Repression of E2F target genes is required for cell cycle arrest in Rb family (Rb, p107, and p130)-deficient cells.
Abstract
The ability of progenitor cells to exit the cell cycle is essential for proper embryonic development and homeostasis, but the mechanisms governing cell cycle exit are still not fully understood. Here, we tested the requirement for the retinoblastoma (Rb) protein and its family members p107 and p130 in G0/G1 arrest and differentiation in mammalian cells. We found that Rb family triple knockout (TKO) mouse embryos survive until days 9–11 of gestation. Strikingly, some TKO cells, including in epithelial and neural lineages, are able to exit the cell cycle in G0/G1 and differentiate in teratomas and in culture. This ability of TKO cells to arrest in G0/G1 is associated with the repression of key E2F target genes. Thus, G1 arrest is not always dependent on Rb family members, which illustrates the robustness of cell cycle regulatory networks during differentiation and allows for the identification of candidate pathways to inhibit the expansion of cancer cells with mutations in the Rb pathway.
Introduction
The retinoblastoma (Rb) protein plays a critical role at the restriction point of the cell cycle (Weinberg, 1995). In mammalian cells, Rb and its family members p107 and p130 are thought to normally ensure cell cycle exit and prevent cells from reentering the cell cycle mainly by binding to E2F transcription factors, inhibiting the expression of E2F target genes, and remodeling chromatin into an inactive state (Classon and Harlow, 2002; Cobrinik, 2005; Gonzalo and Blasco, 2005). In the presence of mitogens, cyclin–Cdk complexes phosphorylate Rb family members, relieving the inhibition of E2F targets and enabling S phase entry. The compromised ability of cells with mutations in the Rb pathway to arrest in G1 is thought to be the major basis of its tumor suppressor activity (Sherr, 2004). However, the Rb family participates in multiple cellular processes, and their functional inactivation may also contribute to genomic instability and altered terminal differentiation; it is also possible that alterations in the Rb pathway have different consequences in different cell types (Classon and Harlow, 2002; Dannenberg and te Riele, 2006; Burkhart and Sage, 2008). A better understanding of the consequences of loss of Rb family function in mammalian cells may help to identify novel therapeutic strategies against many types of human tumors (Knudsen and Knudsen, 2008).
Embryogenesis provides a system to investigate the roles of Rb family proteins at the interface between proliferation and differentiation. Rb−/− embryos die 13.5–15.5 d after fertilization (E13.5–E15.5; Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992). This early embryonic lethality of Rb−/− embryos was shown to be the consequence of hypoxic stress caused by abnormal placental development: in contrast to germline mutant embryos, Rb−/− embryos with wild-type (WT) placentas die at birth from marked defects in muscle differentiation (de Bruin et al., 2003; MacPherson et al., 2003; Wu et al., 2003; Wenzel et al., 2007). p130−/− and p107−/− mice do not have any obvious developmental phenotypes in the 129/Sv and C57BL/6 genetic backgrounds (Cobrinik et al., 1996; Lee et al., 1996). In the same genetic background, p130−/−;p107−/− mice die immediately after birth, with differentiation defects in their bones and cartilage (Cobrinik et al., 1996). Recently, the analysis of Rb−/−;p107−/− mutant embryos with WT placentas showed lethality around E13.5–E14.5, with cardiac differentiation defects and abnormal proliferation of endothelial cells (Berman et al., 2009). These data point to a shared role for Rb family members in cell cycle exit and differentiation during embryonic development.
Although single or double knockout mouse embryonic fibroblasts (MEFs) display a compromised G1 arrest, Rb family triple knockout (TKO) MEFs are unable to arrest in G1 in response to cytostatic signals (Dannenberg et al., 2000; Sage et al., 2000; Peeper et al., 2001). Thus, the TKO strategy may uncover cellular phenotypes that can be masked by the presence of one functional Rb family gene compensating for the loss of the two others. In particular, we surmised that deleting the entire Rb gene family during embryogenesis might reveal the extent to which this gene family is critical for controlling cell cycle exit and differentiation in multiple lineages. We generated embryonic stem cells and mice simultaneously mutated for Rb, p107, and p130. We found that the Rb family is essential for proper embryonic development, but the phenotypes of TKO embryonic cells undergoing differentiation are less severe than expected. Strikingly, some TKO cells are able to arrest in G0/G1 and differentiate in teratomas and in culture. These findings provide evidence for Rb family–independent cellular pathways that can participate in the establishment of cell cycle arrest in G0/G1 in differentiating embryonic cells.
Results
Rb family mutant embryos die at mid-gestation with normal patterning and initial differentiation
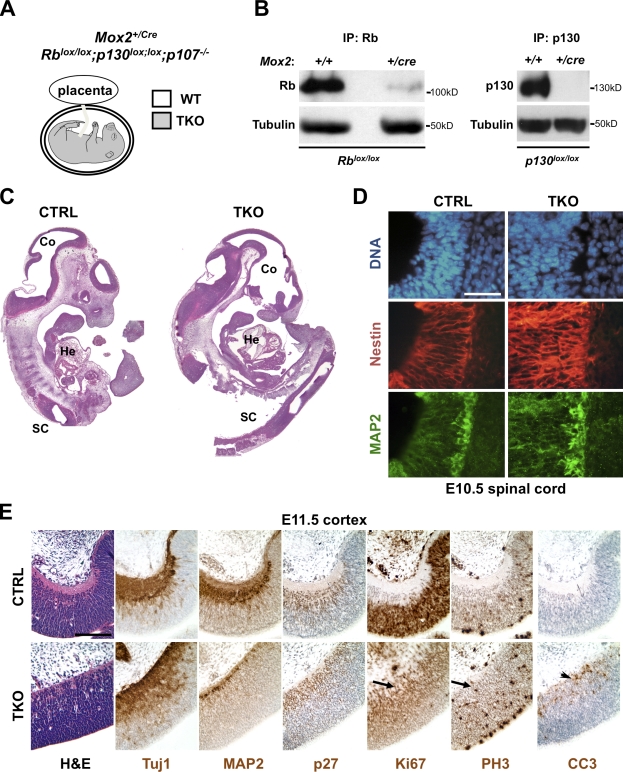
To investigate the composite role of Rb family proteins during embryogenesis, we first sought to generate Rb family TKO mouse embryos with WT placentas to prevent placental defects associated with loss of Rb (Wu et al., 2003). To this end, we bred conditional TKO mice (condTKO, Rblox/lox;p130lox/lox;p107−/−; Viatour et al., 2008) to Mox2+/Cre mice in which Cre expression begins at E5.5 in the embryo but not in the placenta (Fig. 1 A; Tallquist and Soriano, 2000). Survival analysis of embryos collected from condTKO females bred to Mox2+/cre RbΔ/+;p130Δ/+;p107+/− males (where Δ is a deleted conditional allele) supported a model in which TKO embryos die around E11.5 (Table I). PCR and immunoblot analysis showed that the deletion of the Rb and p130 alleles was very efficient although not always complete in this Mox2+/Cre cross (Fig. 1 B and not depicted), potentially explaining the age range of live and dead embryos obtained after E10.5. Strikingly, Mox2+/Cre TKO embryos showed normal dorso–ventral and anterior–posterior axes and development of major organs such as the heart, the spinal cord, the liver, and the brain, with the notable exception of the dorsal musculature (Fig. 1 C and Fig. S1; de Bruin et al., 2003).
Figure 1.
Mox2+/cre TKO embryos develop normally until mid-gestation. (A) Strategy to delete the three Rb family genes in embryos with a WT placenta using Mox2+/Cre mice. (B) Rb (left) and p130 (right) were immunoprecipitated (IP) from lysates of Mox2+/+ and Mox2+/cre E10.5 littermate embryos, and immunoblot analysis was performed with Rb and p130 antibodies, respectively. A direct immunoblot analysis for tubulin serves as a loading control. Protein molecular weights are indicated. (C) Representative sagittal sections from a control (CTRL) and Rb family TKO embryo at E11.5. Counterstain: hematoxylin and eosin (H&E). He, heart; SC, spinal cord; Co, developing cerebral cortex. (D) Immunofluorescence analysis on sagittal sections of the developing spinal cord of an E10.5 TKO embryo (TKO) along with a p107+/− littermate (CTRL) for nestin (red) and MAP2 (green). DAPI stains DNA in blue. (E) Immunostaining (brown signal) of the rostral cortex on control and TKO E11.5 embryos for Tuj1, MAP2, p27, Ki67, PH3 (arrows in the TKO embryo mark ectopically dividing cells), and CC3 (the arrowhead in the TKO embryo marks dying cells). Bars, 100 µm.
Table I.
Generation of Mox2+/Cre TKO and control embryos
| Embryonic stage | 5/6 embryos | TKO embryos | Total number of embryos collected |
| alive/total | alive/total | ||
| E10.5 | 8/8 | 1/1 | 44 |
| E11.5 | 6/7 | 3/4 | 44 |
| E12.5 | 8/9 | 1/2 | 32 |
| E13.5 | 7/8 | 0/0 | 40 |
| E14.5 | 2/5 | 0/0 | 13 |
| E15.5 | 1/1 | 1/2 | 24 |
| E16.5 | 0/4 | 0/0 | 20 |
| Experimental numbers | 32/42 | 6/9 | 281 |
| Expected numbers | 3/16 = 40.68 | 1/16 = 13.56 |
Alive embryos were scored as alive based on a beating heart and blood flowing through their umbilical cord. 5/6 embryos have five mutant alleles and one WT allele among the six alleles for the three Rb family genes. The overall under-representation of TKO embryos after E10.5 is statistically significant in a χ2 test (P = 0.01). All mice analyzed were in a mixed 129Sv/J:C57BL/6 genetic background.
To investigate the consequences of deleting Rb family genes for cell cycle and cell survival in mouse embryos, we performed immunostaining experiments on sections from Mox2+/Cre TKO and control embryos (Mox2+/+ embryos heterozygote for at least one Rb family gene), focusing on the embryonic central nervous system (Fig. 1, D and E). In the spinal cord at E10.5 (Fig. 1 D) and the developing cortex at E11.5 (Fig. 1 E), we found that Mox2+/Cre TKO embryos had a pattern of expression for the postmitotic neuronal marker nestin and for β-tubulin III (clone Tuj1) and MAP2 (microtubule associated protein 2), which was similar to that in control embryos (Fig. 1, D and E). p27, a marker of postmitotic neurons, was up-regulated in both TKO and control differentiating neuronal cells (Fig. 1 E). This analysis confirmed the histopathological observations that loss of Rb family genes in Mox2+/Cre TKO embryos does not prevent the first stages of brain development. However, we also found that, as expected from the analysis of Rb/p107 double mutant embryos (Berman et al., 2009), the brain and other regions of Mox2+/Cre TKO embryos displayed increased proliferation and apoptotic cell death at E11.5, as assayed by immunostaining for Ki67 (a marker of cells that have entered the cell cycle; Gerdes et al., 1984), phospho-histone H3 (PH3; a G2/M marker), and cleaved caspase 3 (CC3; a marker of apoptotic cell death; Fig. 1 E and not depicted). Ectopic proliferation and cell death in critical populations may be responsible for the death of Mox2+/Cre TKO embryos.
These data showing Mox2+/Cre TKO embryos surviving until mid-gestation were surprising given the well-known role of Rb family members in the control of proliferation and differentiation. However, these experiments did not allow us to assess the extent of Rb and p130 deletion in Mox2+/cre TKO embryos at the level of individual cells, and partial or delayed recombination of the conditional alleles in critical cell populations might be the reason why Mox2+/Cre TKO embryos survived so late into gestation and why some TKO cells were able to differentiate.
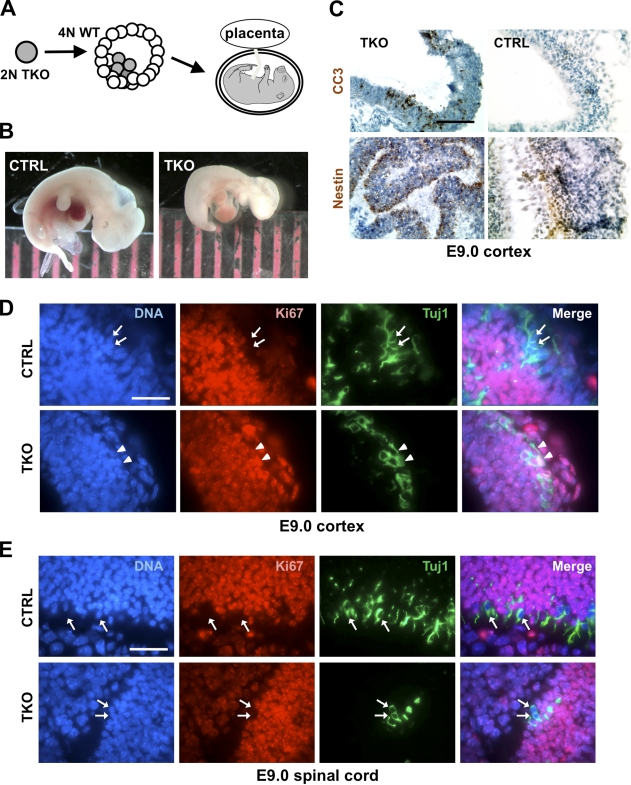
To address this limitation, we next sought to generate embryos composed entirely of TKO cells. To this end, diploid mouse embryonic stem cells (mESCs) with combined mutations in Rb family genes (Dannenberg et al., 2000; Sage et al., 2000) were injected into WT tetraploid blastocysts before reimplantation (Fig. 2 A; Meissner et al., 2007). p107/p130 mutant embryos survive until birth (Cobrinik et al., 1996); accordingly, control p107/p130 double knockout mESCs produced live embryos at 10.5 d of development (E10.5; Fig. 2 B). 7 out of 10 TKO embryos collected at E10.5 were dead. The size and morphological features of these embryos indicated that they were arrested at a developmental stage between embryonic days E8.5 and E9.5 (Table S1). The three remaining TKO embryos were alive but resembled an earlier stage of development (∼E9, or Theiler stage 14; Fig. 2 B). Serial sectioning and histopathological analysis of the three live TKO embryos showed normal development for E9 mouse embryos, with the presence of a well-defined neural tube, a cardiac chamber, and mesenchymal tissue (Fig. S2 A). These data indicated that TKO embryos can develop until E9. The difference between chimeric TKO embryos and Mox2+/Cre TKO embryos may be caused by the activation of Cre; Mox2+/cre TKO embryos begin to express Cre 2 d after the blastocyst stage, when TKO mESCs were introduced into tetraploid WT embryos. It is also possible that the difference between the two experiments is caused by the partial recombination of the conditional alleles in Mox2+/Cre TKO embryos. Nevertheless, these experiments indicate that the Rb gene family is not essential to generate embryos up to mid-gestation.
Figure 2.
Chimeric embryos composed of TKO cells develop until E9. (A) Tetraploid WT blastocysts were injected with TKO diploid mESCs to generate TKO embryos with a WT placenta. (B) Example of a TKO embryo (TKO, right) along with a p107−/−;p130−/− control embryo (CTRL, left). A millimetric ruler is shown for scale. Both embryos were harvested at E10.5. (C) Representative hematoxylin-counterstained neural tube sections from control and TKO embryos at E9 immunostained for CC3 and nestin, as indicated. Bar, 100 µm. (D) Representative sections from control and TKO E9 cortex immunostained for Ki67 (red) and Tuj1 (green). (E) Representative sections from control and TKO E9 spinal cord immunostained with Ki67 (red) and Tuj1 (green). Arrows indicate nuclei staining negatively for Ki67 and positively for Tuj1, and arrowheads indicate nuclei that stain positively for both Ki67 and Tuj1. DAPI stains DNA in blue. Bars: (C) 100 µm; (D and E) 50 µm.
Similar to what we observed in Mox2+/cre TKO embryos, regions of apoptotic cell death could be identified by immunostaining against CC3 in TKO chimeric embryos, including in the developing neural tube; staining of WT embryos at the same stage showed only very few CC3+ cells (Fig. 2 C). We also found that TKO cells were able to develop into specific lineages; in the developing neural tube, TKO cells expressed the neural progenitor marker nestin (Fig. 2 C and Fig. S2 B), whereas TKO cells lining the cardiac chamber expressed sarcomeric myosin (Fig. S2, C and D), similar to WT embryos. Most cells in WT and TKO embryos at E9 were positive for Ki67, as expected for a developing embryo (Fig. S2 B and not depicted). For example, we found that the majority of sarcomeric myosin-positive cells were Ki67+ both in WT and TKO embryos (Fig. S2 D). In contrast, nearly all Tuj1+ neurons were Ki67– in the developing cortex of WT embryos, but most Tuj1+ neurons in the cortex of TKO embryos were Ki67+, which is indicative of a strong cell cycle defect in these embryos (Fig. 2 D).
Rb family mutant mESCs exit the cell cycle and differentiate into multiple cell types in teratomas and in culture
A very small number of TKO cells in the cardiac chamber and in the spinal cord were negative for Ki67 staining but positive for the differentiation markers sarcomeric myosin and Tuj1, respectively (Fig. 2 E and Fig. S2 D). These results suggested that some TKO cells might be capable of cell cycle exit in vivo; however, because of the rarity of these Ki67− TKO cells and the impossibility of following the fate of these cells over extended periods of time in vivo, it was unclear whether these cells represented a true cell cycle arrest independent of the Rb family. To address this question, we turned to systems more amenable to manipulation and quantification.
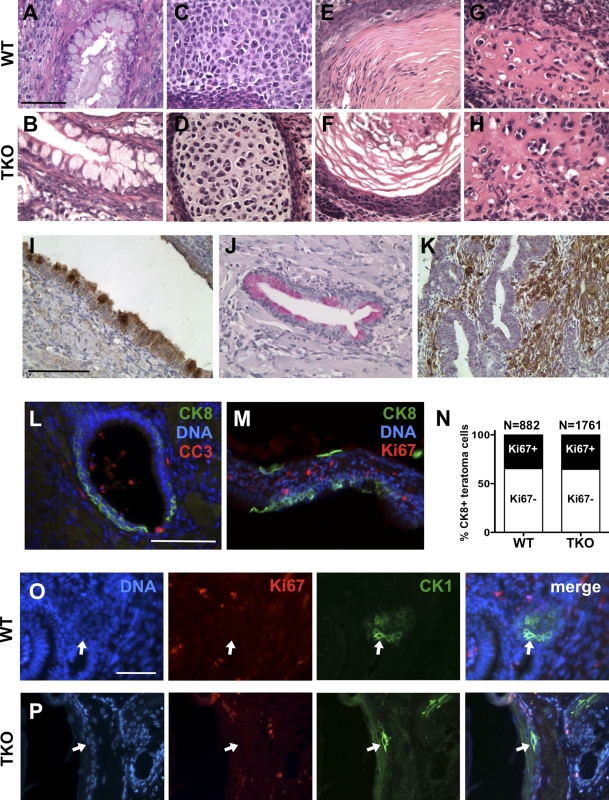
When injected under the skin of immunocompromised mice, WT mESCs formed teratomas comprised of a variety of differentiated endodermal, mesodermal, and ectodermal cell types, as expected (Fig. 3, A, C, E, and G; and not depicted). We used seven independent clones of TKO mESCs: six were generated from independent gene-targeting events as described previously (Sage et al., 2000), and one additional clone came from a different targeting strategy (Dannenberg et al., 2000). All clones produced teratomas that were dissected and analyzed 3 wk after injection. Among the 10 TKO teratomas closely examined, two remained mostly undifferentiated and contained immature cells and cells with a neuroblastic character, as described previously (Dannenberg et al., 2000). However, by histopathological analysis, we also clearly identified multiple differentiated cell types in sections from TKO teratomas, including cartilage, smooth muscle, osteoid, neuronal tissue, and various types of epithelia (Fig. 3, B, D, F, and H; and not depicted). These histological observations were confirmed by immunostaining experiments with markers of differentiation: cells in WT and TKO teratomas were positive for loracrin, smooth muscle actin (SMA), and mucin, confirming the presence of differentiated cells in the ectoderm, mesoderm, and endoderm lineages, respectively (Fig. 3, I–K; and not depicted). Quantification of the expression of these differentiation markers in WT and TKO teratomas did not reveal any significant difference between the two genotypes (Fig. S3, A and B).
Figure 3.
TKO cells generate differentiated cells in teratomas. (A–K) Histopathological analysis of WT and TKO teratomas in paraffin sections. Teratomas formed with WT and TKO mESCs both showed areas of extensive differentiation, including mucin-producing structures (A and B), cartilage (C and D), keratinized epithelium (E and F), and osteoid (G and H). TKO mESCs produce cells that participate in the three major lineages: ectoderm (I; cytokeratin 6, CK6 staining in brown), endoderm (J; mucin staining in pink), and mesoderm (K; SMA staining in brown) in teratomas. (L and M) Representative immunostaining for CK8 and CC3 (L) and for Ki67 and CK8 (M) on sections from a TKO teratoma. (N) Quantification of the proliferative status of CK8+ cells in WT and TKO teratomas. (O and P) Representative immunostaining for Ki67 (red) and CK1 (green) in WT (O) and TKO (P) teratomas. Arrows point to cells that stain negatively for Ki67 and positively for CK1. Bars: (A–M) 100 µm; (O and P) 50 µm.
Double immunostaining for CC3 and the epithelial marker cytokeratin 8 (CK8) also showed that several differentiating epithelial cells in control and in TKO teratomas were not undergoing apoptosis (Fig. 3 L and not depicted). Moreover, immunostaining for the cell cycle marker Ki67 showed that a significant number of CK8+ TKO cells had exited the cell cycle in G0/G1, similar to cells in WT teratomas (Fig. 3, M and N). We confirmed these results using CK1, a well-characterized postmitotic differentiation marker in the skin (Lu et al., 1994): CK1+ Ki67– cells could be readily observed in both WT and TKO teratomas (Fig. 3, O and P). These experiments confirmed that absence of the entire Rb family is compatible with cell fate determination and further indicated that differentiation associated with cell cycle exit is possible in cells with genetic inactivation of the entire Rb gene family.
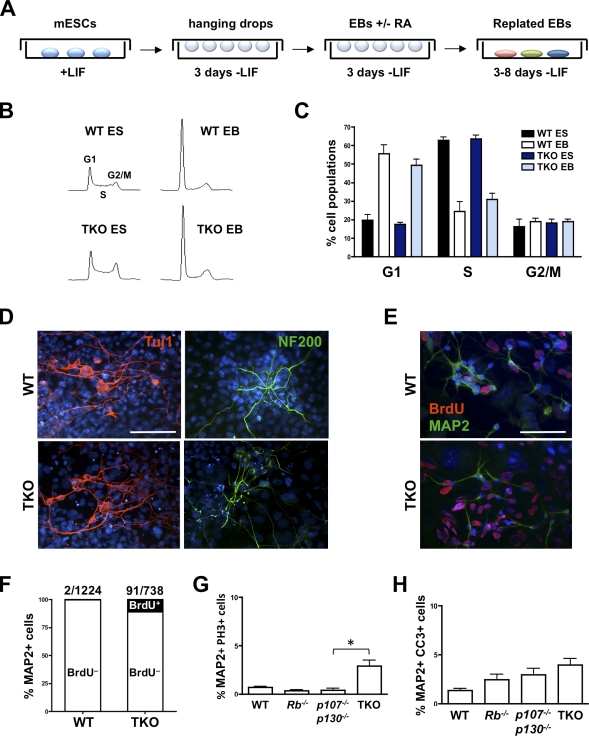
To investigate cell cycle exit and differentiation in TKO cells in a different system, we next produced embryoid bodies (EBs) from four independent TKO mESC clones as well as from control mESCs (Fig. 4 A). In EBs, differentiating mESCs recapitulate some aspects of embryonic development, and retinoic acid treatment has been shown to promote neuronal differentiation (Bain et al., 1995). The number and the size of EBs in the hanging drops were microscopically indistinguishable between the WT, Rb−/−, p107−/−;p130−/−, and TKO cells (unpublished data). To examine changes in gene expression occurring in control and TKO cells during in vitro differentiation, we analyzed the genome-wide expression profiles of WT and TKO mESCs during differentiation. We found that control and TKO undifferentiated mESCs had nearly identical profiles (Fig. S4 A), which supports the idea that Rb family proteins are functionally inactivated in cycling mESCs due to high cyclin–Cdk kinase activities (Ciemerych and Sicinski, 2005; Conklin and Sage, 2009). Although the expression profiles between biological replicates of TKO EB samples were more variable than those of WT EBs (Fig. S4 B), differentiating TKO mESCs clearly showed decreased expression of genes that are associated with undifferentiated mESCs, such as Oct4 and Nanog, and increased expression of genes associated with cellular differentiation, similar to WT cells (Fig. S4 A and not depicted). Thus, loss of Rb family genes is compatible with the early stages of the differentiation process from mESCs in vitro, mimicking the situation observed in embryos.
Figure 4.
TKO embryonic stem cells generate differentiated cells in culture. (A) Generation of EBs from mESCs in culture. mESCs were cultured without LIF in hanging drops for 3 d. Retinoic acid (RA) was added to induce neuronal differentiation. After three additional days of growth in hanging drops, EBs were plated and allowed to differentiate for 1 wk. (B) Cell cycle analysis by propidium iodide staining in WT and TKO mESCs (ES) and EBs. (C) Quantification of the cell cycle analysis (n ≥ 3 for each genotype). (D) Immunostaining for the neuronal markers Tuj1 (red) and NF200 (green) in WT and TKO EBs. Nuclei were visualized with DAPI (blue). Bar, 100 µm. (E) Immunostaining for the neuronal marker MAP2 (green) and BrdU (red) in WT and TKO EBs. Bar, 100 µm. (F) Quantification of MAP2+/BrdU+ cells. A significantly higher number of TKO neurons are BrdU+ compared with WT neurons (P = 0.0073), but the vast majority of TKO neurons are BrdU–. (G) Numbers of MAP2+ cells that are in M phase (stained with PH3) in TKO and control cultures 1 wk after re-plating of EBs (n ≥ 3 for each genotype; *, P = 0.0248). (H) Numbers of CC3+ apoptotic MAP2+ cells in TKO and control EBs 1 wk after re-plating (n = 4 for each genotype). Error bars indicate standard deviation.
Although cells were continuously grown in medium containing 15% serum, we found that both WT and TKO EB populations arrested similarly in G0/G1 upon differentiation (Fig. 4, B and C). Control and mutant cells also expressed markers of the three developmental lineages in culture (Fig. S3 C), as well as markers of postmitotic neuronal cells such as MAP2, Tuj1, and NF200 (Neurofilament 200 kD; Fig. 4 D). Double-staining experiments for MAP2 and BrdU incorporation revealed that the vast majority of TKO cells that have differentiated into mature neurons in culture from EBs did not actively replicate their DNA (Fig. 4, E and F). Co-immunostaining for MAP2 and PH3 confirmed that most TKO differentiated neurons were not going through mitosis (Fig. 4 G). Only a very small number of control and TKO cells expressing markers of terminal neuronal differentiation also stained positive for CC3, which suggests that TKO neurons are largely viable under these differentiation conditions (Fig. 4 H).
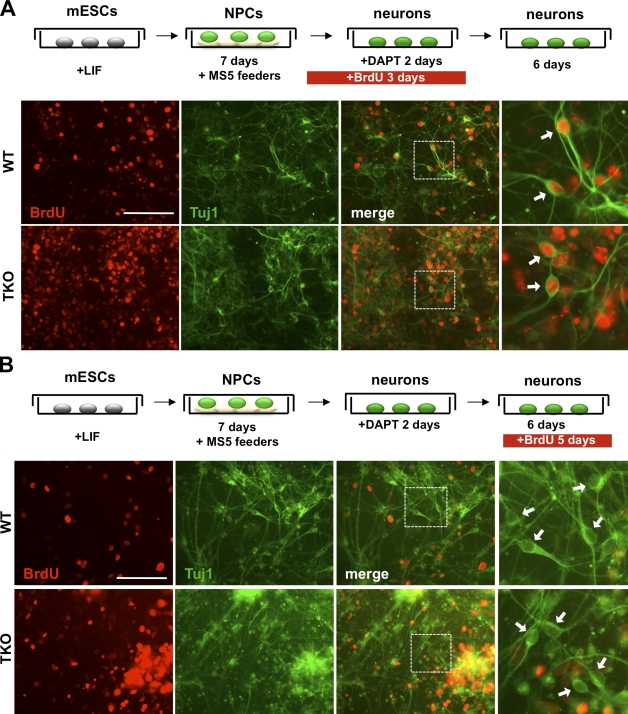
To exclude the possibility that the ability of TKO mESCs to terminally differentiate into neurons was specific to this differentiation protocol, and to study a more homogeneous population of TKO neurons in culture, we turned to an additional differentiation protocol. By differentiating mESCs on a layer of MS5 feeder cells without retinoic acid treatment, mESCs can be directed to efficiently differentiate into cultures of pyramidal neurons (Fig. 5; Ideguchi et al., 2010). Under these conditions, we again found that TKO mESCs efficiently produced neuronal cells expressing markers of terminal differentiation such as Tuj1, MAP2, and NF200 (Fig. 5 and not depicted). We next performed long-term BrdU labeling experiments to investigate whether TKO mESCs were capable of terminally exiting the cell cycle while expressing neuronal markers. First, BrdU was added to cultures of WT and TKO neuronal precursor cells (NPCs) for 3 d, at a time when most WT NPCs exit the cell cycle. 6 d after BrdU was removed from the medium, most WT neurons had retained the BrdU label, which indicated that these cells do not divide after the onset of neuronal differentiation (Fig. 5 A). Under the same conditions, cultures of TKO NPCs were more heterogeneous than WT: some TKO neurons failed to retain BrdU labeling, but we could also readily identify Tuj1+ BrdU+ TKO neurons, which indicates that some TKO cells are capable of terminally exiting the cell cycle upon differentiation (Fig. 5 A). Additionally, we performed labeling experiments on terminally differentiated cultures of WT and TKO neurons by adding BrdU for the last 5 d of differentiation, at a time when the majority of WT cells have completed the final cell division and are terminally differentiated (Fig. 5 B). As expected, WT neurons did not incorporate BrdU under these conditions. Remarkably, we could also identify a large number of TKO neurons that did not incorporate BrdU under the same conditions (Fig. 5 B). Thus, we found that Rb family TKO mESCs are able to stably arrest in G0/G1 and differentiate in teratomas and in culture.
Figure 5.
Postmitotic neurons derived from TKO embryonic stem cells stably exit the cell cycle in culture. (A) Protocol to generate cortical neurons from mESCs in culture and to examine the capacity of these neurons to retain BrdU over time. BrdU was added to NPC cultures that were undergoing the final round of cell division before becoming terminally differentiated. 6 d after the BrdU pulse, WT and TKO cells were immunostained for BrdU incorporation (red) and Tuj1 (green). (B) Long-term BrdU labeling of postmitotic WT and TKO neurons in culture. Neuronal cultures were labeled with BrdU for the last 5 d of culture and then immunostained for BrdU incorporation (red) and Tuj1 (green). White boxes indicate the area magnified in the panels on the right. Individual neurons are shown with arrows. Bars, 100 µm.
Multiple changes in critical cell cycle regulatory networks may explain cell cycle arrest in differentiating TKO mESCs
Several potential models could explain the ability of Rb family TKO cells to arrest in G0/G1. For example, although the Rb family is thought to be essential for proper control of the G1/S transition, some evidence suggests that cyclin E–CDK2 complexes and Myc transcription factors may control this cell cycle transition partly independent of Rb family members (Sears and Nevins, 2002; Hwang and Clurman, 2005; Meyer and Penn, 2008). Another possibility was that in the absence of the Rb family, alternative transcriptional mechanisms could repress the expression of Rb family target genes, including E2F targets.
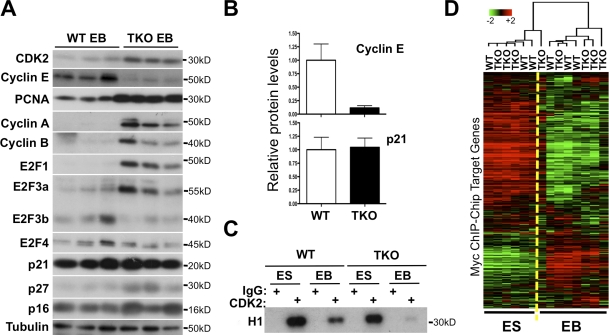
To begin to identify potential mechanisms allowing TKO cells to arrest in G0/G1, we examined the expression of key cell cycle regulators in WT and TKO EBs. Despite the cell cycle arrest observed in both WT and TKO cells, we found that positive regulators of cell cycle progression and E2F targets, including E2F1, E2F3a, cyclin A, cyclin B, CDK2, and proliferating cell nuclear antigen (PCNA), had elevated protein expression in TKO EB samples compared with WT samples (Fig. 6 A). These observations are consistent with the absence of inhibition by Rb family members on E2F activity in these cells. In contrast, however, the expression of another well-known E2F target and critical cell cycle regulator, cyclin E, was significantly lower in TKO cells compared with WT cells (Fig. 6, A and B). The levels of repressor E2Fs (E2F4 and E2F3b) were similar or slightly lower in TKO cells compared with WT cells (Fig. 6 A). The mRNA levels for E2F6, E2F7, and E2F8, three members of the E2F that can repress gene expression independently of Rb family members (Chen et al., 2009; Lammens et al., 2009), were not significantly different between TKO and WT EBs (Fig. S4 C). Levels of the two cell cycle inhibitors p16 and p27 were slightly increased in TKO cells, whereas the levels of p21 were similar in cells from both genotypes (Fig. 6, A and B). In vitro kinase assays showed a down-regulation of CDK2 kinase activity in TKO EBs compared with TKO mESCs (Fig. 6 C), which was similar to the decrease observed in WT cells. Thus, one mechanism by which TKO cells may achieve cell cycle arrest is inhibition of the kinase activity of CDK2 complexes by decreased levels of cyclin E and increased levels of CDK2 inhibitors.
Figure 6.
Low CDK2 and Myc activity in differentiating TKO cells. (A) Immunoblot analysis of cell cycle regulators from three representative WT and three representative Rb family TKO clones. Tubulin serves as a loading control. Protein molecular weights are indicated. (B) Quantification of cyclin E and p21 protein levels in TKO (n = 7) EB extracts relative to the values from WT samples (n = 5) and to tubulin levels. Means ± SEM are shown (error bars). (C) In vitro kinase assay on histone H1 for CDK2 in WT and TKO mESC (ES) and EBs (EB) differentiated following the protocol described in Fig. 5. A similar reduction of CDK2 kinase activity was observed in TKO EBs differentiated following the protocol shown in Fig. 4 compared with TKO mESCs (not depicted). (D) Unsupervised hierarchical clustering of the expression of Myc target genes in WT and TKO mESCs and EBs. Myc target genes were identified from ChIP (ChIP-Chip) data (see Materials and methods for details).
Based on a list of c-Myc target genes identified by whole-genome chromatin immunoprecipitation (ChIP) in mESCs (Kidder et al., 2008), we found that, as expected, the expression of the majority of Myc targets was down-regulated in differentiated WT EB populations compared with mESCs (Fig. 6 D and not depicted). The same trend was observed in TKO populations (Fig. 6 D). The similarity between the gene expression profiles for Myc target genes between WT and TKO cells suggests that Myc activity was similarly reduced as cells from both genotypes differentiate, and provides a second possible mechanism by which TKO cells may be able to exit the cell cycle.
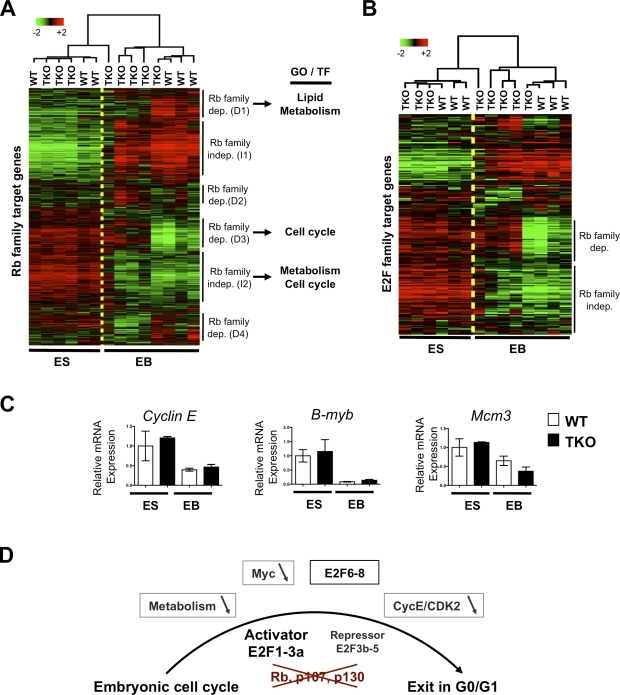
To specifically evaluate the expression of target genes of the Rb family during the differentiation of TKO mESCs in culture, we first used a bioinformatics approach to identify a list of potential direct transcriptional targets of Rb, p107, and p130 based on genome-wide expression profiles and ChIP data (see Materials and methods). When we examined the expression of these 1,095 selected genes during the differentiation of WT and TKO mESCs into EBs, we found four groups of genes whose expression depended on the absence or the presence of Rb family members during differentiation (D1–D4) and two groups whose expression profiles were independent of the Rb family (I1 and I2; Fig. 7 A and Table S6). A gene ontology (GO) term enrichment analysis focusing on the most significant terms (Table S2) indicated that cluster D3 was very significantly enriched for genes with functions classically associated with those of the Rb/E2F axis, such as DNA metabolism (P = 5.7 × 10−14), DNA replication (P = 2.4 × 10−12), M phase (P = 4.4 × 10−10), and cell cycle (P = 2.0 × 10−7); genes in this cluster included classical E2F genes such as Mcm10, Dnmt1, and Tk1. Interestingly, cluster D1 was enriched for lipid metabolism (P = 1.2 × 10−6) and lipid transport (P = 2 × 10−6). I2 genes were associated with biopolymer metabolism (P = 7.6 × 10−6) and intracellular part (P = 4.2 × 10−6). A search for consensus binding sites for transcription factors in the promoters of the genes in these clusters identified E2F/DP as the most significant in cluster D3 (P = 2.4 × 10−5; Table S3). Cluster I2 also showed some potential binding activity for E2F (P = 1.9 × 10−3), but the most significant potential binding activities were for the general transcriptional activator AP2 (P = 1.5 × 10−4) and NRF1 (P = 7.4 × 10−4). Interestingly, NRF1 is a coregulator of a large set of cell cycle and cell metabolism genes with E2F transcription factors (Cam et al., 2004). Thus, a decrease or a lack of activation in the metabolism of lipids and macromolecules is associated with cell cycle arrest in TKO populations. The presence of potential E2F binding sites in the promoter of genes similarly repressed during differentiation in TKO and WT cells also suggested that a subset of E2F target genes was repressed during the differentiation of TKO cells.
Figure 7.
Repression of Rb family and E2F family transcriptional targets in differentiating TKO cells. (A) Unsupervised hierarchical clustering of the expression of 1,095 Rb family target genes in WT and TKO mESCs and EBs. Selected results from the search for GO terms and for potential TFs regulating the six clusters identified are shown on the right (see Table S6 for the list of genes in each cluster and Tables S2 and S3 for the analyses). (B) Unsupervised hierarchical clustering of the expression of E2F target genes in WT and TKO mESCs and EBs. (C) Quantitative RT-PCR analysis of three E2F target genes and three critical cell cycle regulators, Cyclin E (Ccne1), B-myb, and Mcm3, whose repression in EBs is independent of the Rb gene family. Means ± SEM are shown (error bars). (D) Possible mechanisms of G0/G1 arrest in Rb family TKO cells during embryogenesis. Loss of Rb, p107, and p130 results in the activation of activator E2Fs and the loss of repression activity in repressor E2Fs, which is thought to prevent cell cycle exit in G0/G1. Possible mechanisms underlying the cell cycle exit observed in some TKO cells during differentiation include a decreased Myc activity, a decreased CDK2 activity, repression of several E2F targets (possibly by E2F6-8), and low levels of expression of genes involved in general and lipid metabolism.
These observations led us to specifically evaluate E2F activity in TKO cells undergoing differentiation. To this end, we selected a group of E2F target genes based on whole-genome ChIP data for E2Fs in human NTera2 embryonal carcinoma cells (see Materials and methods; Xu et al., 2007), a cell type with similarities to mESCs and EBs. We examined the expression levels of these E2F target genes in WT and TKO mESCs and found that these genes were generally expressed in WT and TKO undifferentiated cycling populations. In differentiating WT mESCs, the expression of most of these E2F target genes decreased, as expected. A subset of these genes remained highly expressed in TKO EBs, confirming that some E2F activity was deregulated in the absence of Rb family genes (Fig. 7 B). Strikingly, however, we found that a significant number of E2F target genes were repressed during differentiation in TKO cells (Fig. 7 B and Table S4), which is indicative of a program in differentiating embryonic cells that leads to the repression of E2F target genes independently of the Rb family. We confirmed these observations by quantitative RT-PCR analysis for the expression of classical E2F target genes using RNA from independent differentiation experiments (Fig. 7 C and Fig. S4, D and E); for instance, we found that the genes coding for cyclin E, B-Myb, and MCM3, three essential regulators of cell cycle progression at the G1–S transition of the cell cycle (Madine et al., 1995; Geng et al., 1996; Le Cam et al., 1999; Hwang and Clurman, 2005; Tarasov et al., 2008), were among the E2F target genes similarly repressed in WT and TKO EB populations (Fig. 7 C and Fig. S4 D). Similar observations were made with RNA from WT and TKO EBs with enriched populations of cortical neurons (Fig. S4 E). Together, these experiments suggest that cells with mutations in Rb family genes are able to arrest in G0/G1 in culture by repressing several E2F target genes involved in cell cycle progression at the G1/S transition of the cell cycle.
Discussion
Retinoblastoma only affects young children and often develops in utero (Lohmann and Gallie, 2004). Loss of Rb function is also implicated in the development of osteosarcoma in teenagers, a time when bone growth is pronounced (Deshpande and Hinds, 2006). These observations raise the possibility that loss of Rb in progenitor cells is a key initiating event in these two cancer types, and possibly in other adult cancers. Indeed, we previously found that loss of Rb family genes in early hematopoietic progenitors was sufficient to initiate a preleukemic disorder (Viatour et al., 2008), and recent observations have corroborated this model in breast cancer (Jiang et al., 2010). These observations led us to investigate the role of Rb family members in embryonic development at a time when many stem/progenitor cells exit the cell cycle and differentiate.
Experiments with viral oncoproteins known to inactivate the function of the three Rb family members have suggested that the Rb family was important but not always essential for several developmental processes (e.g., Kim et al., 1994; Feddersen et al., 1997; Callaghan et al., 1999; McCaffrey et al., 1999; McLear et al., 2006). One limitation of experiments with these viral oncoproteins is the extent to which they inhibit Rb family members (e.g., Casanovas et al., 2005). Data from mice with combined mutations in Rb family genes have further supported a widespread role for Rb, p107, and p130 in the control of cell cycle progression and cellular differentiation in many contexts (for reviews see Khidr and Chen, 2006; Wikenheiser-Brokamp, 2006), including in the formation of skeletal muscle (de Bruin et al., 2003). However, thus far, whether the Rb family is absolutely required for cell cycle exit during differentiation had not been determined. Our original presumption was that mouse embryos with compound genetic inactivation of Rb, p107, and p130 would die from several developmental abnormalities. Indeed, we found that loss of the Rb gene family results in embryonic lethality at mid-gestation and is accompanied by an increase in proliferation and cell death in multiple lineages. This widespread increased apoptosis is probably the cause of death of TKO embryos between E9 and E11; it correlates well with the expression pattern of Rb family genes at this time of development (Jiang et al., 1997; Burkhart et al., 2010). These data demonstrate that the Rb gene family is essential for proper embryonic development, underscoring the role of the Rb pathway in cellular and organismal homeostasis. Surprisingly, however, we did not observe any gross defects in organ size, organ shape, or patterning in TKO embryos. Furthermore, when TKO mESCs are differentiated in culture, we found that some TKO cells can stably arrest in G0/G1 and differentiate, uncovering the existence of a mechanism to arrest cells in G0/G1 independently of the Rb family (Fig. 7 D). Future experiments will explore whether the Rb family–independent cell cycle arrest we observed in culture can also occur in vivo by using mice expressing Cre in specific, well-defined cell types. We will also pursue experiments to determine whether additional and more subtle differentiation phenotypes exist in various TKO cell lineages beyond cell death and cell cycle phenotypes (Chen et al., 2007; McClellan et al., 2007). These experiments will also be critical to determine whether the potential mechanisms of G0/G1 arrest identified in TKO cells in culture are similar or different in vivo.
Because no G1 arrest was observed in TKO MEFs grown under several cytostatic conditions (Dannenberg et al., 2000; Sage et al., 2000), the mechanisms by which TKO embryonic cells arrest in G0/G1 are likely to be cell type dependent. However, recent observations in flies and mammalian cells suggest several models to explain our observations. In Drosophila, feedback mechanisms between E2F and cyclin E–CDK2 ensure that cell cycle arrest is robust, but these mechanisms vary depending on the cell lineage (Buttitta et al., 2007). In mammalian cells, the kinase activity associated with cyclin E–CDK2 is essential for cell cycle progression from G0/G1 (van den Heuvel and Harlow, 1993; Ohtsubo et al., 1995; Aleem et al., 2005; Ciemerych and Sicinski, 2005; Hwang and Clurman, 2005; Geng et al., 2007). Expression of cyclin E is also essential for the enhanced proliferation of Rb mutant cells under senescent conditions (Chicas et al., 2010). Based on these observations, one possibility to explain the G0/G1 arrest observed in TKO cells is a concomitant decrease in E2F transcriptional activity and in cyclin E–associated kinase activity (Fig. 7 D). Indeed, CDK2 activity is decreased in TKO cells, and a subset of E2F targets essential for cell cycle progression, including the gene coding for cyclin E itself, are similarly repressed in WT and TKO differentiating cells. Interestingly, RNA levels for cyclin E are similar in WT and TKO cells, whereas the protein levels are lower in TKO cells, which suggests that additional posttranscriptional mechanisms are at play to down-regulate cyclin E activity in the mutant cells. Nevertheless, the mechanisms that ensure proper cell cycle arrest and prevent cell cycle reentry may be conserved between flies and mammals and may involve dual inhibition of cyclin E–CDK2 activity and key E2F targets.
The mechanisms leading to repression of a large group of E2F target genes in TKO cells undergoing differentiation are still unknown. E2F1-3a, the “activating” members of the E2F family, are not known to have the ability to repress gene transcription in the absence of Rb family members and are thus unlikely to play a role in this process (Trimarchi and Lees, 2002; Chen et al., 2009; Chong et al., 2009; Lammens et al., 2009). Similarly, E2F3b-5 are recruited to the DNA by Rb family proteins to repress their target genes; these “repressor” E2F family members would not be expected to repress E2F targets in TKO cells. Indeed, we found no significant binding of E2F4 at the promoter of E2F target genes in TKO cells (unpublished data). Therefore, the best candidates for the repression of key E2F target genes in TKO cells are “atypical” E2F family members, which can bind DNA and repress transcription independently of Rb family members (Fig. 7 D; Chen et al., 2009; Lammens et al., 2009). Interestingly, E2F6, E2F7, and E2F8 play both cell cycle and developmental roles (Storre et al., 2002; Courel et al., 2008; Li et al., 2008). E2F6 in particular may play a role in quiescent cells (Ogawa et al., 2002), and its repressor functions may be dependent on its interactions with Polycomb group (PcG) protein complexes, which have been involved in embryonic development (Trimarchi et al., 2001; Ogawa et al., 2002; Attwooll et al., 2005; Courel et al., 2008). Although we did not observe increased expression of these atypical E2F family members in our differentiated TKO cells, these factors may become more active or localize to different promoters in the absence of the Rb family members. Although these experiments may be complicated by overlapping functions and compensatory mechanisms between the three atypical E2F family members (Li et al., 2008), genetic crosses between Rb family mutant mice and mice with mutations in atypical E2f genes will help to determine the functional interactions between Rb family members and E2F6-8.
It is also possible that the repression of critical cell cycle genes that are normally E2F targets has become independent of E2F transcription factors in TKO cells and is caused by transcription factors such as YY1 and NRF1. These factors usually partner with E2Fs and may be able to replace their activity in certain circumstances (Cam et al., 2004; Giangrande et al., 2004). Indeed, we see significant enrichment for NRF1 binding sites in the promoters of the Rb-independent target genes in our differentiating TKO cells. We also made the intriguing observation that several genes in metabolic pathways, including lipid metabolism, may be affected by loss of Rb family genes. Interestingly, Rb mutant cells in flies are killed by inactivation of TSC2, in part by inhibition of de novo lipid synthesis (Li et al., 2010). Future experiments will continue to probe this potential link between metabolism and the Rb pathway in mammalian cells.
Rb, p107, and p130 normally promote the differentiation of multiple lineages by controlling the activity of master transcription factors such as MyoD in muscles (Novitch et al., 1999), Runx2 in bones (Thomas et al., 2001), and PGC-1 in adipocytes (Scimè et al., 2005). However, our data demonstrate that Rb family TKO cells are able to fully differentiate, at least in certain lineages in teratomas and in culture. These observations suggest that Rb, p107, and p130 are regulators rather than essential components of the differentiation program. Increasing evidence indicates that cell cycle exit and differentiation programs are not always coupled (Buttitta and Edgar, 2007), and the mechanisms allowing TKO cells to differentiate may or may not be different than those allowing TKO cells to arrest in G0/G1.
Mice with mutations in gene families that were thought to be essential for the G1/S transition of the cell cycle are viable surprisingly often, emphasizing the plasticity of embryonic development in the mouse (Geng et al., 2003; Parisi et al., 2003; Kozar et al., 2004; Malumbres et al., 2004; Aleem and Kaldis, 2006; Berthet et al., 2006; Lee and Sicinski, 2006; Malumbres, 2007; Santamaría et al., 2007). Pursuing these studies will provide novel insights into the mechanisms linking extrinsic signals occurring during development and the intrinsic machinery that governs cell cycle progression and differentiation in normal and cancer cells.
Materials and methods
Mice and generation of embryos
The Rb and p130 conditional knockout alleles and Mox2+/Cre mice have been described previously (Tallquist and Soriano, 2000; Sage et al., 2003; Schaffer et al., 2010). Generation of embryos by tetraploid complementation was performed as described previously (Meissner et al., 2007). In brief, three separate TKO mESC clones and one double knockout control from which the TKO clones were generated were injected into tetraploid blastocysts and implanted. One of the three TKO clones was generated from an independent targeting strategy and provided by J.-H. Dannenberg and H. te Riele (Netherlands Cancer Institute, Amsterdam, Netherlands). Pregnancies were terminated at E10.5–E11.5. All mice were studied in a mixed 129Sv/J:C57BL/6 genetic background. All of the experiments with mice were approved by the Stanford University Institutional Animal Care and Use Committee.
Teratoma formation from mESCs
Teratomas were produced from 106 WT or TKO mESCs injected under the skin of nude mice. Approximately 2–3 wk after the injection, when tumors were visible under the skin, teratomas were dissected and fixed in 4% paraformaldehyde overnight for paraffin embedding and sectioning.
Differentiation of mESCs in culture
mESCs were plated without leukemia inhibitory factor (LIF) in hanging drops at a density of 2 × 105 cells/ml of media. After 3 d in suspension, EBs were fed with 10−7 M retinoic acid in media without LIF. After an additional 3 d of culture in hanging drops, EBs were then collected, washed briefly in PBS, and plated on gelatinized glass coverslips in media without LIF. EBs differentiated for 7 d. A complete protocol for the neuronal differentiation of mESCs on MS5 feeder cells was performed as described previously (Ideguchi et al., 2010), with some modifications. In brief, mESC colonies were grown on a layer of MS5 murine stromal cells (Uzan et al., 1996) for 5 d, then treated with FGF2 for 2 additional days while on feeders. Colonies of NPCs were dissociated into single cells and plated on polyornithine/fibronectin-coated glass slides. On the second day after plating, DAPT was added to the media for 2 d to increase the efficiency of neuronal differentiation. Cells were allowed to differentiate for an additional 8 d, when they were fixed and prepared for immunostaining (Ideguchi et al., 2010). 10 µM BrdU was added to cultures to monitor proliferation.
Immunostaining
For immunohistochemistry, mouse embryos and teratomas were fixed in 4% paraformaldehyde and embedded in paraffin; 5 µm sections were dewaxed and rehydrated in Trilogy buffer (Cell Marque) in a pressure cooker for 15 min. For immunofluorescence to detect YFP, embryos were embedded in OCT media, and 10 µm cryosections were cut. EBs were fixed in 4% paraformaldehyde on glass coverslips, permeabilized in 0.4% Triton X-100, and blocked in 50 mM glycine. All sections were blocked in 5% serum and incubated with primary antibody overnight at 4°C.
The following primary antibodies were used for immunostaining: SMA (Ab15267; Abcam), loracrin (AF62; Covance), NF200 (Ab4680; Abcam), neuronal class III β-tubulin (Tuj1; MMS-43SP; Covance), microtubule-associated protein 2 (Ab5392; Abcam), nestin (MAB353; Millipore), BrdU (347580; BD), PH3 (06-570; Millipore), Ki67 (550609; BD), p27 (610241; BD), cytokeratin 8 (MMS1841; Covance), cytokeratin 1 (PRB-165P; Covance), sarcomeric myosin (MF-20; Developmental Studies Hybridoma Bank), and CC3 (96645; Cell Signaling). After incubation with primary antibody overnight, samples were washed, incubated with secondary antibodies (Alexa Fluor 488 [green] and 594 [red]; Invitrogen), and counterstained with DAPI. Tissues known to express the antibody of interest were used as positive controls, whereas omission of the primary antibodies in identical samples was used as a negative control. BrdU immunostaining was performed as described previously (MacPherson et al., 2003). All images were acquired with a microscope (Eclipse E8000; Nikon) and a camera (Retiga OEM Fast; QImaging) using Osteo II Imagine software (Bioquant). The following Pan Fluor objectives (Nikon) were used at 25°C: 10× differential interference contrast (DIC) L 16 mm, 20× DIC M 2.1 mm, 40× DIC M 0.72 mm, and 100× oil DIC H 0.2 mm. All images within the same panel were processed similarly using Photoshop (Adobe).
Immunoprecipitation and immunoblotting
For immunoprecipitation experiments, embryos were harvested from pregnant females at E11.5 of gestation and snap-frozen on dry ice. Proteins were extracted from the tissues by homogenization in lysis buffer with a pestle. 75 µg of lysate was precleared with protein G beads for 2 h at 4°C and then immunoprecipitated overnight with 20 µg of Rb monoclonal antibody (Rb4.1; Developmental Studies Hybridoma Bank, University of Iowa) or p130 polyclonal antibody (610621; BD). Bound proteins were eluted from the beads with Laemmli buffer at 95°C for 5 min before immunoblot analysis.
Cell pellets for immunoblotting were lysed in whole cell lysis buffer (50 mM Tris, pH 7.6, 1% NP-40, 2 mM EDTA, pH 8, 100 mM NaCl, and protease inhibitors) and 50 µg of protein, run on a 10% acrylamide gel, and transferred to a PVDF membrane (Hybond). Protein concentrations were determined with a standard Bradford protein assay. Membranes were probed with the following antibodies: tubulin (T9026; Sigma-Aldrich), CDK2 (sc-163-G; Santa Cruz Biotechnology, Inc.), cyclin B1 (sc-245; Santa Cruz Biotechnology, Inc.), E2F3 (sc-878; Santa Cruz Biotechnology, Inc.), E2F4 (sc-1082; Santa Cruz Biotechnology, Inc.), p21 (sc-397; Santa Cruz Biotechnology, Inc.), p16 (sc-468; Santa Cruz Biotechnology, Inc.), cyclin E1 (sc-198; Santa Cruz Biotechnology, Inc.), cyclin A2 (sc-751; Santa Cruz Biotechnology, Inc.), E2F1 (sc-193X; Santa Cruz Biotechnology, Inc.), and p27 (610241; BD). Secondary antibodies conjugated to HRP (Jackson ImmunoResearch Laboratories, Inc.) were used at a concentration of 1:5,000, and signal was detected by chemiluminescence. Protein bands for p21 and cyclin E were quantified using the Odyssey Infrared Imaging System (LI-COR Biosciences).
For CDK2 kinase assays, cells were lysed using a whole cell lysis buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 10% glycerol, 0.5% Triton X-100, and protease and phosphatase inhibitors). Two experiments were performed with EB extracts from the general differentiation protocol, and one experiment was performed with the directed neuronal differentiation protocol. Immunoblot analysis for tubulin was used to confirm equal protein amounts. 100 µg of total protein was incubated with 3 µg of a rabbit polyclonal antibody against CDK2 (M2; Santa Cruz Biotechnology, Inc.) overnight at 4°C. Protein A–agarose beads were washed three times with lysis buffer and samples were incubated with 20 µl of beads for 2 h at 4°C. Bead pellets were washed four times with lysis buffer then four times with kinase buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, and 1 mM DTT). Bead pellets were then incubated for 30 min at 37°C with 2 µg of histone H1, 50 µM ATP, and 5 µCi [γ32]ATP in kinase buffer. Samples were then boiled for 5 min with 20 µl of 4× Laemmli buffer and loaded onto a 15% polyacrylamide gel that was subsequently dried for 3 h at 75°C.
Cell cycle and cell death assays
Cells were prepared for cell cycle analysis by fixation in 70% ethanol at −20°C overnight. Cells were rehydrated in PBS and stained with 20 µg/ml propidium iodide in PBS and 20 µg/ml RNase A for 30 min at 37°C, then overnight at 4°C. Cells were filtered with a 100 µm filter to obtain single cell suspensions. Quantification of cell death was performed by AnnexinV-FITC and propidium iodide staining, as described previously (Sage et al., 2000). Samples were analyzed using a FACSCalibur apparatus (BD).
Quantitative RT-PCR
Whole cell populations were collected and mRNA was isolated using TRIZOL (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed using the DyNAmo cDNA Synthesis kit (Finnzymes), with a total of 1 µg of mRNA per synthesis reaction. Quantitative PCR reactions were performed using SYBR GreenER qPCR SuperMix (Invitrogen) according to the manufacturer’s instructions. The samples were analyzed using a 7900HT Detection System with SDS 2.1 software (Applied Biosystems). All samples were normalized to a TATA binding protein (TBP) negative control RT-PCR and performed a minimum of three times. The primer sequences used in these experiments are listed in Table S5.
Microarray analysis
Total RNA was extracted with TRIZOL (Invitrogen) from WT and TKO mESCs and EBs (three independent replicates were isolated for WT mESCs, TKO mESCs, and WT EBs, whereas five replicates were isolated for TKO EBs). The RNA was hybridized to Mouse Genome 430 2.0 arrays (Affymetrix) according to the manufacturer’s instructions. The Invariant Set Normalization method was used to normalize the arrays using dChip software (Li and Hung Wong, 2001). Genes selected for analysis had a raw hybridization value >10 in all samples. The data were converted to log2 values and then mean centered. Cluster (Eisen et al., 1998) was used to carry out unsupervised hierarchical clustering of the genes that changed more than threefold from the mean in four or more samples (out of 14). Primary array data can be accessed through the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession no. GSE13408.
To isolate a set of E2F target genes, we used genome-wide ChIP-chip data from NTera2 human embryonal carcinoma cells (Xu et al., 2007). We isolated the top 200 genes bound by E2F1, E2F4, and E2F6 individually and then combined the lists. We mapped these genes to their corresponding mouse homologues using reciprocal best BLAST hits between human and mouse, resulting in a list of 272 genes (due to overlapping binding of multiple E2Fs to the same genes; Table S4). We then isolated the data corresponding to these genes from our microarray dataset and performed unsupervised hierarchical clustering.
Similarly, to isolate a set of Myc target genes, we used a predefined list of Myc targets identified by genome-wide ChIP in mESCs (Kidder et al., 2008). Using EntrezGene IDs, we then mapped the mouse Myc ESC target genes onto our gene expression data and analyzed it by unsupervised hierarchical clustering.
To identify a list of genes that may be direct targets of Rb family members, we first grouped genes directly bound by both Rb or p130 in human fibroblasts during quiescence and senescence (Chicas et al., 2010), which generated 4,104 potential Rb targets. Separately, we created a list of genes whose expression is dependent on Rb. We did this by isolating genes whose expression changes in mouse cells with mutations in Rb family members in hematopoietic progenitors (Viatour et al., 2008), and upon ectopic expression of Rb in human Saos2 cells (Saddic et al., 2010). For each dataset, we used Significance Analysis of Microarrays (Tusher et al., 2001) to identify genes that are induced or repressed upon mutation of Rb or ectopic expression of Rb as compared with control cells. After combining the three datasets, we identified 4,309 genes whose expression was dependent on Rb. The intersection of these two lists produced a group of 1,095 target genes that are likely to be direct transcriptional targets of Rb family members. We mapped these genes onto our gene expression data and analyzed it by unsupervised hierarchical clustering. We found six clusters of genes whose expression depended on the absence or the presence of Rb family members during differentiation (see Table S6 for a list of the genes). Each cluster was defined as a group of genes that, after unsupervised hierarchical clustering, shared a similar expression patterns across all of the microarray samples (these were readily visible using the TreeView software; http://rana.lbl.gov/EisenSoftware.htm). We performed gene set analysis of these clusters using the “gene module map method” implemented in Genomica (Segal et al., 2004). For each cluster, we looked for a significant enrichment of GO gene sets (P < 0.001; Table S2; Ashburner et al., 2000) and gene sets that share cis-regulatory transcription factor motifs in their promoters (P < 0.01; Table S3; Adler et al., 2007).
Online supplemental material
Fig. S1 shows histopathological analysis of representative control and Mox2+/Cre TKO embryos, and is related to Fig. 1. Fig. S2 shows histopathological analysis of E9 TKO tetraploid embryos, and is related to Fig. 2. Fig. S3 shows immunostaining for markers of each of the three germ cell layers in WT and TKO teratomas, and is related to Figs. 5 and 6. Fig. S4 shows gene expression analysis of WT and TKO mESCs and EBs, and is related to Figs. 6–9. Table S1 shows generation of TKO and control embryos using the tetraploid complementation method. Table S2 shows GO term enrichment analysis (p.001) on genes identified in the clusters shown in Fig. 9 A and Table S6. Table S3 shows a transcription factor (TF) motifs search (p.01) on genes identified in the clusters shown in Fig. 9 A and Table S6. Table S4 lists genes found in the two clusters identified in Fig. 9 B. Table S5 shows primers for quantitative RT-PCR analysis of gene expression. Table S6 lists genes found in the six clusters identified in Fig. 9 A. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003048/DC1.
Acknowledgments
We thank Drs. Hein te Riele, David MacPherson, Vjeko Dulic, Laurent Le Cam, Philipp Kaldis, Laura Attardi, and Anne Brunet for their critical comments on the manuscript; Patrik Asp in the Dynlacht laboratory, Jan-Hermen Dannenberg in the Riele laboratory, and Jackie Lees for sharing unpublished data and providing help with experiments; and Stéphane Vincent for helpful discussions. We thank Chenwei Lin for her help with the microarray analysis.
This work was supported by funding from the American Cancer Society (to H.Y. Chang), the Damon Runyon Cancer Research Foundation and the California Institute for Regenerative Medicine (to J. Sage and H.Y. Chang), the National Institutes of Health (to J. Sage), and the National Science Foundation (to S.E. Wirt). The authors declare that they have no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- CC3
- cleaved caspase 3
- ChIP
- chromatin immunoprecipitation
- CK
- cytokeratin
- EB
- embryoid body
- GO
- gene ontology
- LIF
- leukemia inhibitory factor
- MEF
- mouse embryonic fibroblast
- mESC
- mouse embryonic stem cell
- NPC
- neural precursor cell
- PH3
- phospho-histone H3
- Rb
- retinoblastoma
- SMA
- smooth muscle actin
- TKO
- triple knockout
- WT
- wild type
References
- Adler A.S., Sinha S., Kawahara T.L., Zhang J.Y., Segal E., Chang H.Y. 2007. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 21:3244–3257 10.1101/gad.1588507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleem E., Kaldis P. 2006. Mouse models of cell cycle regulators: new paradigms. Results Probl. Cell Differ. 42:271–328 10.1007/400_023 [DOI] [PubMed] [Google Scholar]
- Aleem E., Kiyokawa H., Kaldis P. 2005. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7:831–836 10.1038/ncb1284 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C., Oddi S., Cartwright P., Prosperini E., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K. 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J. Biol. Chem. 280:1199–1208 10.1074/jbc.M412509200 [DOI] [PubMed] [Google Scholar]
- Bain G., Kitchens D., Yao M., Huettner J.E., Gottlieb D.I. 1995. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168:342–357 10.1006/dbio.1995.1085 [DOI] [PubMed] [Google Scholar]
- Berman S.D., West J.C., Danielian P.S., Caron A.M., Stone J.R., Lees J.A. 2009. Mutation of p107 exacerbates the consequences of Rb loss in embryonic tissues and causes cardiac and blood vessel defects. Proc. Natl. Acad. Sci. USA. 106:14932–14936 10.1073/pnas.0902408106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C., Klarmann K.D., Hilton M.B., Suh H.C., Keller J.R., Kiyokawa H., Kaldis P. 2006. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev. Cell. 10:563–573 10.1016/j.devcel.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Burkhart D.L., Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 8:671–682 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Ngai L.K., Roake C.M., Viatour P., Thangavel C., Ho V.M., Knudsen E.S., Sage J. 2010. Regulation of RB transcription in vivo by RB family members. Mol. Cell. Biol. 30:1729–1745 10.1128/MCB.00952-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L.A., Edgar B.A. 2007. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19:697–704 10.1016/j.ceb.2007.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L.A., Katzaroff A.J., Perez C.L., de la Cruz A., Edgar B.A. 2007. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell. 12:631–643 10.1016/j.devcel.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Callaghan D.A., Dong L., Callaghan S.M., Hou Y.X., Dagnino L., Slack R.S. 1999. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev. Biol. 207:257–270 10.1006/dbio.1998.9162 [DOI] [PubMed] [Google Scholar]
- Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 16:399–411 10.1016/j.molcel.2004.09.037 [DOI] [PubMed] [Google Scholar]
- Casanovas O., Hager J.H., Chun M.G., Hanahan D. 2005. Incomplete inhibition of the Rb tumor suppressor pathway in the context of inactivated p53 is sufficient for pancreatic islet tumorigenesis. Oncogene. 24:6597–6604 10.1038/sj.onc.1208823 [DOI] [PubMed] [Google Scholar]
- Chen D., Opavsky R., Pacal M., Tanimoto N., Wenzel P., Seeliger M.W., Leone G., Bremner R. 2007. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 5:e179 10.1371/journal.pbio.0050179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Z., Tsai S.Y., Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 9:785–797 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R.A., Narita M., Zhang M., Lowe S.W. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 17:376–387 10.1016/j.ccr.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.L., Wenzel P.L., Sáenz-Robles M.T., Nair V., Ferrey A., Hagan J.P., Gomez Y.M., Sharma N., Chen H.Z., Ouseph M., et al. 2009. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 462:930–934 10.1038/nature08677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych M.A., Sicinski P. 2005. Cell cycle in mouse development. Oncogene. 24:2877–2898 10.1038/sj.onc.1208608 [DOI] [PubMed] [Google Scholar]
- Clarke A.R., Maandag E.R., van Roon M., van der Lugt N.M., van der Valk M., Hooper M.L., Berns A., te Riele H. 1992. Requirement for a functional Rb-1 gene in murine development. Nature. 359:328–330 10.1038/359328a0 [DOI] [PubMed] [Google Scholar]
- Classon M., Harlow E. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2:910–917 10.1038/nrc950 [DOI] [PubMed] [Google Scholar]
- Cobrinik D. 2005. Pocket proteins and cell cycle control. Oncogene. 24:2796–2809 10.1038/sj.onc.1208619 [DOI] [PubMed] [Google Scholar]
- Cobrinik D., Lee M.H., Hannon G., Mulligan G., Bronson R.T., Dyson N., Harlow E., Beach D., Weinberg R.A., Jacks T. 1996. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10:1633–1644 10.1101/gad.10.13.1633 [DOI] [PubMed] [Google Scholar]
- Conklin J.F., Sage J. 2009. Keeping an eye on retinoblastoma control of human embryonic stem cells. J. Cell. Biochem. 108:1023–1030 10.1002/jcb.22342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel M., Friesenhahn L., Lees J.A. 2008. E2f6 and Bmi1 cooperate in axial skeletal development. Dev. Dyn. 237:1232–1242 10.1002/dvdy.21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg J.H., te Riele H.P. 2006. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl. Cell Differ. 42:183–225 10.1007/400_002 [DOI] [PubMed] [Google Scholar]
- Dannenberg J.H., van Rossum A., Schuijff L., te Riele H. 2000. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051–3064 10.1101/gad.847700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A., Wu L., Saavedra H.I., Wilson P., Yang Y., Rosol T.J., Weinstein M., Robinson M.L., Leone G. 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. USA. 100:6546–6551 10.1073/pnas.1031853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A., Hinds P.W. 2006. The retinoblastoma protein in osteoblast differentiation and osteosarcoma. Curr. Mol. Med. 6:809–817 [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 95:14863–14868 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddersen R.M., Yunis W.S., O’Donnell M.A., Ebner T.J., Shen L., Iadecola C., Orr H.T., Clark H.B. 1997. Susceptibility to cell death induced by mutant SV40 T-antigen correlates with Purkinje neuron functional development. Mol. Cell. Neurosci. 9:42–62 10.1006/mcne.1997.0601 [DOI] [PubMed] [Google Scholar]
- Geng Y., Eaton E.N., Picón M., Roberts J.M., Lundberg A.S., Gifford A., Sardet C., Weinberg R.A. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 12:1173–1180 [PubMed] [Google Scholar]
- Geng Y., Yu Q., Sicinska E., Das M., Schneider J.E., Bhattacharya S., Rideout W.M., Bronson R.T., Gardner H., Sicinski P. 2003. Cyclin E ablation in the mouse. Cell. 114:431–443 10.1016/S0092-8674(03)00645-7 [DOI] [PubMed] [Google Scholar]
- Geng Y., Lee Y.M., Welcker M., Swanger J., Zagozdzon A., Winer J.D., Roberts J.M., Kaldis P., Clurman B.E., Sicinski P. 2007. Kinase-independent function of cyclin E. Mol. Cell. 25:127–139 10.1016/j.molcel.2006.11.029 [DOI] [PubMed] [Google Scholar]
- Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., Stein H. 1984. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 133:1710–1715 [PubMed] [Google Scholar]
- Giangrande P.H., Zhu W., Rempel R.E., Laakso N., Nevins J.R. 2004. Combinatorial gene control involving E2F and E Box family members. EMBO J. 23:1336–1347 10.1038/sj.emboj.7600134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Blasco M.A. 2005. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 4:752–755 [DOI] [PubMed] [Google Scholar]
- Hwang H.C., Clurman B.E. 2005. Cyclin E in normal and neoplastic cell cycles. Oncogene. 24:2776–2786 10.1038/sj.onc.1208613 [DOI] [PubMed] [Google Scholar]
- Ideguchi M., Palmer T.D., Recht L.D., Weimann J.M. 2010. Murine embryonic stem cell-derived pyramidal neurons integrate into the cerebral cortex and appropriately project axons to subcortical targets. J. Neurosci. 30:894–904 10.1523/JNEUROSCI.4318-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A. 1992. Effects of an Rb mutation in the mouse. Nature. 359:295–300 10.1038/359295a0 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Zacksenhaus E., Gallie B.L., Phillips R.A. 1997. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene. 14:1789–1797 10.1038/sj.onc.1201014 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Deng T., Jones R., Li H., Herschkowitz J.I., Liu J.C., Weigman V.J., Tsao M.S., Lane T.F., Perou C.M., Zacksenhaus E. 2010. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J. Clin. Invest. 120:3296–3309 10.1172/JCI41490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidr L., Chen P.L. 2006. RB, the conductor that orchestrates life, death and differentiation. Oncogene. 25:5210–5219 10.1038/sj.onc.1209612 [DOI] [PubMed] [Google Scholar]
- Kidder B.L., Yang J., Palmer S. 2008. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 3:e3932 10.1371/journal.pone.0003932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Roth K.A., Coopersmith C.M., Pipas J.M., Gordon J.I. 1994. Expression of wild-type and mutant simian virus 40 large tumor antigens in villus-associated enterocytes of transgenic mice. Proc. Natl. Acad. Sci. USA. 91:6914–6918 10.1073/pnas.91.15.6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen E.S., Knudsen K.E. 2008. Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer. 8:714–724 10.1038/nrc2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar K., Ciemerych M.A., Rebel V.I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R.T., et al. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 118:477–491 10.1016/j.cell.2004.07.025 [DOI] [PubMed] [Google Scholar]
- Lammens T., Li J., Leone G., De Veylder L. 2009. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 19:111–118 10.1016/j.tcb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L., Polanowska J., Fabbrizio E., Olivier M., Philips A., Ng Eaton E., Classon M., Geng Y., Sardet C. 1999. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 18:1878–1890 10.1093/emboj/18.7.1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Sicinski P. 2006. Targeting cyclins and cyclin-dependent kinases in cancer: lessons from mice, hopes for therapeutic applications in human. Cell Cycle. 5:2110–2114 [DOI] [PubMed] [Google Scholar]
- Lee E.Y., Chang C.Y., Hu N., Wang Y.C., Lai C.C., Herrup K., Lee W.H., Bradley A. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 359:288–294 10.1038/359288a0 [DOI] [PubMed] [Google Scholar]
- Lee M.H., Williams B.O., Mulligan G., Mukai S., Bronson R.T., Dyson N., Harlow E., Jacks T. 1996. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 10:1621–1632 10.1101/gad.10.13.1621 [DOI] [PubMed] [Google Scholar]
- Li B., Gordon G.M., Du C.H., Xu J., Du W. 2010. Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell. 17:469–480 10.1016/j.ccr.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Hung Wong W. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032 10.1186/gb-2001-2-8-research0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ran C., Li E., Gordon F., Comstock G., Siddiqui H., Cleghorn W., Chen H.Z., Kornacker K., Liu C.G., et al. 2008. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell. 14:62–75 10.1016/j.devcel.2007.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann D.R., Gallie B.L. 2004. Retinoblastoma: revisiting the model prototype of inherited cancer. Am. J. Med. Genet. C. Semin. Med. Genet. 129C:23–28 10.1002/ajmg.c.30024 [DOI] [PubMed] [Google Scholar]
- Lu B., Rothnagel J.A., Longley M.A., Tsai S.Y., Roop D.R. 1994. Differentiation-specific expression of human keratin 1 is mediated by a composite AP-1/steroid hormone element. J. Biol. Chem. 269:7443–7449 [PubMed] [Google Scholar]
- MacPherson D., Sage J., Crowley D., Trumpp A., Bronson R.T., Jacks T. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044–1053 10.1128/MCB.23.3.1044-1053.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine M.A., Khoo C.Y., Mills A.D., Laskey R.A. 1995. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 375:421–424 10.1038/375421a0 [DOI] [PubMed] [Google Scholar]
- Malumbres M. 2007. Cyclins and related kinases in cancer cells. J. BUON. 12:S45–S52 [PubMed] [Google Scholar]
- Malumbres M., Sotillo R., Santamaría D., Galán J., Cerezo A., Ortega S., Dubus P., Barbacid M. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 118:493–504 10.1016/j.cell.2004.08.002 [DOI] [PubMed] [Google Scholar]
- McCaffrey J., Yamasaki L., Dyson N.J., Harlow E., Griep A.E. 1999. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell. Biol. 19:6458–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan K.A., Ruzhynsky V.A., Douda D.N., Vanderluit J.L., Ferguson K.L., Chen D., Bremner R., Park D.S., Leone G., Slack R.S. 2007. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol. 27:4825–4843 10.1128/MCB.02100-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLear J.A., Garcia-Fresco G., Bhat M.A., Van Dyke T.A. 2006. In vivo inactivation of pRb, p107 and p130 in murine neuroprogenitor cells leads to major CNS developmental defects and high seizure rates. Mol. Cell. Neurosci. 33:260–273 10.1016/j.mcn.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Meissner A., Wernig M., Jaenisch R. 2007. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 25:1177–1181 10.1038/nbt1335 [DOI] [PubMed] [Google Scholar]
- Meyer N., Penn L.Z. 2008. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 8:976–990 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- Novitch B.G., Spicer D.B., Kim P.S., Cheung W.L., Lassar A.B. 1999. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9:449–459 10.1016/S0960-9822(99)80210-3 [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro K., Gaubatz S., Livingston D.M., Nakatani Y. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 296:1132–1136 10.1126/science.1069861 [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Theodoras A.M., Schumacher J., Roberts J.M., Pagano M. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi T., Beck A.R., Rougier N., McNeil T., Lucian L., Werb Z., Amati B. 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22:4794–4803 10.1093/emboj/cdg482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeper D.S., Dannenberg J.H., Douma S., te Riele H., Bernards R. 2001. Escape from premature senescence is not sufficient for oncogenic transformation by Ras. Nat. Cell Biol. 3:198–203 10.1038/35055110 [DOI] [PubMed] [Google Scholar]
- Saddic L.A., West L.E., Aslanian A., Yates J.R., Rubin S.M., Gozani O., Sage J. 2010. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J., Mulligan G.J., Attardi L.D., Miller A., Chen S., Williams B., Theodorou E., Jacks T. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037–3050 10.1101/gad.843200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J., Miller A.L., Pérez-Mancera P.A., Wysocki J.M., Jacks T. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 424:223–228 10.1038/nature01764 [DOI] [PubMed] [Google Scholar]
- Santamaría D., Barrière C., Cerqueira A., Hunt S., Tardy C., Newton K., Cáceres J.F., Dubus P., Malumbres M., Barbacid M. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 448:811–815 10.1038/nature06046 [DOI] [PubMed] [Google Scholar]
- Schaffer B.E., Park K.S., Yiu G., Conklin J.F., Lin C., Burkhart D.L., Karnezis A.N., Sweet-Cordero E.A., Sage J. 2010. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 70:3877–3883 10.1158/0008-5472.CAN-09-4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimè A., Grenier G., Huh M.S., Gillespie M.A., Bevilacqua L., Harper M.E., Rudnicki M.A. 2005. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2:283–295 10.1016/j.cmet.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Sears R.C., Nevins J.R. 2002. Signaling networks that link cell proliferation and cell fate. J. Biol. Chem. 277:11617–11620 10.1074/jbc.R100063200 [DOI] [PubMed] [Google Scholar]
- Segal E., Friedman N., Koller D., Regev A. 2004. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 36:1090–1098 10.1038/ng1434 [DOI] [PubMed] [Google Scholar]
- Sherr C.J. 2004. Principles of tumor suppression. Cell. 116:235–246 10.1016/S0092-8674(03)01075-4 [DOI] [PubMed] [Google Scholar]
- Storre J., Elsässer H.P., Fuchs M., Ullmann D., Livingston D.M., Gaubatz S. 2002. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 3:695–700 10.1093/embo-reports/kvf141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M.D., Soriano P. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 26:113–115 [DOI] [PubMed] [Google Scholar]
- Tarasov K.V., Tarasova Y.S., Tam W.L., Riordon D.R., Elliott S.T., Kania G., Li J., Yamanaka S., Crider D.G., Testa G., et al. 2008. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One. 3:e2478 10.1371/journal.pone.0002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.M., Carty S.A., Piscopo D.M., Lee J.S., Wang W.F., Forrester W.C., Hinds P.W. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 8:303–316 10.1016/S1097-2765(01)00327-6 [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Lees J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11–20 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild B., Wen J., Lees J.A. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA. 98:1519–1524 10.1073/pnas.041597698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 98:5116–5121 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan G., Prandini M.H., Rosa J.P., Berthier R. 1996. Hematopoietic differentiation of embryonic stem cells: an in vitro model to study gene regulation during megakaryocytopoiesis. Stem Cells. 14(Suppl 1):194–199 10.1002/stem.5530140725 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S., Harlow E. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 262:2050–2054 10.1126/science.8266103 [DOI] [PubMed] [Google Scholar]
- Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegué E., Sage J. 2008. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 3:416–428 10.1016/j.stem.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. 1995. The retinoblastoma protein and cell cycle control. Cell. 81:323–330 10.1016/0092-8674(95)90385-2 [DOI] [PubMed] [Google Scholar]
- Wenzel P.L., Wu L., de Bruin A., Chong J.L., Chen W.Y., Dureska G., Sites E., Pan T., Sharma A., Huang K., et al. 2007. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 21:85–97 10.1101/gad.1485307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp K.A. 2006. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell. Mol. Life Sci. 63:767–780 10.1007/s00018-005-5487-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., de Bruin A., Saavedra H.I., Starovic M., Trimboli A., Yang Y., Opavska J., Wilson P., Thompson J.C., Ostrowski M.C., et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 421:942–947 10.1038/nature01417 [DOI] [PubMed] [Google Scholar]
- Xu X., Bieda M., Jin V.X., Rabinovich A., Oberley M.J., Green R., Farnham P.J. 2007. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 17:1550–1561 10.1101/gr.6783507 [DOI] [PMC free article] [PubMed] [Google Scholar]