a stop codon triggers degradation of translating mRNAs (reviewed in Stalder and Muhlemann 2008). Splice-dependent EJC deposition can increase the translational efficiency of normal mRNAs while ensuring rapid degradation of aberrant mRNAs.
3'-END PROCESSING
The final step of transcription is endonucleolytic cleavage which occurs 10–30 nucleotides downstream of a signal sequence (conserved AAUAAA sequence in mammals or an AU-rich sequence in yeast), followed by poly(A) addition at the 3′ end (reviewed in Proudfoot 2004). The poly(A) tail is similar to the cap in that it is important for the stability and translational efficiency of the mRNA (Drummond et al. 1985). Cleavage requires multiple proteins, including cleavage/polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), and two cleavage factors (CFIm and CFIIm) in mammals, or cleavage-polyadenylation factor (CPF) and two cleavage factors (CF1A and CF1B) in yeast. Poly(A) polymerase (PAP) then adds the poly(A) tail to the 3′-OH that is exposed on cleavage (reviewed in Proudfoot 2004).
CPSF and CstF are highly conserved between yeast and humans and are required for both cleavage and polyadenylation (reviewed in Shatkin and Manley 2000). CPSF recognizes RNA and facilitates PAP recruitment. The endonuclease responsible for cleavage in mammals is CPSF-73 and Ydh1 in yeast (Ryan et al. 2004; Mandel et al. 2006). CstF recognizes U/GU-rich elements found in the mRNA and is directly involved in polyadenylation. The conserved AAUAAA sequence and downstream U/GU site comprise the core poly(A) element, although additional auxiliary elements can influence polyadenylation efficiency (Gil and Proudfoot 1984; Sadofsky and Alwine 1984; Russnak 1991; Bagga et al. 1995). Cleavage is closely coupled to poly(A) tail synthesis, which also requires PAB1. A number of other factors also participate in 3′-end processing, and many do not share homologs in other systems (reviewed in Shatkin and Manley 2000).
As with splicing, transcripts can be alternatively polyadenylated, thereby altering stability, localization or transport. It is estimated that more than half of the genes in the human genome are subject to such alternative 3′-end processing, generating isoforms that differ in 3′ UTR length or encoding different proteins altogether (Tian et al. 2005). Alternative polyadenylation can be tissue specific, may be coupled to alternative splicing and can have implications for health and disease (Peterson and Perry 1989; Beaudoing and Gautheret 2001; Tian et al. 2005; Lu et al. 2007; reviewed in Danckwardt et al. 2008). In fact, many human genes contain multiple potential 3′-end cleavage sites, and appropriate site selection is achieved by alternate mechanisms, representing an additional layer of complexity in the regulation of gene expression (reviewed in Wilusz and Spector 2010). It is important to note that the mechanism and machinery responsible for alternative polyadenylation remain unclear.
TRANSCRIPTION AND 3′-END PROCESSING INFLUENCE EACH OTHER
Many factors involved in 3′-end processing have been shown to interact with the CTD, including CstF subunits. Purified CTD can stimulate the cleavage step and is needed for processing in reconstituted reactions (Hirose and Manley 1998). As with 5′ capping, the specific CTD phosphorylation pattern is important in 3′-end processing. In this case, loss of serine 2 phosphorylation in Ctk1 (yeast) or Cdk9 (Drosophila) mutants leads to a defect in 3′-end processing, likely resulting from improper or inefficient recruitment of processing factors (reviewed in Hirose and Ohkuma 2007). Furthermore, several yeast proteins involved in 3′-end processing preferentially bind the CTD phosphorylated at serine 2, which may ensure processing occurs as the polymerase reaches the end of a gene. In mammalian cells, unlike yeast, the CTD is also required for cleavage (Licatalosi et al. 2002). Once again, the CTD appears to mediate coupling between transcription and 3′-end processing.
Although transcription elongation continues for quite some distance after the poly(A) signal, transcription termination and 3′-end processing are intimately coupled. This was supported by the finding that termination requires functional poly(A) signals (reviewed in Hirose and Manley 2000). There are multiple ways in which 3′-end processing may be coupled to transcription termination:
In the antiterminator model, extrusion of the poly(A) signal from the polymerase triggers a change in the factors associated, possibly releasing elongation factors or recruiting termination factors (reviewed in Buratowski 2005). The PAF and TREX complexes represent good candidates for such a model, and have been shown to cross-link throughout the gene up to the poly(A) signal, in which they are largely absent (Kim et al. 2004b). The absence of these and other factors may switch the polymerase into a nonprocessive mode, releasing it from the DNA template.
The torpedo model of termination has gained recent support in both yeast and mammals (reviewed in Buratowski 2005). According to this model, the 5′ → 3′ exonucleases Rat1 (Xrn2 in mammals) is recruited to the 3′ end of the mRNA, in which it interacts with Rtt103, which is known to contact the CTD phosphorylated at serine 2. Rat1/Xrn1 degrades the downstream product, eventually catching up with and triggering the release of RNA Pol II (Kim et al. 2004c; West et al. 2004).
Transcriptional pause sites positioned downstream of the poly(A) signal also seem to be important for 3′-end processing and termination of a number of mammalian genes, reestablishing the theme of kinetic coupling between transcription and processing (Yonaha and Proudfoot 2000).
Although 3′-end processing machinery is enriched at the 3′ end of genes, certain factors can be found at or near promoters toward the 5′ end. For example, CPSF can be recruited to promoters through an association with TFIID (Dantonel et al. 1997). Ssu72, a component of yeast CPF, seems to function at many stages, including transcription initiation, 3′-end processing and termination of certain mRNAs and snoRNAs (reviewed in Proudfoot et al. 2002). As described, CTD phosphorylation patterns are linked to processing events, and Ssu72, in conjunction with Pta1 (another component of yeast CPF), also appears to have phosphatase activity specific for serine 5 (Krishnamurthy et al. 2004). Additional genetic and physical interactions have been described between the transcriptional machinery (TFIIB and Sub1) and the 3′-end processing factors Ssu72 and Rna15 (Sun and Hampsey 1996; Wu et al. 1999; Calvo and Manley 2005). Interactions between factors located on opposite ends of genes is likely facilitated by the formation of gene loops, and TFIIB, Ssu72 and Pta1 all appear to have a role in this (Singh and Hampsey 2007). Overall, it seems that the transcriptional machinery, DNA template, nascent mRNA and 3′-end processing machinery are in constant communication for processing.
QUALITY CONTROL OF 3′-END PROCESSING
Evidence in yeast shows that transcripts undergoing aberrant 3′-end processing are disposed of. Defects associated with pap1-1 mutants are suppressed by deletion of the exosomal subunit Rrp6, which is known to interact with both PAP and the export factor Npl3 (Burkard and Butler 2000). Hypoadenylated mRNAs are retained in the nucleus at the site of transcription, and these transcripts are stabilized and exported in the absence of Rrp6 (Hilleren et al. 2001). In rna14 and rna15 mutant strains, defects in termination lead to readthrough transcripts and aberrant polyadenylation. Deletion of Rrp6 stabilizes the aberrantly polyadenylated population whereas depletion of Rrp41 stabilizes the population of long readthrough transcripts (Libri et al. 2002; Torchet et al. 2002). Therefore, different components of the exosome may have evolved specialized roles in mRNA surveillance, ensuring rapid degradation of transcripts that possess aberrant 3′ ends. Additionally, THO complex or sub2 mutants show defects in 3′-end formation, reduced mRNA levels and retention at the site of transcription. Codeletion of Rrp6 and TRAMP components restores mRNA levels, although the retention effect only requires Rrp6 (Libri et al. 2002; Rougemaille et al. 2007). These observations may suggest kinetic competition between 3′ end formation and degradation. Transcripts that do not quickly and efficiently undergo 3′-end processing are exposed to the exosome, and mutation of exosome components may allow more time for defective 3′-end processing machinery to function. Finally, recent evidence suggests that nuclear mRNP assembly factors are involved in releasing the 3′-end processing machinery from the transcript after polyadenylation (Qu et al. 2009). This may be a way to temporally coordinate 3′-end formation, mRNP maturation and export.
EXPORT OF mRNA
Transport through the nuclear pore complex (NPC) represents the link between the nucleus and cytoplasm. Several studies have investigated mRNA mobility in the nucleoplasm and have revealed probabilistic movement of mRNAs with diffusion coefficients between 0.03 µm2/s and 4 µm2/s (Politz et al. 1998; Shav-Tal et al. 2004a; Braga et al. 2007; Siebrasse et al. 2008). In pulse-chase experiments, mRNA was found in the cytoplasm within ∼20 min after transcription (Lewin 1980). However, single-molecule tracking experiments suggest that transit through the NPC is significantly faster, on the order of fractions of a second. Different forms of RNA have been observed in close proximity to the nuclear envelope in electron micrographs (reviewed in Franke and Scheer 1974). Detailed resolution of individual mRNPs and how they move through nuclear pores is mainly derived from work using mRNPs of Balbiani Ring (BR) genes in salivary gland cells of Chironomus (Mehlin et al. 1992; Kiseleva et al. 1998). These large mRNPs, ∼50 nm in diameter, have been visualized interacting with nuclear pores by electron microscopy. Because of their size, these mRNPs unfold at the NPC and show directional translocation through the NPC, beginning with the 5′ end (reviewed in Daneholt 2001). Hypothetical sequences for distinct steps in the export process have been assembled from EM series (Kiseleva et al. 1998). mRNA export likely involves distinct docking, translocation and release steps from the NPC, analogous to a ratchet model that includes a specific function for the mRNA associated DEAD box helicase DBP5 (reviewed in Stewart 2007). Direct interaction of DPB5 with Nup214, a cytoplasmic component of the NPC, has been shown (Napetschnig et al. 2009; von Moeller et al. 2009) and DPB5 is thought to promote export factor release and eventual reorganization of the mRNA (reviewed in Iglesias and Stutz 2008). DBP5 localization, the timing and location of loading onto the transcript, and how many export factors are actually attached to any individual transcript remains unclear (Zhao et al. 2002; Estruch and Cole 2003; Lund and Guthrie 2005; von Moeller et al. 2009). Discrepancy in the number of BR-mRNPs observed on the nuclear and cytoplasmic surfaces of nuclear pores in EM studies have been interpreted as uncoupled asynchronous functions of the export process, which should result in a waiting step during transport. Simultaneously, it was concluded that translocation through the central channel of nuclear pores is probably fast compared to the docking step on the nuclear surface of the pore (Kiseleva et al. 1998). Biochemical studies indicates that mRNA export competence is directly linked to transcription (reviewed in Kohler and Hurt 2007; reviewed in Hurt and Silver 2008; reviewed in Iglesias and Stutz 2008; reviewed in Carmody and Wente 2009; reviewed in Moore and Proudfoot 2009). Recently, a link between actin and transcription has been suggested. Actin (1) can be detected as a component of pre-mRNP complexes, (2) binds transcription factors, (3) is involved in chromatin remodeling, and (4) associates directly to RNA polymerases (reviewed in Miralles and Visa 2006). Interestingly, actin may also have a role in export, as it has been observed to associate cotranscriptionally with BR-mRNPs and remains associated throughout export (Percipalle et al. 2001). Furthermore, the nuclear export receptor exportin-6 shows specificity for profilin-actin, suggesting an additional role as an adaptor for export of certain mRNAs (Stuven et al. 2003; reviewed in Miralles and Visa 2006).
Export is mediated by protein factors associated with the mRNA, and mRNAs that do not carry the necessary adaptor and export factors are retained in the nucleus (reviewed in Iglesias and Stutz 2008). Most mRNAs seem to export via a TAP (Mex67 in yeast)-dependent pathway. TAP is not a member of the karyopherin family and does not rely on the GTPase Ran, which mediates nuclear import (Segref et al. 1997; reviewed in Macara 2001; reviewed in Dreyfuss et al. 2002). Export factors do not recognize the mRNA directly, but rather through adapters such as Aly/REF (Yra1 in yeast), which is necessary for export (Stutz et al. 2000; Zhou et al. 2000; Gatfield et al. 2001; Le Hir et al. 2001). However, CBP80, a component of the CBC, has been implicated in mediating contacts between transport receptors and mRNA (Hamm and Mattaj 1990; Cheng et al. 2006). Aly/REF has been identified to be loaded in a splice-dependent manner as part of the EJC (Le Hir et al. 2001). Complex regulation of Aly/REF links mRNA export to cell cycle progression (Zhou et al. 2000; Okada et al. 2008; reviewed in Okada and Ye 2009) and Yra1, has been linked to S-phase entry (Swaminathan et al. 2007). Conversely, in Drosophila and Caenorhabditis elegans, mRNA export is Aly/REF independent (Gatfield and Izaurralde 2002; Longman et al. 2003).
The transcription-export complex (TREX) exemplifies the tight coupling between transcription and export. Recruitment of the TREX complex is coupled to the transcription machinery in yeast but associated with the splicing machinery in metazoans (Masuda et al. 2005; Cheng et al. 2006). In metazoans, the TREX complex was initially thought to be part of the EJC (Gatfield et al. 2001; Le Hir et al. 2001) but is now known to be recruited to the 5′ end independently, in a splicing and 5′ cap-dependent manner (Masuda et al. 2005; Cheng et al. 2006). In yeast, molecular machinery serves to dock transcribing genes to nuclear pores, resulting in “gene gating.” SAGA, involved in histone modification and DNA remodeling, has also been shown to interact with transcription factors (Grant et al. 1997; Larschan and Winston 2001; reviewed in Daniel and Grant 2007). Components of SAGA and TREX2 complexes are required for gene gating of the GAL locus following activation (Cabal et al. 2006; Wilmes and Guthrie 2009). Sus1, one such component, promotes NPC docking and export (reviewed in Blobel 1985; Jani et al. 2009; Klockner et al. 2009; Wilmes and Guthrie 2009). Sus1 is also involved in transcription elongation and may prevent harmful DNA:RNA hybrids during transcription (Klockner et al. 2009). Interactions have been shown for both TREX and TREX2 complexes with Swt1, an endonuclease that interacts with nuclear pores and is involved in mRNA quality control (Skruzny et al. 2009). Although major pathways for mRNA export have been identified, based on interspecies variations and the number of transport factors and cofactors involved, additional pathways are likely to exist. For example, export of certain viral and other mRNAs depend on CRM1 as an export factor (Ohno et al. 2000; reviewed in Dreyfuss et al. 2002; reviewed in Iglesias and Stutz 2008).
The extent to which processing and export might be regulated in a species- and differentiation-dependent manner currently remains unclear. Dynamic regulation of mRNA processing factors is a relatively new question, and it is still unclear to what degree mRNA export is regulated at the level of individual nuclear pores. A dynamic view of how mRNA transitions through the nuclear pore is lacking, but recent developments in mRNA labeling and imaging technology may provide the opportunity to fill this gap in the near future.
CONCLUDING REMARKS
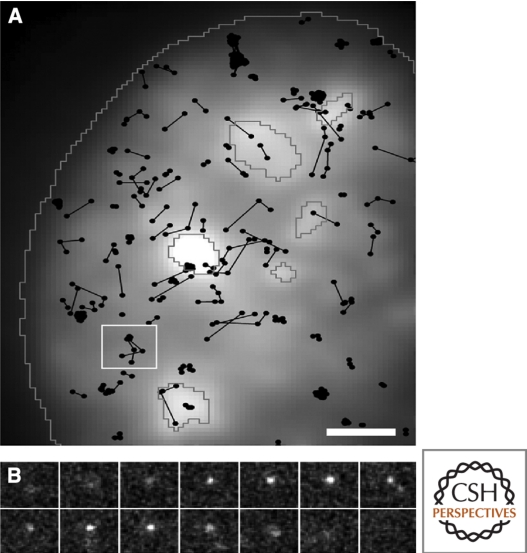
As discussed, a wealth of ensemble biochemical studies has provided great molecular and mechanistic insight into mRNA processing, leading to further questions that will require in vivo imaging approaches. Several recent studies highlight the importance of such techniques in gaining quantitative information on fundamental aspects of mRNA processing events, with spatial and temporal resolution. For example, the nucleus is the site of numerous essential cellular processes that are likely coordinated through highly organized and compartmentalized nuclear bodies with distinct functions, as observed using cellular imaging (reviewed in Misteli 2007). Nuclear bodies lack membranes and are highly dynamic yet steady-state structures. This represents a more advanced view from the original idea of nuclear factories for transcription and replication, in which core components localized to static discrete foci (reviewed in Iborra et al. 1996). This environment facilitates gene expression at multiple levels, including chromatin accessibility, transcriptional control, integration of processing events, stringent quality control, export of mature mRNPs to the cytoplasm and translation. Recent work suggests that Cajal body formation does not require specific gene loci and can initiate from any Cajal body protein, supporting a self-assembling model for nuclear bodies (Kaiser et al. 2008; reviewed in Misteli 2008). Additionally, the long-standing question of whether differentially spliced transcripts recruit distinct sets of basal pre-mRNA splicing factors has recently been addressed. Quantitative single-cell imaging has shown the first in vivo evidence of differential association of pre-mRNA splicing factors with alternatively spliced transcripts, supporting a stochastic model of alternative splicing which would predict that combinatorial sets of splicing factors contribute to splicing outcome (Mabon and Misteli 2005). FRET and FLIM techniques have been applied to investigate interactions between SR proteins and splicing components. Unlike biochemical methods, FRET can be used to study interactions in living cells, with minimal perturbation to the highly structured and dynamic nuclear environment (reviewed in Wouters et al. 2001; Ellis et al. 2008). Such an approach has revealed individual interactions that occur in the presence of RNA Pol II inhibitors, suggesting they are not exclusively cotranscriptional. FRAP analysis suggests that processing factors are highly dynamic and are exchanged between nuclear bodies and other nuclear locations in a matter of seconds (Phair and Misteli 2000; reviewed in Lamond and Spector 2003). Both FRET and FRAP have been used to study the localization and association of SF1 and U2AF. The mobility of these proteins is correlated with their ability to interact with each other, and they are believed to interact in what are described as extraspliceosomal complexes that form before and persist after spliceosome assembly (Rino et al. 2008). The development of real-time, single-molecule imaging techniques provides an especially exciting and promising opportunity to probe in vivo realities, reconciling molecular and mechanistic details within a kinetic and spatial context. One such example involves single particle tracking of U1 snRNP within the nucleus (see Fig. 5), revealing both mobile and transiently immobile states (Grunwald et al. 2006). Single molecule imaging makes the range of mobility states over a population of molecules immediately apparent, and changes in the behavior of any individual molecule during observation can be assessed. A major lesson from these studies is that “the mobility” of a given molecule is more likely a mixed population of different states.
Figure 5.
In vivo trajectories of single U1 snRNPs within the nucleus of HeLa cells. Fluorescently labeled native U1 snRNPs were microinjected to visualize and track single molecules, recorded at 200 Hz. SF2/ASF-GFP was transiently expressed to distinguish mobile and transiently immobilized U1 snRNP particles within the nucleoplasm and speckles, outlined in gray (A). A 8 µm2 area from A is broken down into a short image sequence displaying a single trajectory over time (B). Grunwald et al. 2006, © 2006 by The American Society for Cell Biology.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant EB2060 to R.H.S. and a DFG fellowship (DG 3388) to D.G. The authors would like to thank S. J. Orenstein, Drs. A. Joseph and V. de Turris for critical reading of the manuscript.
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Ares M Jr, Grate L, Pauling MH 1999. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA 5: 1138–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga PS, Ford LP, Chen F, Wilusz J 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res 23: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrass JD, Beggs JD 2003. Splicing goes global. Trends Genet 19: 295–298 [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Gautheret D 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res 11: 1520–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Baumann C, John S, Walker DA, Vigneron M, McNally JG, Hager GL 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep 3: 1188–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ 2008. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol 15: 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr Opin Cell Biol 14: 336–342 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Blobel G 1985. Gene gating: A hypothesis. Proc Natl Acad Sci 82: 8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Braga J, McNally JG, Carmo-Fonseca M 2007. A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys J 92: 2694–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD 1988. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci 85: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2005. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391 [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Calvo O, Manley JL 2005. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J 24: 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR 2009. mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Cho EJ, Rodriguez CR, Takagi T, Buratowski S 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev 12: 3482–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Guthrie C 2000. The question remains: Is the spliceosome a ribozyme? Nat Struct Biol 7: 850–854 [DOI] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708 [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR 1997. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci 94: 11456–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M 2001. Quality control of gene expression in the nucleus. J Cell Mol Med 5: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J 18: 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE 2008. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B 2001. Assembly and transport of a premessenger RNP particle. Proceedings of the National Academy of Sciences of the United States of America 98: 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Grant PA 2007. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res 618: 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell 11: 1279–1289 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. 2009. Imaging transcription in living cells. Annu Rev Biophys 38: 173–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Drummond DR, Armstrong J, Colman A 1985. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res 13: 7375–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Warren SL 1997. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol 136: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 23: 4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JD, Lleres D, Denegri M, Lamond AI, Caceres JF 2008. Spatial mapping of splicing factor complexes involved in exon and intron definition. J Cell Biol 181: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Cole CN 2003. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell 14: 1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci 94: 11893–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933 [DOI] [PubMed] [Google Scholar]
- Franke WW, Scheer U 1974. Pathways of nucleocytoplasmic translocation of ribonucleoproteins. Symp Soc Exp Biol 28: 249–282 [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol 11: 1716–1721 [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Wolffe AP 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell 2: 751–759 [DOI] [PubMed] [Google Scholar]
- Gil A, Proudfoot NJ 1984. A sequence downstream of AAUAAA is required for rabbit β-globin mRNA 3′-end formation. Nature 312: 473–474 [DOI] [PubMed] [Google Scholar]
- Gorski SA, Dundr M, Misteli T 2006. The road much traveled: Trafficking in the cell nucleus. Curr Opin Cell Biol 18: 284–290 [DOI] [PubMed] [Google Scholar]
- Gorski SA, Snyder SK, John S, Grummt I, Misteli T 2008. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell 30: 486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grunwald D, Spottke B, Buschmann V, Kubitscheck U 2006. Intranuclear binding kinetics and mobility of single native U1 snRNP particles in living cells. Mol Biol Cell 17: 5017–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63: 109–118 [DOI] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem 141: 601–608 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Ho CK, Schwer B, Shuman S 1998. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol Cell Biol 18: 5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M Jr 2003. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9: 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA 2005. SRprises along a messenger's journey. Mol Cell 17: 613–615 [DOI] [PubMed] [Google Scholar]
- Hurt JA, Silver PA 2008. mRNA nuclear export and human disease. Dis Model Mech 1: 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, McManus J, Jackson DA, Cook PR 1996. The topology of transcription by immobilized polymerases. Exp Cell Res 229: 167–173 [DOI] [PubMed] [Google Scholar]
- Iglesias N, Stutz F 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett 582: 1987–1996 [DOI] [PubMed] [Google Scholar]
- Inoue K, Ohno M, Sakamoto H, Shimura Y 1989. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev 3: 1472–1479 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668 [DOI] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65: 693–739 [DOI] [PubMed] [Google Scholar]
- Jani D, Lutz S, Marshall NJ, Fischer T, Kohler A, Ellisdon AM, Hurt E, Stewart M 2009. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell 33: 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. 2004. From silencing to gene expression: Real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M 2008. De novo formation of a subnuclear body. Science 322: 1713–1717 [DOI] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J 23: 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, Cho EJ 2004a. mRNA capping enzyme activity is coupled to an early transcription elongation. Mol Cell Biol 24: 6184–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S 2004b. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S 2004c. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Allen TD, Akey CW 1998. Active nuclear pore complexes in Chironomus: Visualization of transporter configurations related to mRNP export. Journal of Cell Science 111: 223–236 [DOI] [PubMed] [Google Scholar]
- Klockner C, Schneider M, Lutz S, Jani D, Kressler D, Stewart M, Hurt E, Kohler A 2009. Mutational uncoupling of the role of Sus1 in nuclear pore complex targeting of an mRNA export complex and histone H2B deubiquitination. J Biol Chem 284: 12049–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA 1984. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell 38: 731–736 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M 2004. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D 2009. A single molecule view of gene expression. Trends Cell Biol 19: 630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28: 215–220 [DOI] [PubMed] [Google Scholar]
- Lewin B 1980. Eucaryotic Chromosomes. in Gene Expression, pp. 730–761 John Wiley & Sons, New York [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol 178: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev 10: 1683–1698 [DOI] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Johnstone IL, Caceres JF 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. Rna 9: 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PJ, Seraphin B 1999. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA 5: 1135–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Huang X, Zhang L, Guo Y, Cheng H, Zhou R 2007. Multiple alternative splicing of mouse Dmrt1 during gonadal differentiation. Biochem Biophys Res Commun 352: 630–634 [DOI] [PubMed] [Google Scholar]
- Lund MK, Guthrie C 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell 20: 645–651 [DOI] [PubMed] [Google Scholar]
- Mabon SA, Misteli T 2005. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol 3: e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG 2001. Transport into and out of the nucleus. Microbiol Mol Biol Rev 65: 570–594, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci 101: 7572–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U 1992. Translocation of a Specific Premessenger Ribonucleoprotein Particle through the Nuclear-Pore Studied with Electron-Microscope Tomography. Cell 69: 605–613 [DOI] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R 2007. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Visa N 2006. Actin in transcription and transcription regulation. Curr Opin Cell Biol 18: 261–266 [DOI] [PubMed] [Google Scholar]
- Misteli T 2007. Beyond the sequence: Cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Misteli T 2008. Cell biology: Nuclear order out of chaos. Nature 456: 333–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P 2002. The yeast capping enzyme represses RNA polymerase II transcription. Mol Cell 10: 883–894 [DOI] [PubMed] [Google Scholar]
- Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel Gn, Hoelz A 2009. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proceedings of the National Academy of Sciences 106: 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim-Rafinia M, Kerem B 2005. The splicing machinery is a genetic modifier of disease severity. Trends Genet 21: 480–483 [DOI] [PubMed] [Google Scholar]
- Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol 12: 144–151 [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Okada M, Ye K 2009. Nuclear phosphoinositide signaling regulates messenger RNA export. RNA Biol 6: 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Jang SW, Ye K 2008. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc Natl Acad Sci 105: 8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Zuccolo M, Loeillet S, Nicolas A, Doye V 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol Biol Cell 16: 5258–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol 153: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Peterson ML, Perry RP 1989. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol 9: 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Jones JC, Greenleaf AL 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: Novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43: 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Browne ES, Wolf DE, Pederson T 1998. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci 95: 6043–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Prasanth KV, Baudendistel N, Fogarty KE, Lifshitz LM, Langowski J, Spector DL, Pederson T 2006. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol Biol Cell 17: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Pyle AM 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C 2009. Assembly of an export-competent mRNP is needed for efficient release of the 3′ end processing complex after polyadenylation. Mol Cell Biol 29: 5327–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rino J, Desterro JM, Pacheco TR, Gadella TW Jr, Carmo-Fonseca M 2008. Splicing factors SF1 and U2AF associate in extraspliceosomal complexes. Mol Cell Biol 28: 3045–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner A, Rinkevich B 2007. The DDX3 subfamily of the DEAD box helicases: Divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem 14: 2517–2525 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak RH 1991. Regulation of polyadenylation in hepatitis B viruses: Stimulation by the upstream activating signal PS1 is orientation-dependent, distance-independent, and additive. Nucleic Acids Res 19: 6449–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M, Alwine JC 1984. Sequences on the 3′ side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol 4: 1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol Cell 13: 377–387 [DOI] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ, Manley JL 2000. The ends of the affair: Capping and polyadenylation. Nat Struct Biol 7: 838–842 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Singer RH, Darzacq X 2004b. Imaging gene expression in single living cells. Nat Rev Mol Cell Biol 5: 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH 2004a. Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol 66: 1–40 [DOI] [PubMed] [Google Scholar]
- Siebrasse JP, Veith R, Dobay A, Leonhardt H, Daneholt B, Kubitscheck U 2008. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Nat Acad Sci 105: 20291–20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J 2005. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12: 645–653 [DOI] [PubMed] [Google Scholar]
- Singh BN, Hampsey M 2007. A transcription-independent role for TFIIB in gene looping. Mol Cell 27: 806–816 [DOI] [PubMed] [Google Scholar]
- Skruzny M, Schneider C, Racz A, Weng J, Tollervey D, Hurt E 2009. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol 7: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Kulkarni S, Pawson T 2004. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol Cell Biol 24: 9274–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2008. The meaning of nonsense. Trends Cell Biol 18: 315–321 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Stark H, Luhrmann R 2006. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct 35: 435–457 [DOI] [PubMed] [Google Scholar]
- Stewart M 2007. Ratcheting mRNA out of the nucleus. Mol Cell 25: 327–330 [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. Rna 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, Gorlich D 2003. Exportin 6: A novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 22: 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Hampsey M 1996. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol 16: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Kile AC, MacDonald EM, Koepp DM 2007. Yra1 is required for S phase entry and affects Dia2 binding to replication origins. Mol Cell Biol 27: 4674–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr, Sachs AB 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D 2002. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell 9: 1285–1296 [DOI] [PubMed] [Google Scholar]
- Valencia P, Dias AP, Reed R 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci 105: 3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP 2000. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol 7: 639–643 [DOI] [PubMed] [Google Scholar]
- Villa T, Rougemaille M, Libri D 2008. Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: Tilting the balance to shape the transcriptome. Biochim Biophys Acta 1779: 524–531 [DOI] [PubMed] [Google Scholar]
- von Moeller H, Basquin C, Conti E 2009. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol 16: 247–254 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N 2000. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 10: 1147–1150 [DOI] [PubMed] [Google Scholar]
- Walther TN, Wittop Koning TH, Schumperli D, Muller B 1998. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4: 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ 2004. Human 5′→ 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Guthrie C 2009. Getting to the gate: Crystallization of a Sac3(CID):Sus1:Cdc31 complex. Mol Cell 33: 671–672 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Spector DL 2010. An unexpected ending: Noncanonical 3′ end processing mechanisms. RNA 16: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters FS, Verveer PJ, Bastiaens PI 2001. Imaging biochemistry inside cells. Trends Cell Biol 11: 203–211 [DOI] [PubMed] [Google Scholar]
- Wu WH, Pinto I, Chen BS, Hampsey M 1999. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28: 978–990 [DOI] [PubMed] [Google Scholar]
- Yao J, Zobeck KL, Lis JT, Webb WW 2008. Imaging transcription dynamics at endogenous genes in living Drosophila tissues. Methods 45: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M, Proudfoot NJ 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J 19: 3770–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci 94: 12898–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci 93: 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR 1994. Localization of pre-mRNA splicing in mammalian nuclei. Nature 372: 809–812 [DOI] [PubMed] [Google Scholar]
- Zhao J, Jin SB, Bjorkroth B, Wieslander L, Daneholt B 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J 21: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL 2004. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res 296: 91–97 [DOI] [PubMed] [Google Scholar]