Abstract
Several affinity resins consisting of ionic metals or metal oxides were investigated for their phosphopeptide enrichment capabilities with subsequent mass spectrometric analyses. Commercially-available enrichment MOAC resins using manufacturer’s and/or published protocols were compared and evaluated for the most efficient and selective method that could be implemented as a standard enrichment procedure. From these comparative analyses using a tryptic digest of casein proteins, it was determined that, in our hands, two of the resins outperformed the others based on a variety of criteria, including the number of phosphorylation sites identified during MS analyses, the lower numbers of non-specifically bound peptides observed, and the limits of detection. Applicability of these enrichment resins to a complex biological mixture was investigated. For this work a mixture of avian histones was digested, subjected to titanium dioxide phosphopeptide enrichment and analyzed by mass spectrometry. Eight phosphorylated tryptic peptides were observed following enrichment and subsequent LC/MS/MS analyses. Of note, seven of the eight phosphopeptides were not observed without titanium dioxide enrichment. From these analyses, four sites of phosphorylation were unequivocally determined; two of which have not been reported in the literature previously. Four additional phosphopeptides were observed, however, the site of phosphorylation could not be distinguished, but was localized to one of two possible amino acids. These methods should aid in the investigation of proteins post-translationally modified with phosphate, especially those present at low concentrations as was demonstrated by successful enrichment at the femtomole level.
INTRODUCTION
One of the most common and important means of biologically regulating protein activity is through reversible phosphorylation and dephosphorylation. Phosphorylation is involved in the regulation of gene expression and protein synthesis, which controls cell growth, division or differentiation. In order to better understand the molecular basis of these regulatory mechanisms, it is useful to identify the specific amino acid residues undergoing phosphorylation.
A number of different approaches to the identification of phosphorylation sites are commonly used, however, mass spectrometry is emerging as the technique of choice (1-3). Even with recent advances in mass spectrometry instrumentation, the detection and determination of sites of phosphorylation remains a challenge since both the amount of phosphorylated protein as well as the stoichiometry of phosphorylation is often times present at low relative abundance. In addition, the suppression of low-level phosphopeptides in complex mixtures and the low sensitivity of phosphorylated peptides under positive ionization conditions due to the negative charges on the phosphate moiety can be difficult obstacles to overcome when analyzing phosphopeptides and proteins by mass spectrometry.
In recent years, the development of enrichment techniques, such as immobilized metal ion affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC), prior to mass spectrometric analyses has proven beneficial for the isolation of phosphopeptides from mixtures with high specificity and for the determination of sites of phosphorylation (reviewed in 3-6). The phosphopeptides can be analyzed by MS following elution from the enrichment resin or can be directly analyzed by MALDI/MS without prior elution (3-10).
Of these enrichment approaches, the metal oxides (e.g. titanium dioxide, zirconium dioxide) show great promise for enhanced selectivity of phosphopeptides. A variety of buffer conditions and/or chemical modification of the phosphate group have been employed to enhance the selectivity of the metal oxide enrichment media (3,5,6,11-23). For example, it has been shown that inclusion of organic displacers (e.g. 2,5-dihydroxybenzoic acid) during binding minimizes the level of non-specifically bound peptides (13). In addition, the methyl esterification of peptides prior to enrichment has been shown to enhance selectivity of the MOAC resins (19).
The goal of the present study is to evaluate several commercially-available enrichment MOAC resins using manufacturer’s and/or published protocols for the most efficient and selective method that could be implemented as a standard enrichment procedure for complex biological samples. A titanium dioxide compact reaction column (CRC) packed with Titansphere® TiO2 resin, NuTips™ comprised of either TiO2, ZrO2, or a TiO2/ZrO2 mixture, as well as magnetic TiO2 beads (Phos-Trap™ Kit) were evaluated and compared to one another as well as to our use of an Fe-nitrilotriacetic acid (Fe-NTA) IMAC CRC (7-10). The parameters used to compare different enrichment methods were based on their selectivity for phosphopeptides, robustness, and reproducibility. In addition, the limits of detection and the applicability of the metal oxides to enrich phosphopeptides from a complex biological sample consisting of core and linker histones from chicken erythrocytes were investigated.
METHODS
Materials
Ethylenediaminetetraaccetic acid (EDTA), α-cyano-4-hydroxycinnamic acid (α-cyano), 2,5-dihydroxybenzoic acid (DHB), ammonium bicarbonate (ABC), formic acid (FA), bovine α-casein, and β-casein (from bovine milk) were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO). The α-cyano was recrystallized from hot methanol and stored in the dark at room temperature. 18 MΩ water was prepared on a model RO 40 water system (Hydro Service and Supplies, Durham, NC). Acetonitrile (ACN) and acetic acid were purchased from Caledon (Georgetown, Ontario, Canada). Trifluoroacetic acid (TFA) was acquired from Pierce (Rockford, IL), phosphoric acid (PA) from EMD Chemicals (Gibbstown, NJ), and ethanol (EtOH) from Warner-Graham (Cockeysville, MD). All solvents were HPLC grade. Ferric chloride (FeCl3) was purchased from Allied Chemical (Brighton, UK). Ni-NTA resin was purchased from Qiagen (Valencia, CA). Compact reaction columns (CRC) and filters (10 μm pore size) were obtained from USB Corporation (Cleveland, OH). Gallus histones were purchased from Upstate Biotechnology (Lake Placid, NY).
Tryptic Digestion Conditions
The proteins were subjected to digestion with sequencing-grade modified porcine trypsin (Promega Corporation, Madison, WI). Enzyme was added at a protein:enzyme ratio of 20:1 and digests were allowed to proceed for at least two hours at 37 °C. For the comparison of the enrichment studies, one microgram each of α- and β-casein was loaded onto the resins. Two independent sets of digests were prepared for the enrichment studies.
Iron NTA IMAC CRC
Fe-NTA IMAC columns were prepared as previously reported (7-10).
Titanium Dioxide CRC
The TiO2 MOAC procedure was adopted from Larsen, et al. (13). Briefly, 1 mg of Titansphere® TiO2 resin (10 μm particle size) was added to 10 μL resin buffer (80% ACN, 19.9% H2O, 0.1% TFA) and loaded into a CRC tube. Titansphere® TiO2 resin was a gift from GL Sciences (Torrance, CA). Binding of phosphopeptides to the TiO2 column was achieved by first draining the buffer, then loading the protein digest mixture with 20 μL protein buffer (100 mg/mL DHB in 80% ACN, 19.9% H2O, 0.1% TFA). The mixture was allowed to incubate at room temperature for 10 minutes, after which, the column was drained, and washed 1 × 30 μL protein buffer and 1 × 30 μL resin buffer. Elution of the bound peptides was accomplished by adding 5 μL of 200 mM ABC solution (pH 10.5) to the column and incubated for 5 minutes at room temperature. The 5 μL eluent was then drained and diluted with 5 μL water. 0.5 μL of the eluent solution was spotted on a MALDI target with 0.5 μL of MALDI matrix (either a saturated solution of α-cyano in 45:45:10 ethanol:water:FA or 20 mg/mL DHB in 50% ACN, 49% H2O, 1% PA).
Phos-Trap™ Kit
The Phos-Trap magnetic titanium beads, binding buffer, and washing buffer (buffers included in kit) were prepared according to the manufacturer’s instructions (Perkin Elmer, Waltham, Massachusetts). Briefly, a 10 μL aliquot of magnetic beads was diluted to 200 μL in de-ionized water and placed in a 96-well plate. The beads were washed 3 times with 200 μL binding buffer. After placing the plate on a separation magnet, which moved the beads to an inner o-ring of the 96-well plate, the supernatant was removed. The sample was diluted 1:10 in binding buffer and incubated with beads for 10 minutes. Three 200 μL washes of binding buffer were followed by a single wash of 100 μL washing buffer. The peptides bound to the magnetic beads were eluted with 20 μL of elution buffer, lyophilized, and re-suspended in 1% FA just prior to MS analyses.
Glygen NuTips™
Enrichment using Glygen Corporation’s (Columbia, MD) NuTips™ was performed similarly to the titanium CRC, with minor variations. The tips, which consist of either titanium dioxide, zirconium dioxide, or a mixture of both, were first equilibrated by washing the resin 10 times with 5 μL of binding solution (80% ACN, 15% H2O, 5% TFA, pH < 3) (13) after which excess buffer was aspirated from the tip. Prior to loading, the digest sample was mixed 1:1 with binding solution. This mixture was loaded onto the tip by taking 2.5 μL aliquots and aspirating and expelling each aliquot ~20 times over several minutes. This was repeated until the desired amount of digest was loaded. Washes consisting of 80% ACN, 19% H2O, 1% TFA (pH < 3) followed (10 × 5 μL), and the bound peptides were eluted with 10 μL of elution solution (2% NH4OH in water, pH=11). To prevent loss of the phosphate moieties under highly basic conditions, the eluent was acidifed with 2 μL FA.
Mass Spectrometry
LC/ESI/MS/MS analyses were performed using an Agilent (Santa Clara, CA) 1100 nanoHPLC interfaced with an Agilent XCT Ultra Ion Trap (IT) equipped with an HPLC Chip Cube MS interface or a Waters Q-Tof Premier hybrid mass spectrometer equipped with a nanoAcquity UPLC system (Manchester, UK). Automated data dependent acquisitions were employed using either CID or ETD fragmentation mechanisms. ETD of parent ions was accomplished using a fluoranthene source for the production of radical anions with a 40 millisecond accumulation time. Positive ion MALDI/MS analyses were performed using either a Voyager Super DE-STR (Applied Biosystems, Framingham, MA) or a Waters Micro MX (Waters Corp., Beverly, MA). Four independent enrichments and subsequent MALDI/MS analyses were performed on all MOAC resins. Additional details regarding the MS analyses can be found in the Supplementary Material.
RESULTS & DISCUSSION
Commercially-available α-casein is a mixture of the S1 variant (major component) which has nine known sites of phosphorylation and the S2 variant (minor component) with eleven known sites of phosphorylation. β-casein contains five sites of phosphorylation. The α- and β-casein tryptic digests were mixed together in solution and analyzed by MALDI/MS (Supplemental Figure S1). The spectrum contains abundant proteolytic peptides, however, no casein phosphopeptides were observed in this analysis.
MALDI/MS Analyses
Fe-NTA IMAC
We have previously shown that phosphopeptides can be directly analyzed by MALDI/MS without prior elution from immobilized metal ion affinity media (IMAC) (7-10) as elution of phosphopeptides from this media can be inefficient (8,10). Figure 1A shows the direct on-resin MALDI-TOF mass spectrum of a casein digest mixture after Fe-NTA bead enrichment. Ions corresponding in mass to several phosphopeptides are observed. Specifically, ions corresponding in mass to eight protonated phosphopeptides representing eight sites of phosphorylation from αS1, nine from αS2, and one from β-casein are observed (Table 1). Additionally, two oxidized phosphopeptides (αS1 T16 and αS1 T7) were also observed; however, we did not determine whether or not oxidation preceded binding to the resin. Surprisingly, ions corresponding to peptides containing four (αS2 T2 and αS2 T7) and five (αS1 T8) phosphate groups were observed. Typically, under positive ion conditions, the MS analysis of peptides containing multiple phosphorylation sites is difficult (24,25). Although at least five nonphosphorylated peptides were observed with the Fe-NTA enrichment (Figure 1A, ions labeled with a filled circle), the abundance of the non-specifically bound peptides is relatively low (<8% relative abundance). The ion of m/z 1832.83 (labeled with an asterisk in Figure 1A) has been previously reported by Larsen, et al. as being a phosphorylated sequence variant and may be attributed to alternative splicing (13). The Fe-NTA resin was capable of enriching phosphopeptides representing 18 of 25 known sites of phosphorylation from a casein digest mixture (Figure 1A and Table 1).
Figure 1.
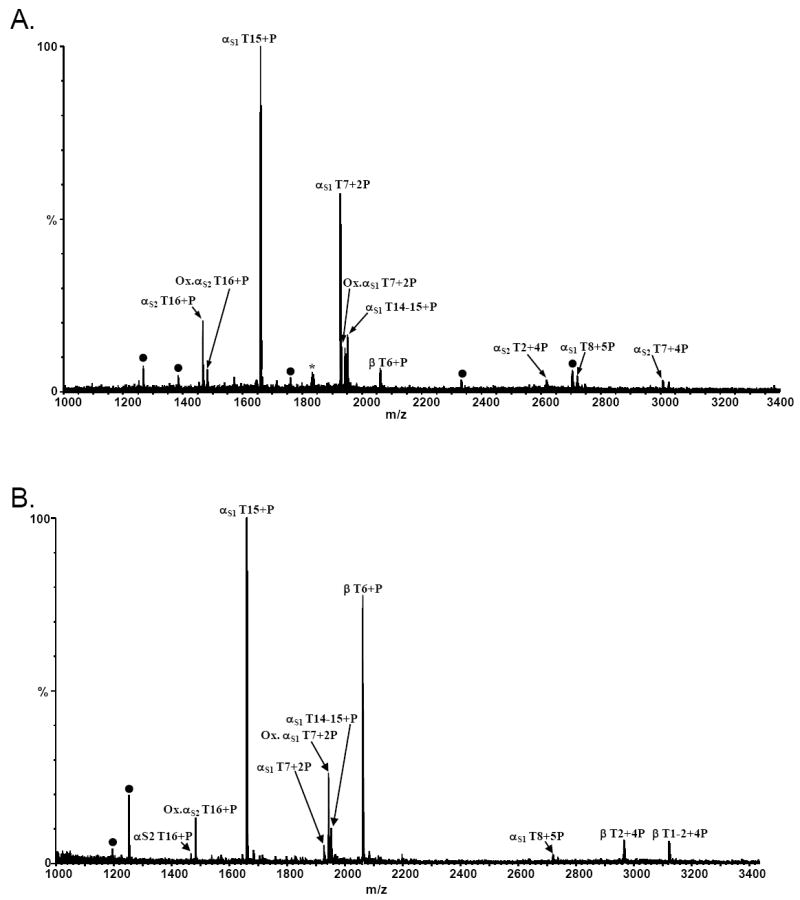
MALDI/MS of an α- and β-casein tryptic digest following A) Fe-NTA IMAC CRC enrichment, and B) titanium dioxide (GL Sciences) MOAC CRC enrichment. The ion labeled with an asterisk (*) corresponds in mass to a previously reported monophosphorylated casein variant (13). Ions labeled with filled circles represent non-specifically bound peptides.
Table 1.
Number of Times Specific Phosphorylation Sites Were Observed by MALDI/MS From Four Independent MOAC Enrichments.
| Tryptic Peptide | Theoretical [M+H]+ | Amino Acid Sequence | Phosphorylated Residue(s) | IMACa Fe-NTA | Titansphere TiO2 CRC | Glygen NuTip |
Phos-Trap TiO2 Kit | ||
|---|---|---|---|---|---|---|---|---|---|
| TiO2 | ZrO2 | TiO2/ZrO2 | |||||||
| αS1 T15/T14-15 | 1660.79 / 1951.95 | (YK)106VPQLEIVPNpSAEER119 | Ser 115 | X | 4 | 4 | 4 | 4 | 4 |
| αS1 T7b | 1927.69 | 43DIGpSEpSTEDQAMEDIK58 | Ser 46,48 | X | 4 | 4 | 4 | 4 | 4 |
| αS1 T8 | 2720.91 | 59QMEAEpSIpSpSpSEEIVPNpSVEQK79 | Ser 64,66,67,68,75 | X | 2 | 4 | 4 | 2 | 2 |
| αS1 T6-7 | 2678.02 | 37VNELpSKDIGpSEpSTEDQAMEDIK58 | Ser 41,46,48 | 0 | 3 | 3 | 2 | 2 | |
| αS2 T14/T14-15 | 1411.50 / 1539.60 | 126EQLpSTpSEENSKK136(K) | Ser 129,131 | 0 | 3 | 2 | 0 | 1 | |
| αS2 T16b/T15-16,T16-17 | 1466.61 / 1594.71 | (K)138TVDMEpSTEVFTK149(K) | Ser 143 | X | 2 | 4 | 4 | 4 | 4 |
| αS2 T2/T1-2 | 2618.91 / 2747.00 | (K)2NTMEHVpSpSpSEESIIpSQETYK21 | Ser 8,9,10,16 | X | 1 | 4 | 4 | 0 | 2 |
| αS2 T7 | 3008.03 | 46NANEEEYSIGpSpSpSEEpSAEVATEEVK7 | Ser 56, 57, 58, 61 | X | 1 | 4 | 3 | 2 | 2 |
| β T6/T5-6 | 2061.83 / 2432.05 | (IEK)33FQpSEEQQQTEDELQDK48 | Ser 35 | X | 4 | 4 | 4 | 4 | 4 |
| β T2 / T1-2 | 2966.17 / 3122.27 | (R)2ELEELNVPGEIVEpSLpSpSpSEESITR25 | Ser 15,17,18,19 | 4 | 4 | 4 | 4 | 4 | |
| # of Unique Sites of Phosphorylation Observed in 4 of 4 MOAC enrichments | 8 | 22 | 17 | 9 | 9 | ||||
Results from a single IMAC enrichment
Observed as both native and oxidized by MALDI/MS
Titanium Dioxide Resin
In 2004, the selectivity of titanium dioxide for phosphopeptides was first reported (12). To investigate the utility, in our hands, of titanium dioxide for phosphopeptide enrichment, casein digest mixtures were loaded onto CRCs packed with a commercially-available TiO2 resin (GL Sciences), eluted, and the resulting MALDI mass spectra obtained. A representative MALDI spectrum from these enrichments is shown in Figure 1B. In this spectrum, ions corresponding in mass to eight unique protonated tryptic phosphopeptides as well as two oxidized forms were observed. These peptides represent fourteen unique sites of phosphorylation. Only two non-specifically bound peptides were observed (T13 and T28-30 from the S2 variant of α-casein) with this resin. As was observed with the Fe-NTA IMAC resin (Figure 1A), peptides containing four or five phosphate groups were observed. Interestingly, the level to which the Fe IMAC or TiO2 MOAC resins enriched various peptides differ; thereby indicating a difference in specificity of binding between the Fe-NTA and titanium resins or elution from the TiO2 resin. For example, the monophosphorylated β-casein T6 peptide is observed at ca. 80% relative abundance in the MALDI spectrum from the TiO2 resin whereas this peptide is observed at ca. 10% relative abundance in the MALDI spectrum from the Fe IMAC resin. Conversely, the doubly phosphorylated α-casein S1 T7 peptide is observed at approximately 60% relative abundance vs 5% relative abundance in the MALDI spectra from the TiO2 resin vs the IMAC resin. The MALDI/MS results of four independent TiO2 enrichments from two independent digestion mixtures are summarized in Table 1. The number of times a specific site of phosphorylation was observed by MALDI/MS from the four independent MOAC enrichments is indicated in the table. Similar to previous reports that organic additives (e.g. DHB and citric acid) can be used to displace non-specifically bound peptides (13,14,16), we observed that the selectivity of the titanium resin was improved using DHB in the loading solution. As was reported by Larsen, et al. (13), we also found that 100 mg/mL DHB was optimal for the casein digest mixture.
Prepacked MOAC Enrichment Tips
Commercially-available prepacked products (Glygen NuTips) were also employed to determine their applicability for enrichment of phosphopeptides. These products contain three different resins (titanium dioxide, zirconium dioxide, or a mixture of both) and were evaluated using the casein digest mixtures. First, the utility of the TiO2 NuTip was evaluated. A representative MALDI/MS spectrum of the casein phosphopeptides enriched using the TiO2 NuTip (Figure 2A) shows ions which correspond in mass to fourteen phosphopeptides and represents twenty-one unique sites of phosphorylation. Moreover, only two non-specifically bound non-phosphorylated peptides (Figure 2A, ions labeled with a filled circle) of relatively low abundance (i.e. <5%) were observed in the eluent from this resin (Figure 2A). The results of four independent enrichments using this resin are shown in Table 1. Interestingly, a comparison of the data obtained from the TiO2 CRCs vs the TiO2 NuTips shows a higher number of unique sites of phosphorylation (8 vs 22) observed with the TiO2 NuTips in four of four enrichments. In addition, the peptides which contain three or more phosphate groups are observed more reproducibly with the TiO2 NuTip than with the TiO2 CRCs. If one includes the phosphopeptides that were observed in three of four independent enrichments, all known sites of phosphorylation were enriched using the TiO2 NuTip protocol.
Figure 2.
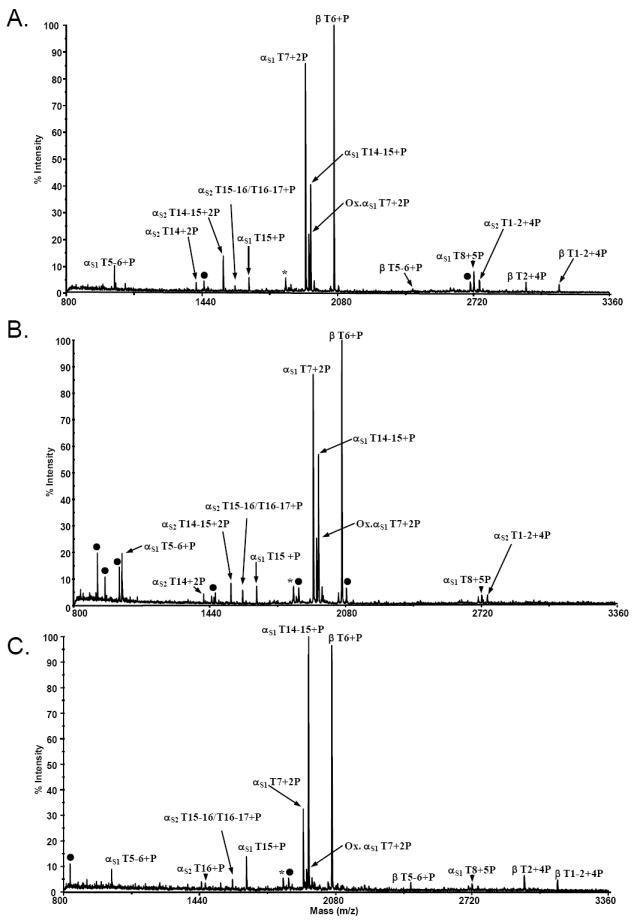
MALDI mass spectra of an α- and β-casein tryptic digest resulting from A) titanium dioxide NuTip enrichment, B) zirconium dioxide NuTip enrichment, and C) a mixed resin tip enrichment consisting of zirconium and titanium dioxide. The ion labeled with an asterisk (*) corresponds in mass to a previously reported monophosphorylated casein variant (13). Ions labeled with filled circles represent non-specifically bound peptides.
Similarly, a representative MALDI-TOF mass spectrum (Figure 2B) acquired from the eluent following enrichment with Glygen’s ZrO2 tip shows ions corresponding in mass to ten unique phosphopeptides representing seventeen sites of phosphorylation. The number of unique sites of phosphorylation observed from four independent enrichments using the ZrO2 resin is seventeen (Table 1). Moreover, many non-phosphorylated casein peptides (e.g. αS1 T10 and T17, and β T10-11) are observed using the ZrO2 resin (Figure 2B, ions labeled with filled circles); thereby, indicating greater non-specific binding compared to either the TiO2 NuTips or the Titansphere TiO2 resin.
Combining different metal oxide resins in one tip has potential for improved recovery of phosphopeptides as well as enhanced selectivity. It has also been reported that zirconium dioxide is more selective for singly phosphorylated peptides and titanium dioxide for multiphosphorylated peptides (15,16). Therefore, a mixed resin tip consisting of titanium dioxide and zirconium dioxide was evaluated using the casein digest mixtures. The MALDI mass spectrum acquired from one eluent from the mixed resin shows eleven ions corresponding in mass to phosphopeptides representing fifteen unique sites of phosphorylation from the digest mixture (Figure 2C). From these data, the number of non-specifically bound peptides observed are lower than with the ZrO2 resin alone and are more similar to the titanium dioxide NuTips. MALDI/MS analyses from four independent mixed resin enrichments are summarized in Table 1. The number of unique sites of phosphorylation observed with the mixed TiO2/ZrO2 resin NuTip in four of four enrichments is similar to that obtained with the TiO2 CRCs (9 and 8, respectively) and is lower than either the single TiO2 resin NuTip (i.e. 22) or the single ZrO2 resin NuTip (i.e. 17). Also of note, in our hands, the mixed resin tips would often leak resin out of the tip when high pH buffers were used; thereby, complicating their analyses. Perhaps this is one reason why the number of unique sites of phosphorylation observed with the mixed resin tips was lower than for either the TiO2 or ZrO2 resin by itself.
It has been reported that the enrichment capabilities of a given resin can vary based on the manner of its manufacture, the method by which it is affixed to the pipette tip, as well as the buffer conditions used during loading and washing steps. Therefore, we investigated the affect of the buffer conditions with the NuTips. Three sets of conditions were tested: 1) binding buffer of 3.3% formic acid and washed with water (15); 2) binding buffer of 80% ACN, 15% H2O, 5% TFA) (16) followed by water washes; and 3) binding buffer of 80% ACN, 15% H2O, 5% TFA followed by a wash solution of 80% ACN, 19% H2O, 1% TFA (16). The use of binding and washing system 3 resulted in the observation of much higher phosphopeptide selectivity (e.g. less than five non-specifically bound peptides observed) and was the protocol used for the analyses shown in Figure 3. Although increased selectivity was observed for the Titansphere titanium dioxide resin when the organic displacer DHB was used, the addition of this modifier did not significantly improve the enrichment capabilities of the NuTips in terms of the number of phosphopeptides observed.
Figure 3.
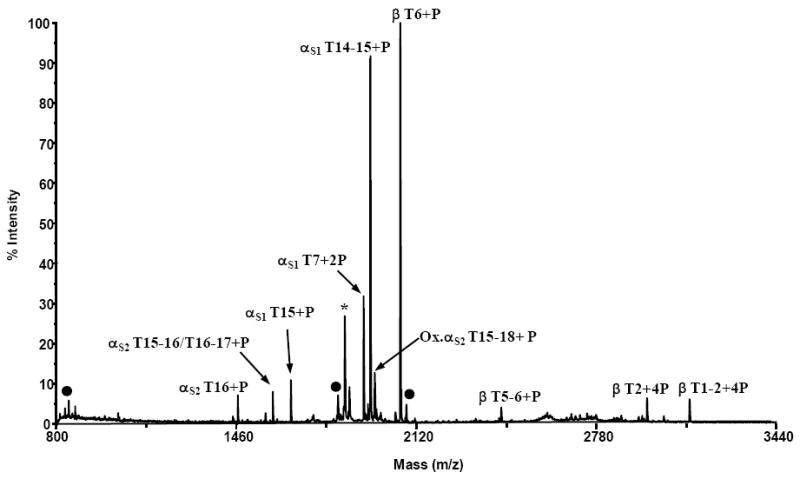
MALDI mass spectrum of an α- and β-casein tryptic digest resulting from enrichment using magnetic titanium dioxide beads (Phos-Trap kit). The ion labeled with an asterisk (*) corresponds in mass to a previously reported monophosphorylated casein variant (13). Ions labeled with filled circles represent non-specifically bound peptides.
To summarize, more unique sites of phosphorylation were reproducibly obtained using titanium dioxide enrichment NuTips than either the zirconium dioxide NuTips or the mixed resin NuTips (Table 1). In addition, the number of non-specifically bound peptides observed with the zirconium dioxide NuTip was, generally, greater in comparison to that of the titanium dioxide resin NuTip (e.g. α-casein S1 T10,α-casein S1 T17, and β-casein T10-11) (Figure 2B vs. Figure 2A). These results are similar to those reported previously (16) in that zirconium dioxide was observed as being less specific for binding of phosphorylated peptides than titanium dioxide. Comparatively, analyses performed with the mixed resin tip (Figure 2C) showed lower levels of non-specific binding than that of the zirconium tip (Figure 2B). In our hands, these enrichment results indicate that, of the three Glygen NuTips evaluated, the titanium dioxide resin is the most reproducibly selective for phosphopeptides with the least non-specific binding under the buffer conditions used in these experiments. In addition, the data obtained from the TiO2 NuTip in terms of the number of unique sites of phosphorylation which could be reproducibly enriched in independent experiments are better, in our hands, than those obtained from either the titanium dioxide CRC.
Magnetic Titanium Beads
Nanoparticles as well as magnetic nanoparticles with a titanium or zirconium surface have also been reported in the literature as being selective for enriching phosphorylated peptides. To explore the phosphopeptide enrichment capabilities of a commercially-available magnetic titanium dioxide particle (Perkin Elmer Phos-Trap™), enrichment of the casein digest mixture was performed using the manufacturer’s protocol and the supplied buffers. A MALDI-TOF mass spectrum (Figure 3) acquired from one casein digest eluent of the magnetic titanium resin enrichment experiments contains nine phosphopeptide ions representing nine of the known sites of phosphorylation. The nonspecific binding observed with this resin was minimal and similar to that observed with the other titanium dioxide resins. It should be noted that the chemical background noise observed in the mass range of m/z 2500-2800 was consistently observed in all eluent analyses from this resin. This most likely inhibited the reproducible observation of several multiply phosphorylated peptides in this region (e.g. αS1 T8,αS1 T6-7, and αS2 T2/T1-2) (Table 1). Without knowing the composition of the manufacturer’s supplied buffers, it is difficult to speculate on the composition of this chemical background. The number of unique sites of phosphorylation observed reproducibly with the Phos-Trap titanium dioxide resin is similar to the Titansphere TiO2 resin and the mixed NuTip resin.
LC/MS/MS Analyses
We also investigated whether or not additional phosphopeptides could be observed following enrichment using LC/ESI/MS/MS. Because it has been reported (13,16) that analyses of the eluent from the titanium dioxide CRC, which contains sizeable amounts of DHB, is not amenable to LC/MS/MS, the subsequent LC/MS/MS analyses were performed on the PhosTrap TiO2 eluent and the three different NuTip eluents. For comparison purposes, a tryptic digest of the casein mixture without any enrichment was also analyzed by LC/MS/MS and structurally diagnostic MS/MS spectra for five phosphopeptides were obtained. These phosphorylated peptides include α-casein S1 T15 and T17, α-casein S2 T16 and T1-2, and β-casein T6. For the mixed resin eluent, no additional phosphopeptides were observed by LC/MS/MS. One additional phosphopeptide was observed from a TiO2 NuTip eluent with the sequence 112VPNpSAEER119 from the S1 variant of α-casein LC/MS/MS analysis. This phosphopeptide was the result of an anomalous cleavage.
Conversely, from the LC/MS/MS analyses of the eluents from the Phos-Trap TiO2 resin, two of the phosphopeptides with molecular ions in the mass-to-charge range of 2500-2800 which were observed above the chemical background in only two of the four MALDI/MS analyses were detected by LC/MS/MS. As an example, the ETD MS/MS data of the [M+4H]4+ ion of m/z 681.1 corresponding in mass to the pentaphosphorylated α-casein S1 T8 tryptic peptide is shown in Figure 4. In this spectrum, a complement of c and z ions are observed due to fragmentations along the peptide backbone chain which allows the determination of the sites of phosphorylation at Ser 64, 66, 67, 68, and 75. Additionally, CID fragmentation was obtained from the MS/MS analyses of the [M+3H]3+ ion of m/z 893.35 which corresponds in mass to the triply phosphorylated α-casein S1 T6-7 tryptic peptide (Supplemental Figure S2). These results illustrate the utility of LC in combination with MS for the analyses of complex mixtures and for obtaining greater sequence coverage of proteins.
Figure 4.
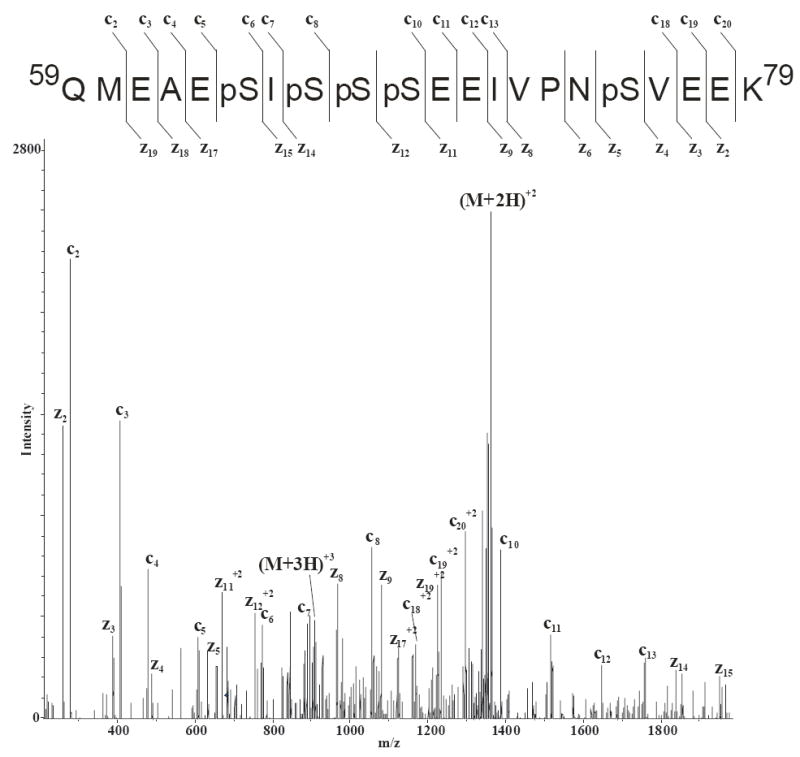
LC/ETD/MS/MS of the [M+4H]4+ ion of m/z 681.1 corresponding in mass to the pentaphosphorylated αS1 tryptic peptide T8 after enrichment with magnetic TiO2 beads.
Under the conditions and MS analyses used in this work, the titanium dioxide NuTip appears to be the most selective phosphopeptide enrichment resin with the least amount of non-specific binding. Although the non-specific binding observed with the Titansphere titanium dioxide resin and the mixed resin NuTip was minimal, the overall number of unique sites of phosphorylation observed reproducibly was less than that observed with the TiO2 NuTip. Additionally, our results indicate that the titanium dioxide NuTip resin exhibits a more reproducible phosphopeptide enrichment capability with lower levels of nonspecific binding than the ZrO2 tips. Even though it has been reported that zirconium dioxide preferentially enriches singly phosphorylated peptides and that titanium dioxide preferentially enriches multiphosphorylated peptides (16), we did not observe this trend.
Additional Figures of Merit
The limits of detection (LOD) for various phosphopeptide enrichment methods have been reported (2,12,14,16,21) and are important figures of merit when evaluating different media. Inefficient elution from affinity media, incomplete phosphopeptide binding upon loading, and sample loss resulting from sample handling are potential causes of reduced limits of detection. Therefore, determining the minimal amount of digest material required to successfully enrich and detect phosphopeptide signals is useful. To assess the lowest levels of phosphopeptides from which enrichment could be performed from the casein digest mixtures, independent replicate enrichments of the TiO2 NuTip and the TiO2 PhosTrap resin were evaluated by MALDI/MS. Employing the criteria of a S/N of 3 or greater and the observation of at least two phosphopeptides, the LOD for the TiO2 NuTip was determined to be ca. 125 fmol for α-casein and 125 fmol for β-casein which showed two monophosphorylated casein peptides, tryptic peptide T6 from β-casein and tryptic peptide T14-15 of α-casein (S1 variant). The limit of detection for the Phos-Trap kit was determined to be ca. 50 fmol for either α- or β-casein. The same two monophosphorylated peptides observed with the NuTip were also observed with the magnetic resin. This is a factor of approximately two lower than the limit of detection determined for the titanium NuTip and similar to the LOD reported for the Titansphere titanium dioxide resin (14).
Reproducibility of a given enrichment method can also play a large part in the number of phosphopeptides recovered run-to-run. In our hands, the MOAC resin exhibiting the highest number of phosphorylation sites that could be reproducibly detected by MALDI/MS from four independent enrichment experiments was the TiO2 NuTip (22 sites) followed by the ZrO2 NuTip (17 sites) (Table 1). Reproducibility when using the titanium CRC methodology and the Phos-Trap kit was lower and could be due to several factors. Some variation in reproducibility when using the titanium CRC may be due to loss of resin (and therefore sample) during enrichment due to the small particle size resin and the pore size of the CRC filter. As discussed previously, the chemical noise observed by MALDI/MS in the Phos-Trap eluents likely inhibited reproducible detection of some phosphopeptides.
Application to Avian Histones
Given the reproducibility results of the NuTip resin and LOD results of the Phos-Trap resin, we chose to further test the capabilities of these two resins by analyzing a more complex biological sample, avian linker and core histones. It is known that histones in avian immature red blood cells undergo many post-translational modifications including phosphorylation, methylation and acetylation (26-29). Although the general phosphorylation of chicken histone H1 has been noted in the literature (27), to our knowledge, only two groups have reported specific sites of phosphorylation of avian histones (27,30). Sung et al. reported the phosphorylation of Ser-3 and Ser-7 of chicken histone H5 using 32P labeling. Dickman et al. recently published the identification of several modifications of the chicken histones, including phosphorylation of histone H5 at Ser-3 and Ser-7 and, possibly, at Thr-1 of this protein. In addition, other sites of phosphorylation that they were unable to unequivocally distinguish include either Ser-22 or Ser-24 of histone H5 and Ser-1 or Thr-3 of histone H1.11L.
LC/MS/MS analyses of the digested and enriched chicken histone sample identified several phosphorylated peptides (Table 2). As an example, the deconvoluted MS/MS data of the [M+2H]2+ ion of m/z 822.2 which corresponds in mass to the N-terminal monophosphorylated acetylated H1.01 tryptic peptide with the sequence SETAPAAAPDAPAPGAK is shown in Figure 5. [Histone nomenclature is according to Coles et al. (31).] These MS/MS data show a series of y ions and b ions which result from fragmentation of the amide bonds in the peptide. In addition, the loss of H3PO4 from several b ions (b7, b8, b11, and b13) and the loss of water plus H3PO4 from the b3, b4 fragments indicating that the phosphate is located at either serine-1 or threonine-3. The observation of the ion corresponding in mass to the loss of water and H3PO4 from the y16 fragment is consistent with a phosphate group located at the Thr-3 residue vs the Ser-1 residue. Thus, these data indicate that the site of phosphorylation in this H1.01 tryptic peptide to Thr-3. Collectively, eight phosphopeptides were observed from the histone enrichment and subsequent LC/MS/MS analyses (Table 2, Figure 5, and Supplemental Figures S3-S9). A similar number of, but not identical, phosphopeptides were observed from the two titanium dioxide resins. Therefore, one should utilize more than one enrichment resin and optimize the conditions according to sample volume and complexity.
Table 2.
Summary of Observed Phosphorylated Peptides following Avian Histone Enrichment.
| Histone Varianta | Accession Number | Theoretical Mass | Observed Mass | Peptide | Sequence with Modificationb | Phosphorylated Residue(s) |
|---|---|---|---|---|---|---|
| H1.10 | AAA48788 | 808.392+ | 808.62+ | 1-17 | Ac-SEpTAPAAAPAVAAPAAK | T-3 |
| H1.01 | NP_001035732 | 822.372+ | 822.22+ | 1-17 | Ac-SEpTAPAAAPDAPAPGAK | T-3 |
| H1.02 | XP_425456 | 823.40+ | 823.72+ | 1-17 | Ac-(pS)E(pT)APVAAPAVSAPGAK | S-1 or T-3 |
| H1.11L | NP_001035733 | 991.472+ | 991.22+ | 1-21 | Ac-(pS)E(pT)APAPAAEAAPAAAPAPAK | S-1 or T-3 |
| H5 | NP_001038138 | 780.412+ | 780.52+ | 1-14 | Ac-(pT)E(pS)LVLSPAPAKPK | T-1 or S-3 |
| H5 | NP_001038138 | 759.402+ | 759.472+ | 1-14 | TEpSLVLSPAPAKPK | S-3 |
| H5 | NP_001038138 | 799.392+ | 799.22+ | 1-14 | TEpSLVLpSPAPAKPK | S-3, S-7 |
| H5 | NP_001038138 | 595.613+ | 595.83+ | 22-37 | (pS)A(pS)HPTYSEMIAAAIR | S-22 or S-24 |
Nomenclature according to Reference 31.
Ac=acetyl; p=phosphorylation
Figure 5.
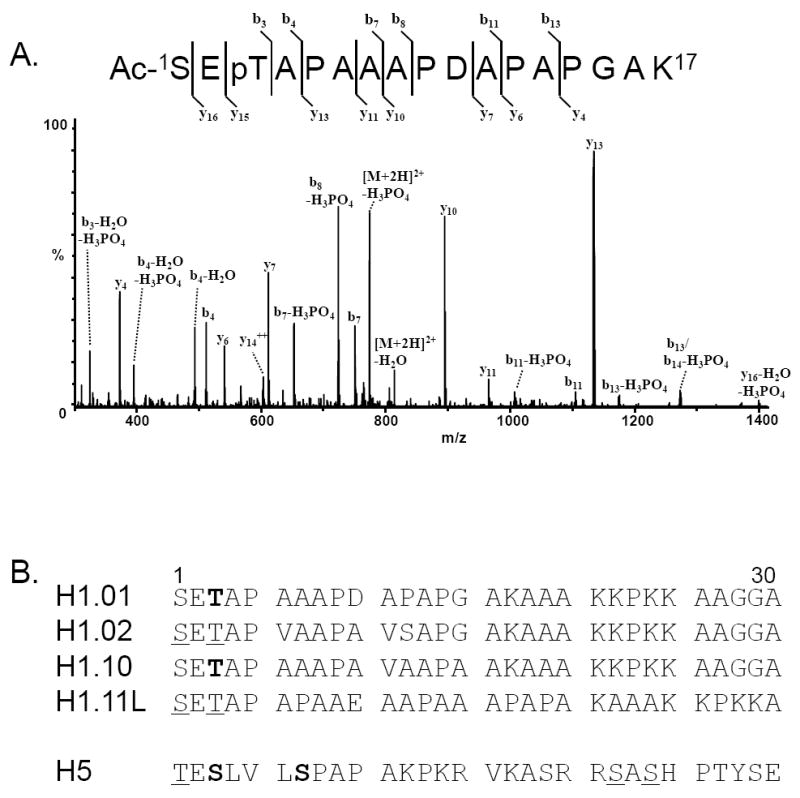
A) LC/CID/MS/MS of the (M+2H)2+ ion of m/z 822.2 corresponding in mass to the N-terminal monophosphorylated acetylated histone H1.01 tryptic peptide. B) N-terminal 30 residues of avian histone proteins for which sites of phosphorylation were determined. Amino acids in bold indicate residues in which a site of phosphorylation was confirmed. Underlined amino acids indicate residues in which the site of phosphorylation could not be unequivocally assigned.
From our MS analyses of phospho-enriched samples, we were able to identify previously unreported sites of phosphorylation. These include Thr-3 of histone H1.01 (Figure 5A) and Thr-3 of histone H1.10 (Supplemental Figure S3). Phosphorylation of histone H1.02 at Ser-1 or Thr-3 was also observed, but could not be unambiguously identified (Supplemental Figure S4). In addition to these novel sites of phosphorylation, the additional sites of phosphorylation of histone H5 (Ser-3, Ser-7, possibly Thr-1, and Ser-22 or Ser-24) and of histone H1.11L (Ser-1 or Thr-3) were identified (Supplemental Figures S5-S9). The N-terminal 30 residues of the chicken histones for which sites of phosphorylation were determined are shown in Figure 5B. Residues shown in bold indicate an amino acid for which a site of phosphorylation was confirmed. Underlined amino acids indicate residues in phosphopeptides for which the site of phosphorylation could not be unequivocally assigned. Of note, only the phosphorylation site at Ser-3 of histone H5 was observed by LC/MS/MS without enrichment. Several of the sites of phosphorylation determined for the chicken histone H1 proteins are homologous to sites of phosphorylation determined for histone H1 proteins from mammals (32,33).
CONCLUSION
All titanium dioxide resins evaluated in this study are capable of enriching phosphopeptides from a casein digest sample, albeit to different extents. These data demonstrate the importance of the experimental conditions employed for the enrichment techniques. Similar to previous reports, organic content in binding and washing solutions proved to be beneficial for the metal oxide resins. Both MALDI and ESI-MS analyses of the titanium dioxide NuTip eluents resulted in the reproducible observation of a greater number of unique sites of phosphorylation with the least amount of nonspecific binding compared to the other MOAC resins. These results demonstrate that enrichment procedures prior to MS analyses dramatically improve detection and sequencing of phosphopeptides compared to analyses without enrichment. In addition, enrichment reduces sample complexity; thereby, reducing the co-elution of peptides during LC/MS/MS analyses.
The applicability of phosphopeptide enrichment of a biological sample was illustrated using titanium dioxide enrichment media. For the avian histone tryptic digest, eight phosphorylated tryptic peptides were observed following enrichment and subsequent LC/MS/MS analyses. Seven of the eight phosphopeptides were not observed without titanium dioxide enrichment. From these analyses, four sites of phosphorylation were unequivocally determined; two of which have not been reported in the literature previously (i.e. Thr-3 of histone H1.10 and Thr-3 of histone H1.01). Four additional phosphopeptides were observed, however, the site of phosphorylation could not be distinguished, but was localized to one of two possible amino acids.
In summary, two of the titanium dioxide resins showed greater selectivity and sensitivity in our hands, however, these analyses were performed on samples of limited complexity. Therefore, depending on sample volume and complexity, it is important to optimize enrichment procedures accordingly. These methods should aid in the investigation of proteins post-translationally modified with phosphate, especially those present at low concentrations as was demonstrated by successful enrichment at the femtomole level.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES050171). The authors would like to thank Dr. Roxana Iacob and Dr. Jeffrey Kuhn for their assistance and thoughtful input, and Dr. Allison Schorzman and Dr. Jason Williams for critical review of this manuscript. NIEHS does not endorse or recommend any commercial products, processes, or services. The views and opinions expressed by authors affiliated with NIEHS do not necessarily state or reflect those of the U.S. Government, and they may not be used for advertising or product endorsement purposes.
Footnotes
Supplemental Material Available: Detailed mass spectrometric methodologies and manual interpretations of MS/MS data showing sites of phosphorylation in casein and avian histone tryptic peptides.
References
- 1.McLachlin DT, Chait BT. Analysis of Phosphorylated Proteins and Peptides by Mass Spectrometry. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 2.Reinders J, Sickmann A. State-of-the-Art in Phosphoproteomics. Proteomics. 2005;5:4052–4061. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 3.Thingholm TE, Jensen ON, Larsen MR. Analytical Strategies for Phosphoproteomics. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 4.Sun X, Chiu JF, He QY. Application of Immobilized Metal Affinity Chromatography in Proteomics. Expert Rev Proteomics. 2005;2:649–657. doi: 10.1586/14789450.2.5.649. [DOI] [PubMed] [Google Scholar]
- 5.Dunn JD, Reid GE, Bruening ML. Techniques for Phosphopeptide Enrichment Prior to Analysis by Mass Spectrometry. Mass Spectrom Rev. 2009 Mar 4; doi: 10.1002/mas.20219. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Thingholm TE, Larsen MR. The Use of Titanium Dioxide Micro-Columns to Selectively Isolate Phosphopeptides from Proteolytic Digests. Methods Mol Biol. 2009;527:57–55. doi: 10.1007/978-1-60327-834-8_5. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Merrick BA, Khaledi MG, Tomer KB. Detection and Sequencing of Phosphopeptides Affinity Bound to Immobilized Metal Ion Beads by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2000;11:273–282. doi: 10.1016/s1044-0305(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 8.Merrick BA, Zhou W, Martin KJ, Jeyarajah S, Parker CE, Selkirk JK, Tomer KB, Borchers CH. Site-Specific Phosphorylation of Human p53 Protein Determined by Mass Spectrometry. Biochemistry. 2001;40:4053–4066. doi: 10.1021/bi002045i. [DOI] [PubMed] [Google Scholar]
- 9.Deterding LJ, Cutalo JM, Tomer KB. In: contribution to “Proteomic Methods for Phosphorylation Site Mapping” in Proteins and Proteomics: A Laboratory Manual. Richard Simpson., editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. p. 597. [Google Scholar]
- 10.Cao H, Deterding LJ, Venable JD, Kennington EA, Yates JR, Tomer KB, Blackshear PJ. Identification of the Anti-Inflammatory Protein Tristetraprolin as a Hyperphosphorylated Protein by Mass Spectrometry and Site-Directed Mutagenesis. Biochem J. 2006;394:285–297. doi: 10.1042/BJ20051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor PA, McQuillan AJ. Phosphate Adsorption onto TiO2 from Aqueous Solutions: An In Situ Internal Reflection Infrared Spectroscopic Study. Langmuir. 1999;15:2916–2921. [Google Scholar]
- 12.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective Isolation at the Femtomole Level of Phosphopeptides from Proteolytic Digests using 2D-NanoLC-ESI-MS/MS and Titanium Oxide Precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 13.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJD. Highly Selective Enrichment of Phosphorylated Peptides from Peptide Mixtures Using Titanium Dioxide Microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly Selective Enrichment of Phosphorylated Peptides using Titanium Dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 15.Kweon HK, Håkansson K. Selective Zirconium Dioxide-Based Enrichment of Phosphorylated Peptides for Mass Spectrometric Analysis. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 16.Jensen SS, Larsen MR. Evaluation of the Impact of Some Experimental Procedures on Different Phosphopeptide Enrichment Techniques. Rapid Commun Mass Spectrom. 2007;21:3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 17.Liang X, Fonnum G, Hajivandi M, Stene T, Kjus NH, Ragnhildstveit E, Amshey JW, Predki P, Pope M. Quantitative Comparison of IMAC and TiO2 Surfaces Used in the Study of Regulated, Dynamic Protein Phosphorylation. J Am Soc Mass Spectrom. 2007;18:1932–1944. doi: 10.1016/j.jasms.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Kweon HK, Håkansson K. Metal Oxide-Based Enrichment Combined with Gas-Phase Ion-Electron Reactions for Improved Mass Spectrometric Characterization of Protein Phosphorylation. J Proteome Res. 2008;7:749–755. doi: 10.1021/pr070386d. [DOI] [PubMed] [Google Scholar]
- 19.Simon ES, Young M, Chan A, Bao ZQ, Andrews PC. Improved Enrichment Strategies for Phosphorylated Peptides on Titanium Dioxide using Methyl Esterification and pH Gradient Elution. Anal Biochem. 2008;377:234–242. doi: 10.1016/j.ab.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y-Q, Fournier J, Gilar M, Gebmler JC. Phosphopeptide Enrichment using Microscale Titanium Dioxide Solid Phase Extraction. J Sep Sci. 2009;32:1189–1199. doi: 10.1002/jssc.200800652. [DOI] [PubMed] [Google Scholar]
- 21.Aryal UK, Ross ARS. Enrichment and Analysis of Phosphopeptides Under Different Experimental Conditions using Titanium Dioxide Affinity Chromatography and Mass Spectrometry. Rapid Commun Mass Spectrom. 2010;24:219–231. doi: 10.1002/rcm.4377. [DOI] [PubMed] [Google Scholar]
- 22.Bai Z, Liu B, Li W, Li P, Wang H, Wang H. The Development of an Improved Simple Titanium Dioxide Enrichment Method for Phosphoproteomic Research. Rapid Commun Mass Spectrom. 2009;23:3013–3017. doi: 10.1002/rcm.4201. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt A, Csaszar E, Ammerer G, Mechtler K. Enhanced Detection and Identification of Multiply Phosphorylated Peptides using TiO2 Enrichment in Combination with MALDI TOF/TOF MS. Proteomics. 2008;8:4577–4592. doi: 10.1002/pmic.200800279. [DOI] [PubMed] [Google Scholar]
- 24.Lee DH, McComb ME, Bromirski M, Jilkine A, Ens W, Standing KG, Perreault H. On-Membrane Digestion of β-casein for Determination of Phosphorylation Sites by Matrix-Assisted Laser Desorption/Ionization Quadrupole/Time-of-Flight Mass Spectrometry. Rapid Commun Mass Spectrom. 2001;15:191–202. doi: 10.1002/1097-0231(20010215)15:3<191::AID-RCM209>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Janek K, Wenschuh H, Bienert M, Krause E. Phosphopeptide Analysis by Positive and Negative Ion Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Rapid Commun Mass Spectrom. 2001;15:1593–1599. doi: 10.1002/rcm.417. [DOI] [PubMed] [Google Scholar]
- 26.Sung MT, Harford J, Bundman M, Vidalakas G. Metabolism of Histones in Avian Erythroid Cells. Biochem. 1977;16:279–285. doi: 10.1021/bi00621a019. [DOI] [PubMed] [Google Scholar]
- 27.Sung MT, Freedlander EF. Sites of in vivo Phosphorylation of Histone H5. Biochem. 1978;17:1884–1890. doi: 10.1021/bi00603a013. [DOI] [PubMed] [Google Scholar]
- 28.Sung MT. Phosphorylation and Dephosphorylation of Histone V (H5): Controlled Condensation of Avian Erythrocyte Chromatin. Biochem. 1977;16:286–290. doi: 10.1021/bi00621a020. [DOI] [PubMed] [Google Scholar]
- 29.Pikaart M, Irving J, Villeponteau B. Decline in Histone H5 Phosphorylation During Erythroid Senescence in Chick Embryos. Mech Of Ageing and Develop. 1991;59:189–195. doi: 10.1016/0047-6374(91)90084-d. [DOI] [PubMed] [Google Scholar]
- 30.Snijders APL, Pongdam S, Lambert SJ, Wood CM, Baldwin JP, Dickman MJ. Characterization of Post-Translational Modifications of the Linker Histones H1 and H5 from Chicken Erythrocytes using Mass Spectrometry. J Proteome Res. 2008;7:4326–4335. doi: 10.1021/pr800260a. [DOI] [PubMed] [Google Scholar]
- 31.Coles LS, Robins AJ, Madley LK, Wells JRE. Characterization of the Chicken Histone H1 Gene Complement. Generation of a Complete Set of Vertebrate H1 Protein Sequences. J Biol Chem. 1987;262:9656–9663. [PubMed] [Google Scholar]
- 32.Garcia BA, Busby SA, Barber CM, Shabanowitz J, Allis CD, Hunt DF. Characterization of Phosphorylation Sites on Histone H1 Isoforms by Tandem Mass Spectrometry. J Prot Res. 2004;3:1219–1227. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- 33.Wisniewski JR, Zougman A, Kruger S, Mann M. Mass Spectrometric Mapping of the Linker Histone H1 Variants Reveals Multiple Acetylations, Methylations, and Phosphorylations as Well as Differences Between Cell Culture and Tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


